Abstract
Hepatitis B virus (HBV) infection is a risk factor for hepatocellular carcinoma (HCC). HBV X protein (HBx) is an important carcinogen for HBV-induced HCC. When the HBx gene is integrated into the host cell genome, it is difficult to eradicate. The identification of an effective target to inhibit the oncogenic function of HBx is therefore critically important. The present study demonstrated that HBx, particularly truncated HBx, was expressed in several HBV-derived cell lines (e.g., Hep3B and SNU423). By analyzing data from The Cancer Genome Atlas, it was revealed that high expression of high mobility group box 1 (HMGB1) was associated with the process and prognosis of HCC. In vitro experiments confirmed that HBx could regulate the expression of HMGB1 and knockdown of HMGB1 could decrease the ability of HBx to promote cellular proliferation. HBx could also upregulate six transcription factors (GATA binding protein 3, Erb-B2 receptor tyrosine kinase 3, heat shock transcription factor 1, nuclear factor κB subunit 1, TATA-box binding protein and Kruppel-like factor 4), which could directly regulate HMGB1. By analyzing genes that are co-expressed with HMGB1, several signaling pathways associated with the development of HCC were identified. HBx and HMGB1 were revealed to be involved in these pathways, which may be the mechanism by which HBx promotes HCC by regulating HMGB1. These findings suggested that HMGB1 may be an effective target for inhibiting HBV-induced HCC.
Keywords: hepatitis B virus-X protein, high mobility group box 1, hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is the most common type of liver tumor, and the mortality rate for HCC is the third highest of all types of cancer, following lung and stomach cancer (1). Hepatitis B virus (HBV) chronic infection is a risk factor for HCC (2), particularly in Asia and South Africa (3). HBV X protein (HBx), a 17-kDa multifunctional protein, serves an important role in HBV-associated HCC (4–6).
During chronic HBV infection, HBV DNA can integrate into the host genome (7). The HBx gene can be maintained and transcribed in human HCC cells, although complete HBV replication does not occur (7). Recent studies have demonstrated that HBx influences the entire process of HBV-associated HCC, including inflammation, cirrhosis and HCC development (8–10). HBx regulates a variety of signaling pathways, including phosphoinositide 3-kinase (PI3K)/protein kinase B (Akt)/mechanistic target of rapamycin, Wnt/β-catenin and P53, through modulating the expression and activity of numerous genes (11–13). These events also include epigenetic modification and mutation. However, it is difficult to eliminate HBx once it has been integrated into the host genome. Our previous study detected HBx, particularly truncated HBx, in HBV-derived HCC cell lines (e.g., Hep3B and SNU423) (14). Therefore, the identification of an effective target is critical for inhibiting the oncogenic function of HBx.
High mobility group box 1 (HMGB1) is a non-histone, nuclear DNA-binding protein (15). HMGB1 is also a multifunctional protein, and its function depends on its location in the cell (15). In the nucleus, as a DNA binding protein, HMGB1 has a role in a number of crucial DNA events (16–20). Under certain conditions, including starvation and reactive oxygen species (ROS), HMGB1 can translocate to the cytoplasm and can transit to the extracellular environment (21). Extra-nuclear HMGB1 regulates several signaling pathways affecting cell functions, including proliferation, autophagy and apoptosis (22–24). Additionally, extracellular HMGB1 can affect the microenvironment by inducing inflammation, fibrosis and angiogenesis (25–27). Our previous study revealed that the deletion of HMGB1 worsened liver ischemia/reperfusion injury (28). Recently, increasing evidence has indicated that HMGB1 has a significant association with HCC. For example, the HMGB1 serum level is increased in patients with chronic HBV and this is associated with the disease stage (29), and HMGB1 can also translocate outside of the nucleus in response to HBV infection (15). These findings suggested that HBV may regulate HMGB1.
The present study demonstrated that the HBx gene was expressed in HBV-derived cell lines. The expression of HMGB1 was associated with the HCC pathological grade and the overall survival of patients. The non-HBV-derived cell line Huh7 was selected to investigate the association between HBx and HMGB1. It was revealed that HBx could regulate the expression of HMGB1 and knockdown of HMGB1 could decrease the ability of HBx to promote cellular proliferation. Next, several transcription factors that had been reported to regulate HMGB1 were investigated using database analysis (e.g., Genecards and Targetscan). Through analysis of these transcription factors, the mechanism for HBx-mediated regulation of HMGB1 was identified. By analyzing the genes co-expressed with HMGB1 in patients with HBV, several key pathways that could promote the formation of HCC were identified. These findings suggested that HMGB1 is involved in the process of HBV-induced HCC.
Materials and methods
Acquisition of human tissue specimens
The three patients were aged 63, 48 and 46 years old (mean was 52.3 years) and comprised of 2 females and 1 male. All these three tissue were obtained in April 2015. Inclusion criteria for the present study were: Each patient was infected by HBV and HBx and HMGB1 had a higher expression in tumor tissue when compared with normal tissue. Paired HCC and adjacent liver tissues were obtained from patients who underwent hepatectomy at the Department of Hepatopancreatobiliary Surgery of the Second Affiliated Hospital of Harbin Medical University (Harbin, China). All human tissues were acquired in accordance with the protocol approved by the Ethics Committee of Harbin Medical University and written informed consent was obtained from all the patients.
