Abstract
Understanding the evolution of Tetraconata or Pancrustacea—the clade that includes crustaceans and insects—requires a well-resolved hypothesis regarding the relationships within and among its constituent taxa. Here, we assembled a taxon-rich phylogenomic dataset focusing on crustacean lineages based solely on genomes and new-generation Illumina-generated transcriptomes, including 89 representatives of Tetraconata. This constitutes, to our knowledge, the first phylogenomic study specifically addressing internal relationships of Malacostraca (with 26 species included) and Branchiopoda (36 species). Seven matrices comprising 81–684 orthogroups and 17 690–242 530 amino acid positions were assembled and analysed under five different analytical approaches. To maximize gene occupancy and to improve resolution, taxon-specific matrices were designed for Malacostraca and Branchiopoda. Key tetraconatan taxa (i.e. Oligostraca, Multicrustacea, Branchiopoda, Malacostraca, Thecostraca, Copepoda and Hexapoda) were monophyletic and well supported. Within Branchiopoda, Phyllopoda, Diplostraca, Cladoceromorpha and Cladocera were monophyletic. Within Malacostraca, the clades Eumalacostraca, Decapoda and Reptantia were well supported. Recovery of Caridoida or Peracarida was highly dependent on the analysis for the complete matrix, but it was consistently monophyletic in the malacostracan-specific matrices. From such examples, we demonstrate that taxon-specific matrices and particular evolutionary models and analytical methods, namely CAT-GTR and Dayhoff recoding, outperform other approaches in resolving certain recalcitrant nodes in phylogenomic analyses.
Keywords: CAT-GTR, Crustacea, Dayhoff recoding, Pancrustacea, phylogenomics, Tetraconata
1. Introduction
The phylogenetic relationships of crustaceans have attracted considerable interest over the years, as the group is diverse in marine and continental environments and many species are of commercial importance. Molecular genetic studies, especially those using phylogenomic datasets, as well as cladistic morphological studies have challenged many traditional hypotheses regarding the relationships of crustacean taxa. Congruence among early molecular studies was scanty (summarized in [1–3]), and despite a slowly emerging consensus regarding the positions of certain taxa, key relationships among and within several major taxa remain highly disputed. Of the traditionally recognized higher taxa, Branchiopoda, Cephalocarida, Malacostraca and Remipedia are clearly monophyletic, whereas Maxillopoda proved to be polyphyletic (e.g. [4–9]).
Certainly unexpected was the paraphyly of crustaceans with regard to Hexapoda (e.g. [10–12]), which is now well established and supported by molecular and morphological data (e.g. [4–9,13–20]). This clade has been termed Tetraconata or Pancrustacea. (We continue to refer to ‘crustaceans’ for all non-hexapod Tetraconata, bearing in mind paraphyly of a proper taxon ‘Crustacea’.) Remipedia are the likely sister group of Hexapoda [6,9,16], but Branchiopoda [13–15,17,18,20] and Xenocarida (Remipedia + Cephalocarida; [4,5]) also have been suggested.
Oligostraca—comprising Branchiura, Mystacocarida, Ostracoda and Pentastomida (previously part of Maxillopoda)—as the sister group to all other Tetraconata (a clade named Altocrustacea) is found in virtually all phylogenomic studies and combined phylogenomic and morphological analyses [4,5,7,9] (but see the morphological result in [6]). The remaining taxa usually fall into two clades, Allotriocarida (comprising Branchiopoda, Cephalocarida, Hexapoda and Remipedia; [7,9,20]) and Multicrustacea (comprising Malacostraca and the previous maxillopodan taxa Copepoda and Thecostraca; [4,5,7,9,13,14]). Other studies grouped Branchiopoda with Multicrustacea (a clade termed Vericrustacea; [4,5]) or Copepoda with Allotriocarida [8]. The most detailed morphological study, including fossils, recovered monophyletic Entomostraca (all Tetraconata, except Hexapoda, Remipedia and Malacostraca) [6]. In addition to these deep tetraconatan relationships, large-scale phylogenomic analyses within the tetraconatan main taxa are largely lacking, except for Ostracoda [7] and Hexapoda [19]. Here, we address this deficiency with a comprehensive analysis of Tetraconata and its two major subclades Malacostraca and Branchiopoda.
More generally, the chosen analytical methods and matrix composition can have a strong effect on the recovered topology (e.g. [8,9,21]). Knowing these limitations is helpful to detect questionable nodes, but identifying the underlying correct topology is intricate. We aim to improve the resolution within the two focal taxa, Branchiopoda and Malacostraca, by constructing taxon-specific matrices for each of these. If congruence among analytical methods is greater for these taxon-specific than for the tetraconatan-wide matrices, we may be able to infer the overall performance of the various analytical approaches.
2. Material and methods
(a). RNA extraction, library preparation and sequencing
Total RNA extraction followed standard TRIzol (Thermo Fisher Scientific) procedures (for details, see [9]). Tissues were fixed in RNAlater and stored at −80°C or fixed directly in TRIzol (figure 1; electronic supplementary material, table S1). mRNA was purified using magnetic oligo(dT)25-coated Dynabeads (Dynabeads mRNA DIRECT Purification Kit, Thermo Fisher Scientific). Fragmented cDNA libraries with 200 bp fragment sizes were constructed with the PrepX Library Preparation Kit (Wafergen Biosystems) on an Apollo 324 (Wafergen Biosystems). Each library was single-indexed with one of 12 indices of the PrepX mRNA Library Preparation Kit for Illumina (electronic supplementary material, table S1). Indexed libraries were PCR-amplified using the KAPA Library Amplification kit (Kapa Biosystems) with 10–20 cycles and cleaned up with magnetic beads (Aline). The mean fragment length was determined with an Agilent 2100 Bioanalyzer HSDNA assay (Agilent Technologies) and concentrations were determined by qPCR (KAPA Library Quantification kits; Kapa Biosystems). Twelve libraries were pooled and sequenced together (150 bp paired-end, on two lanes on an Illumina HighSeq 2500 at the FAS Center for Systems Biology, Harvard). Fifty four transcriptomes were newly sequenced for this study; raw reads of all newly sequenced transcriptomes were deposited with GenBank (electronic supplementary material, table S1).
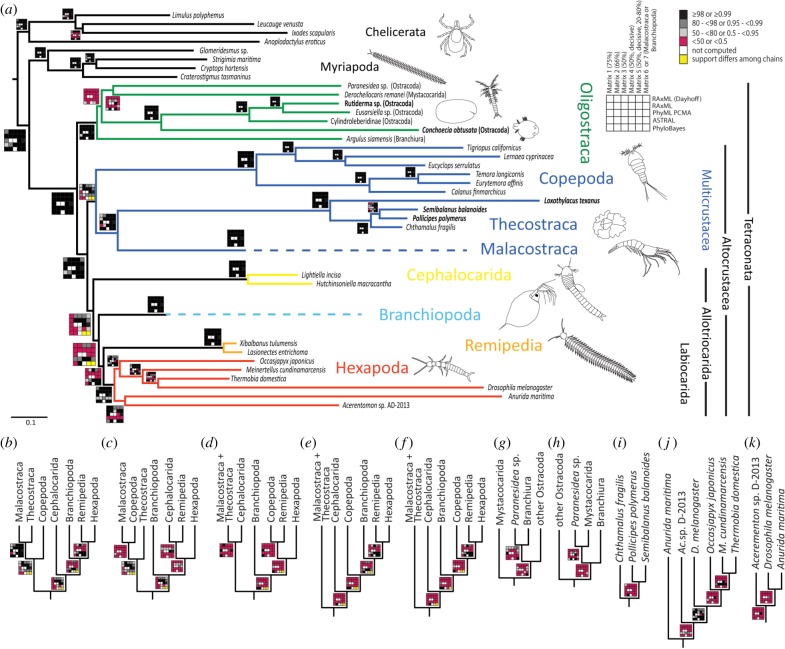
Figure 1.
