Abstract
Non-small cell lung cancer (NSCLC) presents severe threats to the lives of patients. Gefitinib is one of the first-line drugs available for the treatment of NSCLC in the clinical setting. The present study investigated the effects of gefitinib on NSCLC H1650 cell viability and apoptosis via MTT assays and flow cytometry. Western blot analysis was employed to detect tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) expression levels in H1650 cells. In the present study, H1650 cells were treated with TRAIL siRNA or an empty plasmid vector control, followed by gefitinib treatment to investigate apoptosis. Gefitinib treatment markedly inhibited H1650 cell viability, induced apoptosis and reduced TRAIL expression levels. TRAIL interference enhanced H1650 cell apoptosis induced by gefitinib. TRAIL overexpression suppressed gefitinib-induced H1650 cell apoptosis. In addition, gefitinib induced NSCLC H1650 cell apoptosis by downregulating TRAIL expression levels.
Keywords: gefitinib, tumor necrosis factor-related apoptosis-inducing ligand, H1650 cell, apoptosis
Introduction
Non-small cell lung cancer (NSCLC) is one of the most common types of cancer worldwide, and has become the leading cause of cancer-associated mortality in China within the previous 5 years (1). NSCLC includes a variety of cancer types, including large cell carcinoma, squamous cell carcinoma and adenocarcinoma. Compared with SCLC, NSCLC proliferates more slowly and metastasizes later (2,3). NSCLC accounts for ~85% of total lung cancers and >80% of patients diagnosed with NSCLC are in middle- or late-stage disease (4). Therefore, the 5-year survival rate of NSCLC is relatively low (1). As NSCLC seriously threatens the lives of patients, investigation is required to develop a therapeutic method for the treatment of NSCLC (5,6).
As with other types of cancer, chemotherapy, radiotherapy and surgery serve critical roles in the treatment of NSCLC. However, the aforementioned methods are associated with various disadvantages and insufficiencies (7–9). Radiotherapy has been demonstrated to have the best curative effect for NSCLC (10). Radiotherapy mainly targets lymphatic metastasis and primary tumors, with chemotherapy serving an auxiliary role (11,12). Chemotherapy presents marked curative effects on NSCLC; however, bleeding and other side effects have been reported (13). Surgery has the greatest limitation, as it is not applicable for patients with complications or those >70 years old (14,15). Therefore, investigations into the molecular mechanism of NSCLC occurrence and development are required, and may provide valuable information for clinical practice.
Gefitinib exhibits significant curative effects on NSCLC, but its detailed molecular mechanism remains poorly understood (16). The present study aimed to investigate the effects of gefitinib on NSCLC H1650 cell viability and apoptosis. Gefitinib may be used alone or in combination with other chemotherapy drugs for the treatment of NSCLC (17–19). Gefitinib is an antagonist of the tyrosine protein kinase of epidermal growth factor receptor (EGFR). Generally, gefitinib serves an antitumor role by inhibiting EGFR tyrosine protein kinase activity (20). Gefitinib significantly inhibited the proliferation of tumor cells, and also reduced lung cancer tumor growth, invasion and metastasis in a rat model (21,22). However, the knockdown or overexpression of EGFR in vitro or in vivo failed to alter NSCLC cell sensitivity to gefitinib, indicating that gefitinib may have a novel target or molecular mechanism underlying NSCLC (23,24).
Tumor necrosis factor-related apoptotic inducing ligand (TRAIL) is a novel member of the tumor necrosis factor family (25). TRAIL is highly expressed within activated T lymphocytes and may induce apoptosis via interaction with its ligands, including death receptor 4 (DR4) and DR5 (26). By contrast, TRAIL may inhibit apoptosis if not combined with DR4/DR5. A recent study indicated that TRAIL expression levels in NSCLC tissue were altered, indicating that TRAIL may be associated with NSCLC occurrence and development (27).
The present study aimed to investigate the regulatory role and associated mechanism of gefitinib on NSCLC H1650 cell viability and apoptosis in vitro, and may provide valuable information for NSCLC treatment in the clinical setting.
