Abstract
Detection of evolutionary shifts in sensory systems is challenging. By adopting a molecular approach, our earlier study proposed a sensory trade-off hypothesis between a loss of colour vision and an origin of high-duty-cycle (HDC) echolocation in Old World bats. Here, we test the hypothesis in New World bats, which include HDC echolocators that are distantly related to Old World HDC echolocators, as well as vampire bats, which have an infrared sensory system apparently unique among bats. Through sequencing the short-wavelength opsin gene (SWS1) in 16 species (29 individuals) of New World bats, we identified a novel SWS1 polymorphism in an HDC echolocator: one allele is pseudogenized but the other is intact, while both alleles are either intact or pseudogenized in other individuals. Strikingly, both alleles were found to be pseudogenized in all three vampire bats. Since pseudogenization, transcriptional or translational changes could separately result in functional loss of a gene, a pseudogenized SWS1 indicates a loss of dichromatic colour vision in bats. Thus, the same sensory trade-off appears to have repeatedly occurred in the two divergent lineages of HDC echolocators, and colour vision may have also been traded off against the infrared sense in vampire bats.
Keywords: sensory trade-off, pseudogene, opsin, echolocation, vampire bat
1. Introduction
Sensing the ever-changing environment is a fundamental task that all organisms must accomplish to survive in nature. Depending on the species, animals may rely on distinct sets of sensory modalities, with some species lacking one or more basic senses while others develop novel senses [1–3]. Detection of evolutionary shifts in sensory systems is challenging in animals, possibly due to difficulties in observing and measuring sensory functions [4]. By adopting a molecular approach, one group of animals was found to show losses of colour vision—the bats (order Chiroptera) [5], which represent approximately 20% of all living mammal species [6]. Specifically, members of the suborder Yangochiroptera within Chiroptera possess dichromatic colour vision, while some members of the other suborder, Yinpterochiroptera, have lost the ability to distinguish colours [5]. Within Yinpterochiroptera, independent losses of colour vision in several species of the Old World fruit bats (Pteropodidae) may be associated with a behavioural innovation—the choice of caves as roost sites in a clade that otherwise roosts in trees [5]. Loss of colour vision has also occurred independently in another lineage composed of two sister clades of Old World bats, Rhinolophidae and Hipposideridae [5]. This loss apparently coincided with the origin of a novel sensory modality, high-duty-cycle (HDC) echolocation, a sensory specialization that facilitates hunting prey in dense vegetation by separating frequency of emitted call pulses and returning echo [5]. Most echolocating bats use low-duty-cycle (LDC) echolocation and a separate pulse and echo in time, a system that is widely thought to be a primitive in bats [7–9]. As far as is known, the majority of LDC bats also have dichromatic colour vision [5]. As a result of this pattern, a sensory trade-off between the loss of colour vision and the origin of the HDC echolocation in bats has been hypothesized [5]. Under this hypothesis, convergent losses of colour vision with gains of other sensory modalities might be expected to have independently occurred in distantly related lineages. However, other authors have argued that the coincidence of HDC echolocation and loss of colour vision was just a mere coincidence [10]. Addressing these competing hypotheses requires additional data and analyses.
To test the sensory trade-off hypothesis, we turn to the New World bats. These species include the moustached bat (Pteronotus parnellii) and vampire bats. The former has independently evolved HDC echolocation that is similar to the Old World lineages (hipposiderids and rhinolophids) [7,8], while the latter have evolved a unique sensory modality among bats—the infrared sense [2,11]. In mammals, the cone opsins are typically divided into two categories: middle/long wavelength sensitive (M/LWS) and short wavelength sensitive (SWS) [12]. Most mammals have both the two categories of opsin and thus possess dichromatic colour vision, whereas a small proportion of mammals retain only one category of opsin and thus are monochromatic (a condition also known as colour blindness) [10]. Two types of SWS opsin genes are present in mammals: monotremes have SWS2, whereas marsupials and eutherians possess SWS1 [13,14]. Several studies in mammals have demonstrated conservation of M/LWS but extraordinary divergence of SWS1 across taxa [5,15,16], we thus focused on the evolution of SWS1 in this work. Although pseudogenization of coding sequence, transcriptional or translational changes could separately result in functional loss of a gene, we here predict the loss of dichromatic colour vision in New World bats by identifying SWS1 pseudogenes (figure 1; electronic supplementary material, table S1). There is no causal link between pseudogenization and functional loss, as the latter may have preceded the former at transcriptional or translational stage. Thus, pseudogenization of SWS1 can predict a loss of dichromatic colour vision, while an intact SWS1 cannot predict intact colour vision in bats. Through sequencing the short-wavelength opsin gene (SWS1) in 16 species (29 individuals) of New World bats (figure 1; electronic supplementary material, table S1), we inferred losses of dichromatic colour vision to test (i) whether there has been a sensory trade-off between vision and echolocation in Pteronotus species similar to that seen in the distantly-related hipposiderid and rhinolophid lineages, and (ii) whether there has been a sensory trade-off between vision and the infrared sense in vampire bats.
