Abstract
Lung cancer is the leading cause of mortalities among all types of cancer. Therefore, the screening of biomarkers that are related with the progression of lung cancer is crucial for early diagnosis and efficient therapy of lung cancer. In the present study, bioinformatic analysis identified replication factor C 5 (RFC5) as a potential novel oncogene in lung cancer. RFC5 functions as a clamp loader and is involved in DNA replication and repair. Analysis of public databases and reverse transcription-quantitative polymerase chain reaction indicated that RFC5 was significantly increased in tumor tissues compared with adjacent normal tissues. A high RFC5 expression was observed to be associated with more aggressive malignant clinicopathological features, including higher T stage, more advanced regional lymph node metastasis and a higher probability of relapse. Notably, there were notable differences in overall survival (OS), first progression and post-progression survival between the high RFC5 expression group and low RFC5 expression group. Univariate and multivariate Cox regression analyses indicated that RFC5 was an independent risk factor that was associated with poorer OS and disease-free survival. According to GSEA, several gene sets that are associated with cell cycle and DNA damage were enriched in the RFC5 overexpression group, which indicated that RFC5 might promote the proliferation of lung cancer cells. Our finding indicated that RFC5 might be a novel prognostic biomarker of lung cancer, and it might be serve as a potential diagnosis and therapy target for lung cancer in the future.
Keywords: replication factor C 5, lung cancer, prognosis, proliferation, bioinformatic analysis, biomarker
Introduction
Lung cancer is the leading cause of cancer-associated mortality. It was estimated that 155,870 mortalities due to lung cancer occurred in the United States in 2017, which accounted for more than a quarter (26%) of all cancer mortalities (1). Patients with non-small cell lung cancer (NSCLC) approximately account for 80% of lung cancer cases (2). Surgical resection is the best treatment modality for localized lung cancer. However, 79% of lung cancer cases are diagnosed at an advanced stage. The cure rate of patients with advanced lung cancer using conventional chemotherapy is low (3,4).
Tumor-associated genetic alterations have essential roles in the tumorigenesis and progression of lung cancer (5). Extensive study has been focused on finding oncogenes for the early diagnosis or effective therapy for lung cancer. Several drugs that target molecular signaling pathways have been applied to lung cancer treatment, particularly for advanced lung cancer. For example, erlotinib, gefitinib and afatinib worked efficiently for treating lung cancer cases with epidermal growth factor receptor mutations (6–8). Crizotinib is an anaplastic lymphoma kinase inhibitor for treating lung cancer cases with ROS1 translocations (9). However, the molecular pathogenesis of lung cancer has not been fully elucidated. Therefore, finding novel key genes that are associated with tumor progression and prognosis to further understand the molecular mechanisms of lung cancer is necessary.
Replication factor C (RFC), also known as activator 1, was originally purified from the extraction of HeLa cells at the end of 1980s (10). RFC was reported to be a necessary factor for DNA replication of simian virus 40 in vitro (11). One large subunit (RFC1/RFC140) and four small subunits (RFC2/RFC37, RFC3/RFC36, RFC4/RFC40 and RFC5/RFC38) make up the RFC complex and they were commonly found in numerous eukaryotes (12,13). RFC functions as a clamp loader, which has a crucial role in loading proliferating cell nuclear antigen (PCNA) onto primed DNA to elongate the DNA chain (14,15).
Several RFC proteins have been found to be involved in promoting cell proliferation in multiple carcinomas (16–20). Among them, RFC5 subunit has been demonstrated to interact with chromosome transmission fidelity protein 18 homolog (CTF18), This type of interaction not only have a role in sister chromatid cohesion but also stabilize the genome (21,22). Peng et al (23) reported that RFC5 mediated resistance to temozolomide in glioma cells, which was independent of methylguanine-DNA methyltransferase, and its expression was positively regulated by forkhead box M1 (23). The upregulation of the RFC5 gene in multidrug-resistant leukemia cells compared with parental HL-60 cell indicated that RFC5 might participate in drug resistance (24). However, the molecular mechanisms underlying lung cancer and the prognostic value of RFC5 remain to be reported.
The present study aimed to identify whether RFC5 has a key role in promoting lung cancer progression and to investigate its biological function. Furthermore, the prognostic implication was also evaluated by using bioinformatic approaches. Our results showed that RFC5 might serve as an independent predictor for prognosis and a potential therapeutic target for lung cancer.
Materials and methods
Utilization of expression profile datasets
All lung cancer microarray data with large sample sizes were downloaded from the National Center for Biotechnology Information (National Institutes of Health, Bethesda, MD, USA) Gene Expression Omnibus database (GEO; http://www.ncbi.nlm.nih.gov/geo/). The relative expression levels of RFC5 and clinical characteristics as well as follow-up information were extracted for statistical analysis. The probes corresponding to RFC5 were ‘203210_at’ and ‘203209_at’ with which the expression of RFC5 was traced. The basic features of the database were summarized in Table I. In addition, ONCOMINE was used to compare RFC5 expression levels in multiple datasets between cancer specimens and normal specimens. Cluster analysis of RFC5 expression between lung cancer histological subtypes and normal lung tissues was further performed across 6 analyses. The thresholds, two-fold change, P<0.001 and the top 10% gene rank, were used to reduce the false discovery rate (FDR). The ONCOMINE data are available from https://www.oncomine.org.
Table I.
