Abstract
The cerebellum is a brain structure involved in motor and cognitive functions. The development of the cerebellar cortex (the external part of the cerebellum) is under the control of numerous factors. Among these factors, neuropeptides including PACAP or somatostatin modulate the survival, migration and/or differentiation of cerebellar granule cells. Interestingly, such peptides contributing to cerebellar ontogenesis usually exhibit a specific transient expression profile with a low abundance at birth, a high expression level during the developmental processes, which take place within the first two postnatal weeks in rodents, and a gradual decline toward adulthood. Thus, to identify new peptides transiently expressed in the cerebellum during development, rat cerebella were sampled from birth to adulthood, and analyzed by a semi-quantitative peptidomic approach. A total of 33 peptides were found to be expressed in the cerebellum. Among these 33 peptides, 8 had a clear differential expression pattern during development, 4 of them i.e. cerebellin 2, nociceptin, somatostatin and VGF [353–372], exhibiting a high expression level during the first two postnatal weeks followed by a significative decrease at adulthood. A focus by a genomic approach on nociceptin, confirmed that its precursor mRNA is transiently expressed during the first week of life in granule neurons within the internal granule cell layer of the cerebellum, and showed that the nociceptin receptor is also actively expressed between P8 and P16 by the same neurons. Finally, functional studies revealed a new role for nociceptin, acting as a neurotrophic peptide able to promote the survival and differentiation of developing cerebellar granule neurons.
Keywords: Cell biology, Developmental biology, Label-free quantification, Mass Spectrometry, Molecular biology, Neurobiology, Peptides, Peptidomics, Quantification, Tandem Mass Spectrometry, Cerebellum, Development, Nociceptin, Peptidome, Rat
During brain development, neurons originating from germinative zones migrate and establish synaptic connections to generate organized structures and functional networks. This developmental process involves numerous factors which modulate cell proliferation, migration, differentiation and survival. Owing to its postnatal development and its well-organized cell layers, the cerebellum appears to be an excellent model in which to characterize factors controlling neuronal histogenesis. In rodents, during the first two postnatal weeks, immature neurons generated in a secondary germinative zone, i.e. the external granule cell layer (EGL)1, migrate along the radial processes of glial cells through the molecular layer (ML) to reach their destination within the internal granule cell layer (IGL; (1)). In addition to correct cell placement, the formation of functional synapses between parallel fibers of cerebellar granule neurons (CGN) and basket, Golgi and Purkinje cells is essential for establishment of proper motor and cognitive functions of the cerebellum (2). By postnatal day 21 (P21), when CGN have completed their migration, the EGL disappears and only three layers remain i.e. from external to internal side, the molecular layer, the Purkinje cell layer (PL) and the granule cell layer (GL).
The setup of GL requires appropriate spatiotemporal expression of key factors timely expressed throughout development by CGN themselves or by neighboring cells. These factors can be proteases, growth factors, chemokines, neurotransmitters and neuropeptides released in the extracellular matrix to control the various steps of CGN development. For example, sonic hedgehog increases cell proliferation (3), brain-derived neurotrophic factor promotes cell survival (4), vascular endothelial growth factor directs cell migration from the EGL toward the PL (5), glutamate increases neuronal migration speed (6), and serotonin regulates differentiation of CGN (7). Among peptides, pituitary adenylate cyclase-activating polypeptide (PACAP) induces a transient stop of CGN at the level of the PL (8), and promotes their survival (9, 10) and their differentiation in the IGL (11). Concurrently, somatostatin increases CGN migration speed in the EGL but decreases their velocity and induces their final stop in the IGL (12). Although considerable progress has been made toward understanding the guidance systems that regulate development of granule neurons, several molecules regulating proliferation, migration, differentiation, and survival remain to be identified.