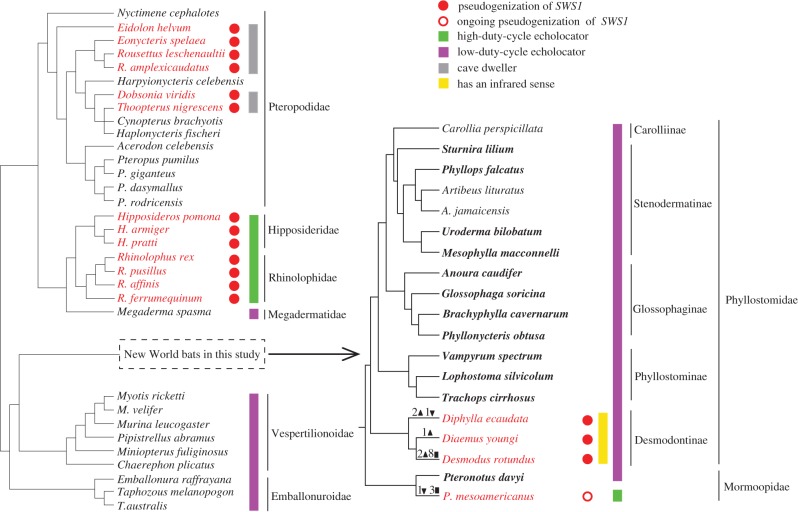
Figure 1.
The species tree depicting the evolution of SWS1 in bats. The tree topology follows previous studies [17–19]. Species shown in the left panel were derived from Zhao et al. [5], while the species detailed in the right panel are the New World bats used in this study; In the New World bats, the 16 species newly sequenced are shown in bold font, whereas the three species previously sequenced are shown in regular font. Numbers of ORF-disrupting insertions (filled inverted triangle), deletions (filled triangle) and premature stops (filled square) are shown on each branch. Species indicated by green bars are high-duty-cycle echolocators, while species with purple bars are low-duty-cycle echolocators; species indicated with the yellow bar are vampire bats that possess infrared sense. Species shown in red possess SWS1 pseudogenes and thus lack typical dichromatic colour vision, and these species were further divided into three categories indicated by three colour bars: green (high-duty-cycle echolocators), yellow (vampire bats) and grey (cave-roosting Old World fruit bats).
2. Material and methods
(a). Taxa and DNA sequencing
Tissues of 16 New World bats (29 individuals) examined in this study were provided by the American Museum of Natural History, and information about these samples is shown in electronic supplementary material, table S1. Our samples contained the HDC echolocator (Pteronotus mesoamericanus) and its congeneric species (Pteronotus davyi) that uses LDC echolocation, and all three extant species of vampire bats (subfamily Desmondontinae within Phyllostomidae) and their related species (electronic supplementary material, table S1). Of note, Pteronotus parnellii was previously thought to be the only bat species in the New World that uses HDC echolocation [7]. However, recent evidence indicates that P. parnellii as traditionally recognized is actually a cryptic species complex comprising at least 9 species [20,21]. Pteronotus mesoamericanus, previously considered a subspecies of P. parnellii, has been elevated to a full species [20,21]. Although most species (n = 13) in our samples have only one individual, three species have either five (Desmodus rotundus and P. davyi) or six individuals (P. mesoamericanus) (electronic supplementary material, table S1). Short-wavelength opsin gene (SWS1) spanning from exon 1 to exon 4 (about 2.2 kb in length) was amplified and directly sequenced in both directions. In addition, we amplified and sequenced the M/LWS opsin gene spanning from exon 1 to exon 6 (about 14 kb in length) in the same five individuals of Desmodus rotundus as those examined SWS1. All primer sequences used in this study are provided in electronic supplementary material, table S2.
