Abstract
The aim of the present study was to investigate the function of the DNA topoisomerase IIα (TOP2A) gene and its associated genes in the progression of papillary renal cell carcinoma (PRCC). Online cancer databases, including cBioportal, Oncomine, OncoLnc and Search Tool for the Retrieval of Interacting Genes/Proteins were used to analyze the TOP2A gene expression profile, function and regulation network in PRCC. The genes that were significantly co-expressed or mutually exclusively expressed with TOP2A were identified. The genes co-expressed with TOP2A were defined as a ‘TOP2A-cancer panel’, which cooperatively promotes PRCC progression. This gene panel performed well in predicting the prognosis of PRCC. In addition, the TOP2A-cancer panel significantly affected the outcome of PRCC compared with clear cell renal cell carcinoma (CCRCC). The protein-protein interaction network of all genes associated with TOP2A was also generated. This interaction network may provide foundation for the additional investigation of TOP2A. Integrative understating of the TOP2A-cancer panel may result in a novel avenue for treatment intervention in PRCC.
Keywords: papillary renal cell carcinoma, DNA topoisomerase IIα, cancer panel
Introduction
Cancer is a multi-gene-associated disease; the genes involved in each malignancy compose a ‘cancer panel’. This ‘cancer panel’ results in a complex protein regulation network that is able to determine the patterns of cancer cell behavior (1,2). Therefore, treatments targeting single genes may result in failure, as there are compensatory effects elicited by other genes that occur when a single pathway is blocked (3–5). Therefore, it is necessary to identify these ‘cancer panels’ in each type of cancer to promote an improved understanding of cell signaling transduction networks and enable the development of higher-efficacy treatments to control cancer cells.
PRCC is the second most common type of kidney cancer. It is also the most malignant type, without any effective therapies (6). Optimal treatment involves surgical removal of the tumor when the disease is in the early stages. However, there remains a lack of treatment options for patients with advanced-stage PRCC (7). Personalized medicine has aimed to distinguish the genetic differences or gene expression pattern alterations in each patient to enable physicians to provide the best treatment for the individual. Cancer is a heterogenetic disease in terms of somatic mutations or gene expression profile alterations in cancer cells (8). Differences in the patterns of gene expression determine the course of treatment to be administered. Therefore, the present study collected data from different cancer databases and integrated the data using a bioinformatics approach to identify a gene panel that affects the progress of PRCC.
DNA topoisomerase IIα (TOP2A) encodes DNA topoisomerase, which is an important enzyme that releases the torsional stress when DNA undergoes DNA replication and transcription (9). TOP2A actively participates in cellular proliferation (10). It is a critical gene in carcinogenesis (11,12). Additionally, mutations in TOP2A are a common cause of the failure of drugs that target the corresponding protein (11). There are numerous data demonstrating that TOP2A is involved in a range of cancer types, including breast, endometrial, colon and ovarian cancer (13–16).
The kidney epitheilal cell is a differentiated cell type. TOP2A is absent or expressed at low levels in kidney epithelial cells (17). A previous study revealed that TOP2A was upregulated in clear cell renal cell carcinoma (CCRCC), and that its expression was predictive of a poor patient outcome (18). Therefore, the present study aimed to identify whether TOP2A was also upregulated in PRCC and its function as a cancer driver, and attempted to mine data online using a bioinformatics approach to examine the cancer panel associated with TOP2A in PRCC.
Materials and methods
Bioinformatics analysis
The Cancer Genome Atlas (TCGA) was joint project launched by the National Cancer Institute and National Human Genome Research Institute, has generated comprehensive, multi-dimensional maps of the key genomic changes in 33 types of human cancer (https://cancergenome.nih.gov/) (19). The present study used the kidney cancer dataset in TCGA. The Oncomine cancer database is a comprehensive cancer database including almost all types of cancer (20). The present study assessed the copy number and gene expression levels of TOP2A in Oncomine (https://www.oncomine.org/resource) by searching the gene symbol and cancer type within the ‘Cancer vs. Normal Analysis’ analysis type filter on 21th March 2017. Furthermore, the outlier analysis tool of Oncomine was used to identify the ‘outlier genes’ that are only expressed in a number of cancer samples on 21th March 2017. The outlier set in the TOP2A positive sample accounted for 5–25% among all samples in the three independent studies (21–23). The survival rate curves were created using OncoLnc (http://www.OncoLnc.org/) on 27th March 2017 (24). The high and low expression groups were set at the upper and lower quartiles, respectively. The high TOP2A expression group was set at >394.46; whilst, the low TOP2A expression group was set at <99.61. Using OncoLnc, the survival curve, the Cox coefficient and the false discovery rate (FDR) were calculated on 27th March 2017. Multiple gene survival analysis was performed using survival tool in cBioPortal for Cancer Genomics (http://www.cbioportal.org) by searching the genes name simultaneously on 1st April 2017 (25,26). The data for the generation of the heat map was downloaded from cBioPortal and hierarchical clustering was performed with MeV software version 4.9.0 developed by GitHub on 11th April 2017 (http://mev.tm4.org/#/welcome). The protein-protein interaction network was completed using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING, version 10.5) by inputting all the genes on 16th April 2017 (http://string-db.org/).
