Keywords: nerve regeneration, cell death, transcription factors, dorsal root ganglia neurons, peripheral nerve injury, sciatic nerve, ingenuity pathway analysis, Cytoscape, bioinformatics analysis, STAT1, IRF7, neural regeneration
Abstract
The peripheral nervous system has the potential to regenerate after nerve injury owing to the intrinsic regrowth ability of neurons and the permissive microenvironment. The regenerative process involves numerous gene expression changes, in which transcription factors play a critical role. Previously, we profiled dysregulated genes in dorsal root ganglion neurons at different time points (0, 3 and 9 hours, and 1, 4 and 7 days) after sciatic nerve injury in rats by RNA sequencing. In the present study, we investigated differentially expressed transcription factors following nerve injury, and we identified enriched molecular and cellular functions of these transcription factors by Ingenuity Pathway Analysis. This analysis revealed the dynamic changes in the expression of transcription factors involved in cell death at different time points following sciatic nerve injury. In addition, we constructed regulatory networks of the differentially expressed transcription factors in cell death and identified some key transcription factors (such as STAT1, JUN, MYC and IRF7). We confirmed the changes in expression of some key transcription factors (STAT1 and IRF7) by quantitative reverse transcription-polymerase chain reaction. Collectively, our analyses provide a global overview of transcription factor changes in dorsal root ganglia after sciatic nerve injury and offer insight into the regulatory transcription factor networks involved in cell death.
Introduction
Peripheral nerve injury affects millions of people every year, resulting in severe disability and a reduction in their quality of life (Wu and Murashov, 2013). Unlike the central nervous system, which is limited in its regenerative capacity, the peripheral nervous system can regenerate following injury owing to the intrinsic growth capacity of neurons and the permissive growth environment established in large part by Schwann cells (Barton et al., 2017; Boerboom et al., 2017). Injuries to the peripheral nerve elicit a complex series of changes at the molecular, cellular and tissue levels involving neurons, glial cells, and the immune and vascular systems (Faroni et al., 2015). However, because the post injury cellular and molecular events are unclear, current therapeutic strategies for peripheral nerve injury are mostly ineffective (Scholz et al., 2009; Muheremu and Ao, 2015). Further investigation of the events and gene alterations after peripheral nerve injury is needed to elucidate the underlying regulatory mechanisms and to identify novel therapeutic targets.
Transcription factors (TFs) regulate gene transcription by binding to specific DNA sequences (Barnes and Adcock, 1995) and are thereby able to switch on or off a large number of genes, playing a critical role in multiple biological processes (Johnston et al., 2003; Palazon et al., 2014; Bhagwat and Vakoc, 2015). Injury signals transmitted from the lesion area trigger the damaged neurons to shift from the neurotransmitter state to a regenerative state (Yu et al., 2013). This process is accompanied by the rapid induction of TFs that trigger a cascade of gene alterations associated with nerve regeneration and a permissive microenvironment (Li et al., 2015). Numerous studies show a pivotal role of TFs in peripheral nerve injury (Patodia and Raivich, 2012; Yoon and Giger, 2016).
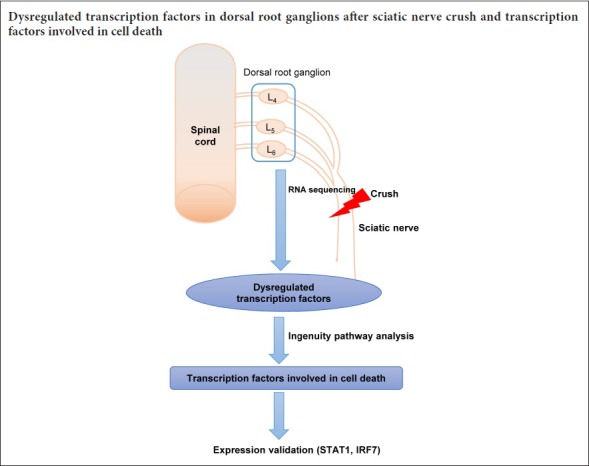
Previously, we performed RNA sequencing to profile dysregulated genes in dorsal root ganglion (DRG) neurons following peripheral nerve injury in rats (Gong et al., 2016). In the present study, we further explore the changes in TFs post nerve injury, and we analyze the enriched molecular and cellular functions of these TFs. In addition, we focus on the TF regulatory networks involved in cell death, which is an important process after nerve injury.