Phylogenetic relationships within Tetraconata. (a) Inferred by PhyloBayes CAT-GTR with matrix 4 (455 decisive genes). All nodes within Malacostraca and Branchiopoda were collapsed (for details, see the electronic supplementary material, table S2 and figures S2 and S3). Support values for all analyses are depicted as rug plots, with each cell representing a specific combination of matrix and analytical method. Nodes supported in only one of two PhyloBayes chains are marked in yellow (the two PhyloBayes chains did not converge for these nodes). Individuals newly sequenced for this study are highlighted in bold. (b–k) All alternative topologies recovered in any analysis with support greater than 50 or 0.5. Alternative relationships within allotriocaridan and multicrustacean taxa (b–k), within Oligostraca (g–h), within Thecostraca (i) and Hexapoda (j–k) are depicted. (Online version in colour.)
(b). Data sanitation and transcriptome assembly
Demultiplexed raw reads were concatenated. Quality trimming to remove adaptor sequences and bases below a Phred score of 30 was performed with Trim Galore! (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/), retaining only reads of 25 bp or longer (corresponding to the minimum k-mer size in transcriptome assembly). We assessed pre- and post-trimming quality of reads with FastQC. All putative rRNA or mtDNA reads were filtered out with Bowtie2 [22] using custom indices built from all relevant rRNA and mtDNA sequences available on GenBank. Following de novo transcriptome assembly with Trinity [23,24] using default parameters (k-mer length 25, path_reinforcement_distance 50), remnant rRNA and mtDNA contigs were filtered out with Bowtie2. For the parasitic Loxothylacus texanus (Thecostraca, Rhizocephala), we also sequenced the transcriptome of the host Callinectes sapidus and eliminated potential host contigs with Bowtie2. The transcriptome of C. sapidus itself was not included in the phylogenomic analyses.
Highly similar isoforms within each assembly were eliminated with CD-HIT-EST [25], with a 95% similarity cut-off to reduce redundancy. Remaining sequences were converted to amino acid sequences with Transdecoder [24] and longest isoform per gene was selected and converted to a single line with two custom python scripts (choose_longest_MOD_v2.py, singleline.pl). Transcriptome data downloaded from NCBI SRA repositories (electronic supplementary material, table S1) were assembled and curated as described above; translated protein sequences from published genomes were only subjected to redundancy reduction with CD-HIT.
Identifying orthologous genes among transcriptome assemblies of all species studied is a crucial step in phylogenomic studies as it provides the foundation for all matrices and subsequent analyses. We used OMA stand-alone v.0.99 [26], which is based on all-by-all graph-based Markov clustering and which was shown to have higher precision in identifying orthologues and in discriminating paralogues than comparable programmes [27]. All computations, with the exception of RAxML analyses (see below), were run on the Odyssey cluster supported by the FAS Division of Science, Research Computing Group at Harvard.
(c). Matrix assembly
Taxon occupancy per orthogroup (as identified by OMA) was assessed with a custom python script (parseoma.py), and for each orthogroup, all amino acid sequences were aligned with MUSCLE v.3.8.31 [28]. Headers were sanitized (replace_header.py) and orthogroups with compositional heterogeneity or potential bacterial contamination were removed. Compositional heterogeneity was assessed with BaCoCa v.1.107 [29] and orthogroups with p < 0.99 in the χ2 test were excluded. Potential bacterial contamination was determined by blasting one sequence per orthogroups against the NCBI nr database (blastp)—selecting the longest available sequence. Four orthogroups with bacterial contamination were identified and removed. This step also allowed the annotation of each retained orthogroup (electronic supplementary material, table S2). In all retained orthogroups, ambiguously aligned positions were masked with Zorro [30] and positions with confidence scores below 5 were removed with a custom Python script (zorry.py).
From the 292 991 unique orthogroups predicted by OMA, seven different amino acid matrices were constructed (table 1), using occupancy as the primary criterion, i.e. the number of species present for each orthogroup [31], using a custom Python script (selectslice.py). The first three matrices were constructed by applying an occupancy threshold of 72 species (75% occupancy; matrix 1; i.e. an orthogroup was selected if 75% of the taxa were represented), 64 species (66% occupancy; matrix 2) and 48 species (50% occupancy; matrix 3), respectively (table 1; electronic supplementary material, tables S2 and S3). Matrices 4 and 5 were specifically aimed at resolving the phylogenetic relationships among tetraconatan taxa and were based on matrix 3, but including only decisive orthogroups (sensu [18]). In addition, the 20% fastest and slowest evolving orthogroups were removed from matrix 5 (see also [9]). Decisive orthogroups are those orthogroups for which at least one representative of each main taxon in question is present, here these were defined as Branchiopoda, Cephalocarida, Hexapoda, Multicrustacea, Oligostraca and Remipedia. We identified decisive orthogroups with a custom Python script (count_taxa_Crustacea.py). Removing orthogroups with the most extreme rates of evolution should reduce biases introduced by heterogeneous evolutionary rates. The evolutionary rate was determined with trimAl [32], calculating the level of conservation (-sct) for each orthogroup. Matrices 6 and 7 are taxon-specific matrices for Malacostraca and Branchiopoda, respectively, aiming to improve occupancy for these taxa, which are well represented with numerous species in our study (see also [7]). Also, artefacts like compositional heterogeneity and heterotachy may be reduced in these taxon-specific matrices, as other taxa with putatively confounding signals are excluded. All non-malacostracans or non-branchiopods were removed from the original OMA output with custom Python scripts (Crustacea_MalacostracaOnly.py, Crustacea_BranchOnly.py). The modified orthogroups were aligned and sanitized as described above for the other matrices. For Malacostraca (matrix 6), all orthogroups with 17 species (64% occupancy) were selected for matrix construction. Speonebalia (Leptostraca) was used as an outgroup, as all analyses employing matrices 1–5 consistently placed Speonebalia as the sister group to all other Malacostraca. For Branchiopoda (matrix 7), all orthogroups with at least 23 species (77% occupancy) were selected (table 1; electronic supplementary material, tables S2 and S3), and the root was placed between Anostraca and Phyllopoda, as supported by all analyses of matrices 1–5. It should be noted that one key branchiopod species (Eoleptestheria cf. ticinensis) became available when all other analyses were well progressed. Therefore, E. cf. ticinensis was included only in the branchiopod-specific matrix 7, but not in the other matrices.
Table 1.
Amino acid matrices for phylogenetic analyses. (Matrices 1–3 were assembled according to minimum occupancy, matrices 4 and 5 are the reduced version of matrix 3, including only decisive orthogroups. In addition, for matrix 5, the orthogroups (OG) with the 20% fastest and slowest evolutionary rate (ER) were removed. Matrices 6 and 7 include only Malacostraca or Branchiopoda, respectively.)
| minimum occupancy per OG (%) | number of OGs | average occupancy per OG (%) | total aa positions | average missing aa positions (%) | |
|---|---|---|---|---|---|
| matrix 1 | 75 | 81 | 80 | 17 690 | 21 |
| matrix 2 | 66 | 232 | 74 | 59 929 | 27 |
| matrix 3 | 50 | 864 | 62 | 242 530 | 40 |
| matrix 4 (decisive) | 50 | 455 | 63 | 122 993 | 38 |
| matrix 5 (decisive, 20–80% ER) | 50 | 267 | 63 | 75 776 | 39 |
| matrix 6 (Malacostraca) | 64 | 277 | 74 | 68 446 | 28 |
| matrix 7 (Branchiopoda) | 77 | 606 | 84 | 167 449 | 18 |
After a first round of analyses were completed, a stricter screening for putative contaminants (sequencing cross-contamination, human or microbial sequences and residual adapters) flagged an additional 195 sequences as putative contaminants and 30 featuring residual adapter sequences (225 out of 53,432 sequences included in our phylogenetic analyses). Their removal (excluding five complete orthogroups and modifying 84 others) had no influence on the recovered topology and negligible effects on support values as tested by ASTRAL and RAxML analyses of matrix 3 (not shown).