Statistical analysis
One-way analysis of variance was conducted to analyze variance among multiple groups and a Student-Newman-Keuls test was used for post-hoc comparisons between the groups. Unpaired Student's t-test was performed for the comparison of mean values of two groups. Pearson's correlation analysis was conducted to test the correlation between genes. All these data analysis were performed using Graphpad Prism 5.0 (GraphPad Software, Inc., La Jolla, CA, USA) and the data were presented as the mean ± the standard deviation. Kaplan Meier analysis was used to compare the survival time between two groups with different expression levels of the genes of interest by OncoLnc. The log-rank test was conducted to compare the survival time distribution of the two groups. Hierarchical clustering was conducted for the generation of the gene expression signature heat map using MeV (Version 4.9.0) developed by GitHub, Inc. (San Francisco, CA, USA). Multivariate Cox regression analysis was used for evaluation of the gene expression of the genes assessed here on the patient's risk of mortality. The Cox coefficient, P-value, FDR and gene rank were calculated using the OncoLnc multivariate Cox regressions model tool (24). P<0.05 was considered to indicate a statistically significant difference.
Results
TOP2A is upregulated, and high expression of TOP2A contributes to poor outcome in PRCC
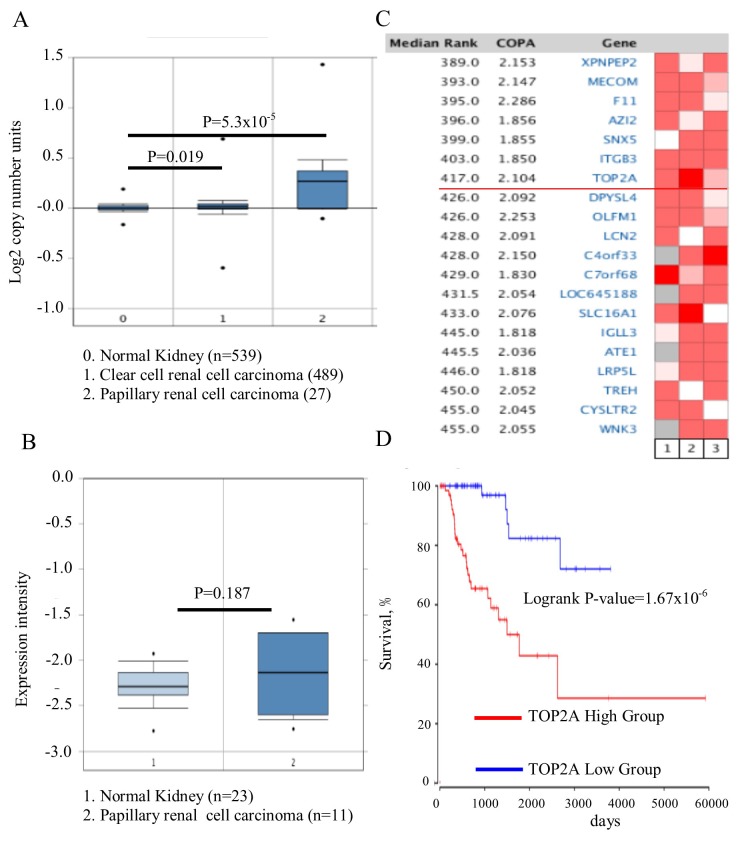
The Cancer Genome Atlas dataset of renal cell carcinoma, which includes 1,017 cases, was analyzed by Oncomine and it was identified that the TOP2A gene copy number in PRCC was significantly increased, compared with kidney or CCRCC (Fig. 1A; P=5.3×10−5). Whether the increase in gene copy number contributed to the upregulation of TOP2A was then assessed. The association between the copy number and its expression level was then analyzed in 23 normal kidney tissues and 11 PRCC tissues. No difference in the expression of TOP2A was observed between the PRCC tissues and normal kidney tissues (Fig. 1B; P=0.187). Owing to the heterogeneity of this cancer type, whether the TOP2A gene was upregulated only in certain patients with a specific genetic background. Therefore, the Oncomine outlier analysis tool was utilized, and 193 patient tumor tissues from 3 independent studies were analyzed (21–23). It was identified that TOP2A was expressed in a subset of patients with high expression of TOP2A (Fig. 1C); its association with the outcome of patients was additionally investigated. The difference in survival rates between the TOP2A high- and low-expression groups was analyzed using the Cox regression model, and it was identified that TOP2A expression was negatively associated with patient outcome (Fig. 1D; P=1.67×10−6). It was concluded that TOP2A was upregulated in one subset of patients with PRCC, and was predictive of poor prognosis.
Figure 1.
TOP2A is dysregulated in a subset of patients with PRCC and predicts poor outcome. (A) The copy number of TOP2A in PRCC was increased, compared with clear cell renal cell carcinoma and normal kidney tissue. (B) There was no significant differential expression of TOP2A in the tissues of PRCC and normal kidney cells. (C) TOP2A was upregulated in a subset of patients with PRCC using outlier analysis. (D) High expression of TOP2A in patients with PRCC predicts a poor survival rate. TOP2A, DNA topoisomerase IIα; COPA, copy number. PRCC, papillary renal cell carcinoma.