Materials and methods
Reagents
High-glucose Dulbecco's modified Eagle's medium (DMEM) was purchased from Gibco; Thermo Fisher Scientific, Inc. (Waltham, MA, USA). Trypsin, EDTA, poly-l-lysine, Hanks buffer, penicillin and streptomycin were obtained from Sigma-Aldrich; Merck KGaA (Darmstadt, Germany). Phosphate-buffered saline (PBS) and dimethyl sulfoxide (DMSO) were purchased from Beijing Dingguo Changsheng Biotechnology Co., Ltd. (Beijing, China). MTT reagent, fluorescein isothiocyanate (FITC)-Annexin V and Caspase 3 Activity Assay kit were obtained from Beyotime Institute of Biotechnology (Haimen, China). TRAIL small interfering RNA (siRNA) and control siRNA were synthetized by Sangon Biotech Co., Ltd. (Shanghai, China). TRAIL plasmids were produced in-house. TRAIL and β-actin antibodies were obtained from Sigma-Aldrich; Merck KGaA.
Cell culture
The H1650 cell line was purchased from the American Type Culture Collection (Manassas, VA, USA) and cultured in high glucose DMEM medium at 37°C and 5% CO2.
Transfection
H1650 cells were cultured at 50% density 1 day prior to transfection. N-[1-(2,3-Dioleoyloxy)propyl]-N,N,N-trimethylammonium methyl-sulfate (DOTAP) Liposomal Transfection Reagent (Sigma-Aldrich; Merck KGaA) was used for transfection. TRAIL siRNA (cat. no. 1027423; Qiagen Sciences, Inc., Gaithersburg, MD, USA) or control siRNA (cat. no. 1027310; Qiagen Sciences, Inc.) (1 µg) were cloned into a TRAIL plasmid with the DOTAP liposomal transfection reagent (Sigma-Aldrich; Merck KGaA) at room temperature for 3 min. The mixture was then added to the cells and maintained for 12 h at room temperature. The cells were cultured for a further 24 h following the replacement of medium and subsequent experimentation.
MTT assay
H1650 cell viability was investigated by colorimetry as previously described (28). Un-transfected H1650 cells were washed in DMEM and treated with 7 µl MTT solution (0.1 M, pH 7.2). The cells were cultured at 37°C and 5% CO2 for 4 h and subsequently washed in DMEM, followed by the addition of 50 µl DMSO for 15 min to dissolve the purple formazan. The plate was analyzed at 540 nm.
Flow cytometry
Flow cytometry was employed to investigate phosphatidylserine expression on the H1650 cell surface as previously described (29). A total of 1×104 H1650 cells were incubated with FITC-Annexin V dye and Annexin Binding Buffer (Thermo Fisher Scientific, Inc.) for 16 min at room temperature. Subsequently, the cells were analyzed using a flow cytometer (BD FACSCalibur™) at 465 (emitted light) and 630 nm (absorbed light) and data were analyzed by BD Cell Quest Pro software, version 5.1 (BD Biosciences, Franklin, NJ, USA).
Caspase-3 activity detection
As previously described, H1650 cell caspase-3 activity was determined using a microplate reader (9). A total of 1×104 H1650 cells were treated with Cell Lysis Buffer (Cell Signaling Technology, Inc., Danvers, MA, USA) for 30 min on ice and incubated with a chromophore p-nitroaniline (pNA) (Included in the caspase-3 activity assay kit) at room temperature for 18 min. Finally, the cells were read on microplate reader at 490 nm.