Materials and Methods
Animals
A total of 18 adult male Sprague-Dawley rats (180–220 g), supplied by the Experimental Animal Center of Nantong University of China, were used for the sciatic nerve crush injury model. Briefly, the sciatic nerve was crushed with a pair of forceps with an applied force of 54 N (three times, 10 seconds each time). The rats were then randomly divided into six groups (n = 3 each) and killed by cervical dislocation at different time points (0, 3 and 9 hours, and 1, 4 and 7 days after surgery). The L4–6 DRGs were then removed and stored at −80°C. All experimental procedures involving rats were conducted in accordance with the Institutional Animal Care Guidelines of Nantong University, China, and were granted ethical approval by the Administration Committee of Experimental Animals, Jiangsu Province, China (approval No. 20150304-005).
TF identification and Venn diagram
After RNA sequencing, differentially expressed genes were screened in DRG tissues at different time points (0, 3 and 9 hours, and 1, 4 and 7 days) following nerve injury (n = 3 per group) (Gong et al., 2016). Genes with false discovery rate < 0.05 and fold change (compared with 0 hours or to the previous time points) > 1.5 or < 0.67 were considered differentially expressed genes. A list of Rattus norvegicus transcription factor genes was obtained from the DBD database [Transcription factor prediction database (Wilson et al., 2008) (http://www.transcriptionfactor.org/index.cgi?Home)]. Differentially expressed genes at each time point were compared with the TF gene list, and common genes were identified as differentially expressed TFs. The Venn diagram online tool (http://bioinformatics.psb.ugent.be/webtools/Venn/) was then used to analyze the intersections of TFs at each time point after nerve injury.
Ingenuity pathway analysis (IPA)
IPA, a powerful search and analysis software, can reveal the significance of research data and identify new targets in biological systems (Kramer et al., 2014). Here, we used Comparison Analysis in IPA to compare TFs at the different time points. We filtered Molecular and Cellular Functions in Diseases and Functions with P < 10–5 and an absolute Z-score value > 1. The results are shown as hierarchical clustering. A gene heat map of certain enriched functions and gene & function networks were also obtained by IPA analysis. The regulatory relationship between TFs was also identified, and the TF network was constructed using Cytoscape (Shannon et al., 2003).
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
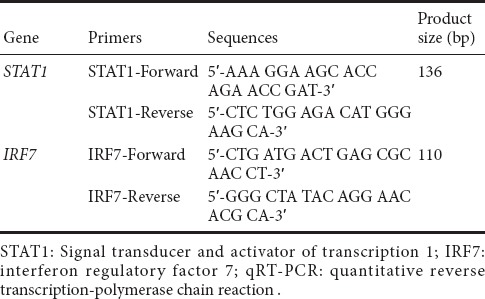
Total RNA was isolated from DRG tissues at different time points after sciatic nerve injury using Trizol reagent (Invitrogen, Carlsbad, CA, USA), and the RNA was reverse transcribed into cDNA with the Prime-Script reagent Kit (TaKaRa, Dalian, China). qRT-PCR was performed for quantification of gene expression with SYBR Premix Ex Taq (TaKaRa) on an ABI StepOne system (Applied Biosystems, Foster City, CA, USA) according to standard protocols. Each sample was assayed in triplicate. Relative expression levels of mRNA were quantified using the comparative 2−ΔΔCt method (Livak and Schmittgen, 2001) and normalized against GAPDH. The primers used are listed in Table 1.
Table 1.
The primers used in qRT-PCR
Statistical analysis
All data are expressed as the mean ± SD. Statistical analyses were performed using Prism 5 software (GraphPad, San Diego, CA, USA). Unpaired two-tailed Student’s t-test was used for comparison between time points after injury and 0 days for qRT-PCR. A value of P < 0.05 was considered statistically significant.