(b). Phylogenetic and evolutionary analysis
Deduced protein sequences were aligned by the BioEdit program [22] with the ClustalW multiple alignment option; the nucleotide sequence alignments were generated according to the protein sequence alignments and carefully checked by eye. A species tree of the bats in our study was derived from previous studies [17–19]. Phylogenetic reconstruction for the SWS1 dataset was conducted using a Bayesian approach implemented in MrBayes (version 3.2.6) [23] with the best-fitting model of sequence evolution predicted by ModelTest (version 2.3) [24]. Phyml (version 3.2) was used to reconstruct the maximum-likelihood (ML) tree of the SWS1 dataset under the HKY+I model that was inferred from jModelTest (version 2.1.10) [25] with 100 bootstrap replicates. Both Kishino–Hasegawa and Shimodaria–Hasegawa tests were performed to test the difference between the ML/Bayesian trees and the established species tree using Tree-puzzle [26].
To visualize the average rates of nonsynonymous (dN) and synonymous (dS) substitutions per site for sequences with open reading frame (ORF)-disrupting indels, the Nei and Gojobori method [27] implemented in SWAAP 1.0.2 [28] was applied. We estimated the variation in ω (the ratio of nonsynonymous to synonymous substitution rates) along the phylogeny and examined differential selective pressures using PAML [29]. We inferred ancestral sequences with a combination of the likelihood-based method in PAML [29,30] and the maximum-parsimony approach [31]. We calculated the selection intensity parameter (k) and detected relaxed selection using RELAX [32] implemented in HyPhy [33]. See also electronic supplementary material.
(c). Pseudogene dating analysis
We investigated when the functional constraint on SWS1 became relaxed in each pseudogenized sequence. We assumed that the functional relaxation on SWS1 started at t million years ago, and the pseudogenization dates were estimated based on two different methods described in Meredith et al. [34] and Zhao et al. [35], respectively. See also electronic supplementary material.
3. Results
(a). Pseudogenization of SWS1 in the New World high-duty-cycle echolocator
Despite the conservation of SWS1 in most New World bats (see also electronic supplementary material), we found frameshifting mutations in the New World HDC echolocator (P. mesoamericanus). We first amplified the SWS1 gene from one individual of this species, and the PCR products were cloned and sequenced. We found that sequences from several clones possess intact ORFs (electronic supplementary material, figure S1a), while one 1 bp insertion was present in the sequences of other clones (electronic supplementary material, figure S1b). The 1 bp insertion in the heterozygous state was further confirmed by direct sequencing in the same individual (electronic supplementary material, figure S1c). The 1 bp insertion created one premature stop codon at the beginning of the fifth transmembrane domain (figure 2; electronic supplementary material, figure S2), and the resulting SWS1 opsin would lack the final three transmembrane domains and thus be nonfunctional. Sliding window analysis also suggested relaxation of functional constraint, because a sudden increase of substitution rates was observed right after the occurrence of the 1 bp insertion (electronic supplementary material, figure S3). We next examined variation in the SWS1 gene using five additional individuals of P. mesoamericanus. A total of six individuals were sequenced: four were found to have the 1 bp insertion in the heterozygous state, one has the 1 bp insertion in the homozygous state, and one was intact for both alleles. Thus, the percentage of this null allele (with the presence of the 1 bp insertion) in six individuals was 50%, indicating that the insertion has not been fixed in populations and the SWS1 gene in P. mesoamericanus is probably under ongoing pseudogenization (electronic supplementary material, figures S2 and S3). For comparison, we additionally sequenced a congeneric species P. davyi that is closely related to P. mesoamericanus but uses the LDC echolocation (figure 1). Five individuals of P. davyi were examined and we found that all of these bats possess an intact SWS1 gene and none contained any frameshifting or nonsense mutations (figure 2; electronic supplementary material, figure S2). These results suggest that the null allele is specific to the New World HDC echolocator (P. mesoamericanus).
Figure 2.
Pseudogenizations of SWS1 in four species of New World bats. Codons containing the first ORF-disrupting mutations are boxed. Dashes indicate alignment gaps and numbers in parentheses indicate the nucleotide positions following human SWS1. The abbreviations of Pdav, Pmes, Drot, Dyou and Deca represent Pteronotus davyi, Pteronotus mesoamericanus, Desmodus rotundus, Diaemus youngi and Diphylla ecaudata, respectively. (Online version in colour.)
