Abstract
T cell signaling is inextricably linked to actin cytoskeletal dynamics at the immunological synapse (IS). This process can be imaged in living T cells expressing GFP actin or fluorescent F-actin binding proteins. Because of its planar nature, the IS provides a unique opportunity to image events as they happen, monitoring changes in actin retrograde flow in T cells interacting with different stimulatory surfaces or after pharmacological treatments. Here, we described the imaging methods and analytical procedures used to measure actin velocity across the IS in T cells spreading on planar stimulatory surfaces.
Keywords: Actin, Cytoskeleton, Kymograph, Immunological synapse, T-cells, Integrin, Planar lipid bilayer, Mobile ligands, Spinning disk, Live cell imaging
1 Introduction
The formation of the immunological synapse (IS) between a T cell and an antigen presenting cell (APC) depends on actin dynamics downstream of T cell receptor (TCR) and integrin engagement [1–4]. TCR signaling activates the Arp2/3 complex-dependent polymerization of branched actin filaments at the edges of the cell- cell contact site, driving initial spreading of the T cell on the APC surface and subsequent centripetal flow of the actomyosin network. This process corresponds to the retrograde actin flow that occurs at the leading edge of a migrating cell. Centripetal actin flow drives the ongoing assembly and function of TCR signaling complexes, and ultimately shuttles these complexes to the center of the IS, where signal extinction takes place [5, 6]. Centripetal flow of the T cell actin network also regulates integrin conformational change, thereby promoting adhesion to ligands on the APC surface, as well as outside-in signals that costimulate T cell activation [7, 8]. Actin dynamics thus function as an essential part of a key feedback loop that coordinates T cell signaling events at the IS. Thus, measuring actin flow at the IS is valuable for understanding the fundamental mechanisms that drive and fine-tune T cell activation.
Much of what is known about actin dynamics at the IS comes from studies of T cells responding to coverslips or planar lipid bilayers coated with stimulatory antibodies or ligands [9]. While these planar stimulatory surfaces do not recapitulate the complex undulations that are formed at a T cell-APC interface [10, 11], they represent a powerful tool because they allow investigators to image the movements of fluorescently tagged cytoskeletal elements and signaling molecules in the microscope’s X–Y plane. In conjunction with these planar stimulatory surfaces, several labs have taken advantage of super resolution approaches such as structured illumination (SIM), stimulated emission depletion (STED), and single molecule localization techniques (PALM/STORM) to examine the molecular architecture at the IS [12–16]. Recently, lattice light sheet technology has also proven to be valuable [17]. Nonetheless, simpler techniques for live cell imaging such as total internal reflection (TIRF) and spinning disk confocal microscopy continue to be the best ways to answer many biological questions. TIRF optics are often used to image movement of signaling molecules at the IS. This modality is favored because it focuses analysis on events occurring at or very near the plasma membrane (within about 100 nm), and offers low background noise. However, the actomyosin network extends more than 1 μm into the cell at early times after TCR engagement [11]. Even at later times when T cells are well spread, the actin-rich lamellipodia are nearly 0.5 μm thick [18]. Thus, only a subset of actin filaments is captured within the TIRF plane. Indeed, it is nearly impossible to gain an overall sense of the actomyosin network using TIRF optics. Because we are interested in the actin cytoskeleton as a functional unit, we prefer to use spinning disk confocal microscopy. As detailed below, we typically capture three planes spanning a total distance of 0.5 μm. This usually captures the entire thickness of the lamellipodium of a spreading T cell. By generating a maximum intensity projection or a 3-dimensional rendering of the three planes, we can analyze the behavior of the lamellipodial actin network as a whole.
Armed with suitable video sequences, it is relatively straightforward to carry out quantitative analysis of actin flow rates by tracking the movement of small structures within the actin network. The most common technique to visualize motion from sequential 2-D imaging is kymographic analysis [19, 20]. To generate a kymograph, one first selects a narrow region of interest and extracts this region from each image in a time series. The selected region is then laid side-by-side for all time points, generating a picture (kymograph) that displays movement of objects within the selected region over time, such that one axis represents space and the other axis represents time. Movement within these space-time plots is seen as diagonal lines of bright or dark features, and the speed of movement can be determined based upon the slope of these lines.
In studies using planar surfaces to analyze protein dynamics at the IS, one important factor is the mobility of stimulatory ligands. Neither glass surfaces bearing immobilized ligands nor planar lipid bilayers bearing freely mobile ligands faithfully recapitulate the biology of bona fide antigen presenting cells, where some ligands exhibit constrained mobility and others are freely mobile [21]. However, these simplified systems provide a means of exploring the ways in which ligand mobility affects actin flow and signaling through actin-coupled receptors.
There are several good protocols in the literature for the preparation of T cell stimulatory surfaces suitable for microscopy [22–24]. Here, we describe our procedures for the preparation of both immobile and mobile stimulatory surfaces, followed by our methods for imaging and measuring lamellipodial actin flow in T cells interacting with these surfaces. We provide details for viewing actin movements in a predetermined plane with optimal spatial and temporal resolution, and testing the effects of ligating specific receptors in the absence of other stimuli and altering actin dynamics using pharmacological agents.
2 Materials
All solutions should be prepared using reverse osmosis (Milli-Q) water and analytical grade reagents. Avoid repeated freeze-thawing of protein solutions and inhibitors; make small aliquots before freezing. Follow institutional safety guidelines in handling and disposing of hazardous chemicals including Hellmanex III and Piranha solution and biohazardous materials such as recombinant viruses and human cells.
2.1 Generation of Stimulatory Surfaces
2.1.1 Preparation of Glass Coverslips
Coverslips for Sticky-Slides (Ibidi): # 1.5H (170 μm ± 5 μm) D 263 M Schott glass, 25 mm × 75 mm.
2% Hellmanex III detergent (Hellma Analytics): dissolve 20 ml of detergent in 980 ml of water. Other alkaline glassware detergents may substitute for Hellmanex III (e.g., Linbro 7×), but optimization of concentration, time, and rinsing requirements is needed.
Piranha solution: Working in a fume hood, mix 300 ml of sulfuric acid with 100 ml of hydrogen peroxide in a glass beaker [7] (see Note 1).
Bath sonicator.
Plasma cleaner.
2.1.2 Preparation of Imaging Chambers
Bottomless Sticky-Slide VI 0.4 6-channel chambers (recommended supplier –Ibidi, Martinsried, Germany). Various home- made flow chambers may be substituted. The desired channel width and length is ~5 × 25 mm with a height of 250 μm.
2.1.3 Coating Surfaces with Immobilized Ligands
Mouse anti-human T cell receptor: OKT3 (recommended supplier—BioXCell, Burlington, VT). Store at 4 °C.
Human VCAM-1: 1 mg/ml in PBS. Available as a soluble Fc fusion from recommended suppliers Sino Biological or R&D Systems. His-tagged protein also works for nonspecific adsorption. Store at −20 to −80 °C in small aliquots.