Co-expressed/mutually exclusively-expressed genes with TOP2A and their role in PRCC
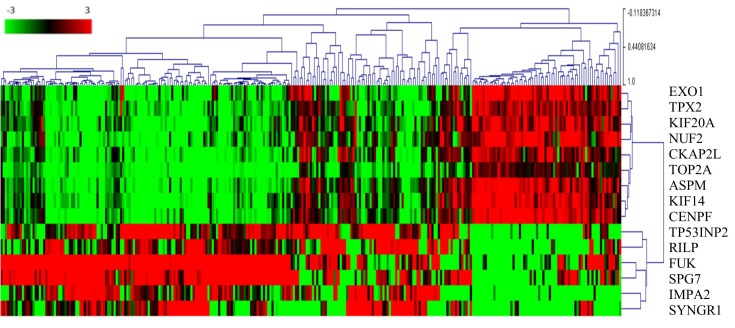
Pearson's correlation analysis was performed, and it was identified that the expression levels of a large number of genes were correlated with TOP2A. Genes with correlation coefficients >0.9 were screened out for additional analysis. The genes co-expressed with the TOP2A were: Abnormal spindle microtubule assembly (ASPM), exonuclease 1 (EXO1), TPX2, microtubule nucleation factor (TPX2), kinesin family member 14 (KIF14), cytoskeleton associated protein 2 like (CKAP2L), KIF20A, NUF, NDC80 kinetochore complex component (NUF2) and centromere protein F (CENPF). The genes that were inversely expressed with TOP2A were also probed. Pearson's correlation analysis was performed, and genes with correlation coefficients <-0.3 were selected. These genes included tumor protein P53 inducible nuclear protein 2 (TP53INP2), Rab interacting lysosomal protein (RILP), fucokinase (FUK), inositol monophosphatase 2 (IMPA2), SPG7, paraplegin matrix AAA peptidase subunit (SPG7) and synaptogyrin 1 (SYNGR1). A heat map for visualizing the association between these genes was generated. The gene expression profiles in the patient tumor tissues were characterized, and the two gene sets of genes that were mutually exclusively-expressed in tumor tissues were identified (Fig. 2). Furthermore, the two sets of genes were compared in CCRCC and PRCC. Not only was the expression level rank but also the Cox regression coefficient was significantly lower in the CCRCC compared with PRCC (Table I).
Figure 2.
TOP2A co-expressed and mutually expressed genes. The heat map depicts the Log2(expression level) of TOP2A-associated genes in patients with papillary renal cell carcinoma (each column represents one case). Green to red transition represents the range from −3 to 3 of gene expression after Log2 transformation. The number next to the branches represents the gene expression correlation with other cases. The higher a correlation value is for a case, the more similar the genes expression profile in the same cluster. EXO1, exonuclease 1; TPX2, TPX2, microtubule nucleation factor; KIF20A, kinesin family member 20A; NUF2, NUF, NDC80 kinetochore complex component; CKAP2L, cytoskeleton associated protein 2 like; TOP2A, DNA topoisomerase IIα; ASPM, abnormal spindle microtubule assembly; KIF14, kinesin family member 14; CENPF, centromere protein F; TP53INP2, tumor protein P53 inducible nuclear protein 2; RILP, Rab interacting lysosomal protein; FUK, fucokinase; SPG7, SPG7, paraplegin matrix AAA peptidase subunit; IMPA2, inositol monophosphatase 2; SYNGR1, synaptogyrin 1.
Table I.