Basic information of the 10 Gene Expression Omnibus datasets.
| Cancer type | Accession no. | Number of samples (tumor/normal) | P-value |
|---|---|---|---|
| Breast carcinoma | GSE10780 | 42/143 | <0.0001 |
| Esophageal carcinoma | GSE23400 | 53/53 | <0.0001 |
| Lung carcinoma | GSE30219 | 293/14 | <0.0001 |
| Gastric cancer | GSE13861 | 71/19 | <0.0001 |
| Acute lymphoblastic leukemia | GSE26713 | 117/7 | 0.1439 |
| Colorectal cancer | GSE32323 | 17/17 | 0.0022 |
| Bladder cancer | GSE3167 | 41/9 | 0.0134 |
| Melanoma | GSE3189 | 45/25 | 0.1813 |
| Cervical cancer | GSE14407 | 33/24 | <0.0001 |
| Nasopharyngeal carcinoma | GSE12452 | 31/10 | <0.0001 |
P-values were acquired using the t-test for the comparison of replication factor C 5 expression between cancer and normal tissues.
Tissue samples and RNA extraction
A total of 26 lung cancer and matched adjacent lung cancer tissues were collected from patients with primary lung cancer surgical resection at the Renmin Hospital of Wuhan University (Hubei, China) from May to July 2017. None of the patients received chemotherapy or radiotherapy prior to operation. The present study was approved by the Ethics Committee of Renmin Hospital of Wuhan University. Written informed consents were obtained from all patients prior to enrollment in the study and anonymity was guaranteed. All samples were immediately cut into pieces and stored in liquid nitrogen. Total RNA was extracted from tissue using TRIzol and quantified by NanoDrop 2000 (Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Reverse transcription was performed using 2 µg total RNA with the RevertAid RT Reverse Transcription kit (Thermo Fisher Scientific, Inc.). According to the manufacturer's protocol, qPCR was performed using the QuantStudio 6 Flex Real-Time PCR system (Thermo Fisher Scientific, Inc.) with SYBR Premix Ex Taq™ II (Takara Bio, Inc., Otsu, Japan). The cycling parameters were 50°C for 30 min, 94.5°C for 15 min, then 40 cycles of 96°C for 30 sec and 59.7°C for 1 min. The sequences of the primers are as follows: RFC5 forward, 5′-GAAGCAGACGCCATGACTCAG-3′ and reverse, 5′-GACCGAACCGAAACCTCGT-3′; β-actin forward, 5′-GAAGAGCTACGAGCTGCCTGA-3′ and reverse, 5′-CAGACAGCACTGTGTTGGCG-3′. The relative expression levels of RFC5 were quantified using RT-qPCR and the 2−ΔΔCq method in triplicate and normalized to β-actin (25).
Gene set enrichment analysis (GSEA)
GSEA was performed using a Java GSEA desktop application that was downloaded from http://www.broad.mit.edu/gsea/. The GSE3141 dataset was analyzed with the GMT file (c5.all.v5.1) gene set, to obtain biological processes enriched by RFC5. The samples were divided into a high RFC5 expression group (top 50%) and a low RFC5 expression group (bottom 50%). A total of four files including expression datasets, gene sets, phenotype labels and chip platforms were loaded for running GSEA according to the manufacturer's specifications. Significant gene sets were confirmed with nominal P-value <0.05 and FDR <0.25.
Kaplan-Meier plotter database analysis
Kaplan-Meier plotter (www.kmplot.com) was used to assess the prognostic significance of RFC5 expression in lung cancer. The database includes gene expression data and clinicopathological features of lung (26), breast (27), ovarian (28) and gastric cancer (29). In order to evaluate the prognostic value of RFC5, the patient samples were divided into two groups, high expression and low expression, on the basis of the median expression of the RFC5 (high expression, top 50%; low expression, bottom 50%). The Kaplan-Meier survival plots were obtained by entering the survival time [overall survival (OS), first progression (FP) and post-progression survival (PPS)] of patients with NSCLC, respectively. The P-values of the log-rank test and hazard ratio (HR) with 95% confidence intervals (CIs) were also calculated.
Statistical analysis
Statistical analyses were performed with GraphPad Prism (version 5.0; GraphPad Software, Inc., La Jolla, CA, USA) and SPSS (version 19.0; SPSS, Inc., Chicago, IL, USA). A paired t-test was used to compare the differences between the expression level of RFC5 in tumor and adjacent normal tissues. An unpaired t-test was used for unpaired comparisons. The comparisons between the experimental groups and the healthy group were performed using one-way ANOVA followed by Dunnett's multiple comparisons test. The P-values were derived from ranked data as variance was unequal among groups (Bartlett's test, P<0.05), and the values were adjusted by Bonferroni's test. The associations between RFC5 expression and clinicopathological parameters were analyzed using the χ2 test. Cox regression was used for univariate and multivariate analysis to determine the independent factors that have a significant effect on patient survival. The HR and 95% CIs of the prognostic factors were calculated. P<0.05 was considered to indicate a statistically significant difference.
Results
RFC5 is upregulated in multiple types of cancer
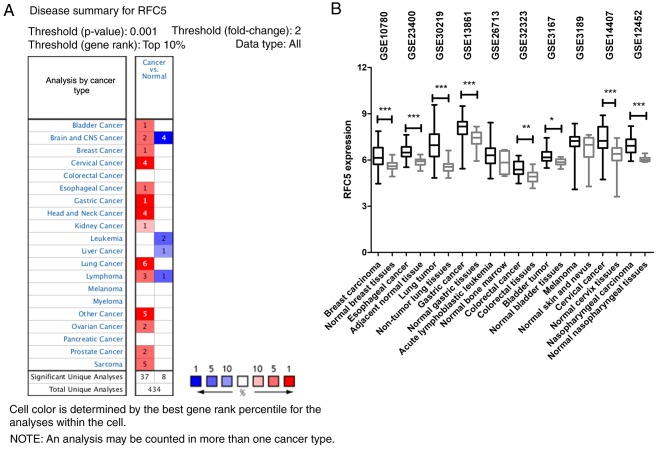
The ONCOMINE database was used to compare the levels of RFC5 expression in cancer and normal samples. The results indicated that RFC5 was overexpressed in multiple cancer types (Fig. 1A). Among 20 common types of cancer, the upregulation of RFC5 was identified in 14 cancer types. The GEO database was searched to analyze the differences in RFC5 expression levels in various tumors. As indicated in Fig. 1B, RFC5 was upregulated in breast carcinoma (P<0.0001), esophageal cancer (P<0.0001), lung tumor (P<0.0001), gastric cancer (P<0.0001), colorectal cancer (P=0.0022), bladder tumor (P=0.0134), cervical cancer (P<0.0001) and nasopharyngeal carcinoma (P<0.0001) tissues compared with normal tissues. These data indicated that RFC5 was upregulated in a variety of tumor types, which suggested it might be relevant to the oncogenesis of these tumors.