Human ICAM-1: 1 mg/ml in PBS. Available as a soluble Fc fusion from recommended suppliers Sino Biological or R&D Systems. His-tagged protein also works for nonspecific adsorption. Store at −20 to −80 °C in small aliquots.
L15 imaging medium: Supplement L15 with 2 mg/ml D-(+) glucose by adding 1 g of D-glucose to 500 ml L15 medium and filtering through a sterile filter unit (0.2 μm pore size). Optionally, use phenol red-free RPMI-1640 enriched with 25 mM HEPES by adding 12.5 ml of sterile 1 M HEPES solution to 500 ml RPMI medium.
Phosphate buffered saline (PBS)- standard formulation with or without D-glucose.
Multichannel pipette.
Poly-L-Lysine hydrobromide (PLL) (mol. wt. 30,000–70,000): Dissolve 5 mg in 10 ml of water. Add 40 μl of 5% NaN3. Freeze in aliquots or store at 4 °C for 1–2 weeks.
2.1.4 Coating Surfaces with Stimulatory Supported Planar Lipid Bilayers
50 ml glass round-bottom flask.
Glass syringes (10, 50, and 500 μl, Hamilton).
2% Hellmanex III detergent: see Subheading 2.1.1, item 2.
Acetone (HPLC grade).
Chloroform (HPLC grade).
Lipids (recommended supplier—Avanti Polar Lipids, see Note 2): DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine) 25 mg/ml in chloroform, DGS-NTA(Ni) (1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl] (nickel salt)) 10 mg/ml in chloroform, DSPE-PEG(2000) Biotin (1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[biotinyl(polyethylene glycol)-2000] (ammonium salt) ) 5 mg/ml in chloroform. Store at −20 °C.
Extruder set (recommended supplier—Avanti Polar Lipids): excluder with two glass syringes and 50 nm pore membranes.
Bath sonicator.
Compressed air (house air is fine, but place a filter on the output to remove contaminants).
Vacuum dessicator.
PBS.
Biotinylated OKT3 (0.5 mg/ml). Store at 4 °C.
Streptavidin, NeutrAvidin or NeutrAvidin –TexasRed conjugate: 1 mg/ml in PBS. Store at −20 °C.
Human VCAM-1-His tagged (recommended supplier—Sino Biological Inc): 1 mg/ml in PBS. Store at −20 to −80 °C in small aliquots.
Human ICAM-1-His tagged (recommended supplier—Sino Biological): 1 mg/ml in PBS. Store at −20 to −80 °C in small aliquots.
L15 imaging medium: L15 supplemented with 2 mg/ml D-(+) glucose.
Multichannel pipette.
2.2 Imaging Actin Dynamics in Living T Cells
2.2.1 Preparation of Cells Expressing Fluorescent Actin Probes for Live-Cell Imaging
Jurkat T-Cells
Jurkat T cell growth medium: RPMI 1640 enriched with 5% fetal bovine serum and 5% newborn calf serum, 2 mM L-alanyl- L-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin (see Note 3).
Jurkat T cells stably expressing fluorescent actin probes (fluorescent protein-labeled actin, Lifeact, or F-tractin) (see Notes 3 and 4 for details on the use of these probes).
Primary Human Peripheral Blood CD4+ T Cells
2.2.2 Fluorescence Microscopy
Below is the description of the setup we use to acquire the images. Other vendors also provide similar systems and there are many options for analysis software.
Inverted microscope (Zeiss Axiovert 200 with Piezo Z-focus).
Yokagawa spinning disk head (PerkinElmer Ultraview ERS6 with Photokinesis unit).
CCD camera: Orca ER camera, Hamamatsu (see Note 7).
Objective: 63× Plan Apo 1.4 NA, oil immersion.
Solent Scientific environmental chamber.
Multi laser module (laser lines 405, 440, 488, 514, 561, 640 nm).
Emission filters (455/60, 485/60, 527/55, 587/125, 615/70, 705/90).
Vibration isolation table (Vibraplane kinetic systems).
Image acquisition software (Volocity v. 6.3, Perkin Elmer).
2.3 Image Analysis
Volocity v. 6.3 imaging software (Perkin Elmer).
Microsoft Excel.
3 Methods
3.1 Generation of Stimulatory Surfaces
3.1.1 Preparation of Glass Coverslips (See Note 8)
Sonicate the glass coverslips in 2% Helmanex detergent for 30 min.
Wash vigorously with water to remove all detergent.
Air dry.
Dip into fresh Piranha solution and incubate for 30 min (see Note 1).
Wash with water.
Air dry.
For studies involving lipid bilayers, we recommend plasma cleaning the glass for 3 min just before the addition of the small unilamellar liposome vesicle (SUV) solution. This step is dispensable for studies where coverslips are coated with stimulatory proteins without lipid bilayers.
3.1.2 Preparation of Imaging Chambers (See Note 9)
Peel the protective paper off the Sticky-Slide 6-channel chambers, revealing the self-adhesive underside. Mount cleaned coverslip, aligning carefully. Take care to touch only the edges of the coverslip.
Press down carefully, using a pen or the back of a pair of tweezers to secure the seal between each well. To prevent leakage, make sure that the tape sticks well to the coverslip. If desired, one can further protect the sides from leaking by applying nail polish around all sides.
3.1.3 Coating Surfaces with Immobile or Mobile Ligands
Two types of stimulatory surfaces can be prepared: ligand can be immobilized by adsorption onto the glass coverslip (Subheading “Coating with Immobile Ligands by Protein Adsorption to the Coverslips”) or ligand can be attached to lipid bilayers where it will have high lateral mobility (Subheading “Coating Surfaces with Stimulatory Supported Planar Lipid Bilayers”).
Coating with Immobile Ligands by Protein Adsorption to the Coverslips
We typically apply OKT3, a monoclonal antibody that reacts with an epitope on the epsilon-subunit within the human CD3 complex [25], in the presence or absence of the adhesion molecules VCAM-1 or ICAM-1, which bind to the integrins VLA-4 and LFA-1, respectively. Depending on the experiment, other stimulatory ligands can be used, and concentrations can be varied (see Note 10).
Dilute OKT3 in PBS to a concentration of 10 μg/ml.
Using the multichannel pipette, add 100 μl of 10 μg/ml OKT3 to each chamber well. To create flow in one direction, it is important always to pipette into one side of the wells (intake ports), and out from the other side (outtake ports). To facilitate this, mark the intake side of the chamber (the side to which OKT3 was added) with an arrow.
Incubate 2 h at RT or overnight at 4 °C.
Wash each well three times with PBS. To do this, use a multi-channel pipette to remove 90% of the solution in the wells (withdraw from the outtake ports). Then pipette in 150 μl of PBS to the marked intake side. Repeat two more times. Never let the wells dry out.