Cell lines
The non-HBV-derived HCC Huh7 cell line was obtained from the Department of Surgery of the University of Pittsburgh Medical Center. The cells were used immediately following receipt and were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum (Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) at 37°C with 5% CO2.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted with TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's protocol. Next, 1 µg total RNA from each sample was reverse transcribed to single-stranded cDNA with RNA to cDNA EcoDry™ Premix (Clontech Laboratories, Inc., Mountainview, CA, USA). cDNA (1 µl) was diluted 50-fold with nuclease-free water and was used as a template for RT-qPCR. All the primers were purchased from Sangon Biotech Co., Ltd. (Shanghai, China). HBx forward, 5′-TTCTTCGTCTGCCGTTCC-3′ and reverse, 5′-TCGGTCGTTGACATTGCT-3′; HMGB1 forward, 5′-TATGGCAAAAGCGGACAAGG-3′ and reverse, 5′-CTTCGCAACATCACCAATGGA-3′; GAPDH forward, 5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse, 5′-GGCTGTTGTCATACTTCTCATGG-3′; GATA3 forward, 5′-GCCCCTCATTAAGCCCAAG-3′ and reverse, 5′-TTGTGGTGGTCTGACAGTTCG-3′; TBP forward, 5′-ACTCCACTGTATCCCTCCCC-3′ and reverse, 5′-TATATTCGGCGTTTCGGGCA-3′; HSF1 forward, 5′-CCATGAAGCATGAGAATGAGGC-3′ and reverse, 5′-CTTGTTGACGACTTTCTGTTGC-3′; ERBB3 forward, 5′-GGTGATGGGGAACCTTGAGAT-3′ and reverse, 5′-CTGTCACTTCTCGAATCCACTG-3′; KLF4 forward, 5′-CCCACATGAAGCGACTTCCC-3′ and reverse, 5′-CAGGTCCAGGAGATCGTTGAA-3′; NFKB1 forward, 5′-AACAGAGAGGATTTCGTTTCCG-3′ and reverse, 5′-TTTGACCTGAGGGTAAGACTTCT-3′. The thermo cycling conditions were as follows: 94°C 20 sec, 60°C 20 sec, 72°C 2 min, 30 cycles. Analysis of relative gene expression data was using real-time quantitative PCR and the 2−ΔΔCq method (30). Gene expression of HBx and HMGB1 were normalized to GAPDH. Gene expression of GATA3, TBP, HSF1, ERBB3, KLF4 and NFKB1 were normalized to the control group. The reverse transcription reactions were prepared using the TaqMan Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.). Each 15-µl multiplex reaction contained 10 ng total RNA as a template.
Western blot analysis
Whole protein was extracted using cell lysis buffer (Cell Signaling Technology, Inc., Danvers, MA, USA). The nuclear protein was extracted as previously described (23). BCA was used as the protein determination method. A total of 20 µg nuclear protein was electrophoresed on 10% SDS-polyacrylamide gels and transferred onto polyvinylidene difluoride membranes. Following blocking with 5% skimmed milk at room temperature for 1 h, the membranes were incubated with a 1:5,000 dilution of the primary antibodies with bovine serum albumin (Thermo Fisher Scientific, Inc.) for HMGB1, HBx and GAPDH (cat. nos. ab79823, ab39716 and ab181602, respectively) at 4°C overnight. Next, the membranes were washed with Tris-buffered saline and Tween-20 (TBST) three times, incubated with a 1:10,000 dilution of the secondary antibody (goat anti-rabbit; HRP; cat. no. ab191866) at room temperature for 1 h, and developed onto X-ray film using a chemiluminescent reagent (cat. no. c510043; Sangon Biotech, Co., Ltd.). All of the antibodies were purchased from Abcam (Cambridge, UK).
Plasmid construct and cell transfection
The PcDNA-3.1-HBx plasmid was purchased from Sangon Biotech Co., Ltd. The control shRNA and HMGB1 shRNA were purchased from Sigma-Aldrich; Merck KGaA (Darmstadt, Germany). The FuGene transfection reagent (Promega Corporation, Madison, WI, USA) was used at a reagent to plasmid ratio of 3:1 and the concentration was 1 µg/ml. Transfection was performed according to the manufacturer's protocol, and cells were harvested after 48 h for the following experiments.
Database analysis and statistical analysis
The Cancer Genome Atlas (TCGA) data were obtained from Synapse TCGA (https://www.synapse.org/). Gene expression data and patient data from 377 patients were analyzed, however 6 patients did not have HBV information. The gene expression association analysis was completed using R2 (http://r2.amc.nl/) and Prism 5. The signaling pathway analysis was performed using Toppgene (https://toppgene.cchmc.org/) and R2. The statistical analysis was performed using Prism 5, and gene information was collected from Genecards (http://www.genecards.org/). Data are presented as the mean ± standard deviation. Two-way analysis of variance with Tukey's multiple comparison test was used for raw data analysis and P<0.05 was considered to indicate a statistically significant difference.