Figure 1.
Analysis of RFC5 expression in various types of human cancer. (A) The ONCOMINE database was queried for RFC5 expression between cancer and normal specimens by using the criteria: Two-fold change for RFC5 expression, top 10% gene rank and P-value<0.001. Red, RFC5 upregulation; blue, RFC5 downregulation. (B) RFC5 expression was visualized by using 10 publicly available Gene Expression Omnibus datasets. RFC5 expression was transformed into log2 (probe intensities) and presented as the mean ± standard error. *P<0.05; **P<0.01; ***P<0.001. CNS, central nervous system; RFC5, replication factor C subunit 5.
RFC5 is significantly overexpressed in lung cancer
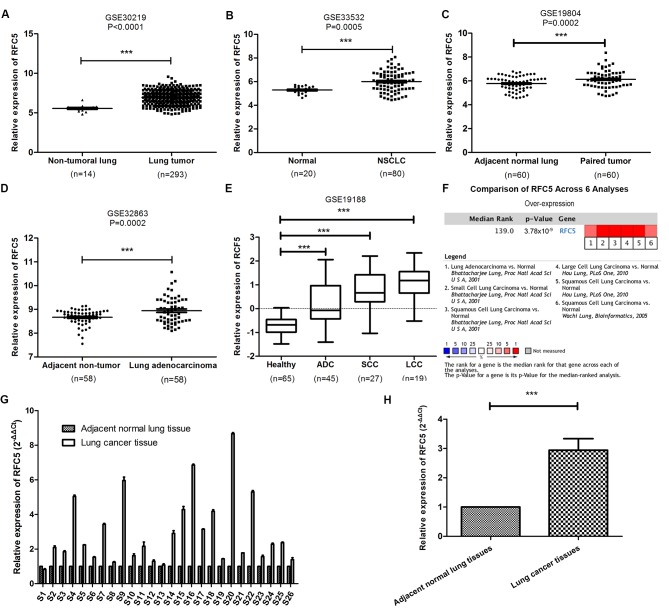
The publicly available GEO datasets were used to analyze the expression levels of RFC5 mRNA in cancer and normal tissues. Compared with normal tissues, the expression levels of RFC5 were significantly higher in lung tumor (Fig. 2A), NSCLC (Fig. 2B), paired tumor (Fig. 2C and D), lung adenocarcinoma (ADC), squamous cell carcinoma (SCC) and large cell carcinoma (LCC) tissues (Fig. 2E). Furthermore, RFC5 was uniformly upregulated in six analyses in ONCOMINE. The level of RFC5 expression was confirmed in 26 paired cancerous and adjacent normal tissues from patients with lung cancer by RT-qPCR. Consistent with the findings of previous bioinformatic analyses, RFC5 was significantly overexpressed in lung cancer tissue compared with adjacent tissues.
Figure 2.
Identification of RFC5 as an overexpressed gene in lung cancer. (A-D) Comparison of the level of RFC5 mRNA expression between cancerous and normal tissue from the datasets (A) GSE30219, (B) GSE33532, (C) GSE19804 (paired samples) and (D) GSE32863 (paired samples). ***P<0.001, paired t-test and unpaired t-test. (E) Expression level of RFC5 in ADC, SCC and LCC in the GSE19188 dataset. ***P<0.001, one-way ANOVA and Dunnett's test. (F) Cluster analysis of RFC5 in different data sets of lung cancer subtypes. Red, significant overexpression. The P-value was calculated using the meta-analysis. (G) Expression levels of RFC5 mRNA in 26-paired lung cancer were assessed by RT-qPCR. (H) Comparison of the expression levels of RFC5 in lung cancer tissues and adjacent tissues. ***P<0.001; RFC5, replication factor C subunit 5; ADC, adenocarcinoma; LCC, large cell carcinoma; SCC, squamous cell carcinoma.
RFC5 expression is associated with the clinicopathological characteristics of lung cancer
In order to elucidate the association between RFC5 expression and the clinicopathological characteristics of lung cancer, the GSE30219 dataset was analyzed by χ2 test. RFC5 expression was associated with sex (P=0.0300), T stage (P<0.0001), N stage (P<0.0001) and relapse (P=0.0010). However, there were no significant associations between RFC5 expression with age (P=0.5580) and M stage (P=0.2820; Table II). These results demonstrated that RFC5 expression was associated with the progression of lung cancer.
Table II.