Results
Altered TFs in the DRG following peripheral nerve injury
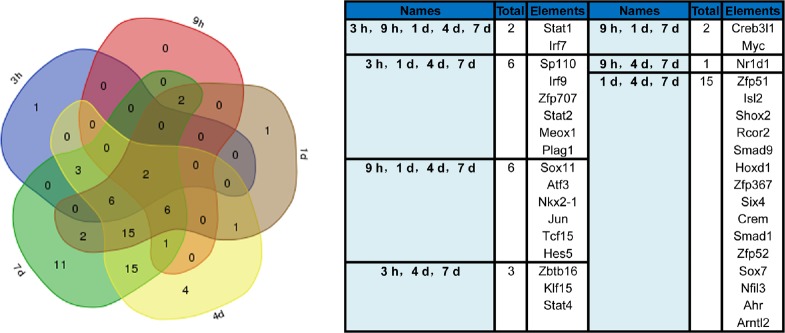
Previously, we investigated the global transcriptome landscape in DRG neurons at 0, 3 and 9 hours, and 1, 4 and 7 days following peripheral nerve injury. Because TFs are activated upon injury to modulate gene expression, we focused on TF changes based on our previous study (Gong et al., 2016). As shown in Table 2 and Additional Table 1 (328.5KB, pdf) , TFs were differentially expressed after nerve injury compared with 0 hours. The number of differential TFs increased as the number of differential genes increased. The percentage of differential TFs among all differential genes peaked at 1 day. The differentially expressed TFs were compared at each time point by Venn diagram analysis. This showed that after nerve injury, two TFs were differentially expressed, signal transducer and activator of transcription 1 (STAT1) and interferon regulatory factor 7 (IRF7). Furthermore, a large number of differentially expressed TFs were common at 1, 4 and 7 days (Figure 1).
Table 2.
Changes in transcription factors following sciatic nerve injury
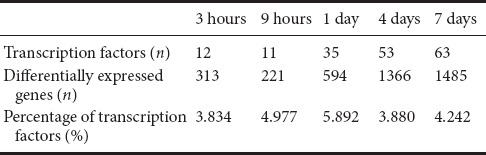
Figure 1.
Venn diagram of significantly differentially expressed transcription factors at 3 and 9 hours, and 1, 4 and 7 days compared with 0 hours.
The transcription factors overlapping the different time points are listed on the right. h: Hour(s); d: day(s).
The differentially expressed TFs at different time points after peripheral nerve injury
Enriched molecular and cellular functions of the differentially expressed TFs
IPA was used for functional analyses of these differentially expressed TFs at each time point, and focusing on Molecular and Cellular Functions in Diseases & Functions. Figure 2 shows the hierarchical clustering of the enriched functions by activation z-score (Additional Table 2 (45.3KB, pdf) ). The blue color indicates a negative regulation of the function, while orange indicates activation. Additionally, from the clustering, we found that functional enrichment of differentially expressed TFs was similar at 4 and 7 days, consistent with the Venn diagram. As shown in Figure 2, at the early stage after nerve injury (3 hours), the differentially expressed TFs mainly regulated cell death and cell viability, which are prerequisites for nerve regeneration. Cell death was inhibited during nerve injury, while cell viability and cell survival were promoted from day 1 after injury. Furthermore, other biological processes were also enhanced from day 1 and day 4, such as differentiation of the nervous system and leukemia cell regulation.
Figure 2.