At this point, another ligand can be added. We use 2 μg/ml of VCAM-1 or ICAM-1 in PBS. Withdraw 100 μl of PBS from the outtake wells and replace with 100 μl of 2 μg/ml VCAM-1 or ICAM-1. Incubate for another 2 h at 37 °C, and wash three times with PBS, all as in steps 2–5 (see Note 11).
Exchange the solution to L-15 imaging medium by washing three times with pre-warmed L-15 medium (37 °C).
Incubate the chamber on the microscope stage at 37 °C for about 10 min before adding cells.
Coating Surfaces with Stimulatory Supported Planar Lipid Bilayers
In order to facilitate specific binding of ligands to the lipid bilayer, functionalized lipid must be incorporated into the lipid mixture during vesicle preparation. Many functionalized lipids are commercially available. We used biotinylated lipids and lipids with a Ni-NTA group, allowing us to attach biotinylated OKT3 (via a Streptavidin bridge) as well as His-tagged ICAM-1 or VCAM-1. The following procedure has two stages: Steps 1–13 describe preparation of lipid vesicles in chloroform in a desired mol% ratio. We use 5 mM DOPC:DSPE-PEG(2000) biotin:DGS-NTA(Ni) (nickel salt) in a 98:1:1 mol% ratio. Steps 14–23 describe the use of these vesicles to generate planar bilayers.
Sonicate the 50 ml glass round-bottom flask and the extruder set in 1% Hellmanex III solution for 10 min.
Thoroughly rinse the flask and extruder set with water to completely remove the residual detergent.
Air dry.
Rinse the flask in acetone and then chloroform, vortexing to be sure to cover all surfaces. It’s fine to leave a little chloroform in the flask.
Wash each glass syringe thoroughly by passing chloroform through it five to ten times.
Use the glass syringes to add 200 μl chloroform to the flask. Again using the glass syringes, add 30.6 μl DOPC, 6 μl DSPE-PEG(2000), and 1 μl DGS-NTA into the flask, pipetting each into the chloroform. Vortex to mix (see Note 12).
Gently dry the lipid solution with compressed air while rotating the round bottle to make a uniform lipid film.
Place the round bottle into a vacuum desiccator and dry for 2 h or overnight.
Rehydrate the lipid film with 200 μl of PBS (or the buffer of your choice) to bring the final lipid concentration to 5 mM, and then sonicate for 5 min to produce micelles.
Assemble the mini-extruder as per Avanti instructions, using the 50 nm pore membrane.
Pass PBS through the extruder a few times to ensure that the assembly does not leak. Monitor the volume that comes across the extruder after five to ten passes. If volume is lost, reassemble the set.
Extrude the lipid solution through the membrane at a constant, steady rate 21 times, creating a lipid vesicle mixture (see Note 13).
Transfer the lipid vesicle mixture to a 1.5 ml conical microcentrifuge tube. This mixture can be kept at 4 °C for 1 week.
For each chamber slide, mix 150 μl of the lipid vesicle mixture with 600 μl PBS to get 1 mM liposome suspension. Vortex to mix (see Note 14).
Using freshly prepared plasma cleaned Sticky-Slide chambers (see Subheading 3.1.2), and a multichannel pipette, add about 100 μl of diluted lipid vesicle mixture to each chamber and incubate for 30 min at RT.
Rinse the wells thoroughly with PBS to remove the excess vesicles. This should be done by sequential addition of PBS to the intake well and removal of flow-through on the other side. About 150 μl of PBS can be added at a time for a total of three to five times. Never allow the wells to dry out or air to enter the channel containing the bilayer.
Remove the final PBS wash and pipette in Streptavidin or fluorescent Neutravidin (1 μg per 100 μl PBS for each well). Incubate for 20 min at RT.
Wash thoroughly as before.
Incubate the chambers with biotinylated OKT3 and His- tagged ICAM-1 or VCAM-1. This should be done sequentially, incubating for 20–30 min and washing three times with PBS after each addition (see Note 15).
Exchange the PBS with three washes of L-15 imaging medium.
Bilayer surfaces should be tested for ligand mobility (see Note 16) and used the same day. We usually transfer them to the microscope environmental chamber (see Subheading 3.2.2). Ligand mobility and surface quality may decrease with longer storage.
Incubate the chamber on the microscope stage at 37 °C for about 10 min before adding cells.
3.2 Imaging Actin Dynamics in Living T Cells
3.2.1 Preparation of Cells Expressing Fluorescent Actin Probes for Live-Cell Imaging
3.2.2 Preparation of the Microscope
Set the environmental chamber on the microscope to 37 °C, and allow it to equilibrate for at least 1 h prior to imaging.
Place all chambers, reagents, etc. into the environmental chamber to allow equilibration.
3.2.3 Preparation of Cells for Live-Cell Imaging
Pipette about 5 ml of cells into a 15 ml tissue culture grade conical tube.
Centrifuge the cell suspension at 250 × g for 5 min at room temperature.
Aseptically aspirate or decant the supernatant without disturbing the cell pellet.
Resuspend the cell pellet in 5 ml of L-15 medium.
Determine the total number of cells using a hemocytometer.
Centrifuge the cells again to remove residual serum.
While the cells are in the centrifuge, calculate the volume of L-15 imaging medium needed to resuspend the cells at the desired density. We typically resuspend the cells at 1 × 106/ml.
Resuspend the washed cells in L-15 medium at the desired final density. Maintain at 37 °C until imaging (see Note 17).
3.2.4 Imaging Cells by Spinning Disk Confocal Microscopy
Open imaging software and set all basic parameters. Configure the time-lapse settings; we usually collect a z-stack of three planes spaced 0.25 μm apart every 0.5–1 s, over a total time of about 4 min.
Inject 50 μl of cell suspension into the intake well of a chamber coated with stimulatory ligands (prepared as described in Subheading 3.1).
Mix and distribute the cells by gently removing about 50 μl of flow-through from the outtake port and adding back to the intake port. Repeat this three to five times.
Place the chamber under the microscope and allow the cells to interact with the stimulatory surface for about 5 min before imaging. In some cases, it is desirable to image the early phases of T cell contact with the coverslip. The easiest way to do this is to add only a few cells initially, allow those cells to settle, and adjusting settings as described in step 5. Then wait for additional cells to touch down, adding more if needed, and analyze that population.
Meanwhile, adjust the focus and set imaging parameters (exposure time and laser power) based on the brightness of cells as they come into contact with the surface. Use the lowest possible intensity and time to minimize photobleaching.
Choose a field with individual cells that are not contacting other cells. If the population of cells is heterogeneous in brightness, take care not to select very bright cells that may overexpress fluorescent actin probes, as overexpression may perturb actin dynamics.
Focus on the bottom of a cell, just above the coverslip. This will be the region where actin-rich lamellipodia form as the cell spreads on the stimulatory surface.