(b). Pseudogenization of SWS1 in all three species of vampire bats
Strikingly, all three extant vampire bats were found to possess ORFs of SWS1 that are disrupted by indels (insertions and/or deletions) (figure 2; electronic supplementary material, figure S4). Specifically, the common vampire bat (Desmodus rotundus) has two 1 bp deletions, one 12 bp deletion and one 3 bp deletion, which together resulted in eight premature stop codons (electronic supplementary material, figure S4). The first premature stop codon is located at the end of the first transmembrane domain, which would lead to the loss of the remaining six transmembrane domains of its protein. Despite multiple attempts, we were only able to sequence a short fragment (298 bp) of the SWS1 gene in the white-winged vampire bat (Diaemus youngi) due to failed amplifications. The sequenced region contains one 1 bp deletion that would result in the disruption of the last transmembrane domain (figure 2; electronic supplementary material, figure S4). The hairy-legged vampire bat (Diphylla ecaudata) has one 1 bp insertion, one 1 bp and one 2 bp deletion, causing frameshifts starting from the second transmembrane domain (figure 2; electronic supplementary material, figure S4). Although premature stop codons were not observed, both vampire bats (Diphylla ecaudata and Diaemus youngi) are likely to have a nonfunctional SWS1 gene, a conclusion that is also supported by our sliding window analysis (electronic supplementary material, figure S3). Indeed, an elevated ω ratio was observed at or downstream of the frameshifting mutations (electronic supplementary material, figure S3), suggesting the strong impact of the ORF-disrupting indels on the gene functionality, even where premature stop codons were not recorded (electronic supplementary material, figure S3b,c). Sliding window analysis also recorded occurrences of ORF-disrupting indels after the ω ratio has increased (electronic supplementary material, figure S3c), suggesting the genetic divergence is at least partially shaped by relaxed selection. Although the sequenced region of SWS1 in Diaemus youngi is short, an apparent high ω ratio was observed ahead of the location of the single frameshifting indel (electronic supplementary material, figure S3c), suggesting that the SWS1 gene in this lineage is under strong relaxed selection and likely nonfunctional. In addition, the relaxed selection on SWS1 in all three vampire bats was further confirmed by the results recovered from the programs PAML and RELAX (electronic supplementary material, table S3). Notably, no shared mutations were found among any pair of the three species, despite the fact that they all possess frame-shifting mutations and share a recent common ancestry [18]. We were unable to examine multiple individuals of Diphylla ecaudata and Diaemus youngi, but we examined five individuals of Desmodus rotundus and found them to each have the same indels in both alleles. Thus, we infer that all ORF-disrupting mutations observed in Desmodus rotundus are likely fixed in its population.
Given the SWS1 gene is pseudogenized in all three vampire bat species, we attempted to determine when the relaxation of functional constraint on SWS1 started in this clade. To this end, we estimated the selective pressures on SWS1 among vampire bats and tested whether there is a signal of relaxation in their most recent common ancestor. The ancestral sequence for vampire bats was inferred with the combination of Bayesian and parsimony approaches [36]. Under the assumption of the same ω for all branches, ω0 was assumed to be 0.16 (model C, electronic supplementary material, table S3), indicative of purifying selection acting on SWS1 across all studied bats. We compared model C with model D (assuming the ancestral branch of vampire bats has ω1 while other non-vampire bats branches have ω0), and found that ω1 is not significantly different from ω0 (p = 1, electronic supplementary material, table S3), suggesting similar levels of selective pressure on SWS1 between the common ancestor of vampire bats and other bats. Similarly, the results recovered from RELAX program also revealed that selective pressure on SWS1 is not relaxed along the common ancestor of vampire bats compared with other lineages (k = 1.128; p = 1, electronic supplementary material, table S3). Together, these findings suggest that SWS1 was under purifying selection on the ancestral branch leading to vampire bats, and that the relaxation of functional constraint leading to pseudogenization of SWS1 in the three extant vampire lineages may have occurred recently.
(c). Dating the SWS1 pseudogenization events in the New World bats
To date the pseudogenization events in New World bats, we estimated when the functional constraint became relaxed for each of the four pseudogenized SWS1 sequences, including three vampire bats (Desmodus rotundus, Diaemus youngi, Diphylla ecaudata) and the null allele of P. mesoamericanus (electronic supplementary material, figure S5). Under the assumption of a complete removal of functional constraints at t million years ago (Ma), we applied two methods to estimate the dates of pseudogenization events [34,35]. In the first method, we estimated a posterior probability distribution of t based on changes in ω (electronic supplementary material, figure S5) [34]. In the second method, we counted the existing ORF-disrupting mutations and calculated the waiting times for generating current number (n) and future number (n + 1) of ORF-disrupting mutations (see also electronic supplementary material). Another posterior probability distribution of t based on the waiting times was estimated (electronic supplementary material, figure S5). Afterwards, we combined the two distributions and obtained a final posterior probability distribution of t (electronic supplementary material, figure S5). Based on these methods, we inferred the dates of SWS1 pseudogenizations in the following lineages as follows: Desmodus rotundus (mode: 4.7 Ma, mean: 5.5 Ma, median: 5.2 Ma, 95% confidence interval: 2.2–10.1 Ma), Diaemus youngi (2.6 Ma, 3.6 Ma, 3.1 Ma, 0.8–8.0 Ma), Diphylla ecaudata (2.5 Ma, 3.9 Ma, 3.5 Ma, 0.9–8.4 Ma) and P. mesoamericanus (1.0 Ma, 2.3 Ma, 1.9 Ma, 0.2–5.9 Ma) (electronic supplementary material, figure S5).