Comparisons of Cox coefficient, P-value, FDR and gene rank between the papillary renal cell carcinoma and clear renal cell carcinoma samples using a Cox regression model.
| A, Co-expressed genes | ||||
|---|---|---|---|---|
| Gene/cancer types | Cox coefficient | P-value | Corrected FDR | Rank |
| TOP2A | ||||
| PRCC | 1.238 | 5.10×10−11 | 2.14×10−7 | 3 |
| CCRCC | 0.259 | 3.00×10−3 | 1.22×10−2 | 4,086 |
| TPX2 | ||||
| PRCC | 1.23 | 4.20×10−10 | 3.21×10−7 | 21 |
| CCRCC | 0.34 | 2.20×10−4 | 1.57×10−3 | 2,332 |
| EXO1 | ||||
| PRCC | 1.039 | 2.90×10−9 | 9.93×10−7 | 48 |
| CCRCC | 0.16 | 6.00×10−2 | 1.27×10−1 | 7,875 |
| KIF14 | ||||
| PRCC | 1.069 | 1.80×10−9 | 7.05×10−7 | 42 |
| CCRCC | 0.317 | 2.50×10−4 | 1.74×10−3 | 2,392 |
| KIF20A | ||||
| PRCC | 1.266 | 9.60×10−11 | 2.25×10−7 | 7 |
| CCRCC | 0.377 | 3.00×10−5 | 3.39×10−4 | 1,471 |
| ASPM | ||||
| PRCC | 1.259 | 1.10×10−10 | 2.26×10−7 | 8 |
| CCRCC | 0.286 | 7.60×10−4 | 4.16×10−3 | 3,044 |
| CKAP2L | ||||
| PRCC | 1.026 | 1.10×10−8 | 2.41×10−6 | 73 |
| CCRCC | 0.173 | 4.00×10−2 | 9.22×10−2 | 7,212 |
| NUF2 | ||||
| PRCC | 1.09 | 5.70×10−10 | 3.46×10−7 | 27 |
| CCRCC | 0.393 | 6.40×10−6 | 1.03×10−4 | 1,033 |
| CENPF | ||||
| PRCC | 1.137 | 3.00×10−10 | 3.21×10−7 | 15 |
| CCRCC | 0.289 | 1.10×10−3 | 5.50×10−3 | 3,283 |
| B, Mutually exclusive genes | ||||
| TP53INP2 | ||||
| PRCC | −0.77 | 2.00×10−6 | 1.30×10−4 | 253 |
| CCRCC | −0.262 | 1.70×10−3 | 7.78×10−3 | 3,644 |
| RILP | ||||
| PRCC | −0.657 | 1.40×10−4 | 2.59×10−3 | 885 |
| CCRCC | 0.099 | 2.20×10−1 | 3.40×10−1 | 10,750 |
| FUK | ||||
| PRCC | −0.688 | 1.40×10−5 | 4.86×10−4 | 467 |
| CCRCC | 0.035 | 6.40×10−1 | 7.43×10−1 | 14,316 |
| IMPA2 | ||||
| PRCC | −0.678 | 8.40×10−6 | 3.41×10−4 | 403 |
| CCRCC | −0.319 | 1.70×10−4 | 1.29×10−3 | 2,167 |
| SPG7 | ||||
| PRCC | −0.917 | 4.60×10−7 | 4.56×10−5 | 165 |
| CCRCC | 0.364 | 1.80×10−6 | 3.98×10−5 | 751 |
| SYNGR1 | ||||
| PRCC | −0.506 | 1.60×10−3 | 1.39×10−2 | 1,891 |
| CCRCC | 0.054 | 5.20×10−1 | 6.42×10−1 | 13,508 |
FDR, false discovery rate; PRCC, papillary renal cell carcinoma; CCRCC, clear cell renal cell carcinoma; ASPM, abnormal spindle microtubule assembly; EXO1, exonuclease 1; TPX2, TPX2, microtubule nucleation factor; KIF14, kinesin family member 14; CKAP2L, cytoskeleton associated protein 2 like; KIF20A, kinesin family member 20A; NUF2, NUF, NDC80 kinetochore complex component; CENPF, centromere protein F; TP53INP2, tumor protein P53 inducible nuclear protein 2; RILP, Rab interacting lysosomal protein; FUK, fucokinase; IMPA2, inositol monophosphatase 2; SPG7, SPG7, paraplegin matrix AAA peptidase subunit; SYNGR1, synaptogyrin 1.
TOP2A cancer panel predicts prognosis in PRCC
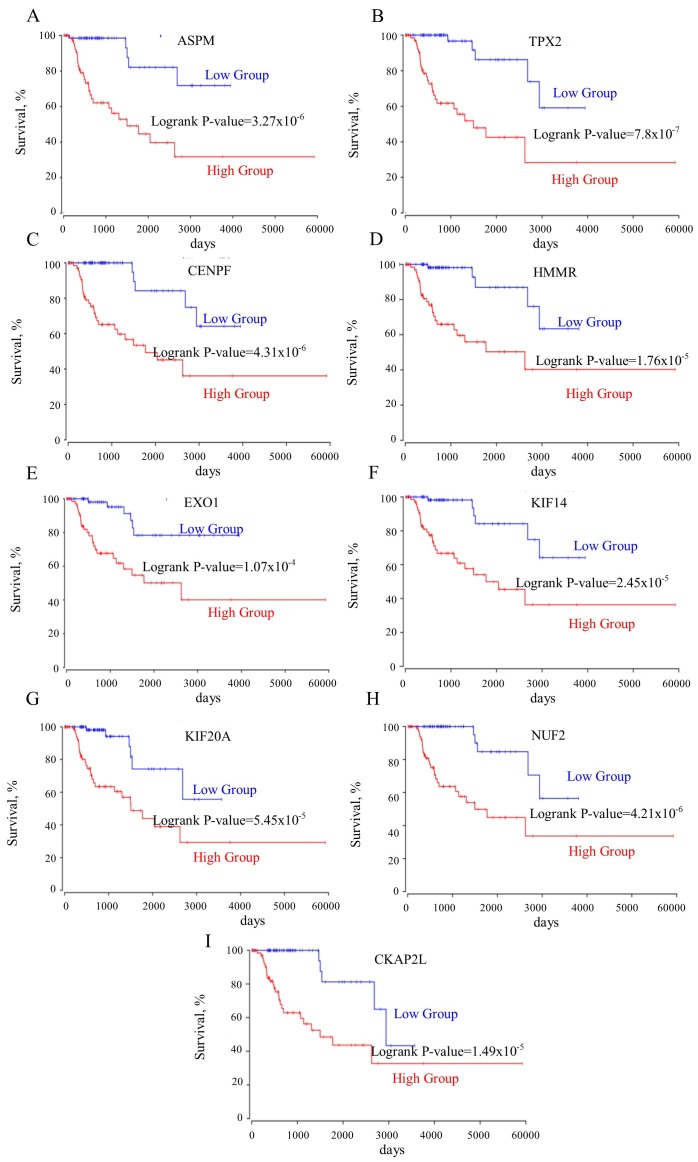
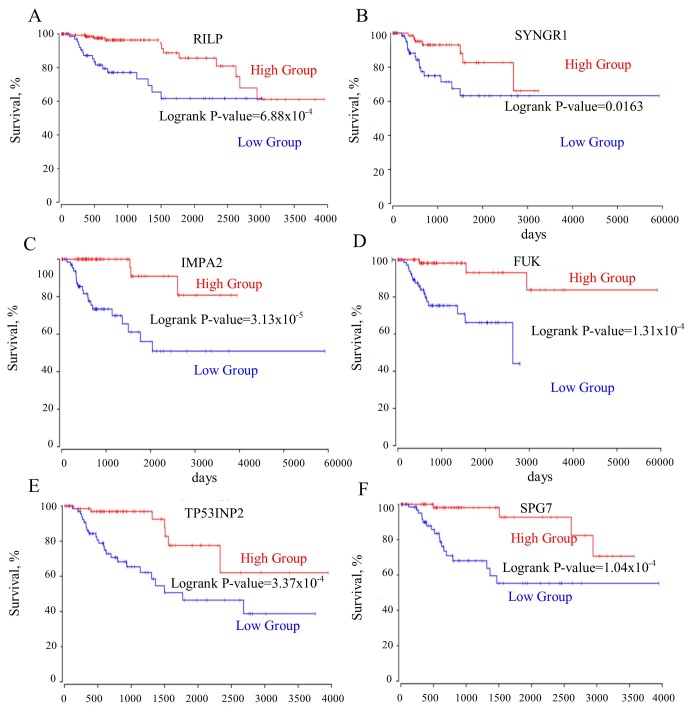
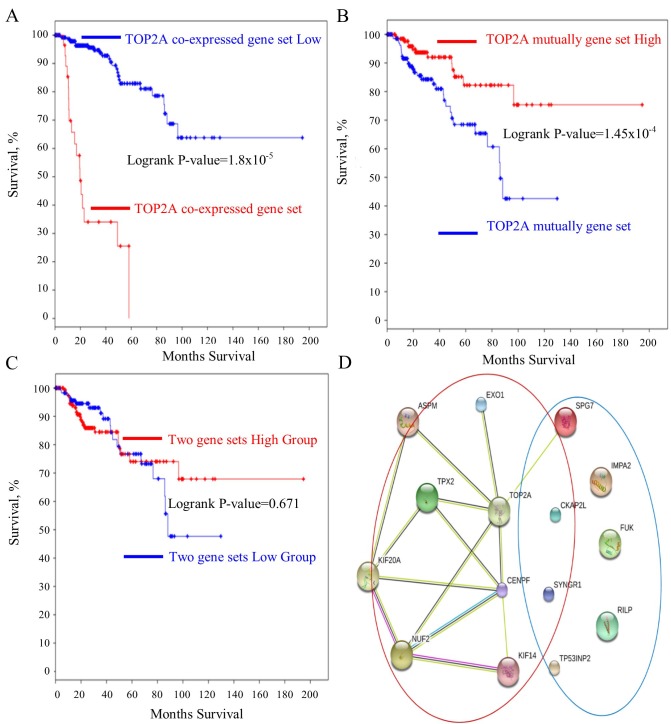