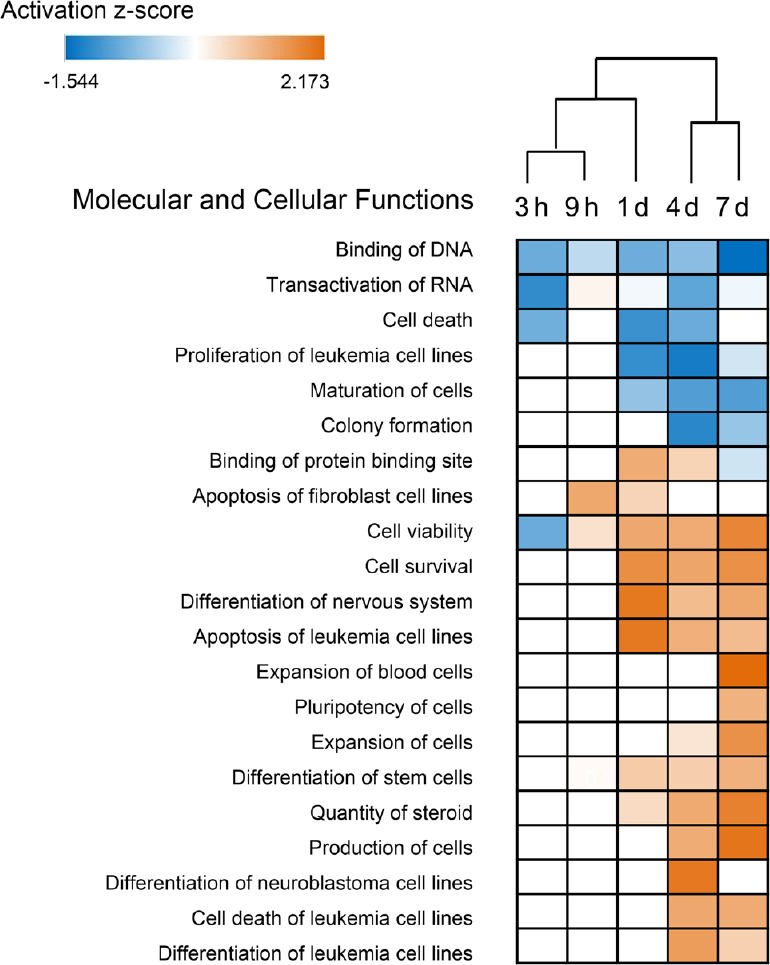
Hierarchical clustering of the enriched molecular and cellular functions of differentially expressed transcription factors at different time points using the Z-score.
Blue indicates negative regulation of the function, while orange indicates activation. The cut-off criterion of enriched functions was P-value < 10–5 and an absolute Z-score value > 1. h: Hours; d: day(s).
The enriched Molecular and Cellular Functions of differentially expressed TFs by Z-score
Regulatory network involved in cell death
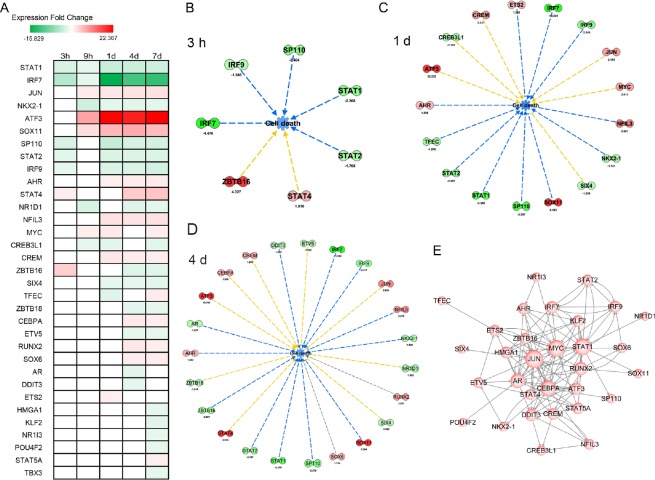
A prerequisite for nerve regeneration is that the DRG neurons survive the injury. Thus, finding effective measures to inhibit cell death is of great importance. We therefore focused on TFs involved in cell death. Among the differential TFs, 33 were involved in cell death. Their expressional fold change trends are given in Figure 3A. STAT1 and IRF7 are included in this list. As shown in Figure 2, cell death was enriched at 3 hours, and 1 and 4 days after the injury. We then constructed gene and function networks of these TFs in cell death at these time points (Figure 3B). The green color indicates that the TF was downregulated, while red indicates increased expression. We also examined the regulatory relationship between the TFs involved in cell death with IPA and constructed a regulatory network with Cytoscape. As shown in Figure 3C, genes at the base of the arrow regulate genes at the tip (target). The more a gene interacts with other genes, the larger the size of the gene circle. Some TFs, such as STAT1, JUN, MYC and IRF7, are particularly critical TFs.