4. Discussion
Sensory trade-offs involve specialization in one sensory modality that may lead to the reduction or absence of other sensory modalities. An earlier study revealed a sensory trade-off between an origin of the HDC echolocation and a reduction of colour vision in the Rhinolophidae + Hipposideridae lineage from one of the two suborders of bats, Yinpterochiroptera [5]. Through identifying SWS1 pseudogenes, the present study revealed the same sensory trade-off in the independently evolved HDC echolocator from the other suborder of bats (Yangochiroptera). In Yangochiroptera, we additionally found a novel sensory trade-off between the gain of the infrared sense and the reduction of colour vision in all three vampire bats.
In contrast to most New World bats (figure 1), the New World HDC bat (Pteronotus mesoamericanus) was found to have a pseudogenized allele of SWS1 in four individuals, while one individual has both pseudogenized alleles and the sixth individual carries both intact alleles (figure 2; electronic supplementary material, figure S4), suggestive of an ongoing pseudogenization. It is unknown whether the intact allele of SWS1 in the four individuals of P. mesoamericanus is functional, but the functional constraint of SWS1 on this species appears to have relaxed. Interestingly, the allelic polymorphism in the SWS1 (one allele is pseudogenized, while the other allele is intact) has not been reported in animals previously. This novel polymorphism prompts us to suggest that multiple individuals of a species should be included in examinations of animal opsin genes. Furthermore, we did not identify any ORF-disrupting mutations in the congeneric species P. davyi that uses the LDC echolocation (figure 2; electronic supplementary material, figure S4), suggesting relaxation of SWS1 should have occurred specifically in the HDC echolocator (P. mesoamericanus) (electronic supplementary material, figure S4). Despite this, we are not able to rule out the possibility that the null allele may have occurred in the common ancestor of subgenus Phyllodia within Pteronotus, which included all species previously recognized as the single species, P. parnellii [21]. One reason is that due to the absence of samples of species belonging to the subgenus Phyllodia within Pteronotus such as P. parnellii and P. rubiginosus, these species may possess shared mutations with P. mesoamericanus. Another reason is that the common ancestor of subgenus Phyllodia diverged very recently [18], the relaxation of functional constraint may have not had enough time to spread the ORF-disruptive mutation in populations. Certainly, it is possible that changes in SWS1 transcription or translation are shared among all species within subgenus Phyllodia, which may have caused loss of function in their common ancestor. The independent origin of the novel sensory modality (HDC echolocation) in the New World bats is broadly coincident with the relaxation of SWS1 evolution within a same timeframe, which parallels with the sensory trade-off between the loss of SWS1 (i.e. loss of colour vision) and an origin of the HDC echolocation in the Old World bats [5]. That the same sensory trade-off is replicated in two different lineages of HDC echolocating bats (figure 1) that evolved this highly sophisticated form of echolocation independently is indicative of convergent evolution resulting from the same selective pressure: HDC echolocation.
The SWS1 opsin gene was found to be pseudogenized in all three vampire bats (figure 2; electronic supplementary material, figure S4), suggesting that they lack colour vision. The absence of common ORF-disrupting mutations in vampire bats and purifying selection on the most recent common ancestor of vampire bats suggest that pseudogenization of SWS1 has occurred recently and independently in the three vampire bat lineages (Desmodus, Diaemus and Diphylla); the ancestral Desmodontinae bats may have retained a functional SWS1. Indeed, this inference was supported by our molecular dating of pseudogenization events (electronic supplementary material, figure S5). However, it remains possible that changes in SWS1 transcription or translation are shared among the three vampire bats, which may have led to functional loss of SWS1 in their common ancestor. Why has SWS1 been lost in all three vampire bats? These bats are well known for their blood-feeding habits, but they are also noteworthy because of the presence of infrared sensors [37]. Vampire bats are the only mammals known to use an infrared sense, which they use to detect the warm skin temperatures of endothermic prey and locate the optimal bite sites [11]. The function of this sensory modality may have rendered the dichromatic vision unimportant, which may have led to relaxation of functional constraints on SWS1. As such, we propose a sensory trade-off between a gain of the heat sense and a loss of colour vision in vampire bats. Certainly, the gain of the heat sense may also lead to losses of other senses such as the sense of taste [36,38,39], possibly due to the huge energy investment devoted to maintenance and use of a particular sense [40].