Interestingly, most mediators controlling granule cell maturation exhibit a transient expression during cerebellar development. For instance, C-X-C motif chemokine ligand 14 (CXCL14) is transiently expressed in the cerebellum between P1 and P8 (13), PACAP expression increases from birth to P14 and then decreases (14), and somatostatin is actively expressed from birth to P21 (15, 16). These observations suggest that factors, notably peptides, displaying an increased expression during the developmental period (i.e. during the first 2 postnatal weeks in rodents) and a subsequent decline of expression toward adulthood are expected to play a key role in cerebellar development. Based on this hypothesis, the aim of the present study was to identify, through omics approaches, peptides exhibiting a transient profile of expression and to subsequently characterize their putative functions.
MATERIALS AND METHODS
Animals
Male and female Wistar rats were born and bred in an accredited animal facility (approved B.76-451-04) in accordance with the French Ministry of Agriculture and the European Community Council Directive 2010/63/UE of September 22.2010 on the protection of animals used for scientific purposes. Tissues were sampled from P2, P4, P8, P16, P21 and P90 animals. For peptidomic experiments only P8, P16, P21 and P90 animals were used whereas for quantitative PCR experiments, all ages were used.
Tissue Sampling for Peptidomic Analysis
Cerebella were quickly removed, placed in saline buffer and cleared of meninges and choroid plexus. Tissues were then rapidly heat-denatured with a Stabilizor™ (Denator, Uppsala, Sweden) to avoid protein and peptide degradation prior to storage at −80 °C. For each stage of development, tissue weight was adjusted by pooling several cerebella to reach the weight of one P90 cerebellum (i.e. 300 mg). For each stage, 10 biological replicates were sampled.
Peptide Extraction
Tissues were sonicated in extraction buffer (Stabilizor Peptide Extraction Kit, Denator) and centrifuged at 14,000 × g, for 30 min at 4 °C. Peptide content in the supernatant was then quantified with the Bradford assay (Bio-Rad, Hercules, CA) and 300 fmol of human serum albumin (HSA; HSA peptide standard mix, Agilent Technologies, Santa Clara, CA) was added as an internal standard in extraction buffer. HSA level was compared between the 4 groups by measuring the area under the curve with Mass Hunter (Agilent Technologies, version B06.00) to assess the quality and reproducibility of the peptide extraction protocol. Samples were filtered with 10 kDa cut-off centrifugal filter units (YM-10 Microcon, Merck Millipore, Darmstadt, Germany) at 14,000 × g for 60 min at 4 °C. The flow through was applied to a solid phase extraction cartridge (Oasis HLB 10 mg, Waters, Milford, MA) and eluted with 60% acetonitrile (ACN)/40% H2O (v/v) and freeze-dried over-night. The peptide extracts were then re-suspended in 300 μl of LC-MS (liquid chromatography-mass spectrometry) solvent (3% ACN/97% H2O(v/v) + 0.1% formic acid (FA)) and concentrated by evaporation to 30 μl.
Mass Spectrometry
Nanoflow liquid chromatography-mass spectrometry (nLC-MS/MS) experiments were performed on an LTQ-Orbitrap Velos (Thermo Fisher Scientific) coupled with a nanoflow liquid chromatography instrument (Easy-nLC II, Thermo Fisher Scientific). A 2 μl volume of each sample was injected onto an enrichment column (C18 PepMap100, Thermo Fisher Scientific). The separation was performed with an analytical column needle (NTCC-360/100–5-153, NikkyoTechnos) in the presence of a mobile phase composed of H2O/0.1% FA (buffer A) and ACN/0.1% FA (buffer B). Elution of peptides was conducted at a flow rate of 300 nL/min, using a two-step linear gradient: from 10 to 40% buffer B over 30 min followed by 10 min at 100% buffer B. The mass spectrometer was operated in positive ionization mode with capillary voltage and source temperature set at 1.5 kV and 275 °C, respectively. The samples were analyzed using collision-induced dissociation method. The first scan (MS spectra) was recorded in the Orbitrap analyzer (r = 60,000) with the mass range m/z 400–1800. Then, the 20 most intense ions were selected for MS2 experiments. Singly charged species were excluded for MS2 experiments. Dynamic exclusion of already fragmented precursor ions was applied for 30 s, with a repeat count of 1, a repeat duration of 30 s and an exclusion mass width of ±10 ppm. Fragmentation occurred in the linear ion trap analyzer with normalized collision energy of 35. All measurements in the Orbitrap analyzer were performed with on-the-fly internal recalibration (lock mass) at m/z 445.12002 (polydimethylcyclosiloxane).