Results
HBx expression is higher in tumor tissue than in adjacent noncancerous tissue, and truncated HBx can be detected in HBV-derived cell lines
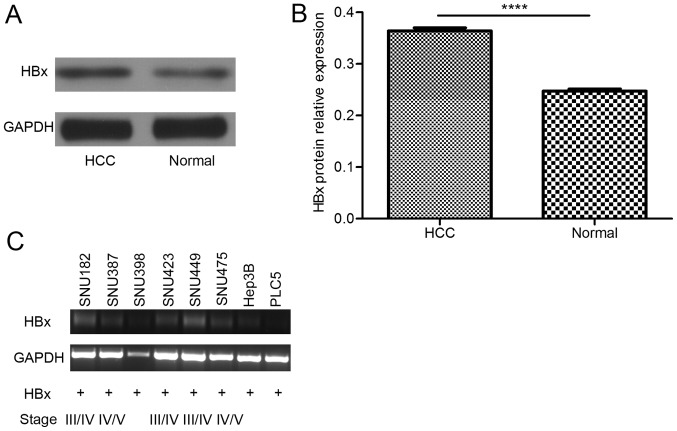
The results of in vivo studies revealed that HBx protein expression was higher in HCC tumor tissue than in the adjacent non-cancerous tissue (Fig. 1A and B). Once the HBx gene is integrated into the host genome, it is difficult to eliminate. Accordingly, in in vitro studies, truncated HBx was detected (Fig. 1C), which is the form of HBx with a greater effect (14). These results suggested that HBx is expressed in liver cells for a long time and contributes to the process of HCC.
Figure 1.
HBx is expressed at a higher level in tumor tissue than in adjacent non-cancerous tissue, and truncated HBx can be detected in hepatitis B virus-derived HCC cell lines. (A) Western blot analysis and (B) relative protein expression of HBx, which was higher in tumor tissues than in adjacent non-cancerous tissues (****P<0.0001). (C) In the HBV-derived cell lines, truncated HBx was detected. HCC, hepatocellular carcinoma; HBx, hepatitis B virus-X protein.
HMGB1 expression is higher in tumor tissue than in adjacent non-cancerous tissue, and a high level of HMGB1 is associated with the pathological grade of HCC and the survival of patients
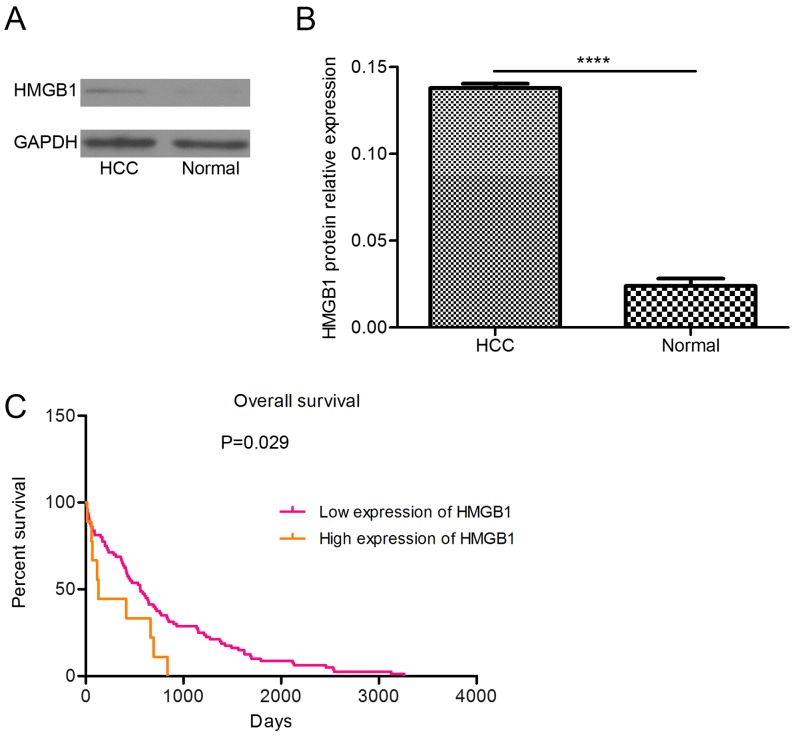
The protein level of HMGB1 was higher in the tumor tissue than in the adjacent non-cancerous tissue (Fig. 2A and B). By analyzing TCGA data, the present study revealed that the prognosis of patients with high-level HMGB1 expression was poor (Fig. 2C). These findings suggested that abnormal expression of HMGB1 can induce HCC.
Figure 2.
HMGB1 is highly expressed in tumor tissue and high HMGB1 expression is associated with patient prognosis. (A) Western blot analysis and (B) relative protein expression of HMGB1, which was higher in HCC tumor tissue than in the adjacent non-cancerous tissue (****P<0.0001). (C) Patients overexpressing HMGB1 exhibited a poor prognosis. (D) High expression of HMGB1 was associated with tumor grade. Gene expression data were retrieved from The Cancer Genome Atlas. HCC, Hepatocellular carcinoma; HMGB1, high mobility group box 1.