Combined with results of earlier studies, our data provide a broader picture of the evolution of vision in bats that are considered sensory specialists. Although dichromatic colour vision is widespread among bats, independent losses of colour vision have now been identified in several lineages (figure 1). These lineages seem to fall into three broad groups. The first group includes the cave-dwelling Old World fruit bats (Pteropodidae), which roost during the day in dark caves in stark contrast with their tree-roosting relatives that retain dichromatic vision (figure 1) [5]. The second group includes two divergent lineages that employ HDC echolocation: an Old World lineage (Hipposideridae + Rhinolophidae), and a New World lineage (P. mesoamericanus within Mormoopidae) (figure 1). A third group comprises the three extant vampire bats (subfamily Desmondontinae within Phyllostomidae) (figure 1). These findings highlight that different taxon-specific ecological reasons may lead to the loss of colour vision in different bat lineages [10]. While each of these observations represents only a correlation between a gain of one sense and a loss of another, rather than direct evidence of causation, together they provide a compelling picture supportive of the sensory trade-off hypothesis due to replicated patterns of evolution of sensory modalities in phylogenetically divergent lineages. Together with previous analyses, our study suggests that evolutionary shifts in animal sensory systems are more frequent than previously thought.
It should be noted that the loss of opsin gene function may have occurred at different stages of protein synthesis: pseudogenization of coding sequences, transcriptional or translational changes. Due to preservation methods, our bat samples from the American Museum of Natural History were only suitable for DNA sequencing, so we were not able to measure mRNA and protein abundances. However, our inference from genetic data was confirmed by mRNA and protein expression data generated by RNA sequencing and immunohistochemical staining [41]. In particular, the SWS1 protein was not detected in the New World high-duty-cycle echolocating bat species (Pteronotus parnellii) [41], which belongs to the same species complex as Pteronotus mesoamericanus examined in our study [20], suggesting a loss of SWS1 function. Both the SWS1 mRNA transcript and protein were not detected in the common vampire bat (Desmodus rotundus), and the SWS1 protein was not found in the other vampire bat (Diaemus youngi) [41], suggesting that both vampire bat species have lost SWS1 function. Despite the consistency between DNA and protein, discordance between DNA and protein was also observed in two species (Trachops cirrhosus and Pteronotus davyi) in which SWS1 protein was not detected but the gene is intact [41]. The discordance suggests that in some cases the loss of SWS1 function was not caused by changes in its coding sequence, and that changes in gene transcription or translation may have preceded changes in coding sequence and also resulted in its functional loss [41]. Thus, we cannot preclude the possibility that SWS1 function may have been lost in other bat species with an intact coding sequence, and call for further investigations involving examination of mRNA and protein levels to address this possibility in the future.
Supplementary Material
Acknowledgements
We are grateful to J. Zhang (University of Michigan) for support in loaning the bat tissues. We thank S. Wang (Wuhan University) for technical assistance in the laboratory.
Data accessibility
DNA sequences: GenBank accessions MG251342–MG251380.
Authors' contributions
H.Z. and Q.L. designed research; H.J. and J.W. performed research; J.W., H.J., Q.L. and H.Z. analysed data; N.B.S. provided bat tissues and revised the manuscript; and J.W., Q.L. and H.Z. wrote the manuscript.
Competing interests
The authors declare that they have no competing interests.
Funding
This work was supported in part by the National Natural Science Foundation of China (91331115 and 31722051).