(d). Phylogenetic analyses
We performed up to five different phylogenetic analyses for each of the inferred matrices. Phylogenetic trees were rooted with Chelicerata, except in matrices 6–7.
Maximum-likelihood analyses were conducted with RAxML [33] using HPC2 on XSEDE of the CIPRES Science Gateway [34]. We performed two different analyses for each matrix, one using the above-described amino acid matrices and one using Dayhoff transformed six-state matrices. The latter is supposed to reduce artefacts of long-branch attraction and taxon-specific compositional heterogeneity [8,35,36] by assigning each amino acid to one of six classes based on their properties: AGPST, FWY, C, HKR, ILMV and EDNQ. For the Dayhoff recoded matrices, a multigamma GTR model was employed (-m MULTIGAMMA -K GTR). For the amino acid matrices, a PROTGAMMALG4X substitution model partitioned by orthogroups was employed. One hundred rapid bootstrap replicates (-f) were computed and the bootstrap (-x) and parsimony random seed (-p) were set to 12345.
A different maximum-likelihood approach was followed with PhyML 3.0 [37]. Here, the amino acid substitution matrices were inferred via a PCMA [38] employing 10 principal components (-pcs 10 -f e -m PCMA -s SPR -n_rand_start 3 -r_seed 123 -d aa -sequential). We did not employ PhyML for matrices 2 and 3, owing to the size of the matrices.
Bayesian inference was conducted with PhyloBayes MPI [39] under the CAT-GTR model [40], which is assumed to be particularly well suited to resolve artefacts caused by long-branch attraction and site-specific compositional heterogeneity [41,42]. We ran two independent chains for each matrix and assessed their convergence with tracecomp and bpcomp. We did not employ PhyloBayes for matrix 3 owing to its large size.
While all of the above phylogenetic methods used a single supermatrix concatenating all orthogroups, ASTRAL 4.10.2 [43] uses a coalescence-based approach. Individual gene trees were computed with RAxML using the PROTGAMMALG4X substitution model, and then the underlying species tree was inferred by ASTRAL, summarizing all gene trees representing the respective matrices. Local posterior probabilities were calculated as an approximation for node support.
3. Results and discussion
(a). Influence of taxon-specific matrices and analytical methods on inferred relationships
Selecting suitable orthogroups to assemble matrices is a crucial step for phylogenomic approaches to balance the number of included inferred orthogroups with the amount of missing data and potential noise. Common approaches for selecting or excluding orthogroups are occupancy thresholds, measures of evolutionary rates, decisiveness and compositional heterogeneity, among others (e.g. [9,18,21,44–47]). The goal of these approaches is to exclude those orthogroups with either too few representatives, too much noise or other artefacts. Taxon-specific matrices represent a very different approach (see also [7]). They aim at maximizing information content for a specific taxon, which is relatively well represented in terms of species numbers in the overall dataset.
With fewer and more closely related species being targeted, taxon-specific matrices increase the information content compared to the full dataset when identical occupancy cut-offs are employed. For example, the tetraconatan-wide matrix 2 and the malacostracan-specific matrix 6 had similar occupancy thresholds at 66 and 64% (table 1), respectively. However, matrix 6 featured an additional 45 orthogroups and the amount of missing data at the orthogroup level was reduced from 31.5% (average for malacostracans in matrix 2) to 26.2%. For Branchiopoda, the positive effect was even higher, featuring more than seven times the number of orthogroups present in matrix 1, which has a similar occupancy threshold. That these matrices improve the resolution for the targeted taxa is clearly shown for the malacostracan-specific matrix (the effects of the branchiopod-specific matrix are less pronounced because conflict was lower for Branchiopoda in matrices 1–5). All analyses of the malacostracan-specific matrix 6 consistently recovered several crucial clades (e.g. Caridoida, Peracarida or Pleocyemata), with little to no conflict among analytical methods (figure 2). By contrast, analyses of the tetraconatan-wide matrices 1–5 showed considerable variation when analysed with different methods.
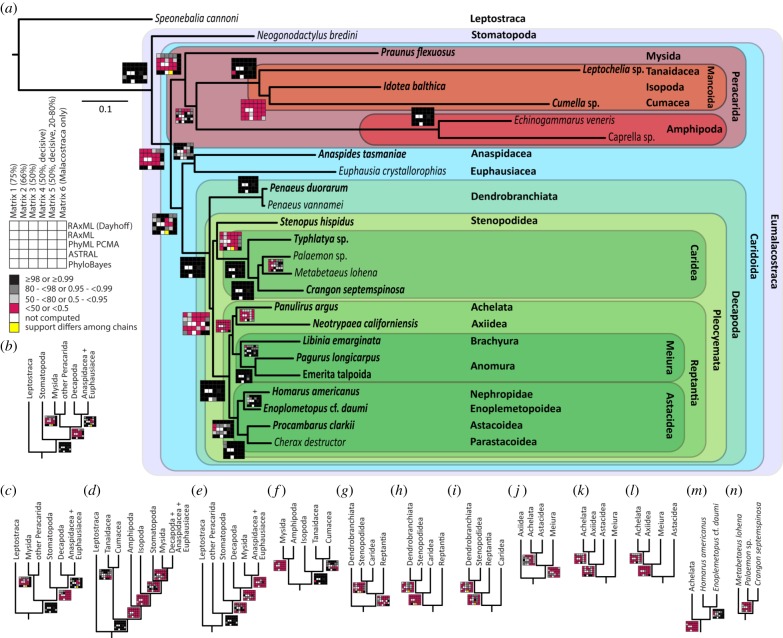
Figure 2.
Phylogenetic relationships within Malacostraca. (a) Topology based on the PhyloBayes CAT-GTR analysis of the malacostracan-specific matrix 6. Support values for all analyses are depicted as rug plots. Nodes supported in only one of two PhyloBayes chains are marked in yellow (the two PhyloBayes chains did not converge for these nodes). Individuals newly sequenced for this study are highlighted in bold. All alternative topologies with support greater than 50 or 0.5 in any analysis are shown: (b–e) position of Stomatopoda and Mysida, (f) relationships within Peracarida, (g–i) Decapoda, (j–m) Reptantia and Astacidea and (n) Caridea. (Online version in colour.)
The taxon-specific matrices may also aid in determining which of the employed analytical approaches were best suited to resolve certain nodes. Although all approaches supported monophyletic Peracarida and Caridoida under the malacostracan-specific matrix 6—a result consistent with morphological studies [48,49], only CAT-GTR (PhyloBayes) and Dayhoff recoding also supported these clades under matrices 1–5. All other approaches supported alternative topologies under these tetraconatan-wide matrices (figure 2). CAT-GTR and Dayhoff recoding reduce artefacts associated with compositional heterogeneity and long-branch attraction [8,35,36,40,42] and, therefore, might have fared better also under the tetraconatan-wide matrices. However, CAT-GTR and Dayhoff recoding also show crucial discrepancies in certain nodes (e.g. regarding relationships within Decapoda; figure 2) and even between certain CAT-GTR chains (see Discussion on the position of Copepoda below).