Collect a time-lapse series.
Image as many fields as needed from one chamber for up to 20 min or when cells start to deform and detach from the surface (see Note 18).
If desired, inhibitors can be added to test effects on actin dynamics (see Note 19, which includes a table of commonly used inhibitors). After imaging the untreated cells for 1–2 min, add the desired inhibitor using a gel-loading tip. Mix gently by pipetting in and out of the intake port, or by removing 50 μl of medium from the outtake port and adding back to the intake port. Take care not to disturb the cells or bump the stage. Resume imaging as soon as possible after adding the inhibitor.
3.3 Image Analysis
Actin flow rates are calculated based on kymographic analysis. Here, we describe the procedure using Volocity v. 6.3. Other software packages have similar capabilities (see Note 20).
Select a video sequence of a cell for analysis.
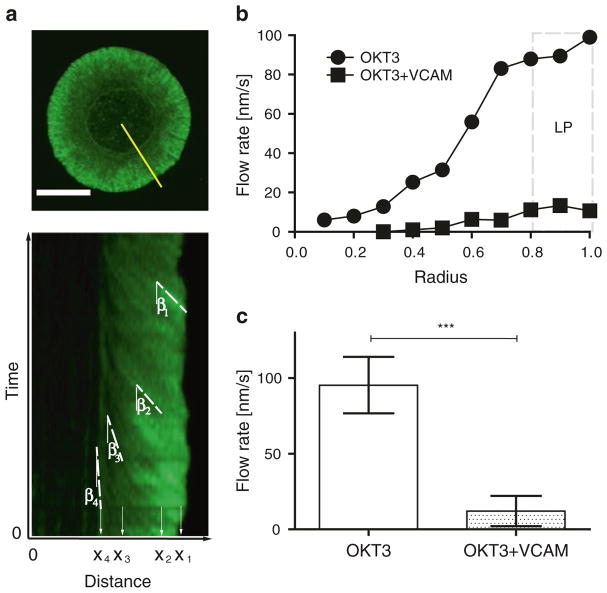
Draw a ray from the center of immunological synapse (IS) to the periphery (see Fig. 1a, yellow line in top panel).
Generate kymograph. (Go to “tool” in Volocity and choose “kymograph”).
In Volocity, you can set the time units so that one pixel equals 1 s. This way, the y axis of the kymograph (displayed in pixels) is equal to time in seconds (see Fig. 1a, bottom panel).
Choose the line tool and draw lines along the diagonal intensity maxima (see the white dashed lines in Fig. 1a, bottom panel) (see Note 21).
Go to the “Measurements” section displayed above the kymograph image. In the Measurements view you will find the length of each of your drawn lines, the location of each line (start position x, start position y, end position x, end position y), and the line angles in degrees. The output you will get is shown in Table 1.
Select all the measurements for lines and copy to the Microsoft Excel folder. You will need angle values (displayed in degrees) to calculate actin flow rates. If you are interested in calculating actin flow rates as a function of position along the cell radius as described in steps 12–19, you will also need end position x.
In Volocity, “angle” ranges from 0 to 180. 0 means pointing “up the screen,” along the y axis. 180 means pointing “down” the screen. Thus, to calculate the angle of the lines you have drawn (see β1 – n in Fig. 1a, bottom panel), you need to subtract the angle obtained from Volocity from 180 (β_degree = 180-Angle[degree]).
Convert each angle β_degree to radians (multiply by π/180°).
Calculate flow rate from the slope: V[μm/s]=tan(β_radian) × (tpixel [pixels]/ts[s]) × Cf[μm/pixel], where Cf is a conversion factor that will convert distance in pixels to metric scale. Note that if you set the time units so that one pixel equals 1 s, in step 4, this conversion is unnecessary because tpixels/ts = 1.
Average the flow rates calculated from all kymographs and calculate standard deviation (SD) and standard error (SE).
Since actin flow decelerates with centripetal movement, it is valuable to calculate actin flow rates as a function of position along the radius of the immunological synapse. To do this, for each line drawn in step 5, record the position (x end point), based upon the distance from IS center (see x1 – n in Fig. 1a, bottom panel). For each cell, record the radius (r) (see Note 22).
Create a table like that shown in Table 2. In addition to measured values from steps 8–10, your table should include normalized distances (Dn = position xn/cell radius r).
Calculate actin flow rates as in step 10.
Create additional kymographs and repeat the analysis of actin slopes and flow rate calculations for 10–30 cells per condition.
Bin the values based on where each measurement was made along the cell radius (after normalizing from 0 to 1). Units of 0.1 radius work well. Average flow rate values for each bin.
Create a graph like that shown in Fig. 1b, showing actin flow rate vs normalized position (from 0 to 1). This graph will show the distribution of F-actin velocity across the immunological synapse, grouped into ten equally spaced bins.
As an alternative to analyzing actin flow across the entire immunological synapse, it is sometimes sufficient to analyze flow where it is fastest, i.e., within the outer lamellipodial region. Based on morphology and localization of actin and myosin, we define this region as the outer 20% of the radius [18]. To obtain this value, bin measurements as described in step 16 and average measurements for all data points in the radius range 0.8–1 (see Fig. 1c).
Calculate statistical significances using Student’s T test for unpaired samples.
Fig. 1.
Characterization of F-actin dynamics in Jurkat T lymphoma cells. T cells were allowed to interact with coverslips coated with anti-CD3 (OKT3) ± VCAM-1 and imaged for 4 min. (a) Single time point of a responding cell stimulated on anti-CD3 (top) and the corresponding kymograph of F-actin dynamics generated along the yellow line (bottom). The dotted lines trace the paths taken by distinct features along the distance xn with their corresponding angles βn. (b) Kymographic analysis of F-actin flow in Jurkat T cells, showing the distribution of F-actin velocity across the immunological synapse. The area marked by the dashed box displays the peripheral lamellipodial region (LP). (c) Actin flow rates within the LP region for Jurkat T cells responding to OKT3 in the absence or presence of VCAM-1. Means ± SD are shown (n = 20–40 cells per condition), ***, P < 0.001. Scale bar 10 μm
Table 1.
Data from the measurements panel in Volocity for the white dashed lines in Fig. 1a
| ID | Item Name | Name | Population | Type | Total line length | Start position x | Start position y | End position x | End position y | Angle (degrees) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Jurkat_Kymograph | Lines 1 | Lines | Line | 28.32 | 98 | 39 | 119 | 58 | 132.14 |
| 2 | Jurkat_Kymograph | Lines 2 | Lines | Line | 24.08 | 77 | 113 | 93 | 131 | 138.37 |
| 3 | Jurkat_Kymograph | Lines 3 | Lines | Line | 29.07 | 63 | 165 | 76 | 191 | 153.43 |
| 4 | Jurkat_Kymograph | Lines 4 | Lines | Line | 28.28 | 54 | 198 | 58 | 226 | 171.87 |
Table 2.