Figure 3.
Differential transcription factors involved in cell death.
(A) Expression trend diagram of transcription factors involved in cell death by expression fold-change. Green indicates downregulation, while red indicates upregulation. (B–D) Gene and function network of differentially expressed transcription factors in cell death at 3 hours (B), and at 1 (C) and 4 (D) days. Green indicates downregulation, while red indicates upregulation. The expressional fold change at certain time points after injury are shown below the symbol for the transcription factor. The blue lines indicate inhibition of the function, while the yellow lines indicate that the regulation was inconsistent with the state of the downstream molecule. The grey line indicates that the effect was unclear. (E) The regulatory network of differentially expressed transcription factors involved in cell death. The circles indicate transcription factors, while the arrows indicate the relationships. The more a transcription factor interacts with another transcription factor, the larger the circle. h: Hour(s); d: day(s).
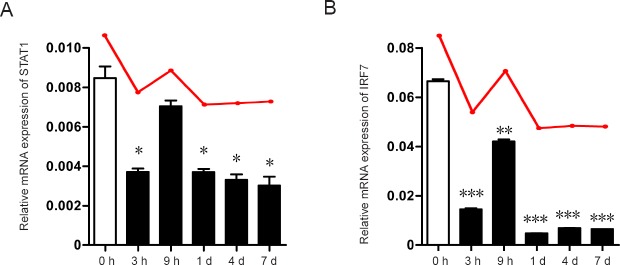
Validation of STAT1 and IRF7 by qRT-PCR
Because STAT1 and IRF7 are key TFs differentially expressed at all time points after peripheral nerve injury, they also serve as key nodes in the cell death regulatory network. qRT-PCR was performed to validate the expressional changes of these TFs. As shown in Figure 4, the bar represents the expression of TFs at the indicated time point post peripheral nerve injury, while the red line represents the expressional changes revealed by RNA sequencing. The qRT-PCR results were highly consistent with previous RNA sequencing data. However, although both STAT1 and IRF7 were downregulated after nerve injury, expression of these TFs was high at 9 hours compared with the other time points. This high expression might be related to the balance of death and survival in DRG neurons. In addition, immunofluorescence for STAT1 and IRF7 was performed in DRG sections, which revealed that these TFs were mainly expressed in DRG neurons, although the expressional changes between 0 and 3 hours were not striking (data not shown).
Figure 4.
STAT1 and IRF7 mRNA expression levels measured by quantitative reverse transcriptionpolymerase chain reaction.
(A, B) Relative STAT1 and IRF7 mRNA expression levels at different time points, compared with 0 hour. The red line above the bar shows the expressional trend revealed by RNA sequencing. *P < 0.05, **P < 0.01, ***P < 0.001, vs. 0 h. Data are expressed as the mean ± SD (n = 3, unpaired two-tailed student’s t-test). STAT1: Signal transducer and activator of transcription 1; IRF7: interferon regulatory factor 7; h: hour(s); d: day(s).
Discussion
Peripheral nerves are commonly damaged after nerve injury, and to provide a permissive environment that facilitates nerve regeneration, multiple cell types undergo changes in gene expression that alter their phenotype (Barton et al., 2017). For example, DRG neurons undergo a switch from the neurotransmitter state to a regenerative state. DRG neurons are primary afferent neurons that transmit sensory information from the periphery to the central nervous system. The sciatic nerve, which is often investigated in peripheral nerve injury models, has its neuronal somas mainly in L4–6 DRGs in rats (Richner et al., 2014).
Previously, we investigated global transcriptional changes in DRGs after sciatic nerve injury by RNA sequencing. Differentially expressed genes were then analyzed according to trends in expressional changes, and two typical clusters were selected for further analysis. However, it remained unclear how these alterations in gene expression are regulated.
