Abstract
Bacillus subtilis is the best described member of the Gram positive bacteria. It is a typical rod shaped bacterium and grows by elongation in its long axis, before dividing at mid cell to generate two similar daughter cells. B. subtilis is a particularly interesting model for cell cycle studies because it also carries out a modified, asymmetrical division during endospore formation, which can be simply induced by starvation. Cell growth occurs strictly by elongation of the rod, which maintains a constant diameter at all growth rates. This process involves expansion of the cell wall, requiring intercalation of new peptidoglycan and teichoic acid material, as well as controlled hydrolysis of existing wall material. Actin-like MreB proteins are the key spatial regulators that orchestrate the plethora of enzymes needed for cell elongation, many of which are thought to assemble into functional complexes called elongasomes. Cell division requires a switch in the orientation of cell wall synthesis and is organised by a tubulin-like protein FtsZ. FtsZ forms a ring-like structure at the site of impending division, which is specified by a range of mainly negative regulators. There it recruits a set of dedicated division proteins to form a structure called the divisome, which brings about the process of division. During sporulation, both the positioning and fine structure of the division septum are altered, and again, several dedicated proteins that contribute specifically to this process have been identified. This chapter summarises our current understanding of elongation and division in B. subtilis, with particular emphasis on the cytoskeletal proteins MreB and FtsZ, and highlights where the major gaps in our understanding remain.
Keywords: Bacillus, B. subtilis, MreB, MreB homologues, Bacterial cell shape, Helical filaments, Circumferential motion, Cell elongation machinery, Peptidoglycan synthesis, PG, Divisome, Min system, MinJ, FtsZ, Z ring, Sporulation, SpoIIE, L-form bacteria
Introduction to B. subtilis
Bacillus subtilis is an aerobic, Gram positive, endospore forming bacterium of the phylum Firmicutes. It is by far the best characterised Gram positive organism and basic knowledge about B. subtilis is frequently used to guide and inform thinking about other Gram positive organisms. Historically, interest in B. subtilis was based largely on three features of its biology: early success in achieving natural transformation with linear DNA, which greatly facilitated genetic analysis of the organism (Anagnostopoulos and Spizizen 1961); its ability to form endospores, which was used as a simple model for cellular development and differentiation (Errington 1993, 2003; Tan and ramamurthi 2014); and industrial interest in its prodigious ability to secrete certain valuable hydrolytic enzymes (e.g. proteases and amylases) directly into the growth medium (Pohl and Harwood 2010).
The biggest driver for study of B. subtilis, at least in the 1960s to 1990s, was probably interest in endospore (spore) formation (Fig. 3.1). Sporulation of B. subtilis is triggered essentially by nutrient stress. The process begins with a modified, highly asymmetric cell division. This generates a small prespore (sometimes called forespore) cell, destined to become the mature endospore, and a much larger mother cell. The mother cell engulfs the prespore, forming a cell within a cell. The two cells then cooperate in a complex developmental process in which the prespore becomes highly differentiated and covered in protective layers. Eventually, the mother cell lyses to release the now dormant endospore. Endospores are incredibly resistant and can remain dormant for extremely long periods of time, before germinating in response to specific chemical signal molecules (germinants). The process of sporulation in B. subtilis is now understood in great detail (Errington 1993, 2003; Tan and Ramamurthi 2014).
Fig. 3.1.
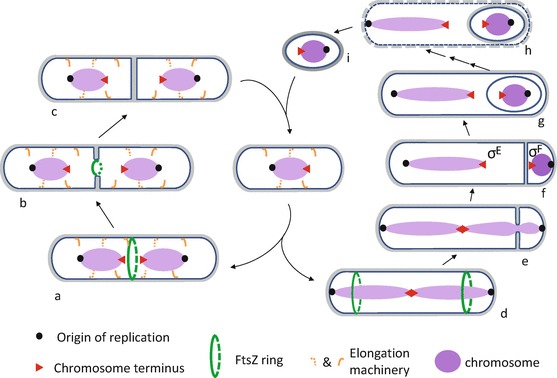
B. subtilis cell cycle. The left half represents the vegetative cycle, where a new born cell (centre) grows in length, controlled by the elongation machinery (orange curved lines), in the meantime the chromosome is replicated and FtsZ (green ring) assembles between segregated chromosomes at mid cell (a). As cell division progresses and septum grows inwards, the Z ring contracts (b). Upon completion of septation, which generates two identical daughter cells, the Z ring and the divisome disassemble (c) and the dividing wall splits to allow separation of the new born cells (centre). Under starvation conditions B. subtilis cells initiate spore development (right half). Instead of segregating the replicated chromosomes to quarter positions, the sister chromosomes undergo a conformational change to form an elongated structure called the axial filament, which extends from pole to pole. FtsZ assembles at two sub-polar positions, one at each cell pole (d). Only one of the two Z rings develops into a septum, which forms over one of the chromosomes (e). Following the completion of asymmetric septation, which generates two unequal sized daughter cells, the small prespore or forespore and the large mother cell, transcription factor Sigma F (σF) is activated in the prespore and the remaining part of the prespore chromosome is segregated. Activation of σF in the prespore leads to the activation of Sigma E ( σE) in the mother cell (f). The different programmes of gene expression in the prespore and the mother cell direct the engulfment of the prespore by the mother cell (g). Finally, the spore undergoes maturation, and the mother cell lyses (h) to release a highly resistant, dormant spore (i), which can germinate and start growing (centre) when nutrients become available
Research on spore formation contributed considerably to the development of methods for studying the sub-cellular distribution of proteins and other important macromolecules in bacteria, laying the foundations for modern bacterial cell biology (Shapiro and Losick 2000; Errington 2003). These imaging methods, together with the exceptionally powerful molecular genetics of B. subtilis, stimulated a new era of studies on the cell cycle and cell morphogenesis. FtsZ, a tubulin homologue that is the key player in bacterial cell division, and MreB, an actin homologue that governs cell shape in many rod shaped bacteria, will be the main topics for discussion in this chapter. As the main focus of the chapter lies on B. subtilis, reference to work on other bacteria will be limited to situations where the contrast or additional information is helpful. For more detail on the E. coli system and on FtsZ and MreB proteins generally, the reader is directed to Chaps. 10.1007/978-3-319-53047-5_2, 10.1007/978-3-319-53047-5_5, 10.1007/978-3-319-53047-5_7 and 10.1007/978-3-319-53047-5_8.
MreB and the Cell Elongation Machinery
Organization of the B. subtilis Cell Wall
Peptidoglycan (PG) is the major component of virtually all bacteria (Typas et al. 2012). It comprises a single huge macromolecule that covers the entire surface of the cell. Lying just outside the cytoplasmic membrane it acts as a protective layer but it also constrains the membrane against the outward turgor pressure imposed by the high osmolarity of the cytoplasm. PG is of considerable practical significance as its synthesis is the target for many useful antibiotics, and fragments of the wall are recognised by innate immune receptors during infection. The PG contributes to the shape of the cell but has no intrinsic 3D shape, so it must be sculpted by synthetic enzymes into the correct form.
PG is composed of long glycan strands with alternating N-acetylglucosamine and N-acetylmuramic acid sugars, cross linked by peptide bridges made up of a mixture of L- and D-amino acids (De Pedro and Cava 2015). The precursor for PG synthesis, called lipid II, is a disaccharide pentapeptide coupled to a C55 isoprenoid lipid (bactoprenol) and is synthesised in the cytosol by a well characterised series of enzymes. Lipid II is flipped to the exterior and assembled into the existing cell wall sacculus by a multiplicity of synthetic enzymes called penicillin-binding proteins (PBPs), which possess the glycosyltransferase and transpeptidase activities needed to extend the glycan strands and create peptide cross bridges (Lovering et al. 2012; Scheffers and Tol 2015). Recently the RodA protein was identified as a possible monofunctional glycosyltransferase (Meeske et al. 2016 and Emami et al. 2017). Extracellular autolytic enzymes are required to allow expansion of the wall by breaking bonds in pre-existing material. Their activities need to be tightly regulated to enable controlled expansion of the wall during growth, while avoiding potentially catastrophic turgor-driven lysis (Vollmer et al. 2008).
Gram positive bacteria lack the outer membrane characteristic of Gram negatives. However, Gram positive walls typically contain a second major class of polymers called teichoic acids (TAs) (Sewell and Brown 2014; Percy and Grundling 2014). In many Gram positives there are two major forms: wall teichoic acids (WTA), which are covalently linked to the PG; and lipoteichoic acids (LTA), which are coupled to a lipid carrier. In B. subtilis WTA and LTA have the same general composition, of poly-[glycerol-phosphate]. TAs have been implicated in many functions. Metal homeostasis is probably a central role – scavenging divalent cations and maintaining surface charge.
B. subtilis cells have a characteristic morphology; in essence, an elongated regular cylindrical tube with hemispherical poles. Growth occurs by elongation along the long axis of the cell, and division occurs approximately each time a doubling in length occurs. The cells have a typical diameter of about 850 nm, irrespective of the growth rate. Changes in growth rate are accommodated by alterations in cell length, with faster growing cells (average up to about 5 μm) being longer than slower growing cells (average minimally about 2 μm) (Sharpe et al. 1998).
Cell shape determination is a central problem in biology. In B. subtilis and many other rod-shaped bacteria, shape is thought to be determined and maintained by the action of “cytoskeletal” proteins of the MreB family (see below). These proteins are structurally and biochemically related to eukaryotic actins. Like actin proteins, they undergo reversible polymerization, which is regulated by binding and hydrolysis of ATP. However, recent work has highlighted certain intrinsic differences, most notably the tight direct association of MreB polymers with the cytoplasmic membrane (Salje et al. 2011). Mutations in the mreB genes of many bacteria abolish proper cell shape control. The possibility that they have a direct role in cell shape determination and or maintenance comes in part from the analogy with shape control by actin filaments in eukaryotes but also from the view that the organization of a large scale (μm) structure requires long range topographical instructions, such as might be provided by elongated MreB filaments. Early experiments examining the localization of MreB proteins in B. subtilis provided strong support for this view through the observation of elongated helical filaments that appeared to wrap around the long axis of the cell (Jones et al. 2001). However, the significance of these elongated filaments and even their existence have been the subject of much recent debate (see below).
B. subtilis Has Three Actin Like MreB Homologues
The mreB gene was first defined by mutations altering cell shape in E. coli (Wachi et al. 1987). Early genome sequence analyses revealed that B. subtilis has three mreB paralogous genes (Levin et al. 1992; Varley and Stewart 1992; Abhayawardhane and Stewart 1995). The gene designated mreB has an equivalent chromosomal location to that of mreB genes of most other bacteria, in lying immediately upstream of homologues of mreC and mreD genes that are also involved in cell elongation. The other homologues – mbl (MreB like) and mreBH (MreB homologue) are located in distant parts of the chromosome. Early mutational studies of mreB and mbl were hampered by the presence of the important mreC and mreD genes downstream from mreB and the lethal nature of the mutations. The latter problem was simplified by the finding that for both paralogues, the viability of null mutants can be rescued by addition of high concentrations (e.g. 20 mM) of Mg2+ to the culture medium, for reasons that are not clear (Formstone and Errington 2005). To summarise the results of several papers, the three paralogues appear to have partially redundant functions, and overexpression of any one of the genes can enable growth and reasonably normal morphology of an otherwise triple null mutant. Single mutations tend to have subtly different effects on morphology: mreB mutants have an increased diameter but remain able to grow in a straight line; mbl mutants are highly twisted with some bulging and lysis; mreBH mutants have a narrow cell phenotype, especially under low Mg2+ conditions (Jones et al. 2001; Carballido-López et al. 2006; Kawai et al. 2009a; Defeu Soufo and Graumann 2006).
Interestingly, Schirner and Errington (2009) found that upregulation of an ECF sigma factor (σI) involved in cell envelope stress was sufficient to suppress the lethality that normally occurs in attempting to construct an mreB, mbl, mreBH triple mutant. The suppressed mutant grew well (though requiring high Mg2+) but with a spherical morphology. Thus, it seems that, as in many other organisms, MreB proteins play a crucial role in rod-shape morphogenesis and cylindrical cell wall extension.
Filaments, Foci and Movement
Early imaging experiments with all three B. subtilis MreB family proteins appeared to reveal extended helical filaments localized close to the cell periphery and thus presumably close to the inner surface of the cytoplasmic membrane (Jones et al. 2001). All three MreB family proteins exhibited roughly similar patterns of localization and seemed to co-localize, at least under some conditions (Carballido-López et al. 2006; Defeu Soufo and Graumann 2006). The localization of various cell elongation proteins (Leaver and Errington 2005; Formstone et al. 2008) and the use of labelling methods designed to identify nascent cell wall synthesis (Daniel and Errington 2003; Tiyanont et al. 2006) also seemed compatible with a helical mode of wall synthesis. Some experiments suggested a degree of remodelling or active movement of the filaments during cell elongation (Carballido-López and Errington 2003; Defeu Soufo and Graumann 2004). The filamentous helical view of MreB localization was exciting because, in principle, the filaments could act as a geometric guide for the synthetic machinery directly defining the morphology of rod-shaped cells.
In 2011, however, three groups described experiments that revealed the circumferential movement of relatively short filaments or foci (rather than long filaments) of MreB proteins in B. subtilis (Garner et al. 2011; Domínguez-Escobar et al. 2011) and E. coli (Van Teeffelen and Gitai 2011). Importantly, the circumferential movement was dependent on active cell wall synthesis, suggesting that MreB follows rather than leads the synthetic process. The problem with the short filament or patchy view of MreB organization lies in the question of what its role is in cell elongation. Domínguez-Escobar et al. (2011) proposed that MreB acts as a scaffold to help assemble the complex of proteins needed to coordinate the synthesis of PG, WTA and other wall associated elements. The complex was proposed to use existing glycan strands as a template for insertion of new material. However, this presumably only works while the template strands have the correct geometry and could not account for the long term fidelity of shape maintenance and especially the restoration of shape in cells with any shape abnormality.
Meanwhile, electron cryotomography failed to detect elongated filaments of native MreB in flash frozen intact E. coli cells (Swulius et al. 2011), and demonstrated that the very prominent helical cytoplasmic filaments made by one particular E. coli MreB/GFP fusion protein were an artefact specific to that fusion protein (Swulius and Jensen 2012). However, Salje et al. (2011) potentially solved the conundrum of the missing filaments of native protein in the cryo-EM experiments through the discovery that MreB protein polymers have a high affinity for membranes and in vivo this tight membrane association would likely hide the filaments from detection in the low contrast cryo-EM images. This also explained the problem with the E. coli GFP-MreB fusion protein mentioned above because the membrane targeting sequence of E. coli MreB is close to the N-terminus and thus probably occluded by the GFP fusion. Meanwhile, Dempwolff et al. (2011) obtained supporting evidence for membrane association of MreB and Mbl, though interestingly not MreBH, by expressing GFP fusions of the 3 proteins in eukaryotic cells.