Calculation of actin velocity across the IS as described in Subheading 3.3
| ID | End position x | Normalized distance D = xn/r | Angle (degrees) | β = 180–Angle (degrees) | β (radian) = β (degree) × π/180° | Slope = tan(βradian) | V [μm/s] = slope × Cf × tpixel/ts |
|---|---|---|---|---|---|---|---|
| 1 | 119 | 0.915 | 132.14 | 47.86 | 0.83 | 1.10 | 0.116 |
| 2 | 93 | 0.715 | 138.37 | 41.63 | 0.73 | 0.89 | 0.093 |
| 3 | 76 | 0.585 | 153.43 | 26.57 | 0.46 | 0.50 | 0.053 |
| 4 | 58 | 0.446 | 171.87 | 8.13 | 0.14 | 0.14 | 0.015 |
| n | xn | Dn | angle | β_degree | β_radian | slope | Vn |
Table 3.
Inhibitors used to perturb dynamics of the acto-myosin network
| Inhibitor name | Mode of action | Dose (μM)a | Effect on T cell actin dynamics | References |
|---|---|---|---|---|
| Cytochalasin D | Inhibits actin polymerization by binding to growing ends of actin nuclei and filaments (F-actin), and preventing addition of monomers (G-actin) to these sites | 10 | Induced the accumulation of disordered F-actin–rich zones | [21] |
| Latrunculin B (Note A) | Inhibits actin polymerization through association with actin monomers | 1 | Led to complete depletion of the F-actin network | [21] |
| Jasplakinolide (Jas) | Disrupts actin filaments and induces polymerization of monomeric actin into amorphous masses. | 1 | Arrested F-actin retrograde flow within 30 s. The network formed a tight band that constricted inward due to myosin II activity | [8, 18, 34] |
| Blebbistatin (Bleb) (Note B) | Inhibits myosin II ATPase activity and slows down phosphate release | 50 | T cell spreading and centripetal actin flow were unaffected by blebbistatin pretreatment. At late time points, myosin-inhibited cells failed to contract normally and became irregularly shaped. Disrupted organization of actin arcs | [8, 18, 34, 37] |
| Y27632 (Y27) | Rho kinase inhibitor, blocks phosphorylation of myosin light chain at S19 and inhibits myosin II filament assembly | 25 | T cell spreading and centripetal actin flow were unaffected. Effects at late time points were similar to blebbistatin | [8, 18] |
| Jas+Bleb / Jas+Y27 | Inhibitor cocktail | See above | Arrested F-actin flow and caused a slow, partial collapse of the actin network | [8, 18] |
| SMIFH2 | Inhibits formin-mediated actin assembly | 2.5–10 | Initially slowed centripetal flow of actin arcs. Eventually arrested arc generation from the cell edge | [38, 39] |
| CK666 | Arp2/3 complex inhibitor; stabilizes the inactive state of Arp2/3 complex and prevents conformational changes required for activation | 100 | Altered lamillipodial actin architecture and slowed centripetal flow. Drove a lamellipodial-to-filopodial shape change. Blocked the formation of TCR- associated actin foci (invadopodia like projections within the central region of the immunological synapse) | [38, 39, 40] |
| CK689 | Inactive homologue of CK666 | 100 | Actin structure was indistinguishable from untreated cells | [39, 40] |
Note A: Latrunculin B is serum sensitive [41]. Thus, a higher dose may be required in the presence of serum
Note B: Blebbstatin is light sensitive. Extended exposure to blue light (450–490 nm) may cause degradation of bleb-bistatin to an inactive product via cytotoxic intermediates. Thus, this drug should be handed in dim lighting and red fluorophores should be used. A photo-stable analog of (−)-blebbistatin, (S)-nitro-blebbistatin, is stable to prolonged irradiation at 450–490 nm. (S)-nitro -blebbstatin is commercially available from Cayman Chemicals and can be used in the same dose as the photo-inactivated form [42].
Final concentration. Inhibitors are usually prepared as 200–500 × stocks, dissolved in DMSO
Footnotes
Piranha solutions are extremely energetic and may result in explosion or skin burns if not handled with extreme caution. Work in a fume hood and wear a lab coat, acid apron, safety goggles, and heavy, nitrile gloves. When preparing Piranha solution, always add the peroxide to the acid. Dispose of Piranha solution in a sturdy container, sitting within a secondary container. Maintain a loose cap; the solution generates gases for several days and could explode if not allowed to vent. It is important to avoid adding any other waste into this container. Mixing with organic compounds like toluene, chloroform, and phenol will result in a violent reaction. The Piranha waste container must be well marked and lab members should be instructed about the associated hazards. We post a prominent sign whenever working with Piranha solution, to minimize traffic and distractions in that area of the laboratory.
Chloroform evaporation can cause inaccuracy in measurement, thus the volumes of lipid solutions should be noted and losses in volume should be replenished before mixing different lipid solutions.
Jurkat cells are grown in suspension, in a 37°C degree incubator containing 5% CO2. The cells should not be allowed to become too dense; ideally, the culture should be kept between 1x105/ml and 2x106/ml. Typically, these cells will need to be split 1:10 every 2 days. Jurkat cells sometimes become unresponsive when passaged multiple times, and we typically thaw a fresh vial about once a month. Thus, it is important to maintain a good supply of frozen vials. Thaw an early freeze and expand again when running low on backup vials. An important caveat to the use of Jurkat T cells is that these cells have known signaling abnormalities, particularly in inositol lipid regulatory pathways [26, 27]. Nonetheless, these cells are a valuable tool because of their large size and the ability to generate stable transfectants. For our studies, we typically use a Jurkat T cell line that was stably transfected with GFP-actin [28]. These cells were screened to rule out changes in TCR signaling, and to ensure that GFP-actin is not grossly overexpressed. Plasmids and recombinant viral particles expressing fluorescent actin are available from Clontech and can be expressed in Jurkat T cells by transfection or lentiviral transduction. Fluorescently tagged-actin can perturb actin dynamics, and can be excluded from lamellar networks and filopodia [29–31]. Moreover, this strategy labels both polymeric and monomeric pools. Thus, we sometimes use cells expressing fluorescent-Lifeact, a 17 amino acid peptide that binds selectively and reversibly to actin filaments (F-actin), and has little or no effect on actin dynamics [32]. Plasmids and recombinant viral particles expressing fluorescent Lifeact are available from Ibidi. Alternatively, transgenic Lifeact mice are available [33] and can be used as a source of T cells; such mice are particularly valuable for studies requiring naive T cells, since activation is typically required for effective viral transduction. Other investigators use F-tractin, the actin-binding domain from rat inositol triphosphate 3-kinase, which facilitates visualization of actin arcs at the lamellar region [34, 35]. Care should be taken in choosing an actin probe, since each has the potential to bias the structures being imaged [30]. Where possible, controls should be performed using multiple approaches.