A number of TFs were dysregulated after injury. The Venn diagram and the enriched molecular and cellular function analyses by IPA revealed that the alterations could be divided into two periods. In the early period following nerve injury (3 and 9 hours), approximately 10 TFs were differentially expressed and mainly regulated cell death and cell viability. In the later period (1, 4 and 7 days), the number of significantly differentially expressed TFs increased. In addition to inhibiting cell death and enhancing cell viability, these TFs also regulate nervous system differentiation and immune cell activity, consistent with a previous study (McLachlan and Hu, 2014). Leukemia cell regulation was also enriched during this period. It has been reported that leukemia inhibitory factor, which induces the terminal differentiation of myeloid leukemia cells and inhibits their proliferation, plays an important role in peripheral nerve regeneration (Nicola and Babon, 2015). However, the relationship between leukemia cell regulation and peripheral nerve injury has not been explored.
Although a favorable microenvironment facilitates axonal regrowth after nerve injury, the capacity of damaged neurons to survive and switch to a regenerative state is critical for nerve regeneration and functional recovery (Navarro et al., 2007). It has been reported that a significant amount of DRG neurons eventually die after peripheral nerve injury (Lin et al., 2011). Therefore, identifying key TFs involved in cell death might provide gene targets for reducing the loss of DRG neurons and improving their survival following peripheral nerve injury.
Here, we identified 33 differentially expressed TFs involved in cell death in DRG tissues after sciatic nerve injury. Consistent with previous studies, ATF3 and JUN, two classical TFs involved in axonal regeneration, were upregulated following nerve injury (Seijffers et al., 2007; Ruff et al., 2012). STAT1 is a key modulator of cell death and has been reported to be associated with neuronal cell death after cerebral ischemia and spinal cord injury (Kim and Lee, 2007; Osuka et al., 2011; Wu et al., 2014). Although STAT1 can directly interact with apoptotic proteins, it promotes cell death mainly through a transcriptional mechanism involving death-promoting gene activation (Kim and Lee, 2007). The downregulation of STAT1 in DRG neurons following sciatic nerve injury might protect against cell death and promote their survival. IRF7, a member of the interferon regulatory transcription factor family, has been shown to participate in the transcriptional activation of virus-induced cellular genes. In the central nervous system, IRF7 is upregulated following axonal transection in the hippocampus and plays an important role in microglial polarization switching (Khorooshi and Owens, 2010; Tanaka et al., 2015). However, in the present study, IRF7 was downregulated after peripheral nerve injury, which might prevent cell death. The roles and mechanisms of action of these TFs deserve further investigation.
In conclusion, numerous TFs undergo changes in expression in DRG tissues after sciatic nerve injury. These differential TFs are enriched in cell death and viability at the early period following nerve injury and later in nervous system differentiation. In addition, we confirmed changes in two key TFs, STAT1 and IRF7, which might serve as potential targets for the treatment of peripheral nerve injuries.
Additional files:
Additional Table 1 (328.5KB, pdf) :: The differentially expressed TFs at different time points after peripheral nerve injury.
Additional Table 2 (45.3KB, pdf) :: The enriched molecular and cellular functions of differentially expressed TFs by Z-score.
Footnotes
Conflicts of interest: The authors declare that there are no conflicts of interest associated with this manuscript.
Financial support: This study was supported by the National Natural Science Foundation of China, No. 31500823; the Natural Science Foundation of Jiangsu Province of China, No. BK20150403; the Natural Science Fund for Colleges and Universities in Jiangsu Province of China, No. 16KJB180024. The funding sources had no role in study conception and design, data analysis or interpretation, paper writing or deciding to submit this paper for publication.
Institutional review board statement: All the experimental procedures involving animals were conducted in accordance with Institutional Animal Care Guidelines of Nantong University of China (20150304-005), and approved ethically by the Administration Committee of Experimental Animals, Jiangsu Province, China.
Copyright license agreement: The Copyright License Agreement has been signed by all authors before publication.
Data sharing statement: Datasets analyzed during the current study are available from the corresponding author on reasonable request.
Plagiarism check: Checked twice by iThenticate.
Peer review: Externally peer reviewed.