Taken together, our analyses underscore that taxon-specific matrices increase overall occupancy, total gene number and resolution for the respective taxon. They also highlight the need for phylogenomic studies to analyse multiple matrices using different analytical methods and models. Not all nodes are equally resolved under each model or with each matrix and problematic nodes can be identified only by comparing multiple analyses, though CAT-GTR and Dayhoff recoding appeared to best suited to resolve problematic nodes. When combined with fast maximum-likelihood algorithms like RAxML, Dayhoff recoding has a much lower computational footprint than CAT-GTR in PhyloBayes. Dayhoff recoding can also be combined with CAT-GTR, reducing site and taxon-specific compositional heterogeneity simultaneously [36].
(b). Phylogenetic relationships of tetraconatan taxa
The heterogeneity of proposed tetraconatan phylogenetic relationships among recent phylogenomic studies (e.g. [4,5,7,9]) is also reflected in our analyses, despite the greatly improved taxon and gene sampling. Overall, Oligostraca, Altocrustacea, Multicrustacea, Communostraca (Malacostraca and Thecostraca), Malacostraca, Thecostraca, Copepoda, Branchiopoda, Cephalocarida, Hexapoda and Remipedia are well supported by our analyses (e.g. [4,5,7,9]) (we use the term ‘well supported’ only for clades with bootstrap support of ≥98 or posterior probabilities ≥0.99 in more than 50% of all analyses and which were rejected in not more than two analyses and not more than one analysis of CAT-GTR, Dayhoff recoding or of matrices 6 and 7, if applicable).
The monophyly of Multicrustacea and of Allotriocarida was supported in several analyses, but in others Cephalocarida clustered with Malacostraca and Thecostraca, while Copepoda clustered within Allotriocarida in most analyses with CAT-GTR (figure 1). Notable are the CAT-GTR analyses of matrices 4 and 5: in one chain, Multicrustacea and Allotriocarida were monophyletic with full support, whereas Copepoda clustered within Allotriocarida with full support in the other chain (the chains of these PhyloBayes analyses did not converge owing to the position of Copepoda). The latter was also observed by Rota-Stabelli et al. [8] in analyses of CAT-GTR and Dayhoff recoding, which they employed to reduce codon usage biases among taxa [8,35]. Our overall strategy to analyse amino acid matrices should already reduce codon usage biases and our analyses using Dayhoff recoding recovered monophyletic Allotriocarida and Multicrustacea. Consequently, we assume monophyletic Multicrustacea and Allotriocarida, and suggest that the non-monophyly of Multicrustacea in some CAT-GTR chains represents an artefact of CAT-GTR, but not necessarily related to codon usage biases. This issue remains to be further tested.
Phylogenetic relationships within Allotriocarida were highly dependent on the employed method and matrix, disregarding the problematic placement of Copepoda. Most analyses recovered either a clade comprising Remipedia and Cephalocarida (Xenocarida) (Miracrustacea; e.g. RAxML analyses of amino acid matrices and PhyML; see also [4,5]) or Remipedia (Labiocarida; e.g. ASTRAL, CAT-GTR with PhyloBayes and RAxML with Dayhoff recoding; see also [7,16,19]) as the sister group to Hexapoda (figure 1; electronic supplementary material, table S2). A similar pattern was observed by Schwentner et al. [9], who attributed these to the respective analyses' power to resolve artefacts like compositional heterogeneity and long-branch attraction (see also [8,35]) and, therefore, rejected Xenocarida as an artefact of compositional heterogeneity and long-branch attraction. Our results are consistent with those findings and interpretations.
Our phylogenetic analyses lend further support to studies that suggested non-monophyly of Ostracoda [48,49], with the Podocopa species (Paranesidea sp.) clustering either with the mystacocarid Derocheilocaris remanei or with the branchiuran Argulus siamensis (figure 1). However, this interpretation remains tentative owing to the overall long branches and low support within Oligostraca. Other phylogenomic studies presented similar results [4,9], and only Oakley et al. [7], who included the largest number of Ostracoda (based mostly on 454-based transcriptomes and thus not included here), recovered monophyletic Ostracoda, but not in analyses that included other tetraconatan representatives. Whether the higher occupancy of our matrices or the denser taxon sampling of Oakley et al. [7] correctly resolved the relationships within Oligostraca remains open. Resolving Oligostraca would benefit from denser taxon sampling and targeted data matrices, as done here for Malacostraca and Branchiopoda.
(c). Phylogenetic relationships within Malacostraca
The current malacostracan classification is largely based on Calman [50,51], who classified Leptostraca as malacostracans and separated four major Eumalacostraca taxa (sensu [52]): Hoplocarida (including Stomatopoda, represented in our analyses by the crown-group Verunipeltata), Syncarida (Bathynellacea and Anaspidacea), Eucarida (Euphausiacea and Decapoda) and Peracarida (Mysida, Amphipoda, Isopoda and allies). Calman rejected the taxon Schizopoda, proposed by Boas [53] and Thomson [54], which comprised Euphausiacea and Mysida and, after the discovery of Anaspides tasmaniae in 1893, Anaspidacea.
The relationships between Calman's eumalacostracan taxa were repeatedly discussed, but the general ‘morphological consensus' was that Hoplocarida is the sister group to the remaining three taxa, collectively known as Caridoida [55–57]. Morphological support for Peracarida is strong [55,58,59]. Most molecular studies so far, however, do not support the monophyly of Peracarida, proposing a closer relationship of Mysida with various combinations of non-peracarid Eumalacostraca [60–64] with the exception of one multilocus study [65]. Our analyses support monophyletic Eumalacostraca (figure 2), but monophyly of Caridoida and Peracarida was not supported by alternative positions of Stomatopoda and Mysida, respectively, in all analyses of ASTRAL, PhyML and RAxML with amino acid matrices of matrices 1–5 (figure 2a–e). Here, Mysida and Stomatopoda largely clustered with Euphausiacea, Anaspidacea and Decapoda. However, Caridoida and Peracarida were monophyletic in all analyses of CAT-GTR, several analyses employing Dayhoff recoding and all analyses with all methods of the malacostracan-specific matrix 6 (figure 2a–e). Based on the latter analyses, especially those employing the malacostracan-specific matrix 6, we propose that Caridoida and Peracarida are monophyletic (see also [55,58,66]) (see below for further discussion).
Within Peracarida, the clade Mancoida (Cumacea, Isopoda and Tanaidacea) is well supported (e.g. [55,59]), rejecting Edriophthalmata (Amphipoda and Isopoda) (e.g. suggested in [66]) (figure 2a,f). The sister group relationship between Cumacea and Tanaidacea, recovered in most of our analyses (figure 2f), contradicts the traditional Tanaidacea–Isopoda clade [55,56], which was not recovered in any of our analyses.
Monophyly of Eucarida was questioned by Richter [67] who suggested a clade of Anaspidacea, Euphausiacea and Peracarida (Xenommacarida; based on the structure of the ommatidia). Our analyses suggest non-monophyly of Eucarida and Xenommacarida, because Euphausiacea clusters consistently with Anaspidacea, but without any member of the Peracarida (figure 2a). Decapoda is potentially the sister group of Anaspidacea and Euphausiacea. Monophyly of Syncarida was not tested because Bathynellacea were not available for transcriptome sequencing.