HBx can regulate the expression of HMGB1
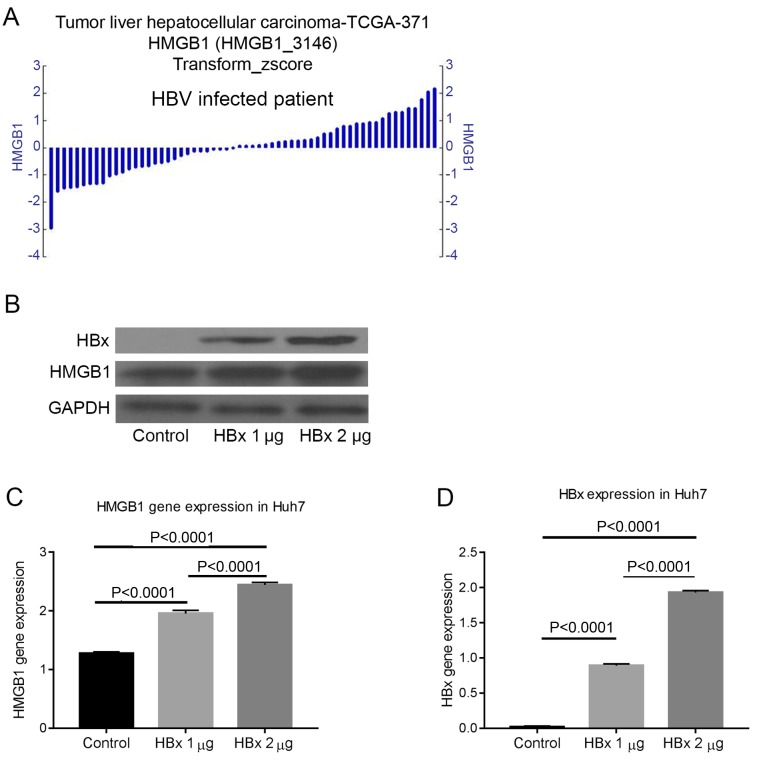
The gene expression of HMGB1 differed in patients with HBV-induced HCC. This finding suggested that HBV may regulate HMGB1 (Fig. 3A). To address this hypothesis, the non-HBV-derived Huh7 cell line was transfected with different quantities of pcDNA-3.1-HBx plasmid and the protein expression was measured by western blot analysis after 72 h. The results confirmed that HBx could regulate the expression of HMGB1 (Fig. 3B-D).
Figure 3.
HBx can regulate the expression of HMGB1. (A) By analyzing data from The Cancer Genome Atlas using R2, it was revealed that HMGB1 was differentially expressed in HBV-associated HCC. (B) Transfecting the HBx plasmid into the non-HBV-derived Huh7 cell line, it was revealed that the protein expression of HMGB1 could be regulated by HBx. The gene expression level of (C) HMGB1 and (D) HBx were also been measured. HBx, hepatitis B virus-X protein; HMGB1, high mobility group box 1; HBV, hepatitis B virus.
HMGB1 is required for HBx to promote the cellular proliferation
To investigate the role of HMGB1 in the carcinogenic process of HBx, a HBx stable expression cell line was created (Fig. 4A and B). HMGB1 was knocked down in the HBx stable expression cell line and it was revealed that the ability of HBx to promote proliferation was decreased (Fig. 4C).
Figure 4.
HMGB1 is required for HBx to promote cellular proliferation. (A) Reverse transcription-quantitative polymerase chain reaction for HMGB1 gene expression in the HBx stable expression cells following transfection for 48 h with control shRNA or HMGB1 shRNA. (B) Knockdown of HMGB1 decreased cellular proliferation in HBx stable expression cells. (C) Western blot analysis for HMGB1 protein expression in the indicated cells following transfection for 72 h with control shRNA or HMGB1 shRNA. HMGB1, high mobility group box 1; HBx, hepatitis B virus-X protein; shRNA, short hairpin RNA; OD, optical density.
HBx increases the expression of HMGB1 by regulating transcription factors associated with HMGB1
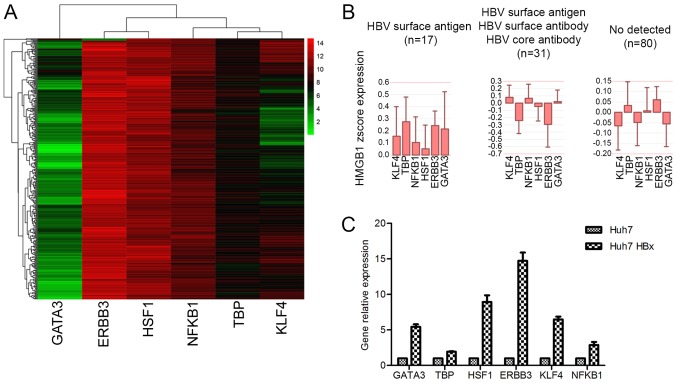
To clarify the molecular mechanism of HBx-mediated regulation of HMGB1, several transcription factors associated with HMGB1 that could be expressed in HCC were investigated. Eventually, six possible transcription factors: GATA3, ERBB3, HSF1, NFKB1, TBP and KLF4 were identified (Fig. 5A). Several of these transcription factors were highly expressed in patients with HBV (Fig. 5B).
Figure 5.