The most recent in vivo imaging experiments on B. subtilis, using various super-resolution methods may have resolved some of the confusion around MreB localization, by demonstrating that the proteins (at least MreB and Mbl) are able to form relatively extended helical filaments, at least under some conditions, but that the whole filament systems undergo overall near circumferential movement, which is dependent on (and presumably driven by) PG synthesis (Reimold et al. 2013; Olshausen et al. 2013). Olshausen et al. (2013) suggested that the elongated filaments could serve to coordinate the synthetic activities of multiple wall synthetic complexes, providing a plausible mechanistic explanation for the function of long MreB filaments.
Two recent lines of work on the E. coli MreB system suggest that MreB may have more than one mode of action in cell shape regulation, which may clarify some of the conflicting data described previously. First, Ursell et al. (2014) and Billings et al. (2014) found that MreB filaments or patches have affinity for regions of a particular (aberrant) curvature. Recruitment of the cell wall machinery to those sites could help to correct the curvature leading to the restoration of shape. Morgenstein et al. (2015) then discovered that in certain mutational backgrounds, MreB motion is not required for maintenance of a rod shape. The authors proposed that MreB can help specify cell shape by two distinct mechanisms: first, by a motion-independent mechanism that relies on recruitment of the synthetic machinery to sites of inappropriate curvature, effectively a “repair” mechanism; and second, by a motion-dependent mechanism that helps distribute synthesis over a greater proportion of the surface and is used when cells are growing rapidly and presumably are relatively unperturbed.
The significance of movement and the possible roles of filaments in shape determination have been reviewed in detail recently (Errington 2015).
A Complex Web of Interactions Between MreB Proteins and Cell Wall Effectors
The models discussed above mainly assume that MreB proteins work by controlling the spatial activity of the enzymes responsible for cell wall expansion. This view is supported by numerous reports describing interactions between MreB and various components of the cell wall machinery. The list of possible MreB interacting proteins identified by various methods as candidate members of the B. subtilis elongation machine or “elongasome” are summarised in Table 3.1. Similar collections of interacting proteins have been identified for several other rod-shaped organisms, particularly E. coli and Caulobacter crescentus (reviewed by (Errington 2015). The methods used to identify these proteins include various indirect approaches, such as co-localization, localization dependence, and genetic epistasis, as well as more direct methods of bacterial or yeast 2-hybrid approaches and biochemical pull downs. Although some of the data are not entirely convincing, for example, the “helix-like” localization patterns, taken together, these methods point to the existence of large elongasome complexes containing multiple proteins involved in synthesis of PG and WTA, as well as potentially extracellular factors, such as autolytic enzymes or their regulators. At the moment, little is known about the stoichiometry of these complexes or about whether they are stable or transient.
Table 3.1.
Possible MreB interacting poteins identified as candidate components of the elongasome
| Proteina | MW (kDa) | Localizationb | Comment | References |
|---|---|---|---|---|
| MreC | 32 | I/E | Cell shape function. Encoded by gene immediately downstream from mreB | Leaver and Errington (2005), Kawai et al. (2009b), Garner et al. (2011) and Domínguez-Escobar et al. (2011) |
| MreD | 19 | I | Cell shape function. Encoded by gene immediately downstream from mreBC | Leaver and Errington (2005), Garner et al. (2011), Domínguez-Escobar et al. (2011) and Muchova et al. (2013) |
| RodZ | 23 | I/C | Required for normal cell shape | Domínguez-Escobar et al. (2011) and Muchova et al. (2013) |
| CwlO | 50 | E | Autolytic enzyme, regulated by FtsEX | Domínguez-Cuevas et al. (2013) |
| LytE | 37 | E | Autolytic enzyme. Export regulated by MreBH? | Carballido-López et al. (2006) |
| FtsE | 25 | C | ABC-transporter (ATP-binding protein). With FtsX regulates CwlO. Controlled specifically by Mbl? | Domínguez-Cuevas et al. (2013) |
| FtsX | 32 | I | ABC-transporter (membrane protein). With FtsE regulates CwlO. Controlled specifically by Mbl? | Domínguez-Cuevas et al. (2013) |
| PBP 1 | 99 | E | Major bifunctional PBP. Important for both cell elongation and division | Van Den Ent et al. (2006) and Kawai et al. (2009a, b) |
| PBP 2A | 79 | E | Major TPase with specific role in elongation. Partially redundant to PBP H | Van Den Ent et al. (2006), Kawai et al. (2009a, b), Garner et al. (2011) and Domínguez-Escobar et al. (2011) |
| PBP 2B | 79 | E | Major TPase with specific role in division | Van Den Ent et al. (2006) and Kawai et al. (2009b) |
| PBP 2C | 79 | E | Bifunctional PBP with unknown function | Van Den Ent et al. (2006) and Kawai et al. (2009b) |
| PBP 2D | 71 | E | Transpeptidase with unknown function | Van Den Ent et al. (2006) and Kawai et al. (2009b) |
| PBP 3 | 74 | E | Accessory TPase that can rescue cell division in the absence of PBP 2B activity | Kawai et al. (2009b) |
| PBP 4 | 70 | E | Bifunctional PBP with unknown function | Kawai et al. (2009a, b) |
| PBP H | 76 | E | Major TPase with specific role in elongation. Partially redundant to PBP 2A | Van Den Ent et al. (2006), Kawai et al. (2009b), Domínguez-Escobar et al. (2011) |
| PBP I | 65 | E | TPase of unknown function. | Van Den Ent et al. (2006) and Kawai et al. (2009b) |
| RodA | 43 | I | PG synthesis. Possible monofunctional GTase | Domínguez-Escobar et al. (2011), Meeske et al. (2016), Emami et al. (2017) |
| DapI | 41 | C | N-acetyl-diaminopimelate deacetylase. PG synthesis | Rueff et al. (2014) |
| TagA | 29 | C | Teichoic acid synthesis. UDP-N-acetyl-D-mannosamine transferase | Formstone et al. (2008) |
| TagB | 44 | C | Teichoic acid synthesis. Putative CDP-glycerol:glycerol phosphate glycerophosphotransferase | Formstone et al. (2008) |
| TagF | 87 | C | Teichoic acid synthesis. CDP-glycerol:polyglycerol phosphate glycero-phosphotransferase | Formstone et al. (2008) |
| TagG | 32 | I | ABC transporter for teichoic acid translocation (permease) | Formstone et al. (2008) |
| TagH | 59 | C | ABC transporter for teichoic acid translocation (ATP-binding protein) | Formstone et al. (2008) |
| TagO | 39 | C | Teichoic acid synthesis. Undecaprenyl-phosphate-GlcNAc-1-phosphate transferase | Formstone et al. (2008) |
| TagT | 35 | E | Transfer of anionic cell wall polymers from lipid-linked precursors to peptidoglycan | Kawai et al. (2011) |
| TagU | 34 | E | Transfer of anionic cell wall polymers from lipid-linked precursors to peptidoglycan | Kawai et al. (2011) |
| YvcK | 34 | C | Required for normal localization of PBP 1 | Foulquier et al. (2011) |
| GpsB | 11 | C | Regulation of PBP 1 localization, especially its switch between elongation and division sites. | Claessen et al. (2008) |
| EF-Tu | 43 | C | Translation elongation factor | Defeu Soufo et al. (2015) |
aIn addition to the above, Kawai et al. (2011) identified many additional MreB-associated proteins by pull-down mass spectrometry
b I integral membrane, E extracellular, C cytoplasmic
The Future
Much remains to be learned about the detailed functions of the MreB family proteins of B. subtilis. It is by no means clear why B. subtilis possesses three paralogous genes. At one level, it reflects the general complexity of the cell wall synthetic machinery of the organism. Thus, it also carries multiple copies of many other synthetic genes, including, for example: 4 class A (TPase and GTase) PBPs (Popham and Setlow 1996), 3 LTA synthases (Grundling and Schneewind 2007a, b; Schirner et al. 2009), 3 WTA transferases (Kawai et al. 2011), and at least 2 families of lipid II flippases (Meeske et al. 2015). The overlapping semi-redundant functions of the 3 MreB proteins may reflect that they interact differentially with subsets of cell envelope proteins in order to adapt cell envelope properties to changing environmental conditions. Perhaps “chemical warfare” between organisms in complex and highly competitive environments such as soil, drives adaptability in cell envelope synthesis and organization. Although some aspects of the differential activities of the 3 MreB proteins are beginning to be worked out (Carballido-López et al. 2006; Domínguez-Cuevas et al. 2013), much more probably remains to be elucidated.
An important related question concerns how the many interactions between MreB proteins and the various other components of the cell envelop synthetic machinery (PG synthases, PBPs, autolysins, WTA synthases, etc) are mediated, particularly whether they are static or dynamic and the extent to which they are hierarchical and mutually permissive or exclusive.
A final major question concerns the localization and dynamic properties of the proteins. What conditions determine the length of the MreB filaments and how do length and movement relate to the various problems associated with cell shape determination, maintenance and repair?
There is a sense that the array of analytical methods we now possess, enabling us to localise proteins with increasing temporal and spatial resolution and to define the components of protein complexes and their stoichiometry, should allow details of the machinery and mechanisms to be resolved. Complexity may be the biggest barrier to progress.
FtsZ and the Cell Division Machinery
Most bacteria with a PG wall divide by directing the ingrowth of a sheet of wall material that eventually forms the new hemispherical poles of the daughter cells. In almost all bacteria, the key cytoskeletal protein involved in defining the site of division and then orchestrating the process is called FtsZ, which is structurally and biochemically homologous to tubulin (Löwe and Amos 1998). In bacteria where the process has been studied in detail, FtsZ appears to form a circumferential ring that defines the site of cell division (Bi and Lutkenhaus 1991). It also serves to recruit, directly or indirectly, multiple protein components of a division machine, sometimes called the “divisome” (Adams and Errington 2009; Egan and Vollmer 2013). Several divisome associated proteins might also be considered as cytoskeletal proteins (e.g. FtsA, DivIVA, MinD; see below), depending on the definition. B. subtilis is an interesting model for the study of bacterial cell division because it has two contrasting modes of division: a “conventional mode”, carried out by vegetatively growing cells; and a modified, highly asymmetric division undertaken by sporulating cells.
Biochemical Properties of FtsZ
The presence of a tubulin GTP-binding signature motif in FtsZ (GGGTGTG) was first reported in the early 1990s (Raychaudhuri and Park 1992; De Boer et al. 1992; Mukherjee and Lutkenhaus 1994). Crystallographic studies confirmed the near congruence of the structures of FtsZ and tubulin proteins (Löwe and Amos 1998). Not surprisingly, the proteins also have similar biochemical properties. Like tubulin, FtsZ assembles in vitro in a head to tail fashion to form single stranded protofilaments, which can further assemble into bundles, sheets or rings. The protofilaments are also highly dynamic and go through cycles of turnover/polymerization, regulated by the binding and hydrolysis of GTP. See Chapter 10.1007/978-3-319-53047-5_5 for a detailed description of FtsZ polymerization dynamics.
FtsZ Visualization During Growth and Sporulation of B. subtilis
The ability of FtsZ to form tubulin-like protofilaments and protofilament bundles raised important questions about the abundance, assembly and dynamics of the protein in vivo. Estimations of protein abundance have suggested about 2000–6000 molecules per cell in B. subtilis (Feucht et al. 2001; Ishikawa et al. 2006; Muntel et al. 2014), giving a protein concentration of about 2–6 μM, well above the in vitro critical concentration for assembly (e.g. 0.72 μM for E. coli FtsZ; Chen et al. 2012). Also, given a protofilament subunit repeat length of 4.3 nm (Oliva et al. 2004), there is enough FtsZ to circumnavigate the cell about 3–10 times.
Several labs have investigated the localization of FtsZ in B. subtilis. Wang and Lutkenhaus (1993) used immunogold electron microscopy to demonstrate association of FtsZ with the leading edge of the invaginating cell division structure. The immunofluorescence studies of (Levin and Losick 1996) confirmed the presence of FtsZ bands (presumed to be rings) at the expected position near mid cell in vegetative cells but also unexpectedly, near both poles of early sporulating cells. This turns out to be a key feature of the mechanism used by B. subtilis to achieve asymmetric cell division during sporulation, which will be discussed in detail below. Several reports have highlighted the possible role of helical FtsZ structures as intermediates in assembly or constriction at cell division sites (Ben-Yehuda and Losick 2002; Feucht and Errington 2005; Peters et al. 2007; Strauss et al. 2012). Helical FtsZ structures are most prominent during the transition from vegetative growth to sporulation in B. subtilis, during which the site of division shifts from mid cell to near the pole. The mid cell Z ring appears to transform into a helix which grows length-wise, before breaking down into two short helices, one near each pole. Each helix then coalesces into a ring (Ben-Yehuda and Losick 2002). Therefore, each sporulating cell assembles two Z rings, one at each pole, but only one develops into a septum (see below). Several mutations in ftsZ have been described that promote a tendency to form spiral Z rings and similarly shaped division events (Feucht and Errington 2005; Michie et al. 2006), suggesting that the helical configuration has functional relevance. Peters et al. (2007) also described helical configurations in vegetative cells, based on modified immunofluorescence imaging methods. On the other hand, several higher resolution imaging methods, including super-resolution fluorescence imaging and cryo-EM of B. subtilis and other organisms have suggested that FtsZ rings may be more complex, and beaded or discontinuous (Jennings et al. 2011; Strauss et al. 2012; Li et al. 2007; Min et al. 2014).
Dynamic movement of FtsZ rings has been observed by time-lapse imaging (Strauss et al. 2012) but more quantitative and perhaps surprising information came from fluorescence recovery after photobleaching (FRAP) experiments. Erickson and colleagues established that FtsZ subunits in Z rings, either pre-constriction or during constriction, turn over with a half time of about 8 seconds (Anderson et al. 2004). This emphasises the likely importance of dynamics in FtsZ function but also, the difficulty in imaging such structures with the need for both high spatial and temporal resolution. Löwe’s lab have recently described compelling evidence for the formation of regular circumferential bands of FtsZ in which the protofilaments are connected by regular lateral contacts for 2 Gram negative bacteria, E. coli and Caulobacter crescentus, as well as in an in vitro system. These observations support a model for constriction involving filament sliding (Szwedziak et al. 2014). It remains to be seen whether this model can be extended to B. subtilis and other Gram positive bacteria, but it seems unlikely that the fundamental features of FtsZ function in bacterial division are not well conserved.