Decapod monophyly is well supported (figure 2a–f). Timm & Bracken-Grissom [68] nicely summarized the numerous relationships recently proposed for decapod taxa, but our results do not resolve all of these outstanding conflicts. The relationships among Dendrobranchiata, Stenopodidea, Caridea and Reptantia remain unresolved (figure 2a,g–i), though most analyses of the malacostracan-specific matrix 6 supported a sister group relationship between Caridea and Stenopodidea (see also [69,70]), with Reptantia as their closest relative (figure 2g). The latter implies monophyletic Pleocyemata (e.g. [55,71]), but is only poorly corroborated by analyses of other matrices. Within Reptantia, Astacidea and Meiura are well-supported clades.
Published molecular phylogenetic analyses covering most malacostracan taxa–even based on one or a few genes– are rare, and phylogenomic approaches included a handful of malacostracan taxa at most. Despite ours being, to our knowledge, the most speciose phylogenomic study to date, several long-standing conflicting phylogenetic hypotheses could not be resolved. The malacostracan-specific matrix appears best suited to resolve at least the backbone of malacostracan relationships. The observed discrepancies among analyses might be attributed to the relatively low representation of some malacostracan species in OMA-derived orthogroups selected for our matrices (electronic supplementary material, table S3). However, the number of contigs per species fed into OMA for Malacostraca and Branchiopoda was similar (electronic supplementary material, table S2), suggesting that the low representation is not based on reduced quality or diversity of the underlying transcriptomes. Potentially, gene duplication events or increased mutation rates in some taxa may have negatively affected orthogroup assignment with OMA.
(d). Phylogenetic relationships within Branchiopoda
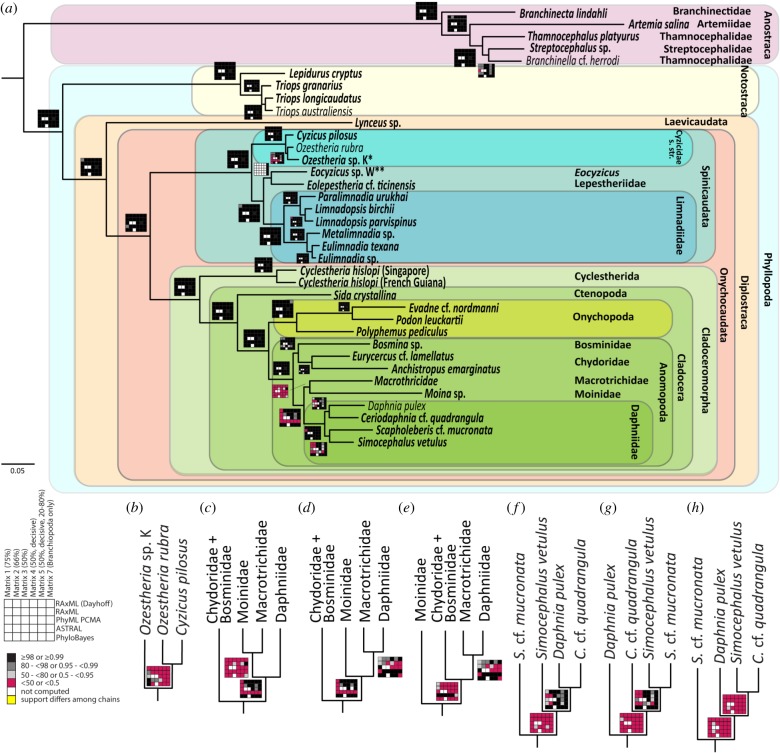
A new era of branchiopod phylogeny started with Fryer's [72] ‘flat’ classification, with the extant Branchiopoda subdivided into eight ‘orders’ (Anostraca, Notostraca, Laevicaudata, Spinicaudata, Anomopoda, Ctenopoda, Haplopoda and Onychopoda). The monophyly of most of Fryer's [72] taxa has later been confirmed [73–76], with the exception of Spinicaudata (as introduced by Linder [77]) from which the aberrant clam shrimp family Cyclestheridae was later removed (see [78]), in agreement with our results (figure 3). Furthermore, Anostraca are well supported as the sister taxon to all other Branchiopoda (Phyllopoda; [75,81,82]) and within Phyllopoda, Notostraca is the sister taxon to a monophyletic Diplostraca, which includes the three ‘clam shrimp’ taxa Laevicaudata, Spinicaudata and Cyclestherida as well as the water fleas (Cladocera) ([75] (morphology only), [81–83]). The sister group relationship of Cyclestherida and Cladocera (Cladoceromorpha) has been well accepted (e.g. [84]) and is well supported in our analyses. With the overall congruence between our detailed phylogenomic analyses, and morphological as well as previous molecular studies, the backbone phylogeny of Branchiopoda can now be considered resolved.
Figure 3.
Phylogenetic relationships within Branchiopoda. (a) Topology based on the PhyloBayes CAT-GTR analysis of the branchiopod-specific matrix 7. Support values are depicted as rug plots. Eoleptestheria cf. ticinensis was included in matrix 7 only. Individuals newly sequenced for this study are highlighted in bold. All alternative topologies with support greater than 50 or 0.5 in any analysis are shown: (b) Ozestheria, (c–e) Anomopoda and (f–h) Daphniidae. Asterisk following Schwentner et al. [79]; double asterisks following Schwentner et al. [80]. (Online version in colour.)
Paraphyly of Cyzicidae has been previously reported [85,86] and is now well supported (figure 3). Eocyzicus is more closely related to Leptestheriidae and Limnadiidae than to remaining Cyzicidae, with the latter being the sister group to all other extant Spinicaudata. This contrasts with previous morphology-based hypotheses of a close relationship of Leptestheriidae and Cyzicidae [87,88].
The cladoceran taxa Ctenopoda, Anomopoda and Onychopoda are monophyletic in most studies (e.g. [74–76]) and further supported here (a single species of Ctenopoda was included, and thus not tested). Ctenopoda is the sister taxon to all other cladocerans (e.g. [89]), a position not recovered by most previous studies [73–76,87,90,91]. Unfortunately, our analyses lack Leptodora kindtii (the only representative of Haplopoda), so we were not able to test whether it is the sister group to Onychopoda (Gymnomera; e.g. [89]) or the remaining Cladocera. Within Anomopoda, a clade comprising Daphniidae and Macrotrichidae is best supported with Moinidae as their sister taxon (figure 3).
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank two anonymous reviewers for their helpful and constructive comments. We acknowledge the FAS Bauer Core and Research Computing Group for their support and technical assistance, including the usage of the Odyssey cluster. We thank Zachary Folz, the late George Hampson, Georg Brenneis, Simone Bourgeois (New Bedford Sea Laboratory) and Alissar Langworthy for their assistance in collecting animals, and David J. Combosch and Rosa Fernández for their insights in analysing the data.
Data accessibility
All resulting matrices, tree files, assemblies with Trinity, orthologues and custom python and perl scripts are avalaible in the Dryad Digital Repository: https://doi:10.5061/dryad.sn35910 [92] and Harvard Dataverse (https://dataverse.harvard.edu/dataverse/Crustacean_Phylogeny). Raw reads were deposited at GenBank SRA (SAMN06174118–SAMN06174176; see the electronic supplementary material, table S1 for details).
Authors' contribution
M.S. carried out laboratory work, analyses and drafted the manuscript. M.S., G.G. and S.R. conceived and designed the study. All authors collected specimens, contributed to and approved the final version of the manuscript.
Competing interests
We declare that we have no competing interests.
Funding
This work was supported by a postdoctoral stipend of the German Science Foundation (Deutsche Forschungsgemeinschaft; DFG SCHW 1810/1-1 to M.S.), the Fenner Chase Fund of the Museum of Comparative Zoology (to M.S.) and internal funds from the Museum of Comparative Zoology and the Faculty of Arts and Sciences to G.G.