Western blot analysis
TRAIL expression levels in H1650 cells were analyzed via western blotting as described previously (17). The cells were lysed on ice for 30 min using Cell Lysis Buffer (Cell Signaling Technology). Following centrifugation at 10,000 × g for 5 min at 4°C, the protein was quantified using Pierce BCA Protein Assay kit (Thermo Fisher Scientific, Inc.) and separated by 10% SDS-PAGE electrophoresis (10 µg/lane). Then the protein was transferred to a polyvinylidene fluoride membrane and blocked with 5% skimmed milk at room temperature for 1 h. Subsequently, the membrane was incubated with a TRAIL primary antibody (cat. no., T9191) (dilution, 1:1,000) and β-actin primary antibody (cat. no., SAB5500001) (dilution, 1:2,000) for 2 h at room temperature. The membrane was then incubated with HRP-conjugated goat anti-rabbit secondary antibody (cat. no., AP187P; dilution, 1:1,000; Sigma-Aldrich; Merck KGaA) for 2 h at room temperature and washed with PBST three times. Finally, the membrane was treated with an enhanced chemiluminescence agent (Thermo Fisher Scientific, Inc.) and analyzed. ImageJ software version 1.51 (National Institutes of Health, Bethesda, MD, USA) was used for data analysis.
Statistical analysis
All statistical analyses were performed using SPSS software and data were presented as mean ± standard deviation (SD) A Levene's test was first performed to detect normal distribution. One-way analysis of variance with Student-Newman-Keuls multiple comparison post-hoc analysis was used for comparison of the means (16). P<0.05 was considered to indicate a statistically significant difference.
Results
Gefitinib inhibits H1650 cell viability
As presented in Fig. 1, an MTT assay revealed that H1650 cell viability was markedly declined following treatment with 1 µg/ml gefitinib (P=0.0067).
Figure 1.
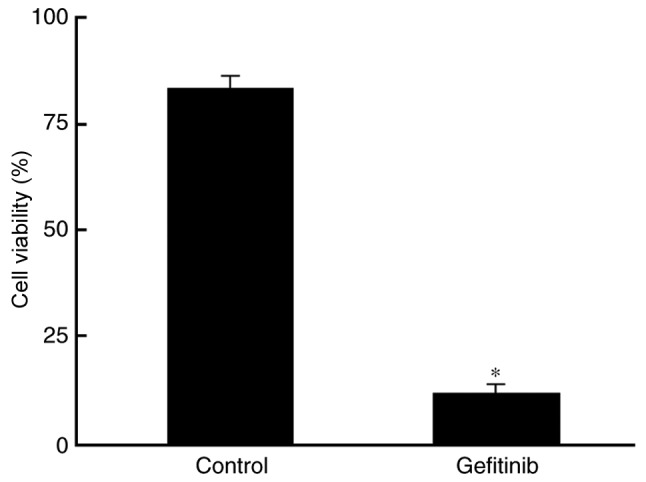
Gefitinib inhibits H1650 cell viability. *P<0.05, compared with the control.
Gefitinib induces H1650 cell apoptosis
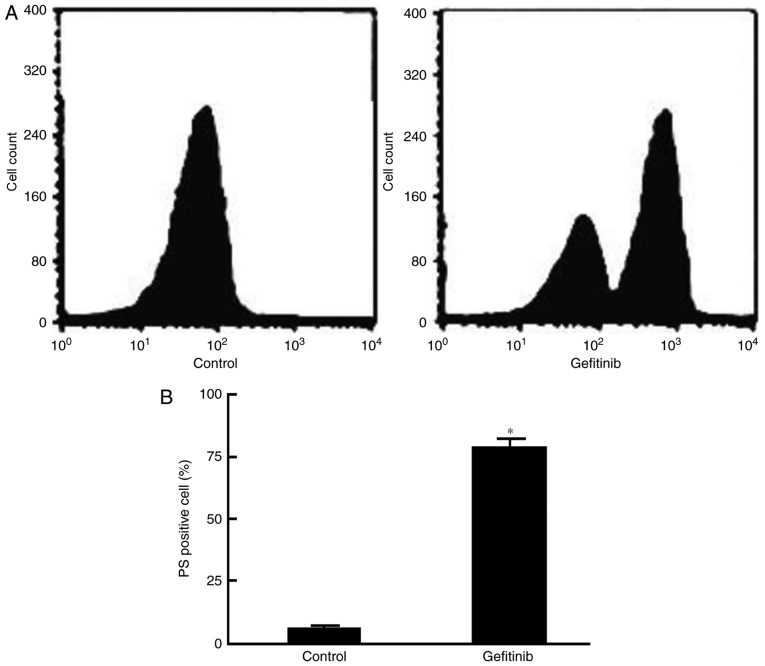
Flow cytometry demonstrated that the phosphatidylserine levels on the H1650 cell surface were significantly increased compared with levels in the control (P=0.023; Figs. 2 and 3).