HBx increases the expression of HMGB1 by regulating transcription factors associated with HMGB1. (A) The gene expression of the six transcription factors in patients with hepatocellular carcinoma. (B) Several of these transcription factors were highly expressed in HBV-infected patients. (C) HBx could upregulate the expression of these transcription factors. Gene data were obtained from The Cancer Genome Atlas. R2 and Prism5 were used to analyze these data. HBx, hepatitis B virus-X protein; HMGB1, high mobility group box 1; HBV, hepatitis B virus; GATA3, GATA binding protein 3; ERBB3, Erb-B2 receptor tyrosine kinase 3; HSF1, heat shock transcription factor 1; NFKB1, nuclear factor κB subunit 1; TBP, TATA-box binding protein; KLF4, Kruppel-like factor 4.
HBx affects a variety of cellular functions through regulation of HMGB1
A number of genes that are co-expressed with HMGB1 were abnormally expressed in HBV-induced HCC. Therefore, several genes that exhibited a significant associated with HMGB1 were further examined. Next, the top 19 genes were selected to analyze the functions associated with HMGB1 (Table I). By analyzing these genes using Toppgene, the top 16 signaling pathways enriched for genes that are co-expressed with HMGB1 were obtained (Table II).
Table I.
Top 19 genes co-expressed with HMGB1 in patients with HBV.
| Gene | Function | R2 | P-value |
|---|---|---|---|
| CDKN1B | Cell cycle, development | 0.303 | 2.5×10−9 |
| PCNA | DNA repair, signaling transduction | 0.289 | 1.5×10−8 |
| CDK2 | Cell cycle, kinase | 0.204 | 7.6×10−5 |
| CREB1 | TF, drug target, signal transduction, transcription regulator activator | 0.199 | 1.2×10−4 |
| CCNE2 | Cell cycle | 0.193 | 1.9×10−4 |
| BIRC5 | Apoptosis, cell cycle | 0.188 | 2.7×10−4 |
| NFKBIA | Apoptosis, signal transduction | 0.186 | 3.1×10−4 |
| CCNA2 | Cell cycle | 0.171 | 9.8×10−4 |
| YWHAQ | Signal transduction | 0.169 | 1.1×10−3 |
| E2F1 | TF, apoptosis, cell cycle, development, transcription regulator activator | 0.168 | 1.2×10−3 |
| CREB3L4 | TF, transcription regulator activator | 0.144 | 5.5×10−3 |
| CDK4 | Cell cycle, kinase | 0.144 | 5.5×10−3 |
| RB1 | TF, cell cycle, drug target, signal transduction, transcription regulator activator | 0.133 | 1.0×10−2 |
| TGFBR1 | Kinase, membrane, signal transduction | 0.123 | 2.0×10−2 |
| E2F2 | TF, cell cycle, transcription regulator activator | 0.117 | 2.0×10−2 |
| SMAD4 | TF, development, signal transduction, transcription regulator activator | 0.113 | 3.0×10−2 |
| STAT4 | TF, signal transduction, transcription regulator activator | 0.109 | 4.0×10−2 |
| APAF1 | Apoptosis, development, drug target | 0.105 | 4.0×10−2 |
| BAX | Apoptosis, cell cycle, development, differentiation, membrane | 0.102 | 5.0×10−2 |
HBV, hepatitis B virus; HMGB1, high expression of high mobility group box 1.
Table II.
Top 16 signaling pathways enriched for genes that were co-expressed with high mobility group box 1.
| Name | Source | P-value | FDR B&H | FDR B&Y | Bonferroni | Gene input |
|---|---|---|---|---|---|---|
| Cell cycle | BioSystems: WikiPathways | 6.16×10−17 | 1.57×10−14 | 1.09×10−13 | 3.6×10−14 | 11 |
| p53 signaling pathway | MSigDB C2 BIOCARTA (v5.1) | 1.07×10−15 | 1.04×10−13 | 7.23×10−13 | 6.25×10−13 | 7 |
| DNA damage response | BioSystems: WikiPathways | 8.61×10−15 | 6.28×10−13 | 4.37×10−12 | 5.03×10−12 | |
| Viral carcinogenesis | BioSystems: KEGG | 1.67×10−13 | 7.49×10−12 | 5.20×10−11 | 9.74×10−11 | 11 |
| Pathways in cancer | BioSystems: KEGG | 8.38×10−13 | 3.49×10−11 | 2.43×10−10 | 4.89×10−10 | 12 |
| PI3K-Akt signaling pathway | BioSystems: KEGG | 1.64×10−10 | 6.26×10−11 | 4.35×10−10 | 9.55×10−10 | 12 |
| E2F transcription factor network | BioSystems: Pathway Interaction Database | 1.82×10−12 | 6.26×10−11 | 4.35×10−10 | 1.06×10−9 | 8 |
| DNA replication | BioSystems: REACTOME | 2.01×10−9 | 4.71×10−8 | 3.27×10−7 | 1.18×10−6 | 7 |
| Regulation of IFNA signaling | BioSystems: REACTOME | 2.10×10−9 | 4.72×10−8 | 3.28×10−7 | 1.23×10−6 | 5 |
| TRAF6-mediated IRF7 activation | BioSystems: REACTOME | 6.38×10−9 | 1.1×10−7 | 7.61×10−7 | 3.73×10−6 | 5 |
| Regulation of autophagy | BioSystems: KEGG | 8.79×10−9 | 1.39×10−7 | 9.64×10−7 | 5.13×10−6 | 5 |
| Integrated cancer pathway | BioSystems: WikiPathways | 1.02×10−8 | 1.57×10−7 | 1.09×10−6 | 5.98×10−6 | 5 |
| Regulation of p27 phosphorylation during cell cycle progression | MSigDB C2 BIOCARTA (v5.1) | 1.27×10−8 | 1.85×10−7 | 1.29×10−6 | 7.41×10−6 | 4 |
| RIG-I/MDA5-mediated induction of IFN-α/β pathways | BioSystems: REACTOME | 1.55×10−8 | 2.21×10−7 | 1.54×10−6 | 9.06×10−6 | 6 |
| IL-7 signaling pathway | BioSystems: WikiPathways | 3.38×10−8 | 4.45×10−7 | 3.09×10−6 | 1.97×10−5 | 5 |
| Regulation of TLR signaling pathway | BioSystems: WikiPathways | 6.74×10−8 | 8.2×10−7 | 5.7×10−6 | 3.94×10−5 | 6 |
PI3K-Akt, phosphoinositide 3-kinase-protein kinase B; IFNA, interferon α; TRAF6, tumor necrosis factor receptor associated factor 6; IRF7, interferon regulatory factor 7; RIG-I/MDA5, retinoic acid-inducible gene I/melanoma differentiation-associated protein 5; IL-7, interleukin 7; TLR, toll-like receptor.