Associations between RFC5 expression and clinicopathological characteristics of patients with lung cancer in the GSE30219 dataset.
| RFC5 expression | |||||
|---|---|---|---|---|---|
| Characteristics | N | High expression | Low expression | χ2 | P-value |
| Age | 0.342 | 0.5580 | |||
| ≤62.0 | 146 | 71 | 75 | ||
| >62.0 | 146 | 76 | 70 | ||
| Sex | 4.711 | 0.0300 | |||
| Female | 43 | 15 | 28 | ||
| Male | 250 | 132 | 118 | ||
| T stage | 29.997 | <0.0001 | |||
| T1 | 166 | 61 | 105 | ||
| T2 | 69 | 42 | 27 | ||
| T3-4 | 52 | 40 | 12 | ||
| N stage | 37.451 | <0.0001 | |||
| Positive | 93 | 71 | 22 | ||
| Negative | 198 | 75 | 123 | ||
| Metastasis | 2.000 | 0.2820a | |||
| Yes | 8 | 6 | 2 | ||
| No | 282 | 140 | 142 | ||
| Relapse | 11.757 | 0.0010 | |||
| No | 164 | 65 | 99 | ||
| Yes | 114 | 69 | 45 | ||
Fisher's exact test. RFC5, replication factor C 5.
High expression of RFC5 indicates a poor prognosis
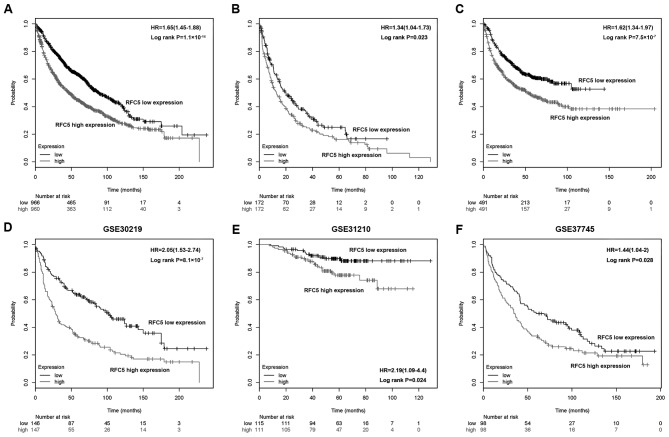
To further investigate the association between RFC5 expression and the outcomes of patients with lung cancer, Kaplan-Meier analysis was applied to assess the OS, FP and PPS in the high RFC5 expression and low RFC5 expression groups. As shown in Fig. 3A-C, a high expression of RFC5 corresponded to a poorer OS (HR, 1.65; 95% CI, 1.45–1.88; P=1.1×10−14), FP (HR, 1.34; 95% CI, 1.04–1.73; P=0.023) and PPS (HR, 1.62; 95% CI, 1.34–1.97; P=7.5×10−7), respectively. Kaplan-Meier analysis was performed in three GEO datasets (GSE30219, GSE31210 and GSE37745, respectively), which consistently indicated that a low RFC5 expression was associated with improved OS (Fig. 3D-F). Furthermore, univariate and multivariate Cox regression analyses were performed to investigate the independent factors that affect patient survival. As shown in Table III, the high RFC5 expression group exhibited a significantly increased risk of OS (HR, 2.027; 95% CI, 1.498–2.742, P<0.0001) compared with the low RFC5 expression group. The characteristics that were significant in the univariate analyses were then incorporated into the multivariate analyses. Age (HR, 2.014; 95% CI, 1.478–2.744, P<0.0001), T stage (HR, 1.363; 95% CI, 1.106–1.678, P<0.0040) and RFC5 expression (HR, 1.557; 95% CI, 1.137–2.132, P<0.0060) were indicated to be significant risk factors in multivariate analysis and were determined as independent prognostic factors of OS. These results also indicated that RFC5 expression (HR, 1.736; 95% CI, 1.143–2.636, P <0.0100); T stage (HR, 1.461; 95% CI, 1.137–1.878, P<0.0030) and N stage (HR, 1.461; 95% CI, 1.016–2.100, P<0.0410) were independent prognostic factors for disease-free survival (Table IV).
Figure 3.
Prognostic values of RFC5 in NSCLC. The associations between RFC5 expression and (A) OS, (B) FP or (C) PPS rate in total samples were analyzed using the Kaplan-Meier Plotter database. (D-F) Overall survival curves for patients in (D) GSE30219, (E) GSE31210 and (F) GSE37745. FP, first progression; HR, hazards ratio; OS, overall survival; PPS, post progression survival; RFC5, replication factor C subunit 5.
Table III.
Univariate and Multivariate analysis of the effect of covariates on overall survival for patients with lung cancer in the GSE30219 dataset.
| A, Univariate analysis | ||
|---|---|---|
| Variables | HR (95% CI) | P-value |
| Age | ||
| ≤62 vs. >62 years | 2.101 (1.544–2.858) | <0.0001 |
| Sex | ||
| Female vs. male | 1.789 (1.098–2.916) | 0.0200 |
| T stage | ||
| T1 vs. T2 vs. T3 vs. T4 | 1.589 (1.368–1.845) | <0.0001 |
| N stage | ||
| N0 vs. N1 | 1.770 (1.431–2.188) | <0.0001 |
| M stage | ||
| M0 vs. M1 | 2.456 (0.908–6.644) | 0.0770 |
| RFC5 expression | ||
| High vs. low | 2.027 (1.498–2.742) | <0.0001 |
| B, Multivariate analysis | ||
| Variables | HR (95% CI) | P-value |
| Age | ||
| ≤62 vs. >62 years | 2.014 (1.478–2.744) | <0.0001 |
| Sex | ||
| Female vs. male | 1.515 (0.926–2.478) | 0.0980 |
| T stage | ||
| T1 vs. T2 vs. T3 vs. T4 | 1.363 (1.106–1.678) | 0.0040 |
| N stage | ||
| N0 vs. N1 | 1.259 (0.935–1.696) | 0.1280 |
| M stage | ||
| M0 vs. M1 | – | – |
| RFC5 expression | ||
| High vs. low | 1.557 (1.137–2.132) | 0.0060 |
CI, confidence interval; HR, hazard ratio; RFC5, replication factor C 5.