Funding: This study was supported by the National Natural Science Foundation of China, No. 31500823; the Natural Science Foundation of Jiangsu Province of China, No. BK20150403; the Natural Science Fund for Colleges and Universities in Jiangsu Province of China, No. 16KJB180024.
(Copyedited by Patel B, Frenchman B, Wang J, Li CH, Qiu Y, Song LP, Zhao M)
References
- Barnes PJ, Adcock IM. Transcription factors. Clin Exp Allergy 25 Suppl. 1995;2:46–49. doi: 10.1111/j.1365-2222.1995.tb00421.x. [DOI] [PubMed] [Google Scholar]
- Barton MJ, John JS, Clarke M, Wright A, Ekberg J. The glia response after peripheral nerve injury: a comparison between schwann cells and olfactory ensheathing cells and their uses for neural regenerative therapies. Int J Mol Sci. 2017;18:E287. doi: 10.3390/ijms18020287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagwat AS, Vakoc CR. Targeting transcription factors in cancer. Trends Cancer. 2015;1:53–65. doi: 10.1016/j.trecan.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerboom A, Dion V, Chariot A, Franzen R. Molecular mechanisms involved in schwann cell plasticity. Front Mol Neurosci. 2017;10:38. doi: 10.3389/fnmol.2017.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faroni A, Mobasseri SA, Kingham PJ, Reid AJ. Peripheral nerve regeneration: experimental strategies and future perspectives. Adv Drug Deliv Rev. 2015;82-83:160–167. doi: 10.1016/j.addr.2014.11.010. [DOI] [PubMed] [Google Scholar]
- Gong L, Wu J, Zhou S, Wang Y, Qin J, Yu B, Gu X, Yao C. Global analysis of transcriptome in dorsal root ganglia following peripheral nerve injury in rats. Biochem Biophys Res Commun. 2016;478:206–212. doi: 10.1016/j.bbrc.2016.07.067. [DOI] [PubMed] [Google Scholar]
- Johnston MV, Alemi L, Harum KH. Learning, memory, and transcription factors. Pediatr Res. 2003;53:369–374. doi: 10.1203/01.PDR.0000049517.47493.E9. [DOI] [PubMed] [Google Scholar]
- Khorooshi R, Owens T. Injury-induced type I IFN signaling regulates inflammatory responses in the central nervous system. J Immunol. 2010;185:1258–1264. doi: 10.4049/jimmunol.0901753. [DOI] [PubMed] [Google Scholar]
- Kim HS, Lee MS. STAT1 as a key modulator of cell death. Cell Signal. 2007;19:454–465. doi: 10.1016/j.cellsig.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Kramer A, Green J, Pollard J, Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Xue C, Yuan Y, Zhang R, Wang Y, Wang Y, Yu B, Liu J, Ding F, Yang Y, Gu X. The transcriptional landscape of dorsal root ganglia after sciatic nerve transection. Sci Rep. 2015;5:16888. doi: 10.1038/srep16888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CR, Yang CH, Huang CE, Wu CH, Chen YS, Sheen-Chen SM, Huang HW, Chen KH. GADD45A protects against cell death in dorsal root ganglion neurons following peripheral nerve injury. J Neurosci Res. 2011;89:689–699. doi: 10.1002/jnr.22589. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- McLachlan EM, Hu P. Inflammation in dorsal root ganglia after peripheral nerve injury: effects of the sympathetic innervation. Auton Neurosci. 2014;182:108–117. doi: 10.1016/j.autneu.2013.12.009. [DOI] [PubMed] [Google Scholar]
- Muheremu A, Ao Q. Past, present, and future of nerve conduits in the treatment of peripheral nerve injury. Biomed Res Int. 2015;2015:237507. doi: 10.1155/2015/237507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro X, Vivo M, Valero-Cabre A. Neural plasticity after peripheral nerve injury and regeneration. Prog Neurobiol. 2007;82:163–201. doi: 10.1016/j.pneurobio.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Nicola NA, Babon JJ. Leukemia inhibitory factor (LIF) Cytokine Growth Factor Rev. 2015;26:533–544. doi: 10.1016/j.cytogfr.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osuka K, Watanabe Y, Usuda N, Atsuzawa K, Yasuda M, Aoshima C, Wakabayashi T, Takayasu M. Activation of STAT1 in neurons following spinal cord injury in mice. Neurochem Res. 2011;36:2236–2243. doi: 10.1007/s11064-011-0547-6. [DOI] [PubMed] [Google Scholar]