Identification of Peptides Expressed in the Cerebellum
Peptides were identified using SpectrumMill (Agilent Technologies, version B04.00.127) implemented with a specific database containing 184 rat neuropeptides collected from the literature (Rat Neuropeptide Database version released on January 2017; supplemental Table S1). Data were also processed using the SwissProt Rat database. For analysis, the following parameters were used: no enzymatic digestion; precursor mass tolerance: 10 ppm; product mass tolerance: 0.5 Da; variable modifications: cysteine sulfoxide, acetylated lysine, oxidized methionine, deaminated, pyroglutamic acid, phosphorylated S, T, and Y. Only peptides with an identification score above 10 (choose from Agilent Technologies recommendations and from the literature (17, 18)) and found in at least 7 samples out of 10 in one or several developmental stages were retained.
Quantification of Regulated Peptides During Development
To measure peptide changes between the four groups (P8, P16, P21 and P90), Progenesis LC-MS software (Nonlinear dynamics, version 4.0.4441.29989) was used. The P8 group was set as the reference to perform two by two comparisons and one P8 sample was chosen to align the LC-MS runs to account for retention time shifts. A minimum of 90% of alignment was required for further analysis. After alignment, statistical analysis was performed with one-way analysis of variance (ANOVA). To highlight peptides differentially expressed between P8 and other groups, a p value < 0.05, a q-value < 0.05 and a power > 0.8 were required. Peptides with a rank > 10 and a score < 20 were excluded. In addition, only peptides with a fold change > 2 in at least one of the 2-by-2 comparison tests were considered differentially expressed. For the identification, Mascot (Matrix Science, Boston, MA, version 2.2.04) was used with the following parameters: no enzymatic digestion; precursor mass tolerance: 10 ppm; product mass tolerance: 0.5 Da; variable modifications: oxidized methionine and carbamidomethylation. Mascot search results were imported into Progenesis.
Targeted Mass Spectrometry
To verify peptide regulations, nanoflow liquid chromatography-mass spectrometry (nLC-MS/MS) experiments were performed on a Q Exactive mass spectrometer (Thermo Scientific) coupled with an Ultimate 3000 nano RSLC (Thermo Scientific). A 2 μl volume of each sample were injected onto an analytical column (PepMap RSLC C18, 75 μm × 25 cm, Thermo Scientific). The separation was performed in the presence of a mobile phase composed of H2O/0.1% FA (buffer A) and 80% ACN/0.1% FA (buffer B). Elution of peptides was conducted at a flow rate of 300 nL/min, using a linear gradient: from 2 to 50% buffer B over 16 min. The mass spectrometer was operated in Parallel Reaction Monitoring (PRM) mode. The resolution was set at 35,000 (at m/z 200), the Automatic Gain Controltarget at 2 × 105 and an isolation window of 1.6 m/z. An inclusion list was generated using the Skyline software (MacCoss Lab, version 4.1.0.11714) based on analytical data (m/z and retention times) obtained with synthetic peptides in the presence of biological matrix VGF [353–372]: 821.0290 m/z, z = 3 from 11 to 13 min and nociceptin: 603.6602 m/z, z = 3 from 12 to 14 min. For quantification, best MS/MS transitions were selected and signals that did not match the retention time were excluded.