Plasmids expressing fluorescent F-tractin are available from Addgene (Cambridge, MA). Regardless of the probe used, cells can be stably transfected by electroporation or lentiviral transduction, and selected with appropriate antibiotics. Stable cell lines should either be screened periodically for expression, or highly stable lines should be generated by two rounds of single cell cloning. Care should always be taken that lines do not change over time, and multiple vials of early passage freezes should be maintained and thawed whenever there is evidence of change.
We obtain primary human peripheral blood CD4+ T cells from the University of Pennsylvania’s Human Immunology Core under an Institutional Review Board approved protocol. However, they can be readily prepared from human peripheral blood using commercial kits based on depletion of other cell types. Purified CD4+ T cells are activated with CD3/CD28 magnetic beads (Dynabeads, Life Technologies) and cultured a humidified 37 °C incubator with 5% CO2. Beads are magnetically removed on day 7 after initial stimulation, and cells are then cultured for an additional day in the presence of 10 U/ml of IL-2. Primary T cells should be used at days 8 and 9 after activation. During this window, the cells are optimally “rested” after the initial activation, and are not yet showing signs of activation-induced cell death.
To generate primary human CD4+ lymphocytes expressing Lifeact-GFP, cells are cultured for 24 h with human T-Activator CD3/CD28 magnetic beads, and then transduced with Lifeact-GFP lentivirus. Lentivirus and 8 μg/ml polybrene (Sigma-Aldrich) is mixed with 5–10×106 T cells in the wells in a 5 ml round-bottom polystyrene tube and centrifuged at 1200 × g for 2 h at 37 °C. Lentivirus-containing media is then replaced with T cell culture media, and the cells are returned to culture. Two days after transduction, the media is supplemented with 2 μg/ml puromycin, and cells are cultured for an additional 4 days before magnetic removal of the activator beads. Cells are cultured for an additional 1–2 days in media with 2 μg/ml puromycin and 10 U/ml IL-2 before use (day 8–9 after activation). A similar approach can be used for expression of other actin probes listed in Note 3.
While many investigators choose a more sensitive EM CCD camera, our system was configured with an ER CCD because it has a smaller pixel size, permitting higher resolution imaging. In addition, the ER camera allows a combination of fluorescence and white light (DIC) imaging. Scientific CMOS cameras also work well for the same reasons, and have a larger field size and improved signal-to-noise over the older ER CCD technology.
Proper cleaning is especially important for the generation of uniform stimulatory surfaces. This is especially true for studies using lipid bilayers, since any impurities may disturb the lipid bilayer and its mobility. Thus, it is important to use a detergent solution followed by an acid bath. We have used both Piranha solution (detailed above), and Nochromix solution successfully. As detailed in Note 1, Piranha solution is quite hazardous. Nochromix, a commercial formulation that contains ammonium persulfate, is a safer alternative. Nochromix solution should be prepared in a fume hood according to package directions. Usually, Nochromix powder is mixed with water and added to sulfuric acid, forming a clear solution. In this case incubate Hellmanex-cleaned glass slides in Nochromix solution overnight. Following cleaning with either Piranha or Nochromix, some investigators plasma clean the glass for 3 min just before the addition of SUV solution.
It is best to prepare chambers just before usage and to use freshly cleaned coverslips. In place of the sticky slides, one can use any high coverslip-bottomed imaging chamber (e.g., Mat-Tek dishes). We prefer the Ibidi sticky slides because they provide a very small controlled volume and access for perfusion of inhibitors. The multi-well format also allows imaging of multiple samples on a single glass surface, thus speeding imaging and promoting reproducibility.
Some investigators recommend coating glass coverslips with poly-L-lysine prior to addition of stimulatory ligands. We have tried the procedure with and without poly-L-lysine, and have not observed a difference in results. However, this may vary depending on the ligand used. To use poly-L-lysine, add 120 μl of poly-L-lysine solution to wells before step 1, incubate for 30 min at RT, and wash three times with water. Remove the water using an aspirator and air dry. Proceed with step 1 as before.
As an alternative to sequential coating with ligands, we have coated the glass with a mixture of OKT3 and VCAM-1 or ICAM-1 overnight at 4 °C. We have not seen a difference in experimental results. Nonetheless, we prefer sequential coating since we want to hold the levels of OKT3 constant, and mixing could affect the amount of OKT3 adsorbed onto the surface.
If desired, the lipid mixture can be prepared in larger quantity, and stored in single use aliquots at −20 °C. Note the volume, and upon thawing, add chloroform as necessary to account for evaporation.
Always do uneven number of passes so that your final micelles are in the opposite syringe than the original, this ensures more uniform size and quality.
Oversaturation of glass surface with lipid vesicles will result in partial fusion of vesicles to glass and diminish the mobility of the bilayers.
Biotinylated and His-tagged ligands should be added in excess to cover the surface completely. We use 1 μg of OKT3-biotin and 2 μg of ICAM-1–His per chamber. Optimal concentrations of ligands should be determined empirically. Saturating all the biotin binding sites of the bilayer bound Neutravidin will also help prevent crosslinking by the multibiotinylated OKT3. If lower densities of anti-CD3 are desired then using monovalent and monobiotinylated UCHT1 Fab′ is an alternative [36].
High concentration of ligand or Streptavidin may cause ligand aggregation and affect its mobility. Thus, it is important to perform control experiments for bilayer quality and ligand mobility. The simplest way to test bilayer or ligand mobility is to perform FRAP experiments. For this, lipids or ligands have to have fluorescent dye. For lipid bilayers we have used Texas Red DHPE (0.3%, about 2 μl of 1 mg/ml stock) but any lipid dye will work. To test ligand mobility we have used Neutravidin- Texas Red, OKT3-FITC, and VCAM-Alexa Fluor 647 that was labeled using a microscale protein labeling kit. When the surface is ready, image the surface and photobleach a small area until the area appears black and next record its recovery. If recovery is observed (the black spot will get brighter over time) then the surface is mobile, but if the bleached area remains dark then that will indicate that the ligands are unable to diffuse and are immobilized on the surface. In this case, the surface has to be prepared again. We have observed ligand immobility even in cases when lipid bilayer was mobile, due to ligand aggregation. Thus, it is the best to test both bilayer and ligand mobility at the same time. We have observed Neutravidin aggregation when higher concentrations of DSPE-PEG(2000) Biotin lipid are used (above 1%) but this may vary with specific lipid composition. Thus, optimal concentrations of ligands should be determined empirically.
Because L-15 medium is buffered to maintain pH in an air environment; cells do not need to be kept in a CO2 incubator during this time. Instead, it is often convenient to keep the working tube of cells in the microscope-mounted environmental chamber. The L-15 imaging medium is serum-free to eliminate molecules that might interfere with cell responses to surface bound ligands. This means that the cells awaiting imaging are undergoing serum deprivation. This can be useful, as it tends to suppress basal levels of cell signaling. However, care should be taken not to maintain cells in L-15 for more than about 2 h. If longer times are needed, it is best to take another batch of cells from tissue culture.