- Palazon A, Goldrath AW, Nizet V, Johnson RS. HIF transcription factors, inflammation, and immunity. Immunity. 2014;41:518–528. doi: 10.1016/j.immuni.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patodia S, Raivich G. Role of transcription factors in peripheral nerve regeneration. Front Mol Neurosci. 2012;5:8. doi: 10.3389/fnmol.2012.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richner M, Ulrichsen M, Elmegaard SL, Dieu R, Pallesen LT, Vaegter CB. Peripheral nerve injury modulates neurotrophin signaling in the peripheral and central nervous system. Mol Neurobiol. 2014;50:945–970. doi: 10.1007/s12035-014-8706-9. [DOI] [PubMed] [Google Scholar]
- Ruff CA, Staak N, Patodia S, Kaswich M, Rocha-Ferreira E, Da Costa C, Brecht S, Makwana M, Fontana X, Hristova M, Rumajogee P, Galiano M, Bohatschek M, Herdegen T, Behrens A, Raivich G. Neuronal c-Jun is required for successful axonal regeneration, but the effects of phosphorylation of its N-terminus are moderate. J Neurochem. 2012;121:607–618. doi: 10.1111/j.1471-4159.2012.07706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz T, Krichevsky A, Sumarto A, Jaffurs D, Wirth GA, Paydar K, Evans GR. Peripheral nerve injuries: an international survey of current treatments and future perspectives. J Reconstr Microsurg. 2009;25:339–344. doi: 10.1055/s-0029-1215529. [DOI] [PubMed] [Google Scholar]
- Seijffers R, Mills CD, Woolf CJ. ATF3 increases the intrinsic growth state of DRG neurons to enhance peripheral nerve regeneration. J Neurosci. 2007;27:7911–7920. doi: 10.1523/JNEUROSCI.5313-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Murakami K, Bando Y, Yoshida S. Interferon regulatory factor 7 participates in the M1-like microglial polarization switch. Glia. 2015;63:595–610. doi: 10.1002/glia.22770. [DOI] [PubMed] [Google Scholar]
- Wilson D, Charoensawan V, Kummerfeld SK, Teichmann SA. DBD--taxonomically broad transcription factor predictions: new content and functionality. Nucleic Acids Res. 2008;36:D88–D92. doi: 10.1093/nar/gkm964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Murashov AK. Molecular mechanisms of peripheral nerve regeneration: emerging roles of microRNAs. Front Physiol. 2013;4:55. doi: 10.3389/fphys.2013.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Yang L, Mei X, Yu Y. Selective inhibition of STAT1 reduces spinal cord injury in mice. Neurosci Lett. 2014;580:7–11. doi: 10.1016/j.neulet.2013.11.055. [DOI] [PubMed] [Google Scholar]
- Yi S, Wang QH, Zhao LL, Qin J, Wang YX, Yu B, Zhou SL. miR-30c promotes Schwann cell remyelination following peripheral nerve injury. Neural Regen Res. 2017;12:1708–1715. doi: 10.4103/1673-5374.217351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon C, Giger RJ. Inside out: core network of transcription factors drives axon regeneration. Neuron. 2016;89:881–884. doi: 10.1016/j.neuron.2016.02.022. [DOI] [PubMed] [Google Scholar]
- Yu B, Zhou S, Hu W, Qian T, Gao R, Ding G, Ding F, Gu X. Altered long noncoding RNA expressions in dorsal root ganglion after rat sciatic nerve injury. Neurosci Lett. 2013;534:117–122. doi: 10.1016/j.neulet.2012.12.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The differentially expressed TFs at different time points after peripheral nerve injury
The enriched Molecular and Cellular Functions of differentially expressed TFs by Z-score