Table IV.
Univariate and multivariate analysis of the effect of covariates on disease-free survival for patients with lung cancer in the GSE30219 dataset.
| A, Univariate analysis | ||
|---|---|---|
| Variables | HR (95% CI) | P-value |
| Age | ||
| ≤62 vs. >62 years | 1.332 (0.902–1.968) | 0.1490 |
| Sex | ||
| Female vs. male | 1.301 (0.740–2.286) | 0.3610 |
| T stage | ||
| T1 vs. T2 vs. T3 vs. T4 | 1.852 (1.541–2.226) | <0.0001 |
| N stage | ||
| N0 vs. N1 | 2.309 (1.786–2.986) | <0.0001 |
| M stage | ||
| M0 vs. M1 | 3.705 (1.359–10.095) | 0.0100 |
| RFC5 expression | ||
| High vs. low | 2.327 (1.563–3.464) | <0.0001 |
| B, Multivariate analysis | ||
| Variables | HR (95% CI) | P-value |
| Age | ||
| ≤62 vs. >62 years | – | – |
| Sex | ||
| Female vs. male | – | – |
| T stage | ||
| T1 vs. T2 vs. T3 vs. T4 | 1.461 (1.137–1.878) | 0.0030 |
| N stage | ||
| N0 vs. N1 | 1.461 (1.016–2.100) | 0.0410 |
| M stage | ||
| M0 vs. M1 | 2.640 (0.960–7.265) | 0.0600 |
| RFC5 expression | ||
| High vs. low | 1.736 (1.143–2.636) | 0.0100 |
CI, confidence interval; HR, hazard ratio; RFC5, replication factor C 5.
RFC5 enhances the proliferation of tumor cells in lung cancer
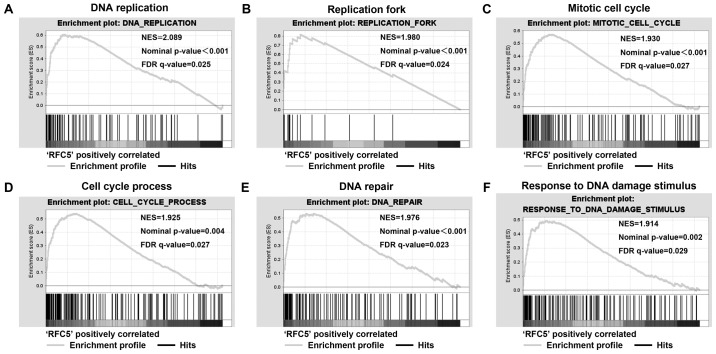
GSEA was used to analyze the biological processes of RFC5 in lung cancer. The GSE3141 dataset was analyzed with GMT file C5 (GO gene set). The first 20 relevant biological processes that met P-value <0.05 and false discovery rates <0.25 are shown in Table V. The results showed that several gene sets that are associated with cell cycle and DNA damage were enriched in the RFC5 overexpression group, which suggested the RFC5 is involved in the proliferation of lung cancer cells (Fig. 4).
Table V.
Enrichment of biological processes in the RFC5 high expression group.
| No. | GS details | Size | ES | NES | NOM P-value | FDR q-value |
|---|---|---|---|---|---|---|
| 1 | DNA replication | 88 | 0.607 | 2.085 | 0.002 | 0.037 |
| 2 | Chromosomal part | 88 | 0.667 | 2.083 | <0.001 | 0.020 |
| 3 | DNA-dependent DNA replication | 47 | 0.672 | 2.072 | 0.002 | 0.015 |
| 4 | Chromosome | 114 | 0.632 | 2.071 | <0.001 | 0.012 |
| 5 | DNA metabolic process | 232 | 0.518 | 2.026 | <0.001 | 0.018 |
| 6 | Double-strand break repair | 22 | 0.739 | 2.013 | 0.002 | 0.019 |
| 7 | Nuclear part | 487 | 0.527 | 1.988 | <0.001 | 0.024 |
| 8 | Nuclear membrane | 47 | 0.651 | 1.985 | <0.001 | 0.022 |
| 9 | Nuclear pore | 28 | 0.761 | 1.982 | <0.001 | 0.020 |
| 10 | Nuclear membrane part | 39 | 0.688 | 1.958 | <0.001 | 0.026 |
| 11 | Nuclear envelope | 67 | 0.607 | 1.951 | <0.001 | 0.026 |
| 12 | Organelle lumen | 391 | 0.512 | 1.940 | <0.001 | 0.027 |
| 13 | Membrane-enclosed lumen | 391 | 0.512 | 1.940 | <0.001 | 0.025 |
| 14 | Mitotic cell cycle | 136 | 0.571 | 1.922 | 0.006 | 0.029 |
| 15 | DNA repair | 117 | 0.533 | 1.918 | <0.001 | 0.029 |
| 16 | Envelope | 158 | 0.587 | 1.912 | <0.001 | 0.030 |
| 17 | Organelle envelope | 158 | 0.587 | 1.912 | <0.001 | 0.028 |
| 18 | Translation factor activity nucleic acid binding | 29 | 0.696 | 1.908 | <0.001 | 0.029 |
| 19 | RNA splicing | 66 | 0.651 | 1.907 | <0.001 | 0.027 |
| 20 | Cell cycle process | 172 | 0.539 | 1.905 | 0.006 | 0.027 |
Statistical analyses were performed using the gene set enrichment analysis software. GS, gene sets; ES, enrichment score; NES, normal enrichment score; NOM, normal; FDR, false discovery rate; RFC5, replication factor C 5.
Figure 4.