References
- 1.Richter S, Møller OS, Wirkner CS. 2009. Advances in crustacean phylogenetics. Arthropod Syst. Phylog. 67, 275–286. [Google Scholar]
- 2.Jenner RA. 2010. Higher-level crustacean phylogeny: consensus and conflicting hypotheses. Arthropod Struct. Dev. 39, 143–153. ( 10.1016/j.asd.2009.11.001) [DOI] [PubMed] [Google Scholar]
- 3.Giribet G, Edgecombe GD. 2013. The Arthropoda: a phylogenetic framework. In Arthropod biology and evolution (eds Minelli A, Boxshall G, Fusco G), pp. 17–40. Berlin, Germany: Springer. [Google Scholar]
- 4.Regier JC, Shultz JW, Zwick A, Hussey A, Ball B, Wetzer R, Martin JW, Cunningham CW. 2010. Arthropod relationships revealed by phylogenomic analysis of nuclear protein-coding sequences. Nature 463, 1079–1083. ( 10.1038/nature08742) [DOI] [PubMed] [Google Scholar]
- 5.Lee MSY, Soubrier J, Edgecombe GD. 2013. Rates of phenotypic and genomic evolution during the Cambrian explosion. Curr. Biol. 23, 1–7. ( 10.1016/j.cub.2013.07.055) [DOI] [PubMed] [Google Scholar]
- 6.Legg DA, Sutton MD, Edgecombe GD. 2013. Arthropod fossil data increase congruence of morphological and molecular phylogenies. Nat. Commun. 4, 2485 ( 10.1038/ncomms3485) [DOI] [PubMed] [Google Scholar]
- 7.Oakley TH, Wolfe JM, Lindgren AR, Zaharoff AK. 2013. Phylotranscriptomics to bring the understudied into the fold: monophyletic Ostracoda, fossil placement, and pancrustacean phylogeny. Mol. Biol. Evol. 30, 215–233. ( 10.1093/molbev/mss216) [DOI] [PubMed] [Google Scholar]
- 8.Rota-Stabelli O, Lartillot N, Philippe H, Pisani D. 2013. Serine codon-usage bias in deep phylogenomics: pancrustacean relationships as a case study. Syst. Biol. 62, 121–133. ( 10.1093/sysbio/sys077) [DOI] [PubMed] [Google Scholar]
- 9.Schwentner M, Combosch DJ, Nelson JP, Giribet G. 2017. A phylogenomic solution to the origin of insects by resolving crustacean-hexapod relationships. Curr. Biol. 27, 1818–1824. ( 10.1016/j.cub.2017.05.040) [DOI] [PubMed] [Google Scholar]
- 10.Giribet G, Richter S, Edgecombe GD, Wheeler WC. 2005. The position of crustaceans within the Arthropoda: evidence from nine molecular loci and morphology. In Crustacean issues 16: crustacea and arthropod relationships. Festschrift for Frederick R. Schram (eds. Koenemann S, Jenner RA), pp. 307–352. Boca Raton, FL: Taylor & Francis. [Google Scholar]
- 11.Richter S. 2002. The Tetraconata concept: hexapod-crustacean relationships and the phylogeny of Crustacea. Org. Divers. Evol. 2, 217–237. [Google Scholar]
- 12.Edgecombe GD. 2010. Arthropod phylogeny: an overview from the perspectives of morphology, molecular data and the fossil record. Arthropod Struct. Dev. 39, 74–87. ( 10.1016/j.asd.2009.10.002) [DOI] [PubMed] [Google Scholar]
- 13.Meusemann K, et al. 2010. A phylogenomic approach to resolve the arthropod tree of life. Mol. Biol. Evol. 27, 2451–2464. ( 10.1093/molbev/msq130) [DOI] [PubMed] [Google Scholar]
- 14.Andrew DR. 2011. A new view of insect–crustacean relationships II. Inferences from expressed sequence tags and comparisons with neural cladistics. Arthropod Struct. Dev. 40, 289–302. ( 10.1016/j.asd.2011.02.001) [DOI] [PubMed] [Google Scholar]
- 15.Rota-Stabelli O, Campbell L, Brinkmann H, Edgecombe GD, Longhorn SJ, Peterson KJ, Pisani D, Philippe H, Telford MJ. 2011. A congruent solution to arthropod phylogeny: phylogenomics, microRNAs and morphology support monophyletic Mandibulata. Proc. R. Soc. B 278, 298–306. ( 10.1098/rspb.2010.0590) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.von Reumont BM, et al. 2012. Pancrustacean phylogeny in the light of new phylogenomic data: support for Remipedia as the possible sister group of Hexapoda. Mol. Biol. Evol. 29, 1031–1045. ( 10.1093/molbev/msr270) [DOI] [PubMed] [Google Scholar]
- 17.Borner J, Rehm P, Schill RO, Ebersberger I, Burmester T. 2014. A transcriptome approach to ecdysozoan phylogeny. Mol. Phylogenet. Evol. 80, 79–87. ( 10.1016/j.ympev.2014.08.001) [DOI] [PubMed] [Google Scholar]
- 18.Dell'Ampio E, et al. 2014. Decisive data sets in phylogenomics: lessons from studies on the phylogenetic relationships of primarily wingless insects. Mol. Biol. Evol. 31, 239–249. ( 10.1093/molbev/mst196) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Misof B, et al. 2014. Phylogenomics resolves the timing and pattern of insect evolution. Science 346, 763–767. ( 10.1126/science.1257570) [DOI] [PubMed] [Google Scholar]
- 20.Lozano-Fernandez J, et al. 2016. A molecular palaeobiological exploration of arthropod terrestrialisation. Phil. Trans. R. Soc. B 371, 20150133 ( 10.1098/rstb.2015.0133) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma PP, Kaluziak S, Pérez-Porro AR, González VL, Hormiga G, Wheeler WC, Giribet G. 2014. Phylogenomic interrogation of Arachnida reveals systemic conflicts in phylogenetic signal. Mol. Biol. Evol. 31, 2963–2984. ( 10.1093/molbev/msu235) [DOI] [PubMed] [Google Scholar]
- 22.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. ( 10.1038/nmeth.1923) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. ( 10.1038/Nbt.1883) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haas BJ, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protocols 8, 1494–1512. ( 10.1038/nprot.2013.084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu LM, Niu BF, Zhu ZW, Wu ST, Li WZ. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152. ( 10.1093/Bioinformatics/Bts565) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altenhoff AM, Gil M, Gonnet GH, Dessimoz C. 2013. Inferring hierarchical orthologous groups from orthologous gene pairs. PLoS ONE 8, e53786 ( 10.1371/journal.pone.0053786) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altenhoff AM, et al. 2016. Standardized benchmarking in the quest for orthologs. Nat. Methods 13, 425–430. ( 10.1038/nmeth.3830) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kück P, Struck TH. 2014. BaCoCa: a heuristic software tool for the parallel assessment of sequence biases in hundreds of gene and taxon partitions. Mol. Phylogenet. Evol. 70, 94–98. ( 10.1016/j.ympev.2013.09.011) [DOI] [PubMed] [Google Scholar]