The roles of the genes in the TOP2A ‘panel’ in PRCC remain elusive. A survival curve analysis for each gene between their respective high- and low-expression groups was performed. It was identified that these genes were good prognostic markers. Notably, these genes performed better in predicting prognosis of the patient in PRCC compared with CCRCC (Table I). ASPM, TPX2, CENPF, hyaluronan mediated motility receptor (HMMR), EXO1, KIF14, KIF20A, NUF2, cytoskeleton associated protein 2 like (CKAP2L) predicted the shortest survival time of patients (Fig. 3A-I). However, the upregulation of the mutually exclusive genes (RILP, SYNGR1, IMPA2, FUK, TP53INP2, SPG7) prolonged the patient survival time (Fig. 4A-F). Furthermore, the mutually exclusive gene expression in the patients with TOP2A high expression may counteract the decreased survival time observed in the TOP2A-cancer panel gene expression analysis (Fig. 5A-C). The gene interaction network of the TOP2A-associated genes was analyzed with the STRING protein interaction analysis tool, and it was observed that the gene co-expressed with TOP2A form a ‘cancer panel’. The downregulated genes may serve as ‘tumor repressor panel’ (Fig. 5D).
Figure 3.
(A-I) All genes co-expressed with TOP2A are negatively associated with the survival rates of patients. ASPM, abnormal spindle microtubule assembly; TPX2, TPX2, microtubule nucleation factor; CENPF, centromere protein F; HMMR, hyaluronan mediated motility receptor; EXO1, exonuclease 1; KIF14, kinesin family member 14; KIF20A, kinesin family member 20A; NUF2, NUF, NDC80 kinetochore complex component; CKAP2L, cytoskeleton associated protein 2 like; TOP2A, DNA topoisomerase IIα.
Figure 4.
(A-F) All mutually exclusively expressed gene with TOP2A are positively associated with the survival rates of patients. RILP, Rab Interacting lysosomal protein; SYNGR1, synaptogyrin 1; IMPA2, Inositol monophosphatase 2; FUK, fucokinase; TP53INP2, tumor protein P53 inducible nuclear protein 2; SPG7, SPG7, paraplegin matrix AAA peptidase subunit; TOP2A, DNA topoisomerase IIα.
Figure 5.
TOP2A mutually exclusively expressed genes counteract the TOP2A co-expressed genes on reducing the survival rates of patients and the protein-protein interaction network between all genes of TOP2A-cancer gene set. (A) The survival rate of the patients with all TOP2A co-expressed gene upregulated. (B) The survival rates of the patients with all TOP2A mutually exclusively expressed gene upregulated. (C) The survival rates of patients with all the TOP2A-associated genes upregulated. (D) The protein-protein interaction network of all genes. Genes co-expressed with TOP2A are included in the red circle. Genes mutually exclusively expressed with TOP2A are included in the blue circle. TOP2A, DNA topoisomerase IIα.