Primary Culture of CGN
Cell cultures were prepared from P7 to P9 rat cerebella, as previously described (19). Briefly, cerebella were removed from skulls and cells were dissociated both enzymatically and mechanically. Cells were plated at a density of 106 cells/ml in costar 24-well culture plates previously coated with poly-d-lysine (12.5 μg/ml, BD Bioscience, Boston, MA). Culture medium consisted of 74% Dubecco's modified Eagle's medium (DMEM), 24% Ham's F-12, 1% antibiotic-antimicotic (100 X, Gibco Life Technologies, Grand Island, NY), 1% N2-supplement (Invitrogen) and 25 mm KCl. Mature neurons used for quantitative PCR experiments were plated in costar 6-well culture plates in the presence of culture medium complemented with fetal bovine serum (10%; Pan Biotech, Wimborne, UK) and treated with cytosine arabinoside (10 μm; Sigma-Aldrich, St. Louis, MO). CGN RNA from 3 wells were harvested every day for 5 days in Tri-Reagent (Sigma-Aldrich). To obtain astrocytes, after 4 days of culture in the presence of fetal bovine serum (10%) without cytosine arabinoside, CGN were killed with hydrogen peroxide (H2O2; 30 μm; Sigma-Aldrich) and 5 days later, proliferating astrocyte RNA was harvested with Tri-Reagent (Sigma-Aldrich).
Cell Treatments
Cells were treated with graded concentrations of nociceptin (10−7 to 10−12 m; ab38198, Abcam, Cambridge, MA). In some experiments H2O2 (10−5 m) was added to promote cell death before treatment and PACAP (10−7 m) was used as a positive control for its stimulatory effect on cell survival (20) and inhibitory action on cell motility (21). PACAP was synthesized by Pr Alain Fournier (INRS-Institut Armand-Frappier, 531 boul. des Prairies, Laval, QC, H7V 1B7, Canada) using the fluorenylmethyloxycarbonyl (Fmoc) chemistry methodology as previously described (22).
Cell Survival Measurement
CGN survival was measured with fluorescein diacetate (FDA) as previously described (23). Culture medium was removed, and cells were incubated with FDA (15 μg/ml) for 8 min at 37 °C in the dark. Cells were then rinsed with PBS and lyzed with a 10-mm Tris-HCl solution containing 1% sodium dodecyl sulfate. To measure cell survival, fluorescence signal was measured at 540 nm with an Infinite 200 microplate reader (Tecan, Hombrechtikon, Germany).
Caspase Activity Measurement
Caspase-3/7 activity in cultured CGN was evaluated with the Caspase-Glo® 3/7 Assay (Promega). Three hours after treatment, cells were rinsed with DMEM, resuspended in DMEM (100 μl) and incubated with 2 X caspase assay buffer containing Z-DEVD-aminoluciferin substrate. To measure enzymatic activity, bioluminescence signal was measured over 1.5 h at 37 °C with an Infinite 200 microplate reader (Tecan).
Cell Motility Measurement
CGN motility was measured using the microexplant technique as previously described (24). Briefly, P7 or P8 rat cerebella were cut into 50 to 100 μm pieces and placed in Costar 24-well culture plates double coated with poly-d-lysine (12.5 μg/ml, BD Bioscience) and laminin (20 μg/ml, Sigma-Aldrich). After plating, the explants were covered with culture medium composed of minimum essential medium (Invitrogen) complemented with 1% N2-supplement and 1% antibiotic-antimicotic (100X, Gibco Life Technologies). After 24 h in a cell culture incubator, dishes were transferred into an incubation chamber (37 °C, 5% CO2 and 95% O2) fixed to the stand of an inverted microscope (Leica DMI 6000B, Rueil Malmaison, France). Images of migrating CGN were acquired with the LAS-AF software every 2 min for 45 min in control conditions and then for another 45 min in treated conditions. Analyses were performed with the MetaMorph software (Roper Scientific).
Cell Differentiation Measurement
Cell differentiation was evaluated by counting and measuring CGN