Figure 2.
Gefitinib induces H1650 cell apoptosis. (A) Representative flow cytometry graph showing PS expression. (B) Quantification of PS positive cells.*P<0.05, compared with the control. PS, phosphatidylserine.
Figure 3.
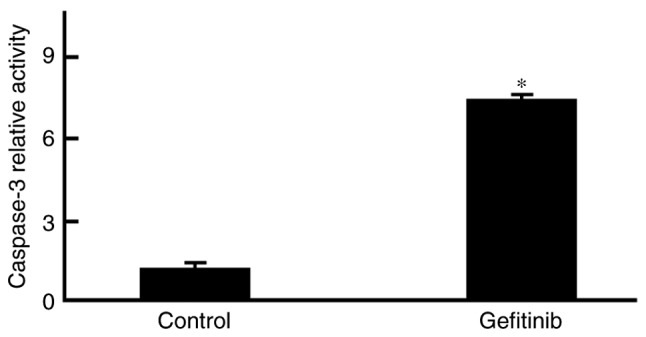
Gefitinib activates caspase-3 within H1650 cells. Analysis of caspase-3 activity demonstrated a marked increase within H1650 cells following treatment with 1 µg/ml gefitinib. *P<0.05, compared with control.
Gefitinib decreases TRAIL protein expression levels
Western blot analysis demonstrated that, compared with the control, TRAIL protein expression levels were markedly declined following treatment with 1 µg/ml gefitinib (P<0.05; Fig. 4).
Figure 4.
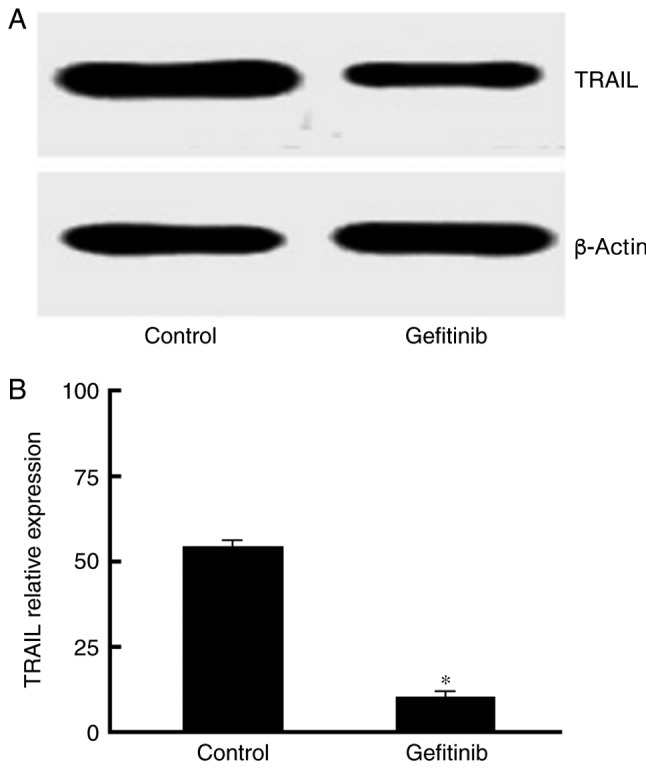
Gefitinib decreases TRAIL protein expression levels. (A) Western blot analysis of TRAIL expression. (B) Quantification of Caspase-3 activity.*P<0.05, compared with the control. TRAIL, tumor necrosis factor-related apoptosis-inducing ligand.
TRAIL knockdown enhances gefitinib-induced H1650 cell apoptosis
H1650 cells were transfected with TRAIL siRNA and treated with gefitinib. As presented in Fig. 5, caspase-3 activity was markedly increased in response to gefitinib treatment (P=0.011).