References
- 1.Au WWL, Simmons JA. 2007. Echolocation in dolphins and bats. Phys. Today 60, 40. [Google Scholar]
- 2.Kurten L, Schmidt U. 1982. Thermoperception in the common vampire bat (Desmodus rotundus). J. Comp. Physiol. A 146, 223–228. ( 10.1007/BF00610241) [DOI] [Google Scholar]
- 3.Feng P, Zheng J, Rossiter SJ, Wang D, Zhao H. 2014. Massive losses of taste receptor genes in toothed and baleen whales. Genome Biol. Evol. 6, 1254–1265. ( 10.1093/gbe/evu095) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones G, Teeling EC, Rossiter SJ. 2013. From the ultrasonic to the infrared: molecular evolution and the sensory biology of bats. Front. Physiol. 4, 117 ( 10.3389/Fphys.2013.00117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao H, Rossiter SJ, Teeling EC, Li CJ, Cotton JA, Zhang S. 2009. The evolution of color vision in nocturnal mammals. Proc. Natl Acad. Sci. USA 106, 8980–8985. ( 10.1073/pnas.0813201106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simmons NB. 2005. Mammal species of the world: a taxonomic and geographic reference. Baltimore, MD: Johns Hopkins University Press. [Google Scholar]
- 7.Jones G, Teeling EC. 2006. The evolution of echolocation in bats. Trends Ecol. Evol. 21, 149–156. ( 10.1016/j.tree.2006.01.001) [DOI] [PubMed] [Google Scholar]
- 8.Arita HT, Fenton MB. 1997. Flight and echlocation in the ecology and evolution of bats. Trends Ecol. Evol. 12, 53–58. ( 10.1016/S0169-5347(96)10058-6) [DOI] [PubMed] [Google Scholar]
- 9.Simmons NB, Geisler JH. 1998. Phylogenetic relationships of Icaronycteris, Archaeonycteris, Hassianycteris, and Palaeochiropteryx to extant bat lineages, with comments on the evolution of echolocation and foraging strategies in Microchiroptera. Bull. Am. Mus. Nat. Hist. 235, 1–182. [Google Scholar]
- 10.Jacobs GH. 2013. Losses of functional opsin genes, short-wavelength cone photopigments, and color vision-a significant trend in the evolution of mammalian vision. Vis. Neurosci. 30, 39–53. ( 10.1017/S0952523812000429) [DOI] [PubMed] [Google Scholar]
- 11.Kunz TH, Fenton MB. 2003. Bat ecology. Chicago, IL: University of Chicago Press. [Google Scholar]
- 12.Peichl L. 2005. Diversity of mammalian photoreceptor properties: adaptations to habitat and lifestyle? Anat. Rec. 287A, 1001–1012. ( 10.1002/ar.a.20262) [DOI] [PubMed] [Google Scholar]
- 13.Cowing JA, Arrese CA, Davies WL, Beazley LD, Hunt DM. 2008. Cone visual pigments in two marsupial species: the fat-tailed dunnart (Sminthopsis crassicaudata) and the honey possum (Tarsipes rostratus). Proc. R. Soc. B 275, 1491–1499. ( 10.1098/rspb.2008.0248) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies WL, Carvalho LS, Cowing JA, Beazley LD, Hunt DM, Arrese CA. 2007. Visual pigments of the platypus: a novel route to mammalian colour vision. Curr. Biol. 17, R161–R163. ( 10.1016/j.cub.2007.01.037) [DOI] [PubMed] [Google Scholar]
- 15.David-Gray ZK, Bellingham J, Munoz M, Avivi A, Nevo E, Foster RG. 2002. Adaptive loss of ultraviolet-sensitive/violet-sensitive (UVS/VS) cone opsin in the blind mole rat (Spalax ehrenbergi). Eur. J. Neurosci. 16, 1186–1194. ( 10.1046/j.1460-9568.2002.02161.x) [DOI] [PubMed] [Google Scholar]
- 16.Tan Y, Yoder AD, Yamashita N, Li W-H. 2005. Evidence from opsin genes rejects nocturnality in ancestral primates. Proc. Natl Acad. Sci. USA 102, 14 712–14 716. ( 10.1073/pnas.0507042102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Datzmann T, von Helversen O, Mayer F. 2010. Evolution of nectarivory in phyllostomid bats (Phyllostomidae Gray, 1825, Chiroptera: Mammalia). BMC Evol. Biol. 10, 165 ( 10.1186/1471-2148-10-165) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rojas D, Warsi OM, Davalos LM. 2016. Bats (Chiroptera: Noctilionoidea) challenge a recent origin of extant neotropical diversity. Syst. Biol. 65, 432–448. ( 10.1093/sysbio/syw011) [DOI] [PubMed] [Google Scholar]
- 19.Dávalos LM, Cirranello AL, Geisler JH, Simmons NB. 2012. Understanding phylogenetic incongruence: lessons from phyllostomid bats. Biol. Rev. 87, 991–1024. ( 10.1111/j.1469-185x.2012.00240.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clare EL, Adams AM, Maya-Simões AZ, Eger JL, Hebert PD, Fenton MB. 2013. Diversification and reproductive isolation: cryptic species in the only New World high-duty cycle bat, Pteronotus parnellii. BMC Evol. Biol. 13, 26 ( 10.1186/1471-2148-13-26) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pavan AC, Marroig G. 2016. Integrating multiple evidences in taxonomy: species diversity and phylogeny of mustached bats (Mormoopidae: Pteronotus). Mol. Phylogenet. Evol. 103, 184–198. ( 10.1016/j.ympev.2016.07.011) [DOI] [PubMed] [Google Scholar]