- 30.Wu M, Chatterji S, Eisen JA. 2012. Accounting for alignment uncertainty in phylogenomics. PLoS ONE 7, e30288 ( 10.1371/journal.pone.0030288) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hejnol A, et al. 2009. Assessing the root of bilaterian animals with scalable phylogenomic methods. Proc. R. Soc. B 276, 4261–4270. ( 10.1098/rspb.2009.0896) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. ( 10.1093/bioinformatics/btp348) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. ( 10.1093/bioinformatics/btu033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees. In Proc. of the Gateway Computing Environments Workshop (GCE), pp. 1–8. New Orleans. [Google Scholar]
- 35.Susko E, Roger AJ. 2007. On reduced amino acid alphabets for phylogenetic inference. Mol. Biol. Evol. 24, 2139–2150. ( 10.1093/molbev/msm144) [DOI] [PubMed] [Google Scholar]
- 36.Feuda R, Dohrmann M, Pett W, Philippe H, Rota-Stabelli O, Lartillot N, Wörheide G, Pisani D. 2017. Improved modeling of compositional heterogeneity supports sponges as sister to all other animals. Curr. Biol. 27, 3864–3870. ( 10.1016/j.cub.2017.11.008) [DOI] [PubMed] [Google Scholar]
- 37.Zoller S, Schneider A. 2013. Improving phylogenetic inference with a semiempirical amino acid substitution model. Mol. Biol. Evol. 30, 469–479. ( 10.1093/molbev/mss229) [DOI] [PubMed] [Google Scholar]
- 38.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. ( 10.1093/sysbio/syq010) [DOI] [PubMed] [Google Scholar]
- 39.Lartillot N, Rodrigue N, Stubbs D, Richer J. 2013. PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst. Biol. 62, 611–615. ( 10.1093/Sysbio/Syt022) [DOI] [PubMed] [Google Scholar]
- 40.Lartillot N, Philippe H. 2004. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol. Biol. Evol. 21, 1095–1109. ( 10.1093/molbev/msh112) [DOI] [PubMed] [Google Scholar]
- 41.Lartillot N, Brinkmann H, Philippe H. 2007. Suppression of long-branch attraction artefacts in the animal phylogeny using a site-heterogeneous model. BMC Evol. Biol. 7 (Suppl 1), S4 ( 10.1186/1471-2148-7-S1-S4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whelan NV, Halanych KM. 2017. Who let the CAT out of the bag? Accurately dealing with substitutional heterogeneity in phylogenomic analyses. Syst. Biol. 66, 232–255. ( 10.1093/sysbio/syw084) [DOI] [PubMed] [Google Scholar]
- 43.Mirarab S, Reaz R, Bayzid MS, Zimmermann T, Swenson MS, Warnow T. 2014. ASTRAL: genome-scale coalescent-based species tree estimation. Bioinformatics 30, i541–i548. ( 10.1093/bioinformatics/btu462) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fernández R, Edgecombe GD, Giribet G. 2016. Exploring phylogenetic relationships within Myriapoda and the effects of matrix composition and occupancy on phylogenomic reconstruction. Syst. Biol. 65, 871–889. ( 10.1093/sysbio/syw041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fernández R, Hormiga G, Giribet G. 2014. Phylogenomic analysis of spiders reveals nonmonophyly of orb weavers. Curr. Biol. 24, 1772–1777. ( 10.1016/j.cub.2014.06.035) [DOI] [PubMed] [Google Scholar]
- 46.Laumer CE, et al. 2015. Spiralian phylogeny informs the evolution of microscopic lineages. Curr. Biol. 25, 2000–2006. ( 10.1016/j.cub.2015.06.068) [DOI] [PubMed] [Google Scholar]
- 47.Kocot KM, et al. 2017. Phylogenomics of Lophotrochozoa with consideration of systematic error. Syst. Biol. 66, 256–282. ( 10.1093/sysbio/syw079) [DOI] [PubMed] [Google Scholar]
- 48.Wakayama N. 2007. Embryonic development clarifies polyphyly in ostracod crustaceans. J. Zool. 273, 406–413. ( 10.1111/j.1469-7998.2007.00344.x) [DOI] [Google Scholar]
- 49.Yamaguchi S, Endo K. 2003. Molecular phylogeny of Ostracoda (Crustacea) inferred from 18S ribosomal DNA sequences: implication for its origin and diversification. Mar. Biol. 143, 23–38. ( 10.1007/s00227-003-1062-3) [DOI] [Google Scholar]
- 50.Calman WT. 1904. On the classification of the Crustacea Malacostraca. Ann. Nat. Hist. 7, 144–158. [Google Scholar]
- 51.Calman WT. 1909. A Treatise on Zoology, 7: Appendiculata, Crustacea. London, UK: Adam and Charles Black. [Google Scholar]
- 52.Grobben K. 1892. Zur Kenntnis des Stammbaumes und des Systems der Crustaceen. Sitzungsberichte Kaiserlichen Akademie Wissenschaften Wien 101, 237–274. [Google Scholar]
- 53.Boas JEV. 1882. Studien über die Verwandtschaftsbeziehungen der Malakostraken. Morphol. Jahr. 8, 485–579. [Google Scholar]
- 54.Thomson GM. 1895. On a freshwater Schizopod from Tasmania. Trans. Linn. Soc. Lond. 6, 285–303. [Google Scholar]
- 55.Richter S, Scholtz G. 2001. Phylogenetic analysis of the Malacostraca (Crustacea). J. Zool. Syst. Evol. Res. 39, 113–136. [Google Scholar]
- 56.Siewing R. 1956. Untersuchungen zur Morphologie der Malacostraca (Crustacea). Zool. Jb. Anat. 75, 39–176. [Google Scholar]
- 57.Hessler RR. 1983. A defense of the caridoid facies: wherein the early evolution of the Eumalacostraca is discussed. In Crustacean phylogeny, crustacean issues, Bd 1 (ed. Schram FR.), pp. 145–164. Rotterdam, The Netherlands: A. A. Balkema. [Google Scholar]
- 58.Wirkner CS, Richter S. 2010. Evolutionary morphology of the circulatory system in Peracarida (Malacostraca; Crustacea). Cladistics 26, 143–167. [DOI] [PubMed] [Google Scholar]
- 59.Wilson GDF. 2009. The phylogenetic position of the Isopoda in the Peracarida (Crustacea: Malacostraca). Arthropod Syst. Phylog. 67, 159–198. [Google Scholar]
- 60.Jenner RA, Dhubhghaill CN, Ferla MP, Wills MA. 2009. Eumalacostracan phylogeny and total evidence: limitations of the usual suspects. BMC Evol. Biol. 9, 21 ( 10.1186/1471-2148-9-21) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meland K, Willassen E. 2007. The disunity of ‘Mysidacea’ (Crustacea). Mol. Phylogenet. Evol. 44, 1083–1104. ( 10.1016/j.ympev.2007.02.009) [DOI] [PubMed] [Google Scholar]
- 62.Spears TR, DeBry W, Abele LG, Chodyla K. 2005. Peracarid monophyly and interordinal phylogeny inferred from nuclear small-subunit ribosomal DNA sequences (Crustacea: Malacostraca: Peracarida). Proc. Biol. Soc. Wash. 118, 117–157. [Google Scholar]
- 63.Watling L. 1999. Toward understanding the relationships of the peracaridan orders: the necessity of determining exact homologies. In Crustaceans and the biodiversity crisis. Proc. of the Fourth Int. Crustacean Congress, Amsterdam, The Netherlands, 20–24 July1998, vol. 1 (eds Schram FR, von Vaupel Klein JC), pp. 73–89. Leiden, The Netherlands: Brill. [Google Scholar]
- 64.Jarman SN, Nicol S, Elliott NG, McMinn A. 2000. 28S rDNA evolution in the Eumalacostraca and the phylogenetic position of krill. Mol. Phylogenet. Evol. 17, 26–36. [DOI] [PubMed] [Google Scholar]