Discussion
HBx serves an important role in the process of HBV-induced HCC (Fig. 1A and B). The carcinogenic mechanisms of HBx have been reported in numerous previous studies (7–13). The HBx gene integrates into the host cell genome during HBV infection (Fig. 1C) (14) and is very difficult to completely remove. Therefore, the best method for inhibiting the function of HBx is to identify an effective target to prevent integration.
HMGB1 is a multifunctional protein, and its function depends on its location. Abnormal expression of HMGB1 in different organs can induce different diseases (15). High expression of HMGB1 can influence several cell functions and the microenvironment of the liver (15–27); accordingly, HMGB1 has been associated with the development and progression of HCC (Fig. 2) and has also been associated with HBV infection (29). In the present study, it was confirmed that HBx could increase the expression of HMGB1 through the regulation of transcription factors (Figs. 3 and 5). Therefore, HMGB1 may be an ideal target for inhibiting the carcinogenic function of HBx.
To study the molecular mechanisms of HBx-induced HCC through the regulation of HMGB1, genes significantly associated with HMGB1 in patients with HBV were investigated (Table I). Next, these genes were sorted into signaling pathways, as presented in Table II. Abnormalities in some of these pathways have been reported to be associated with HBx and HMGB1. For example, the P53 signaling pathway is a classical tumor suppressor pathway, and HBx and HMGB1 can regulate cell proliferation and apoptosis by suppressing P53 (2,31). PI3K/Akt is an important signaling pathway through which HBx can induce autophagy (32), and HMGB1 is involved in the same pathway when responding to oxidative stress (33). Furthermore, the Toll-like receptor (TLR) signaling pathway also contributes to HBx-induced carcinogenesis (34), and a previous study demonstrated that HMGB1 could increase the expression of mitochondria in HCC by binding to TLRs (35).
In conclusion, HBx and HMGB1 have a synergistic effect in promoting HCC. The present study confirmed that HBx could regulate HMGB1. Additionally, several signaling pathways associated with HBx and HMGB1 that may serve as the mechanism whereby HBx promotes HCC through the regulation of HMGB1 were identified. Nevertheless, further investigation is required to identify the precise mechanism through which HBx induces HCC through HMBG1 regulation.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- ERBB3
Erb-B2 receptor tyrosine kinase 3
- GATA3
GATA binding protein 3
- HBx
hepatitis B virus-X protein
- HMGB1
high mobility group box 1
- HSF1
heat shock transcription factor 1
- KLF4
Kruppel-like factor 4
- NFKB1
nuclear factor κB subunit 1
- TBP
TATA-box binding protein
Funding
This study was supported by Heilongjiang Natural Science Foundation (grant no. H2018025).
Availability of data and materials
HCC TCGA data were obtained from http://www.synapse.org/. All other data analyzed during the current study are included in the published article.
Authors' contributions
DW and HL contributed to experiment design, drafting the manuscript and revising. HW, CD, HY and JZ conducted the western blotting and PCR experiments, and data collection. YP, BS, ZS, JW and TC contributed to data analysis. ST contributed to experiment design and drafting the manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Harbin Medical University and written informed consent was obtained from all the patients.