- 22.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98. [Google Scholar]
- 23.Huelsenbeck J, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. ( 10.1093/bioinformatics/17.8.754) [DOI] [PubMed] [Google Scholar]
- 24.Posada D, Crandall K. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14, 817–818. ( 10.1093/bioinformatics/14.9.817) [DOI] [PubMed] [Google Scholar]
- 25.Posada D. 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–1256. ( 10.1093/molbev/msn083) [DOI] [PubMed] [Google Scholar]
- 26.Schmidt HA, Strimmer K, Vingron M, von Haeseler A. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18, 502–504. ( 10.1093/bioinformatics/18.3.502) [DOI] [PubMed] [Google Scholar]
- 27.Nei M, Gojobori T. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3, 418–426. ( 10.1093/oxfordjournals.molbev.a040410) [DOI] [PubMed] [Google Scholar]
- 28.Pride D. 2004. SWAAP 1.0.2: a tool for analyzing substitutions and similarity in multiple alignments. See http://www.bacteriamuseum.org/SWAAP/SwaapPage.htm .
- 29.Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. ( 10.1093/molbev/msm088) [DOI] [PubMed] [Google Scholar]
- 30.Yang Z, Kumar S, Nei M. 1995. A new method of inference of ancestral nucleotide and amino acid sequences. Genetics 141, 1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J, Nei M. 1997. Accuracies of ancestral amino acid sequences inferred by the parsimony, likelihood, and distance methods. J. Mol. Evol. 44, S139–S146. ( 10.1007/Pl00000067) [DOI] [PubMed] [Google Scholar]
- 32.Wertheim JO, Murrell B, Smith MD, Pond SLK, Scheffler K. 2015. RELAX: detecting relaxed selection in a phylogenetic framework. Mol. Biol. Evol. 32, 820–832. ( 10.1093/molbev/msu400) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pond SLK, Frost SDW, Muse SV. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21, 676–679. ( 10.1093/bioinformatics/bti079) [DOI] [PubMed] [Google Scholar]
- 34.Meredith RW, Gatesy J, Murphy WJ, Ryder OA, Springer MS. 2009. Molecular decay of the tooth gene enamelin (ENAM) mirrors the loss of enamel in the fossil record of placental mammals. PLoS Genet. 5, e1000634 ( 10.1371/journal.pgen.1000634) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao H, Yang J-R, Xu H, Zhang J. 2010. Pseudogenization of the umami taste receptor gene Tas1r1 in the giant panda coincided with its dietary switch to bamboo. Mol. Biol. Evol. 27, 2669–2673. ( 10.1093/molbev/msq153) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao H, Zhou Y, Pinto CM, Charles-Dominique P, Galindo-Gonzalez J, Zhang S, Zhang J. 2010. Evolution of the sweet taste receptor gene Tas1r2 in bats. Mol. Biol. Evol. 27, 2642–2650. ( 10.1093/molbev/msq152) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gracheva EO, Cordero-Morales JF, González-Carcacía JA, Ingolia NT, Manno C, Aranguren CI, Weissman JS, Julius D. 2011. Ganglion-specific splicing of TRPV1 underlies infrared sensation in vampire bats. Nature 476, 88–91. ( 10.1038/nature10245) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao H, Xu D, Zhang S, Zhang J. 2012. Genomic and genetic evidence for the loss of umami taste in bats. Genome Biol. Evol. 4, 73–79. ( 10.1093/gbe/evr126) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hong W, Zhao H. 2014. Vampire bats exhibit evolutionary reduction of bitter taste receptor genes common to other bats. Proc. R. Soc. B 281, 20171079 ( 10.1098/rspb.2014.1079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niven JE, Laughlin SB. 2008. Energy limitation as a selective pressure on the evolution of sensory systems. J. Exp. Biol. 211, 1792–1804. ( 10.1242/jeb.017574) [DOI] [PubMed] [Google Scholar]
- 41.Sadier A, et al. 2018. Evidence for multifactorial processes underlying phenotypic variation in bat visual opsins. bioRxiv. ( 10.1101/300301) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequences: GenBank accessions MG251342–MG251380.