Discussion
Oncogene mutations usually drive carcinogenesis, but gene alteration always results in the regulation of the expression of other associated genes. The expression of these associated genes may comprise a network, and their output may affect the behavior of cancer cells. These genes serve a vital role in establishing the properties of different types of cancer usually was defined as ‘cancer panels’ (27). The two most common types of kidney cancer are CCRCC and PRCC (28). CCRCC is sensitive to chemotherapeutics and patients generally exhibit improved outcomes following therapy compared with without therapy (29). However, PRCC often exhibits resistance to current chemotherapeutics (30). It was reported that TOP2A was upregulated in prostate cancer, breast cancer and serves as an indicator of a poor outcome (11,31). Previously, a number of studies have revealed that TOP2A is upregulated in CCRCC and is a poor predicative marker for pgrognosis (18,32). However, to the best of our knowledge, there are no stdies on whether TOP2A is dysregulated in PRCC. The present study identified that TOP2A is upregulated in kidney cancer and is significantly increased in PRCC compared with CCRCC. Additionally, TOP2A functioned well in predicting the prognoses of patients with PRCC compared with in CCRCC. This result indicated that TOP2A may be involved in PRCC.
The ‘cancer panel’ established in the present study included genes that were involved in similar biological functions and contributed to cancer progression. These genes function concomitantly to affect the behavior of cancer cells. Identification of the associations between these genes and their interaction network may enable an improved understanding of how TOP2A causes the development of PRCC. To intervene in cancer progression, the interruption of one driver gene is not sufficient for the complete inhibition of cancer progression. Integral disruption of all of the genes involved in cancer progression is more important for the successful treatment of cancer, than focusing on a single target gene. Therefore, the present study analyzed the genes that were closely associated with TOP2A. Genes that were significantly co-expressed with TOP2A were selected, and it was hypothesized that these genes may be simultaneously involved in PRCC progression. To confirm the function of these genes in PRCC, the survival curves for the high- and low-expression groups of each gene were generated. Notably, it was identified that the expression of these genes significantly reduced the survival time of patients. Therefore, the expression levels of these genes was not only associated with TOP2A, but also upregulation of these genes would reduce the survival rates of the patients. This result indicated that TOP2A may prompt PRCC progression in conjunction with other genes; however, whether TOP2A regulates the expression of these genes requires additional investigation.
Considering the genes in the ‘TOP2A-cancer panel’, the present study aimed to identify the processes they are involved in, and to understand how these genes function together to determine cancer cell properties. The ASPM gene is closely associated with spindle function, which is involved in cell mitosis (33). The TPX2 gene is a spindle assembly factor that servers as a critical role in G2/M transition of cell cycle (34). The HMMR gene encodes a protein that forms a complex with BRCA1/2, which promotes cell proliferation and increases the risk of cancer (35). The CENPF gene is required for chromosome segregation in cell mitosis, which regulates DNA replication and cell cycle progression (36). EXO1 encodes an exonuclease that is responsible for DNA mismatch repair (37). KIF14 contributes to chromosome segregation and spindle formation in the mitosis process (38). KIF20A functions as a spindle assembly mediator, resulting in cell division (39). CKAP2L is involved in spindle organization (40). NUF2 regulates chromosome segregation and centromere function in the cell mitosis (41). Therefore, the genes within the TOP2A-derived cancer panel function in the regulation of cell mitosis. According to the protein-protein network (Fig. 5D), the results of the present study indicated that TOP2A may serve a vital role in the regulation of cell proliferation through interaction with the TOP2A cancer panel.
The present study compared the association between the expression levels of these genes with the survival rates of patients with CCRCC and PRCC. It was identified that these genes that coexpressed with TOP2A significantly increase the survival rate of patients with PRCC compared with patients with CCRCC. However, the genes that inversely expressed with TOP2A decrease the survival rates of patients with PRCC compared with patients with CCRCC, which may provide a method for distinguishing between renal cell carcinoma subtypes by the expression of the TOP2A cancer panel genes. The present study identified that TOP2A was a vital prognostic marker for PRCC, and the genes involved in the network of TOP2A were examined. This network of TOP2A genes may assist in understanding how TOP2A affects cancer cells, and how targeting these genes may provide an avenue for the treatment of PRCC.
Acknowledgements
The results shown here are in whole or part based upon data generated by the TCGA Research Network. The authors also would like to thank Dr. Qingyu Zhang (Faculty of Health Sciences, University of Macau, Macau, China) for providing advice on the data interpretation and critical comments on the discussion.
Funding
This present study was by partially supported by the Zhanjiang scientific research project (grant no. 2016C01005) and Zhanjiang scientific special competition project (grant no. 2016A01011).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
MY and ZH collected data and developed the methodology. DW, ZI and XC interpreted the results. MY and JL designed the work and wrote the manuscript.