Figure 5.
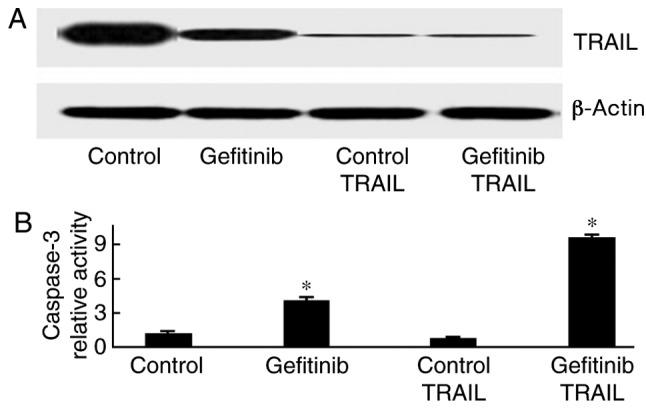
TRAIL knockdown enhances gefitinib-induced H1650 cell apoptosis. *P<0.05, compared with the control. TRAIL, tumor necrosis factor-related apoptosis-inducing ligand.
TRAIL plasmid transfection reduces gefitinib-induced cell apoptosis
H1650 cells were initially transfected with a TRAIL-containing plasmid and were subsequently treated with gefitinib. As presented in Fig. 6, caspase-3 activity was significantly weakened following gefitinib treatment (P=0.013).
Figure 6.
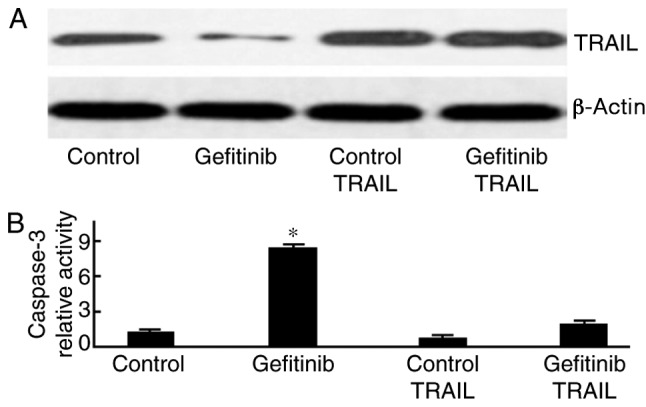
TRAIL-plasmid transfection reduces gefitinib-induced cell apoptosis. (A) Western blot analysis of TRAIL expression. (B) Quantification of Caspase-3 activity. *P<0.05, compared with the control. TRAIL, tumor necrosis factor-related apoptosis-inducing ligand.
Discussion
NSCLC severely threatens the lives of patients, but the molecular mechanism of NSCLC remains to be further investigated (1). A recent study suggested that microRNAs may serve a regulatory role in NSCLC cell proliferation and survival (29,30). The present study aimed to investigate the effects of gefitinib on H1650 cells. The results suggested that gefitinib significantly reduced H1650 cell viability and may have caused H1650 cell apoptosis.
To further analyze the molecular mechanism of gefitinib on NSCLC H1650 cells, various doses of gefitinib (1, 2, 5 and 10 µm) were applied for varying durations (12, 18, 24, 30 and 36 h) to H1650 cells (data not shown). Gefitinib may induce H1650 cell apoptosis in a dose- and time-dependent manner, which is highly consistent with other reports on different types of cancer cells. Additionally, gefitinib exhibited an antitumor effect by inducing apoptosis (3,15,19). Single TRAIL knockdown may not cause apoptosis; however, TRAIL is an important member of the anti-apoptotic proteins, which exhibit anti-apoptotic effects under apoptotic conditions. Its single knockdown did not cause apoptosis, which was the same as the mechanism of other anti-apoptosis proteins, including B-cell lymphoma 2 (Bcl-2) and Bcl-extra large.