Monitor cells during imaging, making focus corrections as needed. This is especially important if your system lacks an autofocus correction mechanism. Even small changes in environmental temperature may cause focus drift. To minimize drift, we use an incubator chamber that encloses the entire stage and objective and preincubate the chamber before imaging. Systems employing a heated dish and an objective heater can be used, but we find that they cause more problems with focus.
Optionally, a syringe pump may be used to introduce an inhibitor into the chamber. Acute treatment with inhibitors by addition during imaging only works for fast-acting inhibitors; some drugs such as Y27632 and blebbistatin require longer to take effect, so cells must be pre-treated prior to adding to the imaging chamber. Conversely, agents that depolymerize actin filaments abolish T cell spreading, so can only be used acutely, after the cells have interacted with stimulatory surfaces. Table 3 lists commonly used inhibitors.
Several software packages offer kymography modules. MetaMorph has been used successfully for studies similar to ours [19, 34]. ImageJ can also be used after installation of plugins that are available online.
In cases when actin speckles are not readily visible, recorded movies can be processed. We used the Smart Sharpen filter in Adobe Photoshop, with 300% amplification of local maxima within a 3 pixel radius. This facilitated identification of individual GFP-actin speckles, which can be used as fiduciary marks for analysis. In control studies, analysis of sharpened and unprocessed movies yielded similar results. As an alternative approach, a portion of the F-actin network can be photo- bleached to induce a synchronous wave of bleached GFP-actin propagating toward the center of the IS. In both cases, a ray was struck from the center of the IS to the periphery, and vertical kymographs were generated in Volocity analyzed as described previously (see Subheading 3.3).
The depletion angles (slopes) are sufficient to calculate rates of actin flow. However, actin flow decelerates as the network moves toward the center of the IS. Thus, we typically calculate actin flow rates as a function of the position within the synapse where each measurement was made. Because the diameter of spread T cells varies somewhat, it is convenient to set the radius of each spread T cell to 1 and normalize all positions accordingly.
References
- 1.Kumari S, Curado S, Mayya V, Dustin ML. T cell antigen receptor activation and actin cytoskeleton remodeling. Biochim Biophys Acta. 2014;1838(2):546–556. doi: 10.1016/j.bbamem.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Comrie WA, Burkhardt JK. Action and traction: cytoskeletal control of receptor triggering at the immunological synapse. Front Immunol. 2016;7:68. doi: 10.3389/fimmu.2016.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Le Floc’h A, Huse M. Molecular mechanisms and functional implications of polarized actin remodeling at the T cell immunological synapse. Cell Mol Life Sci. 2015;72(3):537–556. doi: 10.1007/s00018-014-1760-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beemiller P, Krummel MF. Regulation of T-cell receptor signaling by the actin cytoskeleton and poroelastic cytoplasm. Immunol Rev. 2013;256(1):148–159. doi: 10.1111/imr.12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cemerski S, Das J, Giurisato E, Markiewicz MA, Allen PM, Chakraborty AK, Shaw AS. The balance between T cell receptor signaling and degradation at the center of the immunological synapse is determined by antigen quality. Immunity. 2008;29(3):414–422. doi: 10.1016/j.immuni.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee KH, Dinner AR, Tu C, Campi G, Raychaudhuri S, Varma R, Sims TN, Burack WR, Wu H, Wang J, Kanagawa O, Markiewicz M, Allen PM, Dustin ML, Chakraborty AK, Shaw AS. The immunological synapse balances T cell receptor signaling and degradation. Science. 2003;302(5648):1218–1222. doi: 10.1126/science.1086507. [DOI] [PubMed] [Google Scholar]
- 7.Springer TA, Dustin ML. Integrin inside-out signaling and the immunological synapse. Curr Opin Cell Biol. 2012;24(1):107–115. doi: 10.1016/j.ceb.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Comrie WA, Babich A, Burkhardt JK. F-actin flow drives affinity maturation and spatial organization of LFA-1 at the immunological synapse. J Cell Biol. 2015;208(4):475–491. doi: 10.1083/jcb.201406121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balagopalan L, Sherman E, Barr VA, Samelson LE. Imaging techniques for assaying lymphocyte activation in action. Nat Rev Immunol. 2011;11(1):21–33. doi: 10.1038/nri2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sage PT, Varghese LM, Martinelli R, Sciuto TE, Kamei M, Dvorak AM, Springer TA, Sharpe AH, Carman CV. Antigen recognition is facilitated by invadosome-like protrusions formed by memory/effector T cells. J Immunol. 2012;188(8):3686–3699. doi: 10.4049/jimmunol.1102594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roybal KT, Mace EM, Mantell JM, Verkade P, Orange JS, Wulfing C. Early signaling in primary T cells activated by antigen presenting cells is associated with a deep and transient lamellal actin network. PLoS One. 2015;10(8):e0133299. doi: 10.1371/journal.pone.0133299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown AC, Dobbie IM, Alakoskela JM, Davis I, Davis DM. Super-resolution imaging of remodeled synaptic actin reveals different synergies between NK cell receptors and integrins. Blood. 2012;120(18):3729–3740. doi: 10.1182/blood-2012-05-429977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sherman E, Barr V, Samelson LE. Super-resolution characterization of TCR- dependent signaling clusters. Immunol Rev. 2013;251(1):21–35. doi: 10.1111/imr.12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mace EM, Orange JS. Visualization of the immunological synapse by dual color time- gated stimulated emission depletion (STED) nanoscopy. J Vis Exp. 2014:85. doi: 10.3791/51100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ashdown G, Pandzic E, Cope A, Wiseman P, Owen D. Cortical actin flow in T cells quantified by spatiotemporal image correlation spectroscopy of structured illumination microscopy data. J Vis Exp. 2015;106:e53749. doi: 10.3791/53749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lillemeier BF, Mortelmaier MA, Forstner MB, Huppa JB, Groves JT, Davis MM. TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat Immunol. 2010;11(1):90–96. doi: 10.1038/ni.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ritter AT, Asano Y, Stinchcombe JC, Dieckmann NM, Chen BC, Gawden-Bone C, van Engelenburg S, Legant W, Gao L, Davidson MW, Betzig E, Lippincott-Schwartz J, Griffiths GM. Actin depletion initiates events leading to granule secretion at the immunological synapse. Immunity. 2015;42(5):864–876. doi: 10.1016/j.immuni.2015.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Babich A, Li S, O’Connor RS, Milone MC, Freedman BD, Burkhardt JK. F-actin polymerization and retrograde flow drive sustained PLCgamma1 signaling during T cell activation. J Cell Biol. 2012;197(6):775–787. doi: 10.1083/jcb.201201018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doggett TM, Breslin JW. Study of the actin cytoskeleton in live endothelial cells expressing GFP-actin. J Vis Exp. 2011:57. doi: 10.3791/3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiba K, Shimada Y, Kinjo M, Suzuki T, Uchida S. Simple and direct assembly of kymographs from movies using KYMOMAKER. Traffic. 2014;15(1):1–11. doi: 10.1111/tra.12127. [DOI] [PubMed] [Google Scholar]