Consent for publication
All patients provided consent for the publication of the paper and any associated images.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Hernandez-Gea V, Toffanin S, Friedman SL, Llovet JM. Role of the microenvironment in the pathogenesis and treatment of hepatocellular carcinoma. Gastroenterology. 2013;144:512–527. doi: 10.1053/j.gastro.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu JW, Yang WY, Tsai SM, Lin YM, Chang PH, Chen JR, Wang HD, Wu JL, Jin SL, Yuh CH. Liver-specific expressions of HBx and src in the p53 mutant trigger hepatocarcinogenesis in zebrafish. PLoS One. 2013;8:e76951. doi: 10.1371/journal.pone.0076951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen JY, Chen YJ, Yen CJ, Chen WS, Huang WC. HBx sensitizes hepatocellular carcinoma cells to lapatinib by up-regulating ErbB3. Oncotarget. 2016;7:473–489. doi: 10.18632/oncotarget.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu XY, Tang SH, Wu SL, Luo YH, Cao MR, Zhou HK, Jiang XW, Shu JC, Bie CQ, Huang SM, et al. Epigenetic modulation of insulin-like growth factor-II overexpression by hepatitis B virus X protein in hepatocellular carcinoma. Am J Cancer Res. 2015;5:956–978. [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao J, Wu J, Cai H, Wang D, Yu L, Zhang WH. E3 Ubiquitin ligase Siah-1 is Down-regulated and fails to target natural HBx truncates for degradation in hepatocellular carcinoma. J Cancer. 2016;7:418–426. doi: 10.7150/jca.13019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu JJ, Song W, Zhang SD, Shen XH, Qiu XM, Wu HZ, Gong PH, Lu S, Zhao ZJ, He ML, Fan H. HBx-upregulated lncRNA UCA1 promotes cell growth and tumorigenesis by recruiting EZH2 and repressing p27Kip1/CDK2 signaling. Sci Rep. 2016;6:23521. doi: 10.1038/srep23521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amaddeo G, Cao Q, Ladeiro Y, Imbeaud S, Nault JC, Jaoui D, Gaston Mathe Y, Laurent C, Laurent A, Bioulac-Sage P, et al. Integration of tumour and viral genomic characterizations in HBV-related hepatocellular carcinomas. Gut. 2015;64:820–829. doi: 10.1136/gutjnl-2013-306228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levrero M, Zucman-Rossi J. Mechanisms of HBV-induced hepatocellular carcinoma. J Hepatol. 2016;64(Suppl 1):S84–S101. doi: 10.1016/j.jhep.2016.02.021. [DOI] [PubMed] [Google Scholar]
- 9.Geng M, Xin X, Bi LQ, Zhou LT, Liu XH. Molecular mechanism of hepatitis B virus X protein function in hepatocarcinogenesis. World J Gastroenterol. 2015;21:10732–10738. doi: 10.3748/wjg.v21.i38.10732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tarocchi M, Polvani S, Marroncini G, Galli A. Molecular mechanism of hepatitis B virus-induced hepatocarcinogenesis. World J Gastroenterol. 2014;20:11630–11640. doi: 10.3748/wjg.v20.i33.11630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu M, Guo J, Li W, Xia H, Lu Y, Dong X, Chen Y, Xie X, Fu S, Li M. HBx induced AFP receptor expressed to activate PI3K/AKT signal to promote expression of Src in liver cells and hepatoma cells. BMC Cancer. 2015;15:362. doi: 10.1186/s12885-015-1384-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding SL, Yang ZW, Wang J, Zhang XL, Chen XM, Lu FM. Integrative analysis of aberrant Wnt signaling in hepatitis B virus-related hepatocellular carcinoma. World J Gastroenterol. 2015;21:6317–6328. doi: 10.3748/wjg.v21.i20.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan C, Wang Y, Chow PK, Chung AY, Ooi LL, Lee CG. Altered binding site selection of p53 transcription cassettes by hepatitis B virus X protein. Mol Cell Biol. 2013;33:485–497. doi: 10.1128/MCB.01189-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ng KY, Chai S, Tong M, Guan XY, Lin CH, Ching YP, Xie D, Cheng AS, Ma S. C-terminal truncated hepatitis B virus X protein promotes hepatocellular carcinogenesis through induction of cancer and stem cell-like properties. Oncotarget. 2016;7:24005–24017. doi: 10.18632/oncotarget.8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang R, Chen R, Zhang Q, Hou W, Wu S, Cao L, Huang J, Yu Y, Fan XG, Yan Z, et al. HMGB1 in health and disease. Mol Aspects Med. 2014;40:1–116. doi: 10.2119/molmed.2014.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cato L, Stott K, Watson M, Thomas JO. The interaction of HMGB1 and linker histones occurs through their acidic and basic tails. J Mol Biol. 2008;384:1262–1272. doi: 10.1016/j.jmb.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 17.El Mezayen R, El Gazzar M, Seeds MC, McCall CE, Dreskin SC, Nicolls MR. Endogenous signals released from necrotic cells augment inflammatory responses to bacterial endotoxin. Immunol Lett. 2007;111:36–44. doi: 10.1016/j.imlet.2007.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang C, Peng L, Su J. Two HMGB1 genes from grass carp Ctenopharyngodon idella mediate immune responses to viral/bacterial PAMPs and GCRV challenge. Dev Comp Immunol. 2013;39:133–146. doi: 10.1016/j.dci.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Z, Zhu Y, Lai Y, Wu X, Feng Z, Yu Y, Bast RC, Jr, Wan X, Xi X, Feng Y. Follicle-stimulating hormone inhibits apoptosis in ovarian cancer cells by regulating the OCT4 stem cell signaling pathway. Int J Oncol. 2013;43:1194–1204. doi: 10.3892/ijo.2013.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furuita K, Murata S, Jee JG, Ichikawa S, Matsuda A, Kojima C. Structural feature of bent DNA recognized by HMGB1. J Am Chem Soc. 2011;133:5788–5790. doi: 10.1021/ja2013399. [DOI] [PubMed] [Google Scholar]