There were three main observations of the present study: i) Gefitinib suppressed H1650 cell viability, induced H1650 cell apoptosis and downregulated TRAIL protein expression without affecting the genetic level; ii) TRAIL interference enhanced H1650 apoptosis induced by Gefitinib; and iii) TRAIL overexpression inhibited gefitinib-induced H1650 apoptosis. These results suggested that gefitinib induced the apoptosis of NSCLC H1650 cells by reducing TRAIL expression levels.
The present study also had several drawbacks and limitations: i) How gefitinib regulated TRAIL expression levels was not confirmed; ii) gefitinib-induced H1650 cell apoptosis via the downregulation of TRAIL expression levels was not investigated in an animal model; and iii) clinical cancer and para-carcinoma tissues were not obtained to investigate the association between gefitinib treatment, TRAIL expression levels and potential curative effects. Taken together, the results of the present study confirmed that gefitinib induced the apoptosis of NSCLC H1650 cells by decreasing TRAIL expression levels.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed are included in this published article.
Authors' contributions
HY, SL, HL, PW and HZ performed the experiments and analyzed the data. XC designed the study and wrote the manuscript.
Ethics statement and consent to participate
All experimental procedures involving animals were approved by the Ethnic Committee of Yinzhou Affiliated Hospital to Medical School of Ningbo University (Ningbo, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Cao W, Liu Y, Zhang R, Zhang B, Wang T, Zhu X, Mei L, Chen H, Zhang H, Ming P, Huang L. Homoharringtonine induces apoptosis and inhibits STAT3 via IL-6/JAK1/STAT3 signal pathway in Gefitinib-resistant lung cancer cells. Sci Rep. 2015;5:8477. doi: 10.1038/srep08477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang J, Guo F, Du Y, Liu X, Qin Q, Liu X, Yin T, Jiang L, Wang Y. Continuous exposure of non-small cell lung cancer cells with wild-type EGFR to an inhibitor of EGFR tyrosine kinase induces chemoresistance by activating STAT3. Int J Oncol. 2015;46:2083–2095. doi: 10.3892/ijo.2015.2898. [DOI] [PubMed] [Google Scholar]
- 3.Sudo M, Mori S, Madan V, Yang H, Leong G, Koeffler HP. Short-hairpin RNA library: Identification of therapeutic partners for gefitinib-resistant non-small cell lung cancer. Oncotarget. 2015;6:814–824. doi: 10.18632/oncotarget.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parkin DM, Bray F, Ferlay J, Pisani P. Estimating the world cancer burden: Globocan 2000. Int J Cancer. 2001;94:153–156. doi: 10.1002/ijc.1440. [DOI] [PubMed] [Google Scholar]
- 5.Garofalo M, Romano G, Di Leva G, Nuovo G, Jeon YJ, Ngankeu A, Sun J, Lovat F, Alder H, Condorelli G, et al. EGFR and MET receptor tyrosine kinase-altered microRNA expression induces tumorigenesis and gefitinib resistance in lung cancers. Nat Med. 2011;18:74–82. doi: 10.1038/nm.2577. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Ahn SH, Jeong EH, Lee TG, Kim SY, Kim HR, Kim CH. Gefitinib induces cytoplasmic translocation of the CDK inhibitor p27 and its binding to a cleaved intermediate of caspase 8 in non-small cell lung cancer cells. Cell Oncol (Dordr) 2014;37:377–386. doi: 10.1007/s13402-014-0198-0. [DOI] [PubMed] [Google Scholar]