Ethics approval and consent to publish
This study reinterpreted the data deposited in TCGA and GEO without releasing the information of patients. According to TCGA publication guidelines, there are no restrictions on the use of TCGA data for research purposes.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Dalton E, Thompson J. AB054. Overview of multi-gene panels for hereditary cancer. Ann Transl Med. 2015;3:AB054. [Google Scholar]
- 2.Lincoln SE, Kobayashi Y, Anderson MJ, Yang S, Desmond AJ, Mills MA, Nilsen GB, Jacobs KB, Monzon FA, Kurian AW, et al. A systematic comparison of traditional and multigene panel testing for hereditary breast and ovarian cancer genes in more than 1000 patients. J Mol Diagn. 2015;17:533–544. doi: 10.1016/j.jmoldx.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 3.Housman G, Byler S, Heerboth S, Lapinska K, Longacre M, Snyder N, Sarkar S. Drug resistance in cancer: An overview. Cancers (Basel) 2014;6:1769–1792. doi: 10.3390/cancers6031769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo XE, Ngo B, Modrek AS, Lee WH. Targeting tumor suppressor networks for cancer therapeutics. Curr Drug Targets. 2014;15:2–16. doi: 10.2174/1389450114666140106095151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Q, Madden NE, Wong AST, Chow BKC, Lee LTO. The role of endocrine G protein-coupled receptors in ovarian cancer progression. Front Endocrinol (Lausanne) 2017;8:66. doi: 10.3389/fendo.2017.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shuch B, Hahn AW, Agarwal N. Current treatment landscape of advanced papillary renal cancer. J Clin Oncol. 2017;35:2981–2983. doi: 10.1200/JCO.2017.74.3328. [DOI] [PubMed] [Google Scholar]
- 7.Ross H, Martignoni G, Argani P. Renal cell carcinoma with clear cell and papillary features. Arch Pathol Lab Med. 2012;136:391–399. doi: 10.5858/arpa.2011-0479-RA. [DOI] [PubMed] [Google Scholar]
- 8.Friedman AA, Letai A, Fisher DE, Flaherty KT. Precision medicine for cancer with next-generation functional diagnostics. Nat Rev Cancer. 2015;15:747–756. doi: 10.1038/nrc4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, Darnell J. Molecular Cell Biology. 4th edition. W.H. Freeman; New York, NY: 2000. The Role of topoisomerases in DNA replication. [Google Scholar]
- 10.Chen T, Sun Y, Ji P, Kopetz S, Zhang W. Topoisomerase IIα in chromosome instability and personalized cancer therapy. Oncogene. 2015;34:4019–4031. doi: 10.1038/onc.2014.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Resende MF, Vieira S, Chinen LT, Chiappelli F, da Fonseca FP, Guimarães GC, Soares FA, Neves I, Pagotty S, Pellionisz PA, et al. Prognostication of prostate cancer based on TOP2A protein and gene assessment: TOP2A in prostate cancer. J Transl Med. 2013;11:36. doi: 10.1186/1479-5876-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fountzilas G, Valavanis C, Kotoula V, Eleftheraki AG, Kalogeras KT, Tzaida O, Batistatou A, Kronenwett R, Wirtz RM, Bobos M, et al. HER2 and TOP2A in high-risk early breast cancer patients treated with adjuvant epirubicin-based dose-dense sequential chemotherapy. J Transl Med. 2012;10:10. doi: 10.1186/1479-5876-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ito F, Furukawa N, Nakai T. Evaluation of top2a as a predictive marker for endometrial cancer with taxane-containing adjuvant chemotherapy. Int J Gynecol Cancer. 2016;26:325–330. doi: 10.1097/IGC.0000000000000607. [DOI] [PubMed] [Google Scholar]
- 14.Erriquez J, Becco P, Olivero M, Ponzone R, Maggiorotto F, Ferrero A, Scalzo MS, Canuto EM, Sapino A, Verdun di Cantogno L, et al. TOP2A gene copy gain predicts response of epithelial ovarian cancers to pegylated liposomal doxorubicin: TOP2A as marker of response to PLD in ovarian cancer. Gynecol Oncol. 2015;138:627–633. doi: 10.1016/j.ygyno.2015.06.025. [DOI] [PubMed] [Google Scholar]
- 15.Tarpgaard LS, Qvortrup C, Nygård SB, Nielsen SL, Andersen DR, Jensen NF, Stenvang J, Detlefsen S, Brünner N, Pfeiffer P. A phase II study of epirubicin in oxaliplatin-resistant patients with metastatic colorectal cancer and TOP2A gene amplification. BMC Cancer. 2016;16:91. doi: 10.1186/s12885-016-2124-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, Xu B, Yuan P, Zhang P, Li Q, Ma F, Fan Y. TOP2A amplification in breast cancer is a predictive marker of anthracycline-based neoadjuvant chemotherapy efficacy. Breast Cancer Res Treat. 2012;135:531–537. doi: 10.1007/s10549-012-2167-5. [DOI] [PubMed] [Google Scholar]
- 17.Cheburkin IuV, Kniazeva TG, Peter S, Kniazev IuP, Karelin MI, Shkol'nik MI, Evtushenko VI, Hanson KP, Ullrich A, Kniazev PG. Molecular portrait of human kidney carcinomas: The gene expression profiling of protein-tyrosine kinases and tyrosine phosphatases which controlled regulatory signals in the cells. Mol Biol (Mosk) 2002;36:480–490. doi: 10.1023/A:1016059313254. (In Russian) [DOI] [PubMed] [Google Scholar]
- 18.Chen D, Maruschke M, Hakenberg O, Zimmermann W, Stief CG, Buchner A. TOP2A, HELLS, ATAD2 and TET3 are novel prognostic markers in renal cell carcinoma. Urology. 2017;102:265 e1–265 e7. doi: 10.1016/j.urology.2016.12.050. [DOI] [PubMed] [Google Scholar]