RFC5 enriched cell cycle and DNA damage process in lung cancer. Gene set enrichment analysis was performed by GSEA using the GSE3141 dataset. The gene profile of high RFC5 expression groups (top 50%) and low RFC5 expression groups (bottom 50%) were loaded to the GSEA software, and the ‘c5.all.v5.1’ gene set was selected to process the analysis. (A) RFC5 high expression enriched DNA replication, (B) replication fork, (C) mitotic cell cycle, (D) cell cycle process, (E) DNA repair, (F) response to DNA damage stimulus signature genes. GSEA, gene set enrichment analysis; NES, normal enrichment score; NOM P-value, normal P-value; FDR q-value, false discovery rate q-value.
Discussion
There is no doubt that DNA replication is a crucial process during cell proliferation and enables the ability to proliferate indefinitely, which is a hallmark of tumor cells (5). A number of genes that are associated with DNA replication are deregulated in cancer cells. Therefore, the present study focused on RFC, which is involved in DNA replication and damage repair as well as cell division and proliferation (30–34). In general, RFC is a structurally specific DNA-binding protein that preferentially binds to the 3′ end of the template primer. A study has indicated that RFC catalyzed the formation of a cyclic structure of PCNA around the primers in an ATP-dependent manner (35). Munshi et al (36) observed that cyclin-dependent kinases reduced the stability of RFC, inactivating it in the S phase to regulate DNA replication (36). However despite being an important component of the RFC, the role of RFC5 in lung cancer remains unknown. Therefore, the present study investigated the expression patterns, potential biological functions and the prognostic value of RFC5 in lung cancer.
Bioinformatics is a multidisciplinary research field, and bioinformatic analysis is particularly used for developing methods and software tools to identify candidate genes. Such understanding allows the efficient elucidation of the genetic basis of diseases (37). In the present study, the ONCOMINE database and GEO datasets were applied to analyze RFC5 expression in lung cancer. The results suggested that RFC5 was highly expressed in lung cancer samples compared with normal samples. The expression levels of RFC5 in 26 paired samples were confirmed by RT-qPCR, and consistent results were attained.
To date, conventional surgery and chemotherapy remain the main treatment modalities for treating lung cancer (38). However, numerous lung cancer cases are diagnosed at an advanced stage. When treating patients with advanced lung cancer using conventional surgical resection and chemotherapy, the 5-year relative survival rate is only 4% (3). Therefore, it is important to find a novel target molecule that can be used to diagnose and used for therapy. The prognostic value of RFC5 was rarely investigated in previous studies. In the present study, RFC5 was identified to be a potential independent prognostic factor for patients with lung cancer. A higher expression of RFC5 was significantly associated with higher T stage, more advanced regional lymph node metastasis and a higher probability of relapse. These findings suggested a potential role of RFC5 in the progression of lung cancer, which might contribute to the development of accurate diagnosis and personalized treatment strategies.
A number of studies revealed the association between RFC5 overexpression and DNA replication. The GSEA provides an improved understanding of the biological functional enrichment in the high RFC5 expression groups. The gene sets that were most significant were associated with DNA replication or DNA damage, which suggested that RFC5 might promote the proliferation of lung cancer cells. A study of other RFC subunits found that the interaction between RFC2, RFC3 and c-myc promoted cell division (39). Furthermore, cDNA microarray analysis and meta-analysis confirmed that the RFC4 mRNA was abnormally high in a variety of malignant tumors, including ADC, cervical cancer, head and neck squamous cell carcinoma and colorectal cancer (18,40–43). Chae et al (17) reported that RFC3 might promote cell proliferation, and its promoter directly binds with CAMP responsive element binding protein in acute myeloid leukemia. The copy number of RFC3 also reported to be increased in other types of cancer, and knocking out RFC3 was demonstrated to inhibit the growth of tumor cells. As the five RFC subunits possess similar conserved regions of ATP/GTP-binding proteins, each of the subunit has a remarkable degree of similarity (13); which further indicated that they might be involved in similar biological processes.
In conclusion, this is the first study to systematically demonstrate that RFC5 is an oncogene, which is closely associated with the prognosis of lung cancer. RFC5 was significantly overexpressed in lung cancer tissues compared with normal tissues. Furthermore, a high expression level of RFC5 was associated with poor clinicopathological characteristics and proliferation of tumor cells. In summary, RFC5 might serve as a prognostic biomarker and novel therapeutic target for lung cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (grants no. 81501352, 81270607 and 81541027).
Availability of data and materials
The GEO datasets analyzed during the current study are available from the National Center for Biotechnology Information (National Institutes of Health, Bethesda, MD, USA) (http://www.ncbi.nlm.nih.gov/geo/). The online database Kaplan-Meier Plotter are available from http://kmplot.com/analysis/index.php?p=service&cancer=lung. The ONCOMINE data are available from https://www.oncomine.org.
Authors' contributions
MW and TX conducted the experiments and performed the statistical analysis. YW and QY interpreted the experimental results, wrote and revised the manuscript. SX and QY collected clinical samples and provided critical advice. JX and QZ conceived and designed the study. All authors contributed to this manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Renmin Hospital of Wuhan University (Wuhan, China). Written informed consents were obtained from all patients prior to enrollment in the study, and anonymity was ensured.
Patient consent for publication
Written informed consent were obtained from all examined patients for the publication of their data.