- 7.Chao TT, Wang CY, Lai CC, Chen YL, Tsai YT, Chen PT, Lin HI, Huang YC, Shiau CW, Yu CJ, Chen KF. TD-19, an erlotinib derivative, induces epidermal growth factor receptor wild-type nonsmall-cell lung cancer apoptosis through CIP2A-mediated pathway. J Pharmacol Exp Ther. 2014;351:352–358. doi: 10.1124/jpet.114.215418. [DOI] [PubMed] [Google Scholar]
- 8.Bokobza SM, Jiang Y, Weber AM, Devery AM, Ryan AJ. Combining AKT inhibition with chloroquine and gefitinib prevents compensatory autophagy and induces cell death in EGFR mutated NSCLC cells. Oncotarget. 2014;5:4765–4778. doi: 10.18632/oncotarget.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang H, Zhao PJ, Su D, Feng J, Ma SL. Paris saponin I induces apoptosis via increasing the Bax/Bcl-2 ratio and caspase-3 expression in gefitinib-resistant non-small cell lung cancer in vitro and in vivo. Mol Med Rep. 2014;9:2265–2272. doi: 10.3892/mmr.2014.2108. [DOI] [PubMed] [Google Scholar]
- 10.Kong FM, Zhao J, Wang J, Faivre-Finn C. Radiation dose effect in locally advanced non-small cell lung cancer. J Thorac Dis. 2014;6:336–347. doi: 10.3978/j.issn.2072-1439.2014.01.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee JY, Lee YM, Chang GC, Yu SL, Hsieh WY, Chen JJ, Chen HW, Yang PC. Curcumin induces EGFR degradation in lung adenocarcinoma and modulates p38 activation in intestine: The versatile adjuvant for gefitinib therapy. PLoS One. 2011;6:e23756. doi: 10.1371/journal.pone.0023756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao W, Bao P, Qi H, You H. Resveratrol down-regulates survivin and induces apoptosis in human multidrug-resistant SPC-A-1/CDDP cells. Oncol Rep. 2010;23:279–286. [PubMed] [Google Scholar]
- 13.Zappa C, Mousa SA. Non-small cell lung cancer: Current treatment and future advances. Transl Lung Cancer Res. 2016;5:288–300. doi: 10.21037/tlcr.2016.06.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J, Viallet J, Haura EB. A small molecule pan-Bcl-2 family inhibitor, GX15-070, induces apoptosis and enhances cisplatin-induced apoptosis in non-small cell lung cancer cells. Cancer Chemother Pharmacol. 2008;61:525–534. doi: 10.1007/s00280-007-0499-3. [DOI] [PubMed] [Google Scholar]
- 15.Morgillo F, Kim WY, Kim ES, Ciardiello F, Hong WK, Lee HY. Implication of the insulin-like growth factor-IR pathway in the resistance of non-small cell lung cancer cells to treatment with gefitinib. Clin Cancer Res. 2007;13:2795–2803. doi: 10.1158/1078-0432.CCR-06-2077. [DOI] [PubMed] [Google Scholar]
- 16.Hotta K, Tabata M, Kiura K, Kozuki T, Hisamoto A, Katayama H, Takigawa N, Fujimoto N, Fujiwara K, Ueoka H, Tanimoto M. Gefitinib induces premature senescence in non-small cell lung cancer cells with or without EGFR gene mutation. Oncol Rep. 2007;17:313–317. [PubMed] [Google Scholar]
- 17.Fan XX, Li N, Wu JL, Zhou YL, He JX, Liu L, Leung EL. Celastrol induces apoptosis in gefitinib-resistant non-small cell lung cancer cells via caspases-dependent pathways and Hsp90 client protein degradation. Molecules. 2014;19:3508–3522. doi: 10.3390/molecules19033508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li B, Ren S, Li X, Wang Y, Garfield D, Zhou S, Chen X, Su C, Chen M, Kuang P, et al. MiR-21 overexpression is associated with acquired resistance of EGFR-TKI in non-small cell lung cancer. Lung Cancer. 2014;83:146–153. doi: 10.1016/j.lungcan.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 19.Song JY, Kim CS, Lee JH, Jang SJ, Lee SW, Hwang JJ, Lim C, Lee G, Seo J, Cho SY, Choi J. Dual inhibition of MEK1/2 and EGFR synergistically induces caspase-3-dependent apoptosis in EGFR inhibitor-resistant lung cancer cells via BIM upregulation. Invest New Drugs. 2013;31:1458–1465. doi: 10.1007/s10637-013-0030-0. [DOI] [PubMed] [Google Scholar]