- 19.Grossman RL, Heath AP, Ferretti V, Varmus HE, Lowy DR, Kibbe WA, Staudt LM. Toward a shared vision for cancer genomic data. N Engl J Med. 2016;375:1109–1112. doi: 10.1056/NEJMp1607591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones J, Otu H, Spentzos D, Kolia S, Inan M, Beecken WD, Fellbaum C, Gu X, Joseph M, Pantuck AJ, et al. Gene signatures of progression and metastasis in renal cell cancer. Clin Cancer Res. 2005;11:5730–5739. doi: 10.1158/1078-0432.CCR-04-2225. [DOI] [PubMed] [Google Scholar]
- 22.Lenburg ME, Liou LS, Gerry NP, Frampton GM, Cohen HT, Christman MF. Previously unidentified changes in renal cell carcinoma gene expression identified by parametric analysis of microarray data. BMC Cancer. 2003;3:31. doi: 10.1186/1471-2407-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dannull J, Su Z, Rizzieri D, Yang BK, Coleman D, Yancey D, Zhang A, Dahm P, Chao N, Gilboa E, Vieweg J. Enhancement of vaccine-mediated antitumor immunity in cancer patients after depletion of regulatory T cells. J Clin Invest. 2005;115:3623–3633. doi: 10.1172/JCI25947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anaya J. OncoLnc: Linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Computer Science. 2016;2:e67. doi: 10.7717/peerj-cs.67. [DOI] [Google Scholar]
- 25.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:p11. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cbio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LaDuca H, Stuenkel AJ, Dolinsky JS, Keiles S, Tandy S, Pesaran T, Chen E, Gau CL, Palmaer E, Shoaepour K, et al. Utilization of multigene panels in hereditary cancer predisposition testing: Analysis of more than 2,000 patients. Genet Med. 2014;16:830–837. doi: 10.1038/gim.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Capitanio U, Montorsi F. Renal cancer. Lancet. 2016;387:894–906. doi: 10.1016/S0140-6736(15)00046-X. [DOI] [PubMed] [Google Scholar]
- 29.Escudier B, Eisen T, Stadler WM, Szczylik C, Oudard S, Siebels M, Negrier S, Chevreau C, Solska E, Desai AA, et al. Sorafenib in advanced clear-cell renal-cell carcinoma. N Engl J Med. 2007;356:125–134. doi: 10.1056/NEJMoa060655. [DOI] [PubMed] [Google Scholar]
- 30.Ronnen EA, Kondagunta GV, Ishill N, Spodek L, Russo P, Reuter V, Bacik J, Motzer RJ. Treatment outcome for metastatic papillary renal cell carcinoma patients. Cancer. 2006;107:2617–2621. doi: 10.1002/cncr.22340. [DOI] [PubMed] [Google Scholar]
- 31.Wang J, Xu B, Yuan P, Zhang P, Li Q, Ma F, Fan Y. TOP2A amplification in breast cancer is a predictive marker of anthracycline-based neoadjuvant chemotherapy efficacy. Breast Cancer Res Treat. 2012;135:531–537. doi: 10.1007/s10549-012-2167-5. [DOI] [PubMed] [Google Scholar]
- 32.Parker AS, Eckel-Passow JE, Serie D, Hilton T, Parasramka M, Joseph RW, Wu KJ, Cheville JC, Leibovich BC. Higher expression of topoisomerase II alpha is an independent marker of increased risk of cancer-specific death in patients with clear cell renal cell carcinoma. Eur Urol. 2014;66:929–935. doi: 10.1016/j.eururo.2013.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barr FA, Silljé HH, Nigg EA. Polo-like kinases and the orchestration of cell division. Nat Rev Mol Cell Biol. 2004;5:429–440. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- 34.Kufer TA, Silljé HH, Körner R, Gruss OJ, Meraldi P, Nigg EA. Human TPX2 is required for targeting Aurora-A kinase to the spindle. J Cell Biol. 2002;158:617–623. doi: 10.1083/jcb.200204155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parvin JD, Kais Z, Arora M, Kotian S, Zha A, Ransburgh D, Bozdag D, Catalyurek U, Huang K. Proceedings of the 2009 Ohio Collaborative Conference on Bioinformatics. Bioinformatics. Cleveland, OH: 2009. Identification of a breast cancer associated regulatory network; pp. 71–75. [DOI] [Google Scholar]
- 36.Taylor SS, Scott MI, Holland AJ. The spindle checkpoint: A quality control mechanism which ensures accurate chromosome segregation. Chromosome Res. 2004;12:599–616. doi: 10.1023/B:CHRO.0000036610.78380.51. [DOI] [PubMed] [Google Scholar]
- 37.Kolodner RD, Marsischky GT. Eukaryotic DNA mismatch repair. Curr Opin Genet Dev. 1999;9:89–96. doi: 10.1016/S0959-437X(99)80013-6. [DOI] [PubMed] [Google Scholar]
- 38.Rath O, Kozielski F. Kinesins and cancer. Nat Rev Cancer. 2012;12:527–539. doi: 10.1038/nrc3310. [DOI] [PubMed] [Google Scholar]
- 39.Yu Y, Feng YM. The role of kinesin family proteins in tumorigenesis and progression: Potential biomarkers and molecular targets for cancer therapy. Cancer. 2010;116:5150–5160. doi: 10.1002/cncr.25461. [DOI] [PubMed] [Google Scholar]
- 40.Hong KU, Kim E, Bae CD, Park J. TMAP/CKAP2 is essential for proper chromosome segregation. Cell Cycle. 2009;8:314–324. doi: 10.4161/cc.8.2.7597. [DOI] [PubMed] [Google Scholar]
- 41.DeLuca JG, Dong Y, Hergert P, Strauss J, Hickey JM, Salmon ED, McEwen BF. Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol Biol Cell. 2005;16:519–531. doi: 10.1091/mbc.e04-09-0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.