Competing interests
The authors declare that they have no conflicts of interest.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA A Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, Stein KD, Alteri R, Jemal A. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66:271–289. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 3.Torre LA, Siegel RL, Jemal A. Lung Cancer Statistics. Adv Exp Med Biol. 2016;893:1–19. doi: 10.1007/978-3-319-24223-1_1. [DOI] [PubMed] [Google Scholar]
- 4.Sung WJ, Kim H, Park KK. The biological role of epithelial-mesenchymal transition in lung cancer (Review) Oncol Rep. 2016;36:1199–1206. doi: 10.3892/or.2016.4964. [DOI] [PubMed] [Google Scholar]
- 5.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 6.Kato T, Yoshioka H, Okamoto I, Yokoyama A, Hida T, Seto T, Kiura K, Massey D, Seki Y, Yamamoto N. Afatinib versus cisplatin plus pemetrexed in Japanese patients with advanced non-small cell lung cancer harboring activating EGFR mutations: Subgroup analysis of LUX-Lung 3. Cancer Sci. 2015;106:1202–1211. doi: 10.1111/cas.12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoshi H, Hiyama G, Ishikawa K, Inageda K, Fujimoto J, Wakamatsu A, Togashi T, Kawamura Y, Takahashi N, Higa A, et al. Construction of a novel cell-based assay for the evaluation of anti-EGFR drug efficacy against EGFR mutation. Oncol Rep. 2017;37:66–76. doi: 10.3892/or.2016.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li K, Yang M, Liang N, Li S. Determining EGFR-TKI sensitivity of G719X and other uncommon EGFR mutations in non-small cell lung cancer: Perplexity and solution (Review) Oncol Rep. 2017;37:1347–1358. doi: 10.3892/or.2017.5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Juan O, Popat S. Crizotinib for ROS1 patients: One small step in biomarker testing, one giant leap for advanced NSCLC patients. Lung Cancer. 2017;104:131–133. doi: 10.1016/j.lungcan.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 10.Chen M, Pan ZQ, Hurwitz J. Studies of the cloned 37-kDa subunit of activator 1 (replication factor C) of HeLa cells. Proc Natl Acad Sci USA. 1992;89:5211–5215. doi: 10.1073/pnas.89.12.5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Virshup DM, Kelly TJ. Purification of replication protein C, a cellular protein involved in the initial stages of simian virus 40 DNA replication in vitro. Proc Natl Acad Sci USA. 1989;86:3584–3588. doi: 10.1073/pnas.86.10.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uhlmann F, Cai J, Flores-Rozas H, Dean FB, Finkelstein J, O'Donnell M, Hurwitz J. In vitro reconstitution of human replication factor C from its five subunits. Proc Natl Acad Sci USA. 1996;93:6521–6526. doi: 10.1073/pnas.93.13.6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cullmann G, Fien K, Kobayashi R, Stillman B. Characterization of the five replication factor C genes of Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:4661–4671. doi: 10.1128/MCB.15.9.4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Majka J, Burgers PM. The PCNA-RFC families of DNA clamps and clamp loaders. Prog Nucleic Acid Res Mol Biol. 2004;78:227–260. doi: 10.1016/S0079-6603(04)78006-X. [DOI] [PubMed] [Google Scholar]
- 15.Zhang G, Gibbs E, Kelman Z, O'Donnell M, Hurwitz J. Studies on the interactions between human replication factor C and human proliferating cell nuclear antigen. Proc Natl Acad Sci USA. 1999;96:1869–1874. doi: 10.1073/pnas.96.5.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niu G, Wang D, Pei Y, Sun L. Systematic identification of key genes and pathways in the development of invasive cervical cancer. Gene. 2017;618:28–41. doi: 10.1016/j.gene.2017.03.018. [DOI] [PubMed] [Google Scholar]
- 17.Chae HD, Mitton B, Lacayo NJ, Sakamoto KM. Replication factor C3 is a CREB target gene that regulates cell cycle progression through the modulation of chromatin loading of PCNA. Leukemia. 2015;29:1379–1389. doi: 10.1038/leu.2014.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiang J, Fang L, Luo Y, Yang Z, Liao Y, Cui J, Huang M, Yang Z, Huang Y, Fan X, et al. Levels of human replication factor C4, a clamp loader, correlate with tumor progression and predict the prognosis for colorectal cancer. J Transl Med. 2014;12:320. doi: 10.1186/s12967-014-0320-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lockwood WW, Thu KL, Lin L, Pikor LA, Chari R, Lam WL, Beer DG. Integrative genomics identified RFC3 as an amplified candidate oncogene in esophageal adenocarcinoma. Clin Cancer Res. 2012;18:1936–1946. doi: 10.1158/1078-0432.CCR-11-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arai M, Kondoh N, Imazeki N, Hada A, Hatsuse K, Matsubara O, Yamamoto M. The knockdown of endogenous replication factor C4 decreases the growth and enhances the chemosensitivity of hepatocellular carcinoma cells. Liver Int. 2009;29:55–62. doi: 10.1111/j.1478-3231.2008.01792.x. [DOI] [PubMed] [Google Scholar]
- 21.Murakami T, Takano R, Takeo S, Taniguchi R, Ogawa K, Ohashi E, Tsurimoto T. Stable interaction between the human proliferating cell nuclear antigen loader complex Ctf18-replication factor C (RFC) and DNA polymerase {epsilon} is mediated by the cohesion-specific subunits, Ctf18, Dcc1 and Ctf8. J Biol Chem. 2010;285:34608–34615. doi: 10.1074/jbc.M110.166710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maradeo ME, Garg A, Skibbens RV. Rfc5p regulates alternate RFC complex functions in sister chromatid pairing reactions in budding yeast. Cell Cycle. 2010;9:4370–4378. doi: 10.4161/cc.9.21.13634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng WX, Han X, Zhang CL, Ge L, Du FY, Jin J, Gong AH. FoxM1-mediated RFC5 expression promotes temozolomide resistance. Cell Biol Toxicol. 2017;33:527–537. doi: 10.1007/s10565-017-9381-1. [DOI] [PubMed] [Google Scholar]