- 20.Ansari J, Palmer DH, Rea DW, Hussain SA. Role of tyrosine kinase inhibitors in lung cancer. Anticancer Agents Med Chem. 2009;9:569–575. doi: 10.2174/187152009788451879. [DOI] [PubMed] [Google Scholar]
- 21.Zou M, Xia S, Zhuang L, Han N, Chu Q, Chao T, Peng P, Chen Y, Gui Q, Yu S. Knockdown of the Bcl-2 gene increases sensitivity to EGFR tyrosine kinase inhibitors in the H1975 lung cancer cell line harboring T790M mutation. Int J Oncol. 2013;42:2094–2102. doi: 10.3892/ijo.2013.1895. [DOI] [PubMed] [Google Scholar]
- 22.Chen G, Umelo IA, Lv S, Teugels E, Fostier K, Kronenberger P, Dewaele A, Sadones J, Geers C, De Grève J. miR-146a inhibits cell growth, cell migration and induces apoptosis in non-small cell lung cancer cells. PLoS One. 2013;8:e60317. doi: 10.1371/journal.pone.0060317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee YC, Lee LM, Yang CH, Lin AM, Huang YC, Hsu CC, Chen MS, Chi CW, Yin PH, Kuo CD, et al. Norcantharidin suppresses cell growth and migration with enhanced anticancer activity of gefitinib and cisplatin in human non-small cell lung cancer cells. Oncol Rep. 2013;29:237–243. doi: 10.3892/or.2012.2118. [DOI] [PubMed] [Google Scholar]
- 24.Zhang B, Jiao J, Liu Y, Guo LX, Zhou B, Li GQ, Yao ZJ, Zhou GB. Gefitinib analogue V1801 induces apoptosis of T790M EGFR-harboring lung cancer cells by up-regulation of the BH-3 only protein Noxa. PLoS One. 2012;7:e48748. doi: 10.1371/journal.pone.0048748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan XX, Yao XJ, Xu SW, Wong VK, He JX, Ding J, Xue WW, Mujtaba T, Michelangeli F, Huang M, et al. (Z)3,4,5,4′-trans-tetramethoxystilbene, a new analogue of resveratrol, inhibits gefitinb-resistant non-small cell lung cancer via selectively elevating intracellular calcium level. Sci Rep. 2015;5:16348. doi: 10.1038/srep16348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu T, Wang Z, Liu Y, Mei Z, Wang G, Liang Z, Cui A, Hu X, Cui L, Yang Y, Liu CY. Interleukin 22 protects colorectal cancer cells from chemotherapy by activating the STAT3 pathway and inducing autocrine expression of interleukin 8. Clin Immunol. 2014;154:116–126. doi: 10.1016/j.clim.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 27.Imamura Y, Wang PL, Masuno K, Sogawa N. Salivary protein histatin 3 regulates cell proliferation by enhancing p27(Kip1) and heat shock cognate protein 70 ubiquitination. Biochem Biophys Res Commun. 2016;470:269–274. doi: 10.1016/j.bbrc.2016.01.072. [DOI] [PubMed] [Google Scholar]
- 28.Alam MM, Sohoni S, Kalainayakan SP, Garrossian M, Zhang L. Cyclopamine tartrate, an inhibitor of Hedgehog signaling, strongly interferes with mitochondrial function and suppresses aerobic respiration in lung cancer cells. BMC Cancer. 2016;16:150. doi: 10.1186/s12885-016-2200-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li LH, Wu P, Lee JY, Li PR, Hsieh WY, Ho CC, Ho CL, Chen WJ, Wang CC, Yen MY, et al. Hinokitiol induces DNA damage and autophagy followed by cell cycle arrest and senescence in gefitinib-resistant lung adenocarcinoma cells. PLoS One. 2014;9:e104203. doi: 10.1371/journal.pone.0104203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inamura K, Ishikawa Y. MicroRNA in lung cancer: Novel biomarkers and potential tools for treatment. J Clin Med. 2016;5(pii):E36. doi: 10.3390/jcm5030036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed are included in this published article.