- 21.Tang D, Kang R, Livesey KM, Cheh CW, Farkas A, Loughran P, Hoppe G, Bianchi ME, Tracey KJ, Zeh HJ, III, Lotze MT. Endogenous HMGB1 regulates autophagy. J Cell Biol. 2010;190:881–892. doi: 10.1083/jcb.200911078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sundberg E, Fasth AE, Palmblad K, Harris HE, Andersson U. High mobility group box chromosomal protein 1 acts as a proliferation signal for activated T lymphocytes. Immunobiology. 2009;214:303–309. doi: 10.1016/j.imbio.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 23.Zhu H, Huang L, Zhu S, Li X, Li Z, Yu C, Yu X. Regulation of autophagy by systemic admission of microRNA-141 to target HMGB1 in l-arginine-induced acute pancreatitis in vivo. Pancreatology. 2016;16:337–346. doi: 10.1016/j.pan.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 24.Gdynia G, Keith M, Kopitz J, Bergmann M, Fassl A, Weber AN, George J, Kees T, Zentgraf HW, Wiestler OD, et al. Danger signaling protein HMGB1 induces a distinct form of cell death accompanied by formation of giant mitochondria. Cancer Res. 2010;70:8558–8568. doi: 10.1158/0008-5472.CAN-10-0204. [DOI] [PubMed] [Google Scholar]
- 25.Lin Q, Yang XP, Fang D, Ren X, Zhou H, Fang J, Liu X, Zhou S, Wen F, Yao X, et al. High-mobility group box-1 mediates toll-like receptor 4-dependent angiogenesis. Arterioscler Thromb Vasc Biol. 2011;31:1024–1032. doi: 10.1161/ATVBAHA.111.224048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jia L, Clear A, Liu FT, Matthews J, Uddin N, McCarthy A, Hoxha E, Durance C, Iqbal S, Gribben JG. Extracellular HMGB1 promotes differentiation of nurse-like cells in chronic lymphocytic leukemia. Blood. 2014;123:1709–1719. doi: 10.1182/blood-2013-10-529610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Z, Lin C, Peng L, Ouyang Y, Cao Y, Wang J, Friedman SL, Guo J. High mobility group box 1 activates Toll like receptor 4 signaling in hepatic stellate cells. Life Sci. 2012;91:207–212. doi: 10.1016/j.lfs.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 28.Huang H, Nace GW, McDonald KA, Tai S, Klune JR, Rosborough BR, Ding Q, Loughran P, Zhu X, Beer-Stolz D, et al. Hepatocyte-specific high-mobility group box 1 deletion worsens the injury in liver ischemia/reperfusion: A role for intracellular high-mobility group box 1 in cellular protection. Hepatology. 2014;59:1984–1997. doi: 10.1002/hep.26976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng BQ, Jia CQ, Liu CT, Lu XF, Zhong N, Zhang ZL, Fan W, Li YQ. Serum high mobility group box chromosomal protein 1 is associated with clinicopathologic features in patients with hepatocellular carcinoma. Dig Liver Dis. 2008;40:446–452. doi: 10.1016/j.dld.2007.11.024. [DOI] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Li Q, Li J, Wen T, Zeng W, Peng C, Yan S, Tan J, Yang K, Liu S, Guo A, et al. Overexpression of HMGB1 in melanoma predicts patient survival and suppression of HMGB1 induces cell cycle arrest and senescence in association with p21 (Waf1/Cip1) up-regulation via a p53-independent, Sp1-dependent pathway. Oncotarget. 2014;5:6387–6403. doi: 10.18632/oncotarget.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu M, Guo J, Li W, Xia H, Lu Y, Dong X, Chen Y, Xie X, Fu S, Li M. HBx induced AFP receptor expressed to activate PI3K/AKT signal to promote expression of Src in liver cells and hepatoma cells. BMC Cancer. 2015;15:362. doi: 10.1186/s12885-015-1384-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang FP, Li L, Li J, Wang JY, Wang LY, Jiang W. High mobility group box-1 promotes the proliferation and migration of hepatic stellate cells via TLR4-dependent signal pathways of PI3K/Akt and JNK. PLoS One. 2013;8:e64373. doi: 10.1371/journal.pone.0064373. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Wang Y, Cai J, Zeng X, Chen Y, Yan W, Ouyang Y, Xiao D, Zeng Z, Huang L, Liu A. Downregulation of toll-like receptor 4 induces suppressive effects on hepatitis B virus-related hepatocellular carcinoma via ERK1/2 signaling. BMC Cancer. 2015;15:821. doi: 10.1186/s12885-015-1866-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tohme S, Yazdani HO, Liu Y, Loughran P, van der Windt DJ, Huang H, Simmons RL, Shiva S, Tai S, Tsung A. Hypoxia mediates mitochondrial biogenesis in hepatocellular carcinoma to promote tumor growth through HMGB1 and TLR9 interaction. Hepatology. 2017;66:182–197. doi: 10.1002/hep.29184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
HCC TCGA data were obtained from http://www.synapse.org/. All other data analyzed during the current study are included in the published article.