- 24.Liu SM, Chen W, Wang J. Distinguishing between cancer cell differentiation and resistance induced by all-trans retinoic acid using transcriptional profiles and functional pathway analysis. Sci Rep. 2014;4:5577. doi: 10.1038/srep05577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein D. Quantification using real-time PCR technology: Applications and limitations. Trends Mol Med. 2002;8:257–260. doi: 10.1016/S1471-4914(02)02355-9. [DOI] [PubMed] [Google Scholar]
- 26.Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 28.Gyorffy B, Lanczky A, Szallasi Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr Relat Cancer. 2012;19:197–208. doi: 10.1530/ERC-11-0329. [DOI] [PubMed] [Google Scholar]
- 29.Szasz AM, Lanczky A, Nagy A, Förster S, Hark K, Green JE, Boussioutas A, Busuttil R, Szabó A, Győrffy B. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7:49322–49333. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorbea CM, Marchand P, Jiang W, Copeland NG, Gilbert DJ, Jenkins NA, Bond JS. Cloning, expression and chromosomal localization of the mouse meprin beta subunit. J Biol Chem. 1993;268:21035–21043. [PubMed] [Google Scholar]
- 31.Shimada M, Okuzaki D, Tanaka S, Tougan T, Tamai KK, Shimoda C, Nojima H. Replication factor C3 of Schizosaccharomyces pombe, a small subunit of replication factor C complex, plays a role in both replication and damage checkpoints. Mol Bio Cell. 1999;10:3991–4003. doi: 10.1091/mbc.10.12.3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naiki T, Shimomura T, Kondo T, Matsumoto K, Sugimoto K. Rfc5, in cooperation with rad24, controls DNA damage checkpoints throughout the cell cycle in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:5888–5896. doi: 10.1128/MCB.20.16.5888-5896.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim HS, Brill SJ. Rfc4 interacts with Rpa1 and is required for both DNA replication and DNA damage checkpoints in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:3725–3737. doi: 10.1128/MCB.21.11.3725-3737.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomida J, Masuda Y, Hiroaki H, Ishikawa T, Song I, Tsurimoto T, Tateishi S, Shiomi T, Kamei Y, Kim J, et al. DNA damage-induced ubiquitylation of RFC2 subunit of replication factor C complex. J Biol Chem. 2008;283:9071–9079. doi: 10.1074/jbc.M709835200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shiomi Y, Usukura J, Masamura Y, Takeyasu K, Nakayama Y, Obuse C, Yoshikawa H, Tsurimoto T. ATP-dependent structural change of the eukaryotic clamp-loader protein, replication factor C. Proc Natl Acad Sci USA. 2000;97:14127–14132. doi: 10.1073/pnas.97.26.14127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Munshi A, Cannella D, Brickner H, Salles-Passador I, Podust V, Fotedar R, Fotedar A. Cell cycle-dependent phosphorylation of the large subunit of replication factor C (RF-C) leads to its dissociation from the RF-C complex. J Biol Chem. 2003;278:48467–48473. doi: 10.1074/jbc.M309349200. [DOI] [PubMed] [Google Scholar]
- 37.Qi S, Wang CH, Li CL, Wang P, Liu MH. Candidate genes investigation for severe nonalcoholic fatty liver disease based on bioinformatics analysis. Medicine (Baltimore) 2017;96:e7743. doi: 10.1097/MD.0000000000007743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arnold BN, Thomas DC, Rosen JE, Salazar MC, Detterbeck FC, Blasberg JD, Boffa DJ, Kim AW. Effectiveness of local therapy for stage I non-small-cell lung cancer in nonagenarians. Surgery. 2017;162:640–651. doi: 10.1016/j.surg.2017.04.025. [DOI] [PubMed] [Google Scholar]
- 39.Koch HB, Zhang R, Verdoodt B, Bailey A, Zhang CD, Yates JR, III, Menssen A, Hermeking H. Large-scale identification of c-MYC-associated proteins using a combined TAP/MudPIT approach. Cell cycle. 2007;6:205–217. doi: 10.4161/cc.6.2.3742. [DOI] [PubMed] [Google Scholar]
- 40.Erdogan E, Klee EW, Thompson EA, Fields AP. Meta-analysis of oncogenic protein kinase Ciota signaling in lung adenocarcinoma. Clin Cancer Res. 2009;15:1527–1533. doi: 10.1158/1078-0432.CCR-08-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang BY, You H, Bandyopadhyay S, Agrawal N, Melchert RB, Basnakian AG, Liu Y, Hermonat PL. Cervical cancer isolate PT3, super-permissive for adeno-associated virus replication, over-expresses DNA polymerase delta, PCNA, RFC and RPA. BMC Microbiol. 2009;9:79. doi: 10.1186/1471-2180-9-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Narayan G, Bourdon V, Chaganti S, Arias-Pulido H, Nandula SV, Rao PH, Gissmann L, Dürst M, Schneider A, Pothuri B, et al. Gene dosage alterations revealed by cDNA microarray analysis in cervical cancer: Identification of candidate amplified and overexpressed genes. Genes chromosomes cancer. 2007;46:373–384. doi: 10.1002/gcc.20418. [DOI] [PubMed] [Google Scholar]
- 43.Slebos RJ, Yi Y, Ely K, Carter J, Evjen A, Zhang X, Shyr Y, Murphy BM, Cmelak AJ, Burkey BB. Gene expression differences associated with human papillomavirus status in head and neck squamous cell carcinoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2006;12:701–709. doi: 10.1158/1078-0432.CCR-05-2017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The GEO datasets analyzed during the current study are available from the National Center for Biotechnology Information (National Institutes of Health, Bethesda, MD, USA) (http://www.ncbi.nlm.nih.gov/geo/). The online database Kaplan-Meier Plotter are available from http://kmplot.com/analysis/index.php?p=service&cancer=lung. The ONCOMINE data are available from https://www.oncomine.org.