The B. subtilis Divisome
The B. subtilis divisome has been studied in considerable detail: some properties of the proteins thought to contribute directly or indirectly to divisome function in this organism are described in Table 3.2. These proteins have been identified through homology to known division proteins in other organisms, by biochemical pull downs or through various genetic screens.
Table 3.2.
Proteins of the B. subtilis divisome and its regulators
| Protein | MW (kDa) | Locationa | Comments | Key references |
|---|---|---|---|---|
| FtsZ | 40 | C | Tubulin-like protein. Assembles into protofilaments and higher order structures to generate the “Z ring” at the division site. Recruits other divisome proteins to the ring. | Beall et al. (1988), Beall and Lutkenhaus (1991), and Wang and Lutkenhaus (1993) |
| FtsA | 48 | C | Actin / HSP70 superfamily ATPase. Dimerises and can form higher order structures. C-terminal amphipathic helix promotes membrane association. Direct interaction with FtsZ, which contributes to membrane association of the Z ring. | Beall and Lutkenhaus (1991), Feucht et al. (2001), Jensen et al. (2005) and Ishikawa et al. (2006) |
| SepF | 17 | C | Forms regular 50 nm diameter rings in vitro and interacts directly with FtsZ in vitro, promoting FtsZ bundling. Membrane targeting domain contributes to membrane association of the Z ring. | Hamoen et al. (2006) and Gündoğdu et al. (2011) |
| ZapA | 9.0 | C | Widely conserved protein that promotes Z ring formation by direct interaction with FtsZ. | Gueiros-Filho and Losick (2002) |
| EzrA | 65 | C | N-terminal transmembrane anchor. Cytosolic domain has a spectrin-like fold. Interacts with FtsZ, contributing to membrane association of the Z ring. Additional role in cell elongation via interactions with PBP 2B and GpsB. | Levin et al. (1999), Haeusser et al. (2004), Claessen et al. (2008), and Cleverley et al. (2014) |
| GpsB | 11 | C | DivIVA-related protein involved in both cell elongation and cell division. Interacts with the major PG synthase, PBP 1, and thought to be involved in shuttling of this protein between elongation and division complexes. Synthetic lethal in combination with ftsA mutation. Synthetic “sick” in combination with ezrA. EzrA-SepF interaction probably important for shuttling. | Claessen et al. (2008) and Tavares et al. (2008) |
| FtsL | 13 | E | Bitopic membrane protein with short extracytoplasmic coiled-coil-like domain. Target of several cell division regulatory mechanisms. Unstable protein subject to degradation by a regulated intramembrane proteolysis (RIP) process involving YluC protease. Stability also regulated by interactions with DivIC and DivIB. | Daniel et al. (1998), Daniel and Errington (2000), Sievers and Errington (2000a, b), Kawai and Ogasawara (2006), Bramkamp et al. (2006) and Daniel et al. (2006) |
| DivIB | 30 | E | Bitopic membrane protein with large extracellular domain. Structural data from other organisms suggests two domains, one of which resembles the POTRA domain often involved in protein protein interactions. Complex pattern of interactions with FtsL and DivIC. Homologue called FtsQ in E. coli. | Beall and Lutkenhaus (1989), Harry and Wake (1989, 1997), Katis and Wake (1999), Katis et al. (2000), Daniel and Errington (2000) and Daniel et al. (2006) |
| DivIC | 15 | E | Bitopic membrane protein with short extracytoplasmic coiled-coil-like domain. Interacts with FtsL and DivIB. Likely homologue confusingly called FtsB in E. coli. | Katis et al. (1997), Katis and Wake (1999), Katis et al. (2000), Sievers and Errington (2000b), Robson et al. (2002) and Daniel and Errington (2000) |
| FtsW | 44 | I | Integral membrane protein closely related to RodA involved in cell elongation. | Lu et al. (2007) |
| Pbp2B | 79 | E | Penicillin binding protein. Monofunctional (class B) transpeptidase specifically required for cell division. | Yanouri et al. (1993), Daniel et al. (1996) and Daniel and Errington (2000) |
| DivIVA | 19 | C | Coiled coil protein with weak similarity to eukaryotic tropomyosins. Targeted to division sites and cell poles at least in part by sensing membrane curvature. Membrane interaction through conserved N-terminal domain containing essential tryptophan residue. Involved in a range of cell pole associated functions in Gram positive bacteria. | Cha and Stewart (1997), Edwards and Errington (1997), Hamoen and Errington (2003) and Lenarcic et al. (2009), Ramamurthi and Losick (2009) and Van Baarle et al. (2013) |
| MinC | 25 | C | Widely conserved division inhibitor acting on FtsZ and possibly other steps in division. | Reeve et al. (1973), Levin et al. (1992), Marston and Errington (1999) and Gregory et al. (2008) |
| MinD | 29 | C | Widely conserved indirect division inhibitor that works by spatial regulation of MinC protein. Poorly characterised additional role in chromosome segregation during sporulation. | Reeve et al. (1973), Levin et al. (1992), Marston et al. (1998) and Marston and Errington (1999), Kloosterman et al. (2016) |
| MinJ | 44 | I / C | PDZ-domain protein targeted to cell poles by interaction with DivIVA (at least). Required for correct spatial localization of the MinCD complex and thus the regulation of cell division. | Patrick and Kearns (2008), Bramkamp et al. (2008) and Van Baarle and Bramkamp (2010) |
| Noc | 33 | C | Site-specific DNA binding protein. Inhibitor of division. Major factor effecting nucleoid occlusion. | Wu and Errington (2004), Wu et al. (2009) and Adams et al. (2015) |
| WhiA | 36 | C | Enigmatic nucleoid associated factor. whiA mutation causes severe filamentation when combined with zapA, ezrA or various regulatory proteins of cell division. | Surdova et al. (2013) |
| SpoIIE | 92 | C/I | Bifunctional sporulation-specific protein. C-terminal kinase domain regulates prespore-specific gene expression. C-terminal domain required for efficient switch in cell division position from mid cell to sub-polar position, probably via a direct interaction with FtsZ. | Arigoni et al. (1995), Feucht et al. (1996), Wu et al. (1998), Lucet et al. (2000), Carniol et al. (2005) and Bradshaw and Losick (2015) |
| MciZ | 4.0 | C | Mother cell-specific inhibitor of FtsZ assembly. Caps FtsZ protofilaments at the “minus” end. | Handler et al. (2008) and Bisson-Filho et al. (2015) |
| RefZ | 24 | C | Site-specific DNA-binding protein that contributes to precise relative positioning of chromosome and asymmetric division site during sporulation. | Wagner-Herman et al. (2012) and Miller et al. (2015) |
a C cytosolic, I integral membrane, E extracytoplasmic
Imaging experiments suggest that the divisome assembles in at least two distinct steps (Gamba et al. 2009). In the first step, which seems to involve mainly cytosolic factors, a “ring” of FtsZ protein assembles, in parallel with the recruitment of “early” divisome proteins FtsA, SepF, ZapA and EzrA. After a delay representing about 20% of the cell cycle, the second step of assembly takes place, in which the “late” proteins are recruited. These are mainly proteins with major extracellular domains or integral membrane proteins. Various regulatory proteins, including GpsB, DivIVA, MinJ, MinD and MinC arrive at about the same time or slightly later, possibly being dependent on initiation of membrane or PG ingrowth. Ishikawa et al. (2006) detected interactions between the various early proteins in a series of biochemical pull-down experiments.
Three “early” cytosolic proteins appear to promote the formation of a functional Z ring in B. subtilis – FtsA, SepF and ZapA. FtsA was identified by its conserved location immediately upstream of and adjacent to FtsZ (Beall et al. 1988). Unlike E. coli, ftsA mutants of B. subtilis are viable, though they are substantially deficient in division (Beall and Lutkenhaus 1992). FtsZ still localizes at regular intervals but most of the Z rings are abnormal, often appearing as multiple diffuse bands rather than one clear, strong band (Jensen et al. 2005). Purified B. subtilis FtsA binds and hydrolyses ATP (Feucht et al. 2001) but little more work has been done on this protein so far. Using the Thermotoga maritima protein, Löwe and colleagues have demonstrated that even though FtsA has a different subdomain architecture to actin, the protein can form canonical actin-like protofilaments in vitro (Van Den Ent and Löwe 2000; Szwedziak et al. 2012). FtsA interacts specifically with the C-terminal domain of FtsZ. Despite a great deal of work over nearly 2 decades, little is known about the precise function of ftsA other than that it can form high MW dynamic complexes of various kinds with FtsZ (e.g., Loose and Mitchison 2014). Perhaps its best defined function lies in membrane association, which occurs through a C-terminal amphipathic helix (Pichoff and Lutkenhaus 2005) and enables the protein to anchor the Z ring to the membrane (Szwedziak et al. 2014). Interestingly, this interaction with the membrane is strongly dependent on the membrane potential (Strahl and Hamoen 2010).
Genetic and biochemical experiments suggest that SepF protein provides a second membrane anchor for the Z ring in Gram positive bacteria. sepF was discovered simultaneously in two labs by different methods. Ishikawa et al. (2006) identified the SepF protein in pull-down experiments using FtsZ, FtsA, EzrA and ZapA as bait. Yeast 2-hybrid experiments detected the formation of a SepF-SepF self interaction, as well as an interaction with FtsZ. Meanwhile, Hamoen et al. (2006) identified sepF as a candidate cell division gene from its conserved position (in Gram positive bacteria) between ftsZ and divIVA. Deletion of the gene gives a mild reduction in division frequency but the division septa formed are thick and morphologically abnormal. Mutation of sepF turned out to be lethal in the presence of mutations in ftsA or another division associated gene, ezrA (Ishikawa et al. 2006; Hamoen et al. 2006). In vitro, SepF protein assembles into large and regular protein rings with a diameter of about 50 nm: these rings are able to bundle FtsZ protofilaments into long tubular structures (Gündoğdu et al. 2011). Detailed structural analysis of the protein (Duman et al. 2013) suggests that the N-terminal region, like FtsA, contains a membrane associating amphipathic helix, whereas the C-terminal domain is globular and responsible for both the formation of SepF rings and association with FtsZ. Duman et al. (2013) suggest that the amphipathic helices of FtsA and SepF both serve to promote association of the Z ring with the leading edge of the septum, since this region contains positively curved (convex) membrane into which the helices can readily insert. Duman et al. (2013) proposed a model in which SepF polymers bind as arc onto the convex leading edge of the nascent division septum and maintain the Z ring in this position by bundling FtsZ protofilaments. However, this model is slightly unsatisfactory in leaving open the question of why SepF makes complete rings in vitro.
ZapA is a low MW (9 kDa) positive regulator of FtsZ assembly. It was identified in a screen for genes which, when overexpressed, could overcome the cell division block caused by overproduction of MinD (Gueiros-Filho and Losick 2002). Absence of ZapA gives no discernible phenotype under normal conditions but causes a severe division block in cells producing lower than normal levels of FtsZ, or lacking the ezrA, divIVA or whiA genes (Gueiros-Filho and Losick 2002; Surdova et al. 2013). ZapA interacts directly with FtsZ and, in vitro, it promotes FtsZ polymerisation as well as lateral association, yielding both single and bundled filaments (Gueiros-Filho and Losick 2002; Low et al. 2004). A temperature-sensitive mutant of FtsZ (FtsZ(Ts1)), defective in lateral association between FtsZ protofilaments at high temperatures, can be rescued by overexpressing ZapA. This supports the proposed function of ZapA as a promoter of FtsZ bunding (Monahan et al. 2009). ZapA of Pseudomonas aeruginosa forms dimers or tetramers in solution but oligomerizes at high concentrations; a property that could support its function as an effective FtsZ cross-linker (Low et al. 2004).
The ezrA gene, which is present only in Gram positive bacteria, was identified by mutations suppressing the division phenotype of a thermosensitive ftsZ allele (Levin et al. 1999). ezrA mutants can tolerate reduced levels of active FtsZ and the gene name derives from the observation that the ezrA single mutant makes extra Z-rings. The protein has an unusual topology with a single N-terminal transmembrane span followed by a major domain that is cytosolic. Curiously, one protein that shares this unusual topology is E. coli ZipA, which is an essential FtsZ-interacting protein in E. coli (Hale and De Boer 1997), but it seems that EzrA and ZipA are otherwise unrelated in sequence. ezrA mutants also have a slightly reduced cell diameter, indicating a mild defect in cell elongation. Genetic experiments suggest that this may be due to incorrect regulation of the activity of the major PBP (PBP 1) involved in synthesis of PG during both elongation and division (Claessen et al. 2008). The results of detailed mutational analysis of the gene suggest that different regions of the large protein contribute in different ways to the regulation of Z ring dynamics (Haeusser et al. 2007; Land et al. 2014). The crystal structure of the cytoplasmic domain of EzrA was recently solved (Cleverley et al. 2014) and shown to be similar to that of eukaryotic spectrins, comprising multiple, connected repeats of antiparallel α-helices, forming a complete semi-circle of 12 nm diameter. Spectrins are cytoskeletal proteins that can form two-dimensional polygonal networks lining the membrane, and they help maintain plasma membrane integrity and cytoskeletal structure in eukaryotic cells. The formation of a semi-circle could enable both the C-terminal four-helix bundle and the N-terminal transmembrane domain to interact with the membrane at the same time. Structural modelling indicates that an antiparallel dimer of EzrA molecules, as found in some crystal structure forms, could trap a paired FtsA-FtsZ protofilament inside the arch. In principle, this could serve to both anchor the protofilaments to the membrane and or locally prevent the formation of protofilament bundles.
Because FtsZ protein is thought to be indirectly associated with the cell membrane, through its interactions with FtsA, SepF and possible EzrA, it seems likely that most of the remaining divisome proteins, which are largely integral membrane or extracytoplasmic proteins (summarised in Table 3.2), do not interact directly with FtsZ. Their functions are probably concerned mainly with membrane dynamics, or peptidoglycan synthesis and turnover and will not be discussed in detail here.