- 65.Bybee SM, Bracken-Grissom H, Haynes BD, Hermansen RA, Byers RL, Clement MJ, Udall JA, Wilcox ER, Crandall KA. 2011. Targeted amplicon sequencing (TAS): a scalable next-gen approach to multilocus, multitaxa phylogenetics. Genome Biol. Evol. 3, 1312–1323. ( 10.1093/Gbe/Evr106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Poore GCB. 2005. Peracarida: monophyly, relationships and evolutionary success. Nauplius 13, 1–27. [Google Scholar]
- 67.Richter S. 1999. The structure of the ommatidia of the Malacostraca (Crustacea): a phylogenetic approach. Verhandlungen des naturwissenschaftlichen Vereins in Hamburg 38, 161–204. [Google Scholar]
- 68.Timm L, Bracken-Grissom HD. 2015. The forest for the trees: evaluating molecular phylogenies with an emphasis on higher-level Decapoda. J. Crustacean Biol. 35, 577–592. ( 10.1163/1937240x-00002371) [DOI] [Google Scholar]
- 69.Dixon CJ, Ahyong ST, Schram FR. 2003. A new hypothesis of decapod phylogeny. Crustaceana 76, 935–975. [Google Scholar]
- 70.Tsang LM, Lin F-J, Chu KH, Chan T-Y. 2008. Phylogeny of Thalassinidea (Crustacea, Decapoda) inferred from three rDNA sequences: implications for morphological evolution and superfamily classification. J. Zool. Syst. Evol. Res. 46, 216–223. [Google Scholar]
- 71.Shen H, Braband A, Scholtz G. 2013. Mitogenomic analysis of decapod crustacean phylogeny corroborates traditional views on their relationships. Mol. Phylogenet. Evol. 66, 776–789. [DOI] [PubMed] [Google Scholar]
- 72.Fryer G. 1987. A new classification of the branchiopod Crustacea. Zool. J. Linn. Soc. 91, 357–383. [Google Scholar]
- 73.Braband A, Richter S, Hiesel R, Scholtz G. 2002. Phylogenetic relationships within the Phyllopoda (Crustacea, Branchiopoda) based on mitochondrial and nuclear markers. Mol. Phylogenet. Evol. 25, 229–244. [DOI] [PubMed] [Google Scholar]
- 74.Stenderup JT, Olesen J, Glenner H. 2006. Molecular phylogeny of the Branchiopoda (Crustacea): multiple approaches suggest a 'diplostracan' ancestry of the Notostraca. Mol. Phylogenet. Evol. 41, 182–194. [DOI] [PubMed] [Google Scholar]
- 75.Richter S, Olesen J, Wheeler WC. 2007. Phylogeny of Branchiopoda (Crustacea) based on a combined analysis of morphological data and six molecular loci. Cladistics 23, 301–336. ( 10.1111/j.1096-0031.2007.00148.x) [DOI] [PubMed] [Google Scholar]
- 76.de Waard JR, Sacherova V, Cristescu ME, Remigio EA, Crease TJ, Hebert PDN. 2006. Probing the relationships of the branchiopod crustaceans. Mol. Phylogenet. Evol. 39, 491–502. [DOI] [PubMed] [Google Scholar]
- 77.Linder F. 1945. Affinities within the Branchiopoda with notes on some dubious fossils. Arkiv för Zoologi 37A, 1–28. [Google Scholar]
- 78.Martin JW, Davis GE. 2001. An updated classification of the recent Crustacea. Contrib. Sci. 39, 1–124. [Google Scholar]
- 79.Schwentner M, Just F, Richter S. 2015. Evolutionary systematics of the Australian Cyzicidae (Crustacea, Branchiopoda, Spinicaudata) with the description of a new genus. Zool. J. Linn. Soc. 173, 271–295. ( 10.1111/zoj.12209) [DOI] [Google Scholar]
- 80.Schwentner M, Timms BV, Richter S. 2014. Evolutionary systematics of the Australian Eocyzicus fauna (Crustacea: Branchiopoda: Spinicaudata) reveals hidden diversity and phylogeographic structure. J. Zool. Syst. Evol. Res. 52, 15–31. [Google Scholar]
- 81.Olesen J. 2007. Monophyly and phylogeny of Branchiopoda, with focus on morphology and homologies of branchiopod phyllopodous limbs. J. Crustacean Biol. 27, 165–183. [Google Scholar]
- 82.Olesen J. 2009. Phylogeny of Branchiopoda (Crustacea): character evolution and contribution of uniquely preserved fossils. Arthropod Syst. Phylog. 67, 3–39. [Google Scholar]
- 83.Olesen J, Richter S. 2013. Onychocaudata (Branchiopoda: Diplostraca), a new high-level taxon in branchiopod systematics. J. Crustacean Biol. 33, 62–65. [Google Scholar]
- 84.Ax P. 1999. Das System der Metazoa II. Ein Lehrbuch der phylogenetischen Systematik. Stuttgart: Gustav Fischer Verlag. [Google Scholar]
- 85.Weeks SC, Chapman EG, Rogers DC, Senyo DM, Hoeh WR. 2009. Evolutionary transitions among dioecy, androdioecy and hermaphroditism in limnadiid clam shrimp (Branchiopoda: Spinicaudata). J. Evol. Biol. 22, 1781–1799. [DOI] [PubMed] [Google Scholar]
- 86.Schwentner M, Timms BV, Bastrop R, Richter S. 2009. Phylogeny of Spinicaudata (Branchiopoda, Crustacea) based on three molecular markers: an Australian origin for Limnadopsis. Mol. Phylogenet. Evol. 53, 716–725. [DOI] [PubMed] [Google Scholar]
- 87.Olesen J. 1998. A phylogenetic analysis of the Conchostraca and Cladocera (Crustacea, Branchiopoda, Diplostraca). Zool. J. Linn. Soc. 122, 491–536. [Google Scholar]
- 88.Astrop TI, Hegna TA. 2015. Phylogenetic relationships between living and fossil spinicaudatan taxa (Branchiopoda Spinicaudata): reconsidering the evidence. J. Crustacean Biol. 35, 339–354. [Google Scholar]
- 89.Martin JW, Cash-Clark CE. 1995. The external morphology of the onychopod ‘cladoceran’ genus Bythotrephes (Crustacea, Branchiopoda, Onychopoda, Cercopagididae), with notes on the morphology and phylogeny of the order Onychopoda. Zool. Scr. 24, 61–90. [Google Scholar]
- 90.Taylor DJ, Crease TJ, Brown WM. 1999. Phylogenetic evidence for a single long-lived clade of crustacean cyclic parthenogens and its implications for the evolution of sex. Proc. R. Soc. Lond. B 266, 791–797. [Google Scholar]
- 91.Olesen J. 2000. An updated phylogeny of the Conchostraca–Cladocera clade (Branchiopoda, Diplostraca). Crustaceana 73, 869–886. [Google Scholar]
- 92.Schwentner M, Richter S, Rogers DC, Giribet G. 2018. Data from: Tetraconatan phylogeny with specific focus on Malacostraca and Branchiopoda: highlighting the strength of taxon-specific matrices in phylogenomics Dryad Digital Repository. ( 10.5061/dryad.sn35910) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Schwentner M, Richter S, Rogers DC, Giribet G. 2018. Data from: Tetraconatan phylogeny with specific focus on Malacostraca and Branchiopoda: highlighting the strength of taxon-specific matrices in phylogenomics Dryad Digital Repository. ( 10.5061/dryad.sn35910) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
All resulting matrices, tree files, assemblies with Trinity, orthologues and custom python and perl scripts are avalaible in the Dryad Digital Repository: https://doi:10.5061/dryad.sn35910 [92] and Harvard Dataverse (https://dataverse.harvard.edu/dataverse/Crustacean_Phylogeny). Raw reads were deposited at GenBank SRA (SAMN06174118–SAMN06174176; see the electronic supplementary material, table S1 for details).