- 21.Comrie WA, Li S, Boyle S, Burkhardt JK. The dendritic cell cytoskeleton promotes T cell adhesion and activation by constraining ICAM-1 mobility. J Cell Biol. 2015;208(4):457–473. doi: 10.1083/jcb.201406120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bunnell SC, Barr VA, Fuller CL, Samelson LE. High-resolution multicolor imaging of dynamic signaling complexes in T cells stimulated by planar substrates. Sci STKE. 2003;2003(177):PL8. doi: 10.1126/stke.2003.177.pl8. [DOI] [PubMed] [Google Scholar]
- 23.Vardhana S, Dustin M. Supported planar bilayers for the formation of study of immunological synapses and kinapse. J Vis Exp. 2008:19. doi: 10.3791/947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dustin ML, Starr T, Varma R, Thomas VK. Supported planar bilayers for study of the immunological synapse. Curr Protoc Immunol. 2007;Chapter 18(Unit 18):13. doi: 10.1002/0471142735.im1813s76. [DOI] [PubMed] [Google Scholar]
- 25.Nguyen K, Sylvain NR, Bunnell SC. T cell costimulation via the integrin VLA-4 inhibits the actin-dependent centralization of signaling microclusters containing the adaptor SLP-76. Immunity. 2008;28(6):810–821. doi: 10.1016/j.immuni.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 26.Abraham RT, Weiss A. Jurkat T cells and development of the T-cell receptor signalling paradigm. Nat Rev Immunol. 2004;4(4):301–308. doi: 10.1038/nri1330. [DOI] [PubMed] [Google Scholar]
- 27.Bartelt RR, Cruz-Orcutt N, Collins M, Houtman JC. Comparison of T cell receptor-induced proximal signaling and downstream functions in immortalized and primary T cells. PLoS One. 2009;4(5):e5430. doi: 10.1371/journal.pone.0005430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gomez TS, McCarney SD, Carrizosa E, Labno CM, Comiskey EO, Nolz JC, Zhu P, Freedman BD, Clark MR, Rawlings DJ, Billadeau DD, Burkhardt JK. HS1 functions as an essential actin-regulatory adaptor protein at the immune synapse. Immunity. 2006;24(6):741–752. doi: 10.1016/j.immuni.2006.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spracklen AJ, Fagan TN, Lovander KE, Tootle TL. The pros and cons of common actin labeling tools for visualizing actin dynamics during Drosophila oogenesis. Dev Biol. 2014;393(2):209–226. doi: 10.1016/j.ydbio.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Belin BJ, Goins LM, Mullins RD. Comparative analysis of tools for live cell imaging of actin network architecture. Bioarchitecture. 2014;4(6):189–202. doi: 10.1080/19490992.2014.1047714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doyle T, Botstein D. Movement of yeast cortical actin cytoskeleton visualized in vivo. Proc Natl Acad Sci U S A. 1996;93(9):3886–3891. doi: 10.1073/pnas.93.9.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, Sixt M, Wedlich-Soldner R. Lifeact: a versatile marker to visualize F-actin. Nat Methods. 2008;5(7):605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riedl J, Flynn KC, Raducanu A, Gartner F, Beck G, Bosl M, Bradke F, Massberg S, Aszodi A, Sixt M, Wedlich-Soldner R. Lifeact mice for studying F-actin dynamics. Nat Methods. 2010;7(3):168–169. doi: 10.1038/nmeth0310-168. [DOI] [PubMed] [Google Scholar]
- 34.Yi J, Wu XS, Crites T, Hammer JA., 3rd Actin retrograde flow and actomyosin II arc contraction drive receptor cluster dynamics at the immunological synapse in Jurkat T cells. Mol Biol Cell. 2012;23(5):834–852. doi: 10.1091/mbc.E11-08-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson HW, Schell MJ. Neuronal IP3 3-kinase is an F-actin-bundling protein: role in dendritic targeting and regulation of spine morphology. Mol Biol Cell. 2009;20(24) doi: 10.1091/mbc.E09-01-0083E09-01-0083. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schubert DA, Gordo S, Sabatino JJ, Jr, Vardhana S, Gagnon E, Sethi DK, Seth NP, Choudhuri K, Reijonen H, Nepom GT, Evavold BD, Dustin ML, Wucherpfennig KW. Self-reactive human CD4 T cell clones form unusual immunological synapses. J Exp Med. 2012;209(2):335–352. doi: 10.1084/jem.20111485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X, Moore SW, Iskratsch T, Sheetz MP. N-WASP-directed actin polymerization activates Cas phosphorylation and lamellipodium spreading. J Cell Sci. 2014;127(Pt 7):1394–1405. doi: 10.1242/jcs.134692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henson JH, Yeterian M, Weeks RM, Medrano AE, Brown BL, Geist HL, Pais MD, Oldenbourg R, Shuster CB. Arp2/3 complex inhibition radically alters lamellipodial actin architecture, suspended cell shape, and the cell spreading process. Mol Biol Cell. 2015;26(5):887–900. doi: 10.1091/mbc.E14-07-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vitriol EA, McMillen LM, Kapustina M, Gomez SM, Vavylonis D, Zheng JQ. Two functionally distinct sources of actin monomers supply the leading edge of lamellipodia. CellRep. 2015;11(3):433–445. doi: 10.1016/j.celrep.2015.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumari S, Depoil D, Martinelli R, Judokusumo E, Carmona G, Gertler FB, Kam LC, Carman CV, Burkhardt JK, Irvine DJ, Dustin ML. Actin foci facilitate activation of the phospholipase C-gamma in primary T lymphocytes via the WASP pathway. eLife. 2015:4. doi: 10.7554/eLife.04953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wakatsuki T, Schwab B, Thompson NC, Elson EL. Effects of cytochalasin D and latrunculin B on mechanical properties of cells. J Cell Sci. 2001;114(Pt 5):1025–1036. doi: 10.1242/jcs.114.5.1025. [DOI] [PubMed] [Google Scholar]
- 42.Ponsaerts R, D’Hondt C, Bultynck G, Srinivas SP, Vereecke J, Himpens B. The myosin II ATPase inhibitor blebbistatin prevents thrombin-induced inhibition of intercellular calcium wave propagation in corneal endothelial cells. Invest Ophthalmol Vis Sci. 2008;49(11):4816–4827. doi: 10.1167/iovs.07-1533. [DOI] [PubMed] [Google Scholar]