Regulation of Z Ring Formation and Cell Division
Cell division needs to be tightly coordinated with other cell cycle events, particularly chromosome replication and segregation. Recently, the field of bacterial cell cycle regulation has been invigorated by the unexpected discovery that cell size homeostasis is achieved by an “adder” process in which new born cells grow by a relatively fixed length increment before dividing again, rather than by measuring a “division mass”, according to a decades old dogma (Campos et al. 2014; taheri-araghi et al. 2015). The key questions now concern how the length increment is measured by the cell and used to regulate divisome function. In B. subtilis the intracellular concentration of FtsZ stays constant throughout the cell cycle and, although the frequency of Z ring formation varies with growth rate, the levels of FtsZ are unaffected. Artificially increasing the level of FtsZ in B. subtilis cells only leads to a small increase in Z ring frequency (Weart and Levin 2003).
One factor that could be involved in buffering the levels of available FtsZ is the two-component ATP-dependent protease, ClpXP. ClpX is a member of the AAA+ (ATPases associated with various cellular activities) family of ATPases. It recognizes and unfolds specific protein substrates and transfers the unfolded protein to the serine protease ClpP for degradation (Sauer et al. 2004). ClpXP is thought to participate in the regulation of FtsZ assembly by maintaining the pool of subunits available for ring formation (Weart et al. 2005; Camberg et al. 2009; Dziedzic et al. 2010). In B. subtilis ClpX inhibits FtsZ polymerization in vivo and in vitro in an ATP-and ClpP-independent manner but does not degrade it, though ATP hydrolysis has been shown to be required for maximum inhibition (Weart et al. 2005; Haeusser et al. 2009). This is in contrast to E. coli in which ClpX modulates the level of FtsZ by degrading FtsZ (Camberg et al. 2009).
Several spatial and or temporal regulators of divisome function have been identified, mainly acting by negative regulatory mechanisms.
Nucleoid Occlusion (NO)
The fact that cell division tends not ever to bisect the nucleoid, even in cells with perturbations in chromosome replication or organization, led Woldringh and colleagues to postulate a negative regulation exerted by the nucleoid, potentially by the DNA itself (Mulder and Woldringh 1989; Woldringh et al. 1991), which Rothfield later termed “nucleoid occlusion” (Cook et al. 1989). About 10 years ago, protein factors contributing to NO were identified almost simultaneously in B. subtilis (noc) and E. coli (slmA) (Wu and Errington 2004; Bernhardt and De Boer 2005): surprisingly, they were unrelated proteins that turned out to have different modes of division inhibition. B. subtilis noc was identified serendipitously as a factor that had a synthetic lethal division phenotype when combined with mutations affecting the Min system (which is described below). Three lines of evidence suggested that Noc protein was a NO factor: first, the protein had a classical helix-turn-helix motif and bound tightly to DNA; second, overexpression of Noc inhibited division; third, noc mutants had an increased frequency of nucleoid “guillotining” when DNA replication or segregation was perturbed (Wu and Errington 2004). Later work established that Noc is a site-specific DNA-binding protein with recognition sites (Noc binding sites; NBS) distributed all over the chromosome, except in the replication terminus region, where binding sites are scarce (Wu and Errington 2004; Wu et al. 2009). Noc appears to have been derived by gene duplication and divergence from the ParB (Spo0J) protein in Firmicutes, and like Spo0J it spreads from its primary binding sites to form arrays which are important for its function. While the N-terminal domain of Spo0J (ParB) is involved in interaction with its partner protein Soj, the N-terminal domain of Noc contains an amphipathic helix, which enables it to associate with the cell membrane (Adams et al. 2015). Both overexpression and deletion of Noc affect division at the level of FtsZ assembly (Wu and Errington 2004). It seems that the formation of Noc arrays around NBSs enhances membrane association and that recruitment of these DNA-Noc arrays to the membrane prevents the local formation of FtsZ ring assemblies (Adams et al. 2015). It is possible that Noc works by enhancing a natural NO system, akin to that originally described by Woldringh, in which the presence of chromosomal DNA excludes accumulation or formation of high MW divisome complexes.
The Min System
NO prevents division from occurring in the vicinity of the nucleoid, but it cannot protect the nucleoid free regions at the nascent and old cell poles. The system that prevents polar division is called Min and was first discovered via E. coli mutants that frequently divide near to the cell pole, giving small anucleate cells called minicells (Adler et al. 1967). The B. subtilis MinC and MinD proteins were identified by sequence homology to their E. coli counterparts (Levin et al. 1992). The B. subtilis Min system is now known to consist of at least four proteins: MinC, MinD, MinJ and DivIVA. MinC is an FtsZ inhibitor which interacts directly with FtsZ. In vitro studies have shown that like E. coli MinC, the B. subtilis protein inhibits FtsZ bundle formation by disrupting lateral interactions between protofilaments (Dajkovic et al. 2008; Scheffers 2008; Blasios et al. 2013), though E. coli MinC has been shown to also destabilize FtsZ protofilaments (Hu et al. 1999; Shen and Lutkenhaus 2010). MinD is a membrane-associated activator of MinC. Both MinC and MinD are relatively well conserved among bacteria. The two proteins form a heterotetrameric complex and in B. subtilis are recruited to the division site and the cell poles by MinJ, which in turn associates with the “topological specificity” determinant DivIVA (Edwards and Errington 1997; Marston et al. 1998; Bramkamp et al. 2008; Patrick and Kearns 2008). E. coli lacks counterparts of the MinJ and DivIVA proteins and instead uses an amazing oscillating MinCD mechanism to prevent division at the cell poles (Lutkenhaus 2007).
The key feature of DivIVA that enables it to spatially control the Min inhibitory effect lies in its targeted localization to division sites and cell poles. DivIVA oligomers have affinity for high negative membrane curvature, which normally occurs only at invaginating division septa or recently completed cell poles (Lenarcic et al. 2009; Ramamurthi and Losick 2009; Eswaramoorthy et al. 2011). It is probably recruited to the site of division as soon as membrane invagination begins, due to divisome constriction. Therefore, accumulation of DivIVA at the division site is dependent on the presence of a functional divisome but once curvature has been generated, the rings of DivIVA, one on each side of the growing septum, are no longer affected by contraction of the divisome (Eswaramoorthy et al. 2011). Upon completion of septation, the divisome disassembles and the septum splits to generate new cell poles for the two daughter cells. In cells that are not dividing, DivIVA-GFP is concentrated at the hemispherical cell poles (Eswaramoorthy et al. 2011) but in dividing cells, DivIVA is remodelled and a portion of the DivIVA molecules remain at the pole, while some protein migrates to the new division site (Eswaramoorthy et al. 2011; Bach et al. 2014). The main structural feature of DivIVA is a parallel coiled coil, similar to the yeast tropomyosin Cdc8, a eukaryotic cytoskeletal protein involved in cytokinesis (Edwards et al. 2000; Oliva et al. 2010). This major C-terminal portion of DivIVA resembles the crescent shape of eukaryotic BAR domains normally found at the interface between the actin cytoskeleton and lipid membranes, which bind to curved membranes and also introduce curvature (Oliva et al. 2010). This raises the possibility that DivIVA senses membrane curvature using a mechanism similar to the Bar domain proteins. Structural and genetic evidence suggest that membrane interaction occurs via a hairpin structure with conserved exposed basic and hydrophobic residues in the N-terminal domain of the protein (Oliva et al. 2010).
MinJ is presently the least well characterised component of the Min system. It has 6 transmembrane helices with both N- and C-termini in the cytoplasm. The C-terminal globular portion of the protein comprises a classical PDZ domain; a protein fold often involved in protein-protein interactions (Van Baarle and Bramkamp 2010). MinJ can interact with both DivIVA and MinD, based on 2-hybrid experiments (Patrick and Kearns 2008; Bramkamp et al. 2008), suggesting that MinJ is the immediate polar target for recruitment of MinD, rather than DivIVA (Bramkamp et al. 2008).
As originally described by Marston et al. (1998) and reinforced by subsequent papers (Gregory et al. 2008; Bramkamp et al. 2008; Van Baarle and Bramkamp 2010), DivIVA and presumably now MinJ are recruited to mid cell soon after the initiation of division. MinJ, in turn, recruits the MinCD complex, which has no effect on the ongoing division but is poised to disassemble the divisome as division is completed, and or prevent the assembly of a new division complex. Although in general it appears that little, if any, of these proteins are retained at completed “old” cell poles, some activity is probably retained to prevent inappropriate minicell divisions from occurring there.
The mechanism of action of the MinCD inhibitor is not yet fully understood, despite over 20 years of study. MinD is a member of the Walker A cytoskeletal ATPase (WACA) family, a group of cytoskeletal proteins thought to be unique to bacteria (Löwe and Amos 2009; Shih and Rothfield 2006; Michie et al. 2006; Pilhofer and Jensen 2013). Characteristic of this family is a ‘deviant’ Walker A motif – KGGXXGKT’ containing two conserved lysines, both important for binding and hydrolysis of ATP (Lutkenhaus 2012). As in E. coli, the ATPase activity of B. subtilis MinD is required for membrane binding and activation of MinC (Karoui and Errington 2001), but biochemical details of how the inhibitory activity of the MinCD complex is regulated remain elusive. One interesting recent development has been the report that E. coli MinC and MinD form alternating copolymeric cytomotive filaments with structural similarity to septins (Ghosal et al. 2014). Septins are a group of eukaryotic GTP-binding cytoskeletal proteins that polymerize into hetero-oligomeric protein complexes and play many important roles, including serving as membrane scaffolds for protein recruitment and as diffusion barriers for subcellular compartmentalization (Mostowy and Cossart 2012). It is not clear whether the B. subtilis homologues behave similarly or what the functional significance of the copolymer organization is.
Nutritional Regulation of Cell Division
In B. subtilis and probably many other bacteria, cell size is regulated according to the growth rate, such that fast growing cells are, on average, larger than slow growing cells (Sharpe et al. 1998). Weart et al. (2007) identified the UgtP protein as a key metabolic regulator of cell division in B. subtilis. UgtP is responsible for synthesis of glucolipids using UDP-glucose as a substrate. Mutations in the ugtP gene, or genes upstream in the UDP glucose synthetic pathway (pgsC or gtaB) had a small cell phenotype, in which both FtsZ ring formation and cell division occur at a smaller average cell size than in the wild type. UgtP turned out to interact directly with FtsZ in vitro and in vivo, and its inhibitory effect on FtsZ assembly is stimulated by UDP-Glucose (Weart et al. 2007). Under nutrient rich conditions UgtP levels are increased, as is the availability of its UDP-Glucose substrate, leading to an inhibition/delay in assembly of the FtsZ ring.
Monahan et al. (2014) identified a similar but quite distinct regulatory effect on cell division involving central carbon metabolism. They showed that a temperature sensitive ftsZ mutant could be rescued by mutations in genes encoding pyruvate kinase (pyk) or phosphoglycerate kinase (pgk) and established that these mutations work by limiting the supply of pyruvate from glycolysis. They identified the E1α subunit of pyruvate dehydrogenase, which uses pyruvate as a substrate in generating acetyl-CoA. Localization of E1α was found to shift between nucleoid associated and nucleoid excluded depending on the availability of nutrients (high vs low, respectively). Various genetic tests were consistent with a model in which E1α is a positive regulator of FtsZ ring formation helping to couple this to sensing of nutrient availability. Molecular details of the putative interaction between the various players in this process remain to be worked out.
Z Rings and Cell Division During Sporulation
Early in sporulation the cell division cycle is substantially modified to pave the way for generation of the distinct prespore and mother cell progeny and their subsequent differentiation. As mentioned above, this involves a repositioning of FtsZ rings away from the normal mid cell position to the cell poles (Errington 2003). The following section focuses mainly on events involving FtsZ. The key cell cycle changes, which are controlled by global changes in transcription in response to starvation, are as follows. First, medial division is blocked by a mechanism that is presently unclear, but a major change in chromosome configuration – the formation of a structure called the axial filament (Ryter 1965; Bylund et al. 1993) – probably contributes, through a nucleoid occlusion effect. Instead FtsZ rings are directed to sub-polar positions at each end of the cell (Levin and Losick 1996). Formation of these polar Z-rings requires both a small upregulation of FtsZ synthesis and the synthesis of a sporulation-specific protein SpoIIE (Ben-Yehuda and Losick 2002; Feucht et al. 1996) (see below). The upregulation of ftsZ occurs via a promoter controlled by the σH form of RNA polymerase, which is active only in stationary phase and sporulation (Gholamhoseinian et al. 1992; Gonzy-Tréboul et al. 1992).
Mutations in several key regulatory genes of sporulation give rise to an interesting phenotype called “disporic”, in which prespore-like cells form at both poles of the cell (Piggot and Coote 1976). Lewis et al. (1994) showed that in these cells the asymmetric division events occurred sequentially, with the first preceding the second by about 20 min. This suggested that the Z rings at the two poles develop at different rates, ultimately contributing to the generation of asymmetry – whichever potential divisomes matures first defines the pole which the prespore cell forms (Lewis et al. 1994). The polar sporulation septum differs from a normal vegetative septum in having a much thinner layer of PG (Ryter 1965; Illing and Errington 1991; Tocheva et al. 2013). This thinning may be related to the fact that a little while later, the PG needs to be hydrolysed to enable the remarkable process of prespore engulfment to occur: the small prespore is engulfed by the mother cell to produce a cell within a cell, similar to eukaryotic phagocytosis (Illing and Errington 1991; Tocheva et al. 2013). In addition to FtsZ (Beall and Lutkenhaus 1991), FtsA is probably also required for sporulation division (Beall and Lutkenhaus 1992), though curiously, the latter protein only appears to accumulate at one of the polar potential division sites – presumably the one that goes on to support division (Feucht et al. 2001). DivIB, DivIC and FtsL, at least, are also required for formation of the sporulation septum (Levin and Losick 1994; Daniel et al. 1998; Feucht et al. 1999) but whether the other vegetative divisome proteins are also required for the sporulation septum has not been systematically studied. How the polar septum is formed despite continued presence of the Noc, MinCDJ and DivIVA proteins is also not clear.
The large (92 kDa) SpoIIE protein plays two distinct critical roles in the prespore developmental programme. In addition to its role in asymmetric septation, it is also essential for activation of the first compartment-specific transcription factor, σF, in the prespore (Duncan et al. 1995; Arigoni et al. 1996; Feucht et al. 1996). The N-terminal domain of SpoIIE contains 10 predicted transmembrane spans. This is followed by a central regulatory domain and a C-terminal PP2C phosphatase domain. SpoIIE is recruited to both polar Z rings sequentially (Arigoni et al. 1995; Levin et al. 1997; Wu et al. 1998), probably via a direct interaction with FtsZ (Lucet et al. 2000). The precise role of SpoIIE in FtsZ assembly is still not clear. Absence of SpoIIE causes a delayed and reduced frequency of polar divisions, as well as a vegetative-like thickening of the septa that are formed (Illing and Errington 1991; Barák and Youngman 1996, Feucht et al. 1996; Khvorova et al. 1998; Ben-Yehuda and Losick 2002; Carniol et al. 2005). Crucially, after septation, the SpoIIE protein from the septum is sequestered into the smaller prespore compartment (Wu et al. 1998; Campo et al. 2008; Bradshaw and Losick 2015), where the phosphatase domain helps to drive the activation of σF specifically in that compartment. Regulation of SpoIIE is highly complex and involves association with and release from the divisome, recruitment at the adjacent cell pole by interaction with DivIVA, oligomerization and controlled proteolysis (Bradshaw and Losick 2015).
At the transcriptional level, formation of a polar septum, through the localized action of SpoIIE phosphatase, triggers the prespore localized activation of σF, which then turns on the early prespore programme of gene expression. One of the newly expressed genes, spoIIR, encodes a factor that triggers the activation of a different sigma factor, σE, in the mother cell compartment. Among the proteins made as a result of transcription by σE-RNA polymerase is a specific inhibitor of ftsZ assembly, called MciZ (Handler et al. 2008) that appears to work as a protofilament capping protein (Bisson-Filho et al. 2015). MciZ helps to block the utilization of the second polar FtsZ ring located in the mother cell compartment. Three other σE-dependent proteins (SpoIID, SpoIIM and SpoIIP) also facilitate the formation of a second polar division, but apparently by working downstream on PG synthesis (Eichenberger et al. 2001).
FtsZ Inhibitors as Potential Antibiotics
FtsZ of B. subtilis and closely related Gram positive bacteria, including Staphylococcus aureus, is susceptible to inhibition by a family of related benzamide compounds, with potential for use as antibiotics. These compounds bind to an allosteric site in the protein and apparently trap the protein in the “open” state, which promotes protofilament assembly (Haydon et al. 2008; Tan et al. 2012). In vivo, this results in the formation of multiple discrete foci of FtsZ, which recruit all tested downstream divisome proteins (4 early and 4 late) (Haydon et al. 2008; Adams et al. 2011). However, productive Z rings are not formed, leading to a complete division block. The potential clinical use of these compounds has not yet been evaluated.
L-Form (Cell Wall Deficient) Bacteria
Despite the extraordinary complexity of the wall, its various important functions, and its role as the target for many powerful antibiotics, it is surprisingly easy for B. subtilis to lose its wall. Only one or two mutations are needed to enable B. subtilis (and many other organisms; Mercier et al. (2014)) to switch into a wall deficient mode called L-form (Leaver et al. 2009; Mercier et al. 2013; Kawai et al. 2015). L-forms require an osmoprotective medium to prevent them from incurring osmotic lysis and they have pleomorphic shapes, due to lack of the rigid cell wall. Remarkably, L-forms can tolerate the complete deletion of many genes that are normally essential for growth and division, including both FtsZ and the complete set of MreB homologues (Leaver et al. 2009; Mercier et al. 2012). They divide by a blebbing mechanism that requires only an increase in membrane synthesis (Mercier et al. 2013). Apart from interest in L-forms as potential biotechnological devices or possibly as agents responsible for certain infectious diseases, they are likely to be useful in basic studies of cell wall elongation and division through their ability to tolerate the disruption of many genes that are normally essential (Kawai et al. 2014).
The Future
Some of the open questions about the FtsZ system are similar to those of MreB. The advent of cryo-EM tomography is beginning to resolve the nature of the FtsZ ring (Szwedziak et al. 2014) but resolution of the detailed structure in vivo remains probably the biggest impediment to understand divisome function. Although various temporal and spatial regulators are now known and have been subjected to detailed study, again, understanding of their precise mechanism of action will probably await resolution of the Z-ring structure problem. Furthermore, even in the absence of the key negative regulators (MinCD and Noc), residual cell divisions still tend to occur between replicated chromosomes (Wu and Errington 2004; Rodrigues and Harry 2012), indicating the existence of as yet unidentified regulatory factors.
Once the structure of the Z ring has been resolved many details of the division process will need to be worked out, including the enigmatic roles of several membrane associated divisome proteins, such as DivIB, DivIC, FtsL and FtsW. Finally, it will be interesting to resolve how the division machinery is modified in order to bring about the various subtle changes associated with the asymmetric division of sporulating cells. How is mid cell division blocked? How is polar division promoted: in particular, how are the normal activities of the NO and Min systems overridden? Finally, how does the cell regulate the differential thickness of the vegetative vs sporulation septa?
Acknowledgements
Work in the Errington lab is funded by a Wellcome Trust Senior Investigator Award (WT098374AIA) and a European Research Council Advanced Investigator Award (GA 670980 ELFBAD).
References
- Abhayawardhane Y, Stewart GC. Bacillus subtilis possesses a second determinant with extensive sequence similarity to the Escherichia coli mreB morphogene. J Bacteriol. 1995;177:765–773. doi: 10.1128/jb.177.3.765-773.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams DW, Errington J. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol. 2009;7:642–653. doi: 10.1038/nrmicro2198. [DOI] [PubMed] [Google Scholar]
- Adams DW, Wu LJ, Czaplewski LG, Errington J. Multiple effects of benzamide antibiotics on FtsZ function. Mol Microbiol. 2011;80:68–84. doi: 10.1111/j.1365-2958.2011.07559.x. [DOI] [PubMed] [Google Scholar]
- Adams DW, Wu LJ, Errington J. Nucleoid occlusion protein Noc recruits DNA to the bacterial cell membrane. EMBO J. 2015;34:491–501. doi: 10.15252/embj.201490177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler HI, Fisher WD, Cohen A, Hardigree AA. Miniature Escherichia coli cells deficient in DNA. Proc Natl Acad Sci U S A. 1967;57:321–326. doi: 10.1073/pnas.57.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anagnostopoulos C, Spizizen J. Requirements for transformation in Bacillus subtilis. J Bacteriol. 1961;81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DE, Gueiros-Filho FJ, Erickson HP. Assembly dynamics of FtsZ rings in Bacillus subtilis and Escherichia coli and effects of FtsZ-regulating proteins. J Bacteriol. 2004;186:5775–5781. doi: 10.1128/JB.186.17.5775-5781.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arigoni F, Pogliano K, Webb CD, Stragier P, Losick R. Localization of protein implicated in establishment of cell type to sites of asymmetric division. Science. 1995;270:637–640. doi: 10.1126/science.270.5236.637. [DOI] [PubMed] [Google Scholar]
- Arigoni F, Duncan L, Alper S, Losick R, Stragier P. SpoIIE governs the phosphorylation state of a protein regulating transcription factor σF during sporulation in Bacillus subtilis. Proc Natl Acad Sci U S A. 1996;93:3238–3242. doi: 10.1073/pnas.93.8.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach JN, Albrecht N, Bramkamp M. Imaging DivIVA dynamics using photo-convertible and activatable fluorophores in Bacillus subtilis. Front Microbiol. 2014;5:59. doi: 10.3389/fmicb.2014.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barák I, Youngman P. SpoIIE mutants of Bacillus subtilis comprise two distinct phenotypic classes consistent with a dual functional role for the SpoIIE protein. J Bacteriol. 1996;178:4984–4989. doi: 10.1128/jb.178.16.4984-4989.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall B, Lutkenhaus J. Nucleotide sequence and insertional inactivation of a Bacillus subtilis gene that affects cell division, sporulation, and temperature sensitivity. J Bacteriol. 1989;171:6821–6834. doi: 10.1128/jb.171.12.6821-6834.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall B, Lutkenhaus J. FtsZ in Bacillus subtilis is required for vegetative septation and for asymmetric septation during sporulation. Genes Dev. 1991;5:447–455. doi: 10.1101/gad.5.3.447. [DOI] [PubMed] [Google Scholar]
- Beall B, Lutkenhaus J. Impaired cell division and sporulation of a Bacillus subtilis strain with the ftsA gene deleted. J Bacteriol. 1992;174:2398–2403. doi: 10.1128/jb.174.7.2398-2403.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall B, Lowe M, Lutkenhaus J. Cloning and characterization of Bacillus subtilis homologs of Escherichia coli cell division genes ftsZ and ftsA. J Bacteriol. 1988;170:4855–4864. doi: 10.1128/jb.170.10.4855-4864.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yehuda S, Losick R. Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell. 2002;109:257–266. doi: 10.1016/S0092-8674(02)00698-0. [DOI] [PubMed] [Google Scholar]
- Bernhardt TG, de Boer PAJ. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over Chromosomes in E. coli. Mol Cell. 2005;18:555–564. doi: 10.1016/j.molcel.2005.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi EF, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- Billings G, Ouzounov N, Ursell T, Desmarais SM, Shaevitz J, Gitai Z, Huang KC. De novo morphogenesis in L-forms via geometric control of cell growth. Mol Microbiol. 2014;93:883–896. doi: 10.1111/mmi.12703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisson-Filho AW, Discola KF, Castellen P, Blasios V, Martins A, Sforca ML, Garcia W, Zeri AC, Erickson HP, Dessen A, Gueiros-Filho FJ. FtsZ filament capping by MciZ, a developmental regulator of bacterial division. Proc Natl Acad Sci U S A. 2015;112:E2130–E2138. doi: 10.1073/pnas.1414242112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasios V, Bisson-Filho AW, Castellen P, Nogueira ML, Bettini J, Portugal RV, Zeri AC, Gueiros-Filho FJ. Genetic and biochemical characterization of the MinC-FtsZ interaction in Bacillus subtilis. PLoS One. 2013;8:e60690. doi: 10.1371/journal.pone.0060690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw N, Losick R (2015) Asymmetric division triggers cell-specific gene expression through coupled capture and stabilization of a phosphatase. Elife 4 pii: e08145, doi: 10.7554/eLife.08145. [DOI] [PMC free article] [PubMed]
- Bramkamp M, Weston L, Daniel RA, Errington J. Regulated intramembrane proteolysis of FtsL protein and the control of cell division in Bacillus subtilis. Mol Microbiol. 2006;62:580–591. doi: 10.1111/j.1365-2958.2006.05402.x. [DOI] [PubMed] [Google Scholar]
- Bramkamp M, Emmins R, Weston L, Donovan C, Daniel RA, Errington J. A novel component of the division-site selection system of Bacillus subtilis and a new mode of action for the division inhibitor MinCD. Mol Microbiol. 2008;70:1556–1569. doi: 10.1111/j.1365-2958.2008.06501.x. [DOI] [PubMed] [Google Scholar]
- Bylund JE, Haines MA, Piggot PJ, Higgins ML. Axial filament formation in Bacillus subtilis: induction of nucleoids of increasing length after addition of chloramphenicol to exponential-phase cultures approaching stationary phase. J Bacteriol. 1993;175:1886–1890. doi: 10.1128/jb.175.7.1886-1890.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camberg JL, Hoskins JR, Wickner S. ClpXP protease degrades the cytoskeletal protein, FtsZ, and modulates FtsZ polymer dynamics. Proc Natl Acad Sci U S A. 2009;106:10614–10619. doi: 10.1073/pnas.0904886106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campo N, Marquis KA, Rudner DZ. SpoIIQ anchors membrane proteins on both sides of the sporulation septum in Bacillus subtilis. J Biol Chem. 2008;283:4975–4982. doi: 10.1074/jbc.M708024200. [DOI] [PubMed] [Google Scholar]
- Campos M, Surovtsev IV, Kato S, Paintdakhi A, Beltran B, Ebmeier SE, Jacobs-Wagner C. A constant size extension drives bacterial cell size homeostasis. Cell. 2014;159:1433–1446. doi: 10.1016/j.cell.2014.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballido-López R, Errington J. The bacterial cytoskeleton: in vivo dynamics of the actin-like protein Mbl of Bacillus subtilis. Dev Cell. 2003;4:19–28. doi: 10.1016/S1534-5807(02)00403-3. [DOI] [PubMed] [Google Scholar]
- Carballido-López R, Formstone A, Li Y, Ehrlich SD, Noirot P, Errington J. Actin homolog MreBH governs cell morphogenesis by localization of the cell wall hydrolase LytE. Dev Cell. 2006;11:399–409. doi: 10.1016/j.devcel.2006.07.017. [DOI] [PubMed] [Google Scholar]
- Carniol K, Ben-Yehuda S, King N, Losick R. Genetic dissection of the sporulation protein SpoIIE and its role in asymmetric division in Bacillus subtilis. J Bacteriol. 2005;187:3511–3520. doi: 10.1128/JB.187.10.3511-3520.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha J-H, Stewart GC. The divIVA minicell locus of Bacillus subtilis. J Bacteriol. 1997;179:1671–1683. doi: 10.1128/jb.179.5.1671-1683.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Milam SL, Erickson HP. SulA inhibits assembly of FtsZ by a simple sequestration mechanism. Biochemistry. 2012;51:3100–3109. doi: 10.1021/bi201669d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claessen D, Emmins R, Hamoen LW, Daniel RA, Errington J, Edwards DH. Control of the cell elongation-division cycle by shuttling of PBP1 protein in Bacillus subtilis. Mol Microbiol. 2008;68:1029–1046. doi: 10.1111/j.1365-2958.2008.06210.x. [DOI] [PubMed] [Google Scholar]
- Cleverley RM, Barrett JR, Basle A, Bui NK, Hewitt L, Solovyova A, Xu ZQ, Daniel RA, Dixon NE, Harry EJ, Oakley AJ, Vollmer W, Lewis RJ. Structure and function of a spectrin-like regulator of bacterial cytokinesis. Nat Commun. 2014;5:5421. doi: 10.1038/ncomms6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook WR, De Boer PA, Rothfield LI. Differentiation of the bacterial cell division site. Int Rev Cytol. 1989;118:1–31. doi: 10.1016/S0074-7696(08)60871-2. [DOI] [PubMed] [Google Scholar]
- Dajkovic A, Lan G, Sun SX, Wirtz D, Lutkenhaus J. MinC spatially controls bacterial cytokinesis by antagonizing the scaffolding function of FtsZ. Curr Biol. 2008;18:235–244. doi: 10.1016/j.cub.2008.01.042. [DOI] [PubMed] [Google Scholar]
- Daniel RA, Errington J. Intrinsic instability of the essential cell division protein FtsL of Bacillus subtilis and a role for DivIB protein in FtsL turnover. Mol Microbiol. 2000;36:278–289. doi: 10.1046/j.1365-2958.2000.01857.x. [DOI] [PubMed] [Google Scholar]
- Daniel RA, Errington J. Control of cell morphogenesis in bacteria. Cell. 2003;113:767–776. doi: 10.1016/S0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]
- Daniel RA, Williams AM, Errington J. A complex four-gene operon containing essential cell division gene pbpB in Bacillus subtilis. J Bacteriol. 1996;178:2343–2350. doi: 10.1128/jb.178.8.2343-2350.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel RA, Harry EJ, Katis VL, Wake RG, Errington J. Characterization of the essential cell division gene ftsL (yIID) of Bacillus subtilis and its role in the assembly of the division apparatus. Mol Microbiol. 1998;29:593–604. doi: 10.1046/j.1365-2958.1998.00954.x. [DOI] [PubMed] [Google Scholar]
- Daniel RA, Noirot-Gros MF, Noirot P, Errington J. Multiple interactions between the transmembrane division proteins of Bacillus subtilis and the role of FtsL instability in divisome assembly. J Bacteriol. 2006;188:7396–7404. doi: 10.1128/JB.01031-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boer P, Crossley R, Rothfield L. The essential bacterial cell-division protein FtsZ is a GTPase. Nature. 1992;359:254–256. doi: 10.1038/359254a0. [DOI] [PubMed] [Google Scholar]
- De Pedro MA, Cava F. Structural constraints and dynamics of bacterial cell wall architecture. Front Microbiol. 2015;6:449. doi: 10.3389/fmicb.2015.00449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defeu Soufo HJ, Graumann PL. Dynamic movement of actin-like proteins within bacterial cells. EMBO Rep. 2004;5:789–794. doi: 10.1038/sj.embor.7400209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defeu Soufo HJ, Graumann PL. Dynamic localization and interaction with other Bacillus subtilis actin-like proteins are important for the function of MreB. Mol Microbiol. 2006;62:1340–1356. doi: 10.1111/j.1365-2958.2006.05457.x. [DOI] [PubMed] [Google Scholar]
- Defeu Soufo HJ, Reimold C, Breddermann H, Mannherz HG, Graumann PL. Translation elongation factor EF-Tu modulates filament formation of actin-like MreB protein in vitro. J Mol Biol. 2015;427:1715–1727. doi: 10.1016/j.jmb.2015.01.025. [DOI] [PubMed] [Google Scholar]
- Dempwolff F, Reimold C, Reth M, Graumann PL. Bacillus subtilis MreB orthologs self-organize into filamentous structures underneath the cell membrane in a heterologous cell system. PLoS One. 2011;6:e27035. doi: 10.1371/journal.pone.0027035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez-Cuevas P, Porcelli I, Daniel RA, Errington J. Differentiated roles for MreB-actin isologues and autolytic enzymes in Bacillus subtilis morphogenesis. Mol Microbiol. 2013;89:1084–1098. doi: 10.1111/mmi.12335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez-Escobar J, Chastanet A, Crevenna AH, Fromion V, Wedlich-Soldner R, Carballido-López R. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science. 2011;333:225–228. doi: 10.1126/science.1203466. [DOI] [PubMed] [Google Scholar]
- Duman R, Ishikawa S, Celik I, Strahl H, Ogasawara N, Troc P, Lowe J, Hamoen LW. Structural and genetic analyses reveal the protein SepF as a new membrane anchor for the Z ring. Proc Natl Acad Sci U S A. 2013;110:E4601–E4610. doi: 10.1073/pnas.1313978110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan L, Alper S, Arigoni F, Losick R, Stragier P. Activation of cell-specific transciption by a serine phosphatase at the site of asymmetric division. Science. 1995;270:641–644. doi: 10.1126/science.270.5236.641. [DOI] [PubMed] [Google Scholar]
- Dziedzic R, Kiran M, Plocinski P, Ziolkiewicz M, Brzostek A, Moomey M, Vadrevu IS, Dziadek J, Madiraju M, Rajagopalan M. Mycobacterium tuberculosis ClpX interacts with FtsZ and interferes with FtsZ assembly. PLoS One. 2010;5:e11058. doi: 10.1371/journal.pone.0011058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards DH, Errington J. The Bacillus subtilis DivIVA protein targets to the division septum and controls the site specificity of cell division. Mol Microbiol. 1997;24:905–915. doi: 10.1046/j.1365-2958.1997.3811764.x. [DOI] [PubMed] [Google Scholar]
- Edwards DH, Thomaides HB, Errington J (2000) Promiscuous targeting of Bacillus subtilis cell division protein DivIVA to division sites in Escherichia coli and fission yeast [DOI] [PMC free article] [PubMed]
- Egan AJ, Vollmer W. The physiology of bacterial cell division. Ann N Y Acad Sci. 2013;1277:8–28. doi: 10.1111/j.1749-6632.2012.06818.x. [DOI] [PubMed] [Google Scholar]
- Eichenberger P, Fawcett P, Losick R. A three-protein inhibitor of polar septation during sporulation in Bacillus subtilis. Mol Microbiol. 2001;42:1147–1162. doi: 10.1046/j.1365-2958.2001.02660.x. [DOI] [PubMed] [Google Scholar]
- Emami K et al (2017) RodA as the missing glycosyltransferase in Bacillus subtilis and antibiotic discovery for the peptidoglycan polymerase pathway. Nat Microbiol 2:16253. doi:10.1038/nmicrobiol.2016.253 [DOI] [PMC free article] [PubMed]
- Errington J. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol Rev. 1993;57:1–33. doi: 10.1128/mr.57.1.1-33.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington J. Regulation of endospore formation in Bacillus subtilis. Nat Rev Microbiol. 2003;1:117–126. doi: 10.1038/nrmicro750. [DOI] [PubMed] [Google Scholar]
- Errington J. Bacterial morphogenesis and the enigmatic MreB helix. Nat Rev Microbiol. 2015;13:241–248. doi: 10.1038/nrmicro3398. [DOI] [PubMed] [Google Scholar]
- Eswaramoorthy P, Erb ML, Gregory JA, Silverman J, Pogliano K, Pogliano J, Ramamurthi KS (2011) Cellular architecture mediates DivIVA ultrastructure and regulates min activity in Bacillus subtilis. MBio 2 (b). pii: e00257-11. doi: 10.1128/mBio.00257-11 [DOI] [PMC free article] [PubMed]
- Feucht A, Errington J. ftsZ mutations affecting cell division frequency, placement and morphology in Bacillus subtilis. Microbiology. 2005;151:2053–2064. doi: 10.1099/mic.0.27899-0. [DOI] [PubMed] [Google Scholar]
- Feucht A, Magnin T, Yudkin MD, Errington J. Bifunctional protein required for asymmetric cell division and cell-specific transcription in Bacillus subtilis. Genes Dev. 1996;10:794–803. doi: 10.1101/gad.10.7.794. [DOI] [PubMed] [Google Scholar]
- Feucht A, Daniel RA, Errington J. Characterization of a morphological checkpoint coupling cell-specific transcription to septation in Bacillus subtilis. Mol Microbiol. 1999;33:1015–1026. doi: 10.1046/j.1365-2958.1999.01543.x. [DOI] [PubMed] [Google Scholar]
- Feucht A, Lucet I, Yudkin MD, Errington J. Cytological and biochemical characterization of the FtsA cell division protein of Bacillus subtilis. Mol Microbiol. 2001;40:115–125. doi: 10.1046/j.1365-2958.2001.02356.x. [DOI] [PubMed] [Google Scholar]
- Formstone A, Errington J. A magnesium-dependent mreB null mutant: implications for the role of mreB in Bacillus subtilis. Mol Microbiol. 2005;55:1646–1657. doi: 10.1111/j.1365-2958.2005.04506.x. [DOI] [PubMed] [Google Scholar]
- Formstone A, Carballido-López R, Noirot P, Errington J, Scheffers DJ. Localization and interactions of teichoic acid synthetic enzymes in Bacillus subtilis. J Bacteriol. 2008;190:1812–1821. doi: 10.1128/JB.01394-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulquier E, Pompeo F, Bernadac A, Espinosa L, Galinier A. The YvcK protein is required for morphogenesis via localization of PBP1 under gluconeogenic growth conditions in Bacillus subtilis. Mol Microbiol. 2011;80:309–318. doi: 10.1111/j.1365-2958.2011.07587.x. [DOI] [PubMed] [Google Scholar]
- Gamba P, Veening JW, Saunders NJ, Hamoen LW, Daniel RA. Two-step assembly dynamics of the Bacillus subtilis divisome. J Bacteriol. 2009;191:4186–4194. doi: 10.1128/JB.01758-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner EC, Bernard R, Wang W, Zhuang X, Rudner DZ, Mitchison T. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science. 2011;333:222–225. doi: 10.1126/science.1203285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gholamhoseinian A, Shen Z, Wu J-J, Piggot P. Regulation of transcription of the cell division gene ftsA during sporulation of Bacillus subtilis. J Bacteriol. 1992;174:4647–4656. doi: 10.1128/jb.174.14.4647-4656.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosal D, Trambaiolo D, Amos LA, Lowe J. MinCD cell division proteins form alternating copolymeric cytomotive filaments. Nat Commun. 2014;5:5341. doi: 10.1038/ncomms6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzy-Tréboul G, Karmazyn-Campelli C, Stragier P. Developmental regulation of transcription of the Bacillus subtilis ftsAZ operon. J Mol Biol. 1992;224:967–979. doi: 10.1016/0022-2836(92)90463-T. [DOI] [PubMed] [Google Scholar]
- Gregory JA, Becker EC, Pogliano K. Bacillus subtilis MinC destabilizes FtsZ-rings at new cell poles and contributes to the timing of cell division. Genes Dev. 2008;22:3475–3488. doi: 10.1101/gad.1732408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundling A, Schneewind O. Genes required for glycolipid synthesis and lipoteichoic acid anchoring in Staphylococcus aureus. J Bacteriol. 2007;189:2521–2530. doi: 10.1128/JB.01683-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundling A, Schneewind O. Synthesis of glycerol phosphate lipoteichoic acid in Staphylococcus aureus. Proc Natl Acad Sci U S A. 2007;104:8478–8483. doi: 10.1073/pnas.0701821104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueiros-Filho FJ, Losick R. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 2002;16:2544–2556. doi: 10.1101/gad.1014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gündoğdu ME, Kawai Y, Pavlendova N, Ogasawara N, Errington J, Scheffers DJ, Hamoen LW. Large ring polymers align FtsZ polymers for normal septum formation. EMBO J. 2011;30:617–626. doi: 10.1038/emboj.2010.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusser DP, Schwartz RL, Smith AM, Oates ME, Levin PA. EzrA prevents aberrant cell division by modulating assembly of the cytoskeletal protein FtsZ. Mol Microbiol. 2004;52:801–814. doi: 10.1111/j.1365-2958.2004.04016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusser DP, Garza AC, Buscher AZ, Levin PA. The division inhibitor EzrA contains a seven-residue patch required for maintaining the dynamic nature of the medial FtsZ ring. J Bacteriol. 2007;189:9001–9010. doi: 10.1128/JB.01172-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusser DP, Lee AH, Weart RB, Levin PA. ClpX inhibits FtsZ assembly in a manner that does not require its ATP hydrolysis-dependent chaperone activity. J Bacteriol. 2009;191:1986–1991. doi: 10.1128/JB.01606-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CA, De Boer PAJ. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell. 1997;88:175–185. doi: 10.1016/S0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- Hamoen LW, Errington J (2003) Polar targeting of DivIVA in Bacillus subtilis is not directly dependent on FtsZ or PBP 2B. J Bacteriol 185 [DOI] [PMC free article] [PubMed]
- Hamoen LW, Meile JC, De Jong W, Noirot P, Errington J. SepF, a novel FtsZ-interacting protein required for a late step in cell division. Mol Microbiol. 2006;59:989–999. doi: 10.1111/j.1365-2958.2005.04987.x. [DOI] [PubMed] [Google Scholar]
- Handler AA, Lim JE, Losick R. Peptide inhibitor of cytokinesis during sporulation in Bacillus subtilis. Mol Microbiol. 2008;68:588–599. doi: 10.1111/j.1365-2958.2008.06173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harry EJ, Wake RG. Cloning and expression of a Bacillus subtilis division initiation gene for which a homolog has not been identified in another organism. J Bacteriol. 1989;171:6835–6839. doi: 10.1128/jb.171.12.6835-6839.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harry EJ, Wake RG. The membrane-bound cell division protein DivIB is localized to the division site in Bacillus subtilis. Mol Microbiol. 1997;25:275–283. doi: 10.1046/j.1365-2958.1997.4581822.x. [DOI] [PubMed] [Google Scholar]
- Haydon DJ, Stokes NR, Ure R, Galbraith G, Bennett JM, Brown DR, Baker PJ, Barynin VV, Rice DW, Sedelnikova SE, Heal JR, Sheridan JM, Aiwale ST, Chauhan PK, Srivastava A, Taneja A, Collins I, Errington J, Czaplewski LG. An inhibitor of FtsZ with potent and selective anti-staphylococcal activity. Science. 2008;321:1673–1675. doi: 10.1126/science.1159961. [DOI] [PubMed] [Google Scholar]
- Hu Z, Mukherjee A, Pichoff S, Lutkenhaus J. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc Natl Acad Sci U S A. 1999;96:14819–14824. doi: 10.1073/pnas.96.26.14819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illing N, Errington J. Genetic regulation of morphogenesis in Bacillus subtilis: roles of σE and σF in prespore engulfment. J Bacteriol. 1991;173:3159–3169. doi: 10.1128/jb.173.10.3159-3169.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa S, Kawai Y, Hiramatsu K, Kuwano M, Ogasawara N. A new FtsZ-interacting protein, YlmF, complements the activity of FtsA during progression of cell division in Bacillus subtilis. Mol Microbiol. 2006;60:1364–1380. doi: 10.1111/j.1365-2958.2006.05184.x. [DOI] [PubMed] [Google Scholar]
- Jennings PC, Cox GC, Monahan LG, Harry EJ. Super-resolution imaging of the bacterial cytokinetic protein FtsZ. Micron. 2011;42:336–341. doi: 10.1016/j.micron.2010.09.003. [DOI] [PubMed] [Google Scholar]
- Jensen SO, Thompson LS, Harry EJ. Cell division in Bacillus subtilis: FtsZ and FtsA association is Z-ring independent, and FtsA is required for efficient midcell Z-Ring assembly. J Bacteriol. 2005;187:6536–6544. doi: 10.1128/JB.187.18.6536-6544.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones LJF, Carballido-López R, Errington J. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell. 2001;104:913–922. doi: 10.1016/S0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- Karoui ME, Errington J. Isolation and characterization of topological specificity mutants of minD in Bacillus subtilis. Mol Microbiol. 2001;42:1211–1221. doi: 10.1046/j.1365-2958.2001.02710.x. [DOI] [PubMed] [Google Scholar]
- Katis VL, Wake RG. Membrane-bound division proteins DivIB and DivIC of Bacillus subtilis function solely through their external domains in both vegetative and sporulation division. J Bacteriol. 1999;181:2710–2718. doi: 10.1128/jb.181.9.2710-2718.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katis VL, Harry EJ, Wake RG. The Bacillus subtilis division protein DivIC is a highly abundant membrane-bound protein that localizes to the division site. Mol Microbiol. 1997;26:1047–1055. doi: 10.1046/j.1365-2958.1997.6422012.x. [DOI] [PubMed] [Google Scholar]
- Katis VL, Wake RG, Harry EJ. Septal localization of the membrane-bound division proteins of Bacillus subtilis DivIB and DivIC is codependent only at high temperatures and requires FtsZ. J Bacteriol. 2000;182:3607–3611. doi: 10.1128/JB.182.12.3607-3611.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai Y, Ogasawara N. Bacillus subtilis EzrA and FtsL synergistically regulate FtsZ ring dynamics during cell division. Microbiology. 2006;152:1129–1141. doi: 10.1099/mic.0.28497-0. [DOI] [PubMed] [Google Scholar]
- Kawai Y, Asai K, Errington J. Partial functional redundancy of MreB isoforms, MreB, Mbl and MreBH, in cell morphogenesis of Bacillus subtilis. Mol Microbiol. 2009;73:719–731. doi: 10.1111/j.1365-2958.2009.06805.x. [DOI] [PubMed] [Google Scholar]
- Kawai Y, Daniel RA, Errington J. Regulation of cell wall morphogenesis in Bacillus subtilis by recruitment of PBP1 to the MreB helix. Mol Microbiol. 2009;71:1131–1144. doi: 10.1111/j.1365-2958.2009.06601.x. [DOI] [PubMed] [Google Scholar]
- Kawai Y, Marles-Wright J, Cleverley RM, Emmins R, Ishikawa S, Kuwano M, Heinz N, Bui NK, Hoyland CN, Ogasawara N, Lewis RJ, Vollmer W, Daniel RA, Errington J. A widespread family of bacterial cell wall assembly proteins. EMBO J. 2011;30:4931–4941. doi: 10.1038/emboj.2011.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai Y, Mercier R, Errington J. Bacterial cell morphogenesis does not require a preexisting template structure. Curr Biol. 2014;24:863–867. doi: 10.1016/j.cub.2014.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai Y, Mercier R, Wu LJ, Dominguez-Cuevas P, Oshima T, Errington J. Cell growth of wall-free L-form bacteria is limited by oxidative damage. Curr Biol. 2015;25:1613–1618. doi: 10.1016/j.cub.2015.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Zhang L, Higgins ML, Piggot PJ. The spoIIE locus is involved in the Spo0A-dependent switch in the location of FtsZ rings in Bacillus subtilis. J Bacteriol. 1998;180:1256–1260. doi: 10.1128/jb.180.5.1256-1260.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman TG, Lenarcic R, Willis CR, Roberts DM, Hamoen LW, Errington J, Wu LJ (2016) Complex polar machinery required for proper chromosome segregation in vegetative and sporulating cells of Bacillus subtilis. Mol Microbiol 101(2):333–350 [DOI] [PMC free article] [PubMed]
- Land AD, Luo Q, Levin PA. Functional domain analysis of the cell division inhibitor EzrA. PLoS One. 2014;9:e102616. doi: 10.1371/journal.pone.0102616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaver M, Errington J. Roles for MreC and MreD proteins in helical growth of the cylindrical cell wall in Bacillus subtilis. Mol Microbiol. 2005;57:1196–1209. doi: 10.1111/j.1365-2958.2005.04736.x. [DOI] [PubMed] [Google Scholar]
- Leaver M, Dominguez-Cuevas P, Coxhead JM, Daniel RA, Errington J. Life without a wall or division machine in Bacillus subtilis. Nature. 2009;457:849–853. doi: 10.1038/nature07742. [DOI] [PubMed] [Google Scholar]
- Lenarcic R, Halbedel S, Visser L, Shaw M, Wu LJ, Errington J, Marenduzzo D, Hamoen LW. Localisation of DivIVA by targeting to negatively curved membranes. EMBO J. 2009;28:2272–2282. doi: 10.1038/emboj.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin PA, Losick R. Characterization of a cell division gene from Bacillus subtilis that is required for vegetative and sporulation septum formation. J Bacteriol. 1994;176:1451–1459. doi: 10.1128/jb.176.5.1451-1459.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin PA, Losick R. Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes Dev. 1996;10:478–488. doi: 10.1101/gad.10.4.478. [DOI] [PubMed] [Google Scholar]
- Levin PA, Margolis PS, Setlow P, Losick R, Sun D. Identification of Bacillus subtilis genes for septum placement and shape determination. J Bacteriol. 1992;174:6717–6728. doi: 10.1128/jb.174.21.6717-6728.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin PA, Losick R, Stragier P, Arigoni F. Localization of the sporulation protein SpoIIE in Bacillus subtilis is dependent upon the cell division protein FtsZ. Mol Microbiol. 1997;25:839–846. doi: 10.1111/j.1365-2958.1997.mmi505.x. [DOI] [PubMed] [Google Scholar]
- Levin PA, Kurtser IG, Grossman AD. Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis. Proc Natl Acad Sci U S A. 1999;96:9642–9647. doi: 10.1073/pnas.96.17.9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PJ, Partridge SR, Errington J. Sigma factors, asymmetry, and the determination of cell fate in Bacillus subtilis. Proc Natl Acad Sci U S A. 1994;91:3849–3853. doi: 10.1073/pnas.91.9.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Trimble MJ, Brun YV, Jensen GJ. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J. 2007;26:4694–4708. doi: 10.1038/sj.emboj.7601895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loose M, Mitchison TJ. The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns. Nat Cell Biol. 2014;16:38–46. doi: 10.1038/ncb2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovering AL, Safadi SS, Strynadka NC. Structural perspective of peptidoglycan biosynthesis and assembly. Annu Rev Biochem. 2012;81:451–478. doi: 10.1146/annurev-biochem-061809-112742. [DOI] [PubMed] [Google Scholar]
- Low HH, Moncrieffe MC, Löwe J. The crystal structure of ZapA and its modulation of FtsZ polymerisation. J Mol Biol. 2004;341:839–852. doi: 10.1016/j.jmb.2004.05.031. [DOI] [PubMed] [Google Scholar]
- Löwe J, Amos LA. Crystal structure of the bacterial cell-division protein FtsZ. Nature. 1998;391:203–206. doi: 10.1038/34472. [DOI] [PubMed] [Google Scholar]
- Löwe J, Amos LA. Evolution of cytomotive filaments: the cytoskeleton from prokaryotes to eukaryotes. Int J Biochem Cell Biol. 2009;41:323–329. doi: 10.1016/j.biocel.2008.08.010. [DOI] [PubMed] [Google Scholar]
- Lu Z, Takeuchi M, Sato T. The LysR-type transcriptional regulator YofA controls cell division through the regulation of expression of ftsW in Bacillus subtilis. J Bacteriol. 2007;189:5642–5651. doi: 10.1128/JB.00467-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucet I, Feucht A, Yudkin MD, Errington J. Direct interaction between the cell division protein FtsZ and the cell differentiation protein SpoIIE. EMBO J. 2000;19:1467–1475. doi: 10.1093/emboj/19.7.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu Rev Biochem. 2007;76:539–562. doi: 10.1146/annurev.biochem.75.103004.142652. [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J. The ParA/MinD family puts things in their place. Trends Microbiol. 2012;20:411–418. doi: 10.1016/j.tim.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston AL, Errington J. Selection of the midcell division site in Bacillus subtilis through MinD-dependent polar localization and activation of MinC. Mol Microbiol. 1999;33:84–96. doi: 10.1046/j.1365-2958.1999.01450.x. [DOI] [PubMed] [Google Scholar]
- Marston AL, Thomaides HB, Edwards DH, Sharpe ME, Errington J. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev. 1998;12:3419–3430. doi: 10.1101/gad.12.21.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeske AJ, Sham LT, Kimsey H, Koo BM, Gross CA, Bernhardt TG, Rudner DZ. MurJ and a novel lipid II flippase are required for cell wall biogenesis in Bacillus subtilis. Proc Natl Acad Sci U S A. 2015;112:6437–6442. doi: 10.1073/pnas.1504967112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeske AJ et al (2016) SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature. doi:10.1038/nature19331 [DOI] [PMC free article] [PubMed]
- Mercier R, Domínguez-Cuevas P, Errington J. Crucial role for membrane fluidity in proliferation of primitive cells. Cell Rep. 2012;1:417–423. doi: 10.1016/j.celrep.2012.03.008. [DOI] [PubMed] [Google Scholar]
- Mercier R, Kawai Y, Errington J. Excess membrane synthesis drives a primitive mode of cell proliferation. Cell. 2013;152:997–1007. doi: 10.1016/j.cell.2013.01.043. [DOI] [PubMed] [Google Scholar]
- Mercier R, Kawai Y, Errington J (2014) General principles for the formation and proliferation of a wall-free (L-form) state in bacteria. Elife 3 [DOI] [PMC free article] [PubMed]
- Michie KA, Monahan LG, Beech PL, Harry EJ. Trapping of a spiral-like intermediate of the bacterial cytokinetic protein FtsZ. J Bacteriol. 2006;188:1680–1690. doi: 10.1128/JB.188.5.1680-1690.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AK, Brown EE, Mercado BT, Herman JK. A DNA-binding protein defines the precise region of chromosome capture during Bacillus sporulation. Mol Microbiol. 2015;99(1):111–122. doi: 10.1111/mmi.13217. [DOI] [PubMed] [Google Scholar]
- Min J, Holden SJ, Carlini L, Unser M, Manley S, Ye JC. 3D high-density localization microscopy using hybrid astigmatic/ biplane imaging and sparse image reconstruction. Biomed Opt Express. 2014;5:3935–3948. doi: 10.1364/BOE.5.003935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan LG, Robinson A, Harry EJ. Lateral FtsZ association and the assembly of the cytokinetic Z ring in bacteria. Mol Microbiol. 2009;74:1004–1017. doi: 10.1111/j.1365-2958.2009.06914.x. [DOI] [PubMed] [Google Scholar]
- Monahan LG, Liew AT, Bottomley AL, Harry EJ. Division site positioning in bacteria: one size does not fit all. Front Microbiol. 2014;5:19. doi: 10.3389/fmicb.2014.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstein RM, Bratton BP, Nguyen JP, Ouzounov N, Shaevitz JW, Gitai Z. RodZ links MreB to cell wall synthesis to mediate MreB rotation and robust morphogenesis. Proc Natl Acad Sci U S A. 2015;112:12510–12515. doi: 10.1073/pnas.1509610112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowy S, Cossart P. Septins: the fourth component of the cytoskeleton. Nat Rev Mol Cell Biol. 2012;13:183–194. doi: 10.1038/nrm3284. [DOI] [PubMed] [Google Scholar]
- Muchova K, Chromikova Z, Barak I. Control of Bacillus subtilis cell shape by RodZ. Environ Microbiol. 2013;15:3259–3271. doi: 10.1111/1462-2920.12200. [DOI] [PubMed] [Google Scholar]
- Mukherjee A, Lutkenhaus J. Guanine nucleotide-dependent assembly of FtsZ into filaments. J Bacteriol. 1994;176:2754–2758. doi: 10.1128/jb.176.9.2754-2758.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder E, Woldringh CL. Actively replicating nucleoids influence positioning of division sites in Escherichia coli filaments forming cells lacking DNA. J Bacteriol. 1989;171:4303–4314. doi: 10.1128/jb.171.8.4303-4314.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntel J, Fromion V, Goelzer A, Maabeta S, Mader U, Buttner K, Hecker M, Becher D. Comprehensive absolute quantification of the cytosolic proteome of Bacillus subtilis by data independent, parallel fragmentation in liquid chromatography/mass spectrometry (LC/MS(E)) Mol Cell Proteomics. 2014;13:1008–1019. doi: 10.1074/mcp.M113.032631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva MA, Cordell SC, Löwe J. Structural insights into FtsZ protofilament formation. Nat Struct Mol Biol. 2004;11:1243–1250. doi: 10.1038/nsmb855. [DOI] [PubMed] [Google Scholar]
- Oliva MA, Halbedel S, Freund SM, Dutow P, Leonard TA, Veprintsev DB, Hamoen LW, Löwe J. Features critical for membrane binding revealed by DivIVA crystal structure. EMBO J. 2010;29:1988–2001. doi: 10.1038/emboj.2010.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olshausen PV, Defeu Soufo HJ, Wicker K, Heintzmann R, Graumann PL, Rohrbach A. Superresolution imaging of dynamic MreB filaments in B. subtilis--a multiple-motor-driven transport? Biophys J. 2013;105:1171–1181. doi: 10.1016/j.bpj.2013.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick JE, Kearns DB. MinJ (YvjD) is a topological determinant of cell division in Bacillus subtilis. Mol Microbiol. 2008;70:1166–1179. doi: 10.1111/j.1365-2958.2008.06469.x. [DOI] [PubMed] [Google Scholar]
- Percy MG, Grundling A. Lipoteichoic acid synthesis and function in gram-positive bacteria. Annu Rev Microbiol. 2014;68:81–100. doi: 10.1146/annurev-micro-091213-112949. [DOI] [PubMed] [Google Scholar]
- Peters PC, Migocki MD, Thoni C, Harry EJ. A new assembly pathway for the cytokinetic Z ring from a dynamic helical structure in vegetatively growing cells of Bacillus subtilis. Mol Microbiol. 2007;64:487–499. doi: 10.1111/j.1365-2958.2007.05673.x. [DOI] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol Microbiol. 2005;55:1722–1734. doi: 10.1111/j.1365-2958.2005.04522.x. [DOI] [PubMed] [Google Scholar]
- Piggot PJ, Coote JG. Genetic aspects of bacterial endospore formation. Bacteriol Rev. 1976;40:908–962. doi: 10.1128/br.40.4.908-962.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilhofer M, Jensen GJ. The bacterial cytoskeleton: more than twisted filaments. Curr Opin Cell Biol. 2013;25:125–133. doi: 10.1016/j.ceb.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl S, Harwood CR. Heterologous protein secretion by Bacillus species. Adv Appl Microbiol. 2010;73:1–25. doi: 10.1016/S0065-2164(10)73001-X. [DOI] [PubMed] [Google Scholar]
- Popham DL, Setlow P. Phenotypes of Bacillus subtilis mutants lacking multiple class a high-molecular-weight penicillin-binding proteins. J Bacteriol. 1996;178:2079–2085. doi: 10.1128/jb.178.7.2079-2085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthi KS, Losick R. Negative membrane curvature as a cue for subcellular localization of a bacterial protein. Proc Natl Acad Sci U S A. 2009;106:13541–13545. doi: 10.1073/pnas.0906851106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri D, Park JT. Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature. 1992;359:251–254. doi: 10.1038/359251a0. [DOI] [PubMed] [Google Scholar]
- Reeve JN, Mendelson NH, Coyne SI, Hallock LL, Cole RM. Minicells of Bacillus subtilis. J Bacteriol. 1973;114:860–873. doi: 10.1128/jb.114.2.860-873.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimold C, Defeu Soufo HJ, Dempwolff F, Graumann PL. Motion of variable-length MreB filaments at the bacterial cell membrane influences cell morphology. Mol Biol Cell. 2013;24:2340–2349. doi: 10.1091/mbc.E12-10-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson SA, Michie KA, Mackay JP, Harry E, King GF. The Bacillus subtilis cell division proteins FtsL and DivIC are intrinsically unstable and do not interact with one another in the absence of other septasomal components. Mol Microbiol. 2002;44:663–674. doi: 10.1046/j.1365-2958.2002.02920.x. [DOI] [PubMed] [Google Scholar]
- Rodrigues CD, Harry EJ. The Min system and nucleoid occlusion are not required for identifying the division site in Bacillus subtilis but ensure its efficient utilization. PLoS Genet. 2012;8:e1002561. doi: 10.1371/journal.pgen.1002561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueff AS, Chastanet A, Domínguez-Escobar J, Yao Z, Yates J, Prejean MV, Delumeau O, Noirot P, Wedlich-Soldner R, Filipe SR, Carballido-López R. An early cytoplasmic step of peptidoglycan synthesis is associated to MreB in Bacillus subtilis. Mol Microbiol. 2014;91:348–362. doi: 10.1111/mmi.12467. [DOI] [PubMed] [Google Scholar]
- Ryter A. Etude morphologique de la sporulation de Bacillus subtilis. Annales de l’Institut Pasteur. 1965;108:40–60. [PubMed] [Google Scholar]
- Salje J, Van Den Ent F, De Boer P, Löwe J. Direct membrane binding by bacterial actin MreB. Mol Cell. 2011;43:478–487. doi: 10.1016/j.molcel.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer RT, Bolon DN, Burton BM, Burton RE, Flynn JM, Grant RA, Hersch GL, Joshi SA, kenniston JA, Levchenko I, Neher SB, Oakes ES, Siddiqui SM, Wah DA, Baker TA. Sculpting the proteome with AAA(+) proteases and disassembly machines. Cell. 2004;119:9–18. doi: 10.1016/j.cell.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffers DJ. The effect of MinC on FtsZ polymerization is pH dependent and can be counteracted by ZapA. FEBS Lett. 2008;582:2601–2608. doi: 10.1016/j.febslet.2008.06.038. [DOI] [PubMed] [Google Scholar]
- Scheffers DJ, Tol MB. LipidII: just another brick in the wall? PLoS Pathog. 2015;11:e1005213. doi: 10.1371/journal.ppat.1005213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirner K, Errington J. The cell wall regulator σI specifically suppresses the lethal phenotype of mbl mutants in Bacillus subtilis. J Bacteriol. 2009;191:1404–1413. doi: 10.1128/JB.01497-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirner K, Marles-Wright J, Lewis RJ, Errington J. Distinct and essential morphogenic functions for wall- and lipo-teichoic acids in Bacillus subtilis. EMBO J. 2009;28:830–842. doi: 10.1038/emboj.2009.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewell EW, Brown ED. Taking aim at wall teichoic acid synthesis: new biology and new leads for antibiotics. J Antibiot (Tokyo) 2014;67:43–51. doi: 10.1038/ja.2013.100. [DOI] [PubMed] [Google Scholar]
- Shapiro L, Losick R. Dynamic spatial regulation in the bacterial cell. Cell. 2000;100:89–98. doi: 10.1016/S0092-8674(00)81686-4. [DOI] [PubMed] [Google Scholar]
- Sharpe ME, Hauser PM, Sharpe RG, Errington J. Bacillus subtilis cell cycle as studied by fluorescence microscopy: constancy of the cell length at initiation of DNA replication and evidence for active nucleoid partitioning. J Bacteriol. 1998;180:547–555. doi: 10.1128/jb.180.3.547-555.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Lutkenhaus J. Examination of the interaction between FtsZ and MinCN in E. coli suggests how MinC disrupts Z rings. Mol Microbiol. 2010;75:1285–1298. doi: 10.1111/j.1365-2958.2010.07055.x. [DOI] [PubMed] [Google Scholar]
- Shih YL, Rothfield L. The bacterial cytoskeleton. Microbiol Mol Biol Rev. 2006;70:729–754. doi: 10.1128/MMBR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers J, Errington J. Analysis of the essential cell division gene ftsL of Bacillus subtilis by mutagenesis and heterologous complementation. J Bacteriol. 2000;182:5572–5579. doi: 10.1128/JB.182.19.5572-5579.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers J, Errington J. The Bacillus subtilis cell division protein FtsL localizes to sites of septation and interacts with DivIC. Mol Microbiol. 2000;36:846–855. doi: 10.1046/j.1365-2958.2000.01895.x. [DOI] [PubMed] [Google Scholar]
- Strahl H, Hamoen LW. Membrane potential is important for bacterial cell division. Proc Natl Acad Sci U S A. 2010;107:12281–12286. doi: 10.1073/pnas.1005485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss MP, Liew AT, Turnbull L, Whitchurch CB, Monahan LG, Harry EJ. 3D-SIM super resolution microscopy reveals a bead-like arrangement for FtsZ and the division machinery: implications for triggering cytokinesis. PLoS Biol. 2012;10:e1001389. doi: 10.1371/journal.pbio.1001389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surdova K, Gamba P, Claessen D, Siersma T, Jonker MJ, Errington J, Hamoen LW. The conserved DNA-binding protein WhiA is involved in cell division in Bacillus subtilis. J Bacteriol. 2013;195:5450–5460. doi: 10.1128/JB.00507-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swulius MT, Jensen GJ. The helical MreB cytoskeleton in Escherichia coli MC1000/pLE7 is an artifact of the N-Terminal yellow fluorescent protein tag. J Bacteriol. 2012;194:6382–6386. doi: 10.1128/JB.00505-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swulius MT, Chen S, Jane Ding H, Li Z, Briegel A, Pilhofer M, Tocheva EI, Lybarger SR, Johnson TL, Sandkvist M, Jensen GJ. Long helical filaments are not seen encircling cells in electron cryotomograms of rod-shaped bacteria. Biochem Biophys Res Commun. 2011;407:650–655. doi: 10.1016/j.bbrc.2011.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szwedziak P, Wang Q, Freund SM, Löwe J. FtsA forms actin-like protofilaments. EMBO J. 2012;31:2249–2260. doi: 10.1038/emboj.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szwedziak P, Wang Q, Bharat TA, Tsim M, Löwe J. Architecture of the ring formed by the tubulin homologue FtsZ in bacterial cell division. Elife. 2014;3:e04601. doi: 10.7554/eLife.04601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taheri-Araghi S, Bradde S, Sauls JT, Hill NS, Levin PA, Paulsson J, Vergassola M, Jun S. Cell-size control and homeostasis in bacteria. Curr Biol. 2015;25:385–391. doi: 10.1016/j.cub.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan IS, Ramamurthi KS. Spore formation in Bacillus subtilis. Environ Microbiol Rep. 2014;6:212–225. doi: 10.1111/1758-2229.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan CM, Therien AG, Lu J, Lee SH, Caron A, Gill CJ, Lebeau-Jacob C, Benton-Perdomo L, Monteiro JM, Pereira PM, Elsen NL, Wu J, Deschamps K, Petcu M, Wong S, Daigneault E, Kramer S, Liang L, Maxwell E, Claveau D, Vaillancourt J, Skorey K, Tam J, Wang H, Meredith TC, Sillaots S, Wang-Jarantow L, Ramtohul Y, Langlois E, Landry F, Reid JC, Parthasarathy G, Sharma S, Baryshnikova A, Lumb KJ, Pinho MG, Soisson SM, Roemer T. Restoring methicillin-resistant Staphylococcus aureus susceptibility to β-lactam antibiotics. Sci Transl Med. 2012;4:126ra35. doi: 10.1126/scitranslmed.3003592. [DOI] [PubMed] [Google Scholar]
- Tavares JR, De Souza RF, Meira GL, Gueiros-Filho FJ. Cytological characterization of YpsB, a novel component of the Bacillus subtilis divisome. J Bacteriol. 2008;190:7096–7107. doi: 10.1128/JB.00064-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiyanont K, Doan T, Lazarus MB, Fang X, Rudner DZ, Walker S. Imaging peptidoglycan biosynthesis in Bacillus subtilis with fluorescent antibiotics. Proc Natl Acad Sci U S A. 2006;103:11033–11038. doi: 10.1073/pnas.0600829103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocheva EI, Lopez-Garrido J, Hughes HV, Fredlund J, Kuru E, Vannieuwenhze MS, Brun YV, Pogliano K, Jensen GJ. Peptidoglycan transformations during Bacillus subtilis sporulation. Mol Microbiol. 2013;88:673–686. doi: 10.1111/mmi.12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 2012;10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursell TS, Nguyen J, Monds RD, Colavin A, Billings G, Ouzounov N, Gitai Z, Shaevitz JW, Huang KC. Rod-like bacterial shape is maintained by feedback between cell curvature and cytoskeletal localization. Proc Natl Acad Sci U S A. 2014;111:E1025–E1034. doi: 10.1073/pnas.1317174111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Baarle S, Bramkamp M. The MinCDJ system in Bacillus subtilis prevents minicell formation by promoting divisome disassembly. PLoS One. 2010;5:e9850. doi: 10.1371/journal.pone.0009850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Baarle S, Celik IN, Kaval KG, Bramkamp M, Hamoen LW, Halbedel S. Protein-protein interaction domains of Bacillus subtilis DivIVA. J Bacteriol. 2013;195:1012–1021. doi: 10.1128/JB.02171-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Ent F, Löwe J. Crystal structure of the cell division protein FtsA from Thermotoga maritima. EMBO J. 2000;19:5300–5307. doi: 10.1093/emboj/19.20.5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Ent F, Leaver M, Bendezu F, Errington J, De Boer P, Löwe J. Dimeric structure of the cell shape protein MreC and its functional implications. Mol Microbiol. 2006;62:1631–1642. doi: 10.1111/j.1365-2958.2006.05485.x. [DOI] [PubMed] [Google Scholar]
- Van Teeffelen S, Gitai Z. Rotate into shape: MreB and bacterial morphogenesis. EMBO J. 2011;30:4856–4857. doi: 10.1038/emboj.2011.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varley AW, Stewart GC. The divIVB region of the Bacillus subtilis chromosome encodes homologs of Escherichia coli septum placement (MinCD) and cell shape (MreBCD) determinants. J Bacteriol. 1992;174:6729–6742. doi: 10.1128/jb.174.21.6729-6742.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer W, Joris B, Charlier P, Foster S. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol Rev. 2008;32:259–286. doi: 10.1111/j.1574-6976.2007.00099.x. [DOI] [PubMed] [Google Scholar]
- Wachi M, Doi M, Tamaki S, Park W, Nakajima-Iijima S, Matsuhashi M. Mutant isolation and molecular cloning of mre genes, which determine cell shape, sensitivity to mecillinam, and amount of penicillin-binding proteins in Escherichia coli. J Bacteriol. 1987;169:4935–4940. doi: 10.1128/jb.169.11.4935-4940.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner-Herman JK, Bernard R, Dunne R, Bisson-Filho AW, Kumar K, Nguyen T, Mulcahy L, Koullias J, Gueiros-Filho FJ, Rudner DZ. RefZ facilitates the switch from medial to polar division during spore formation in Bacillus subtilis. J Bacteriol. 2012;194:4608–4618. doi: 10.1128/JB.00378-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lutkenhaus J. The FtsZ protein of Bacillus subtilis is localized at the division site and has GTPase activity that is dependent upon FtsZ concentration. Mol Microbiol. 1993;9:435–442. doi: 10.1111/j.1365-2958.1993.tb01705.x. [DOI] [PubMed] [Google Scholar]
- Weart RB, Levin PA. Growth rate-dependent regulation of medial FtsZ ring formation. J Bacteriol. 2003;185:2826–2834. doi: 10.1128/JB.185.9.2826-2834.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weart RB, Nakano S, Lane BE, Zuber P, Levin PA. The ClpX chaperone modulates assembly of the tubulin-like protein FtsZ. Mol Microbiol. 2005;57:238–249. doi: 10.1111/j.1365-2958.2005.04673.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weart RB, Lee AH, Chien AC, Haeusser DP, Hill NS, Levin PA. A metabolic sensor governing cell size in bacteria. Cell. 2007;130:335–347. doi: 10.1016/j.cell.2007.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woldringh CL, Mulder E, Huls PG, Vischer N. Toporegulation of bacterial division according to the nucleoid occlusion model. Res Microbiol. 1991;142:309–320. doi: 10.1016/0923-2508(91)90046-D. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell. 2004;117:915–925. doi: 10.1016/j.cell.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Feucht A, Errington J. Prespore-specific gene expression in Bacillus subtilis is driven by sequestration of SpoIIE phosphatase to the prespore side of the asymmetric septum. Genes Dev. 1998;12:1371–1380. doi: 10.1101/gad.12.9.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LJ, Ishikawa S, Kawai Y, Oshima T, Ogasawara N, Errington J. Noc protein binds to specific DNA sequences to coordinate cell division with chromosome segregation. EMBO J. 2009;28:1940–1952. doi: 10.1038/emboj.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanouri A, Daniel RA, Errington J, Buchanan CE. Cloning and sequencing of the cell division gene pbpB, which encodes penicillin-binding protein 2B in Bacillus subtilis. J Bacteriol. 1993;175:7604–7616. doi: 10.1128/jb.175.23.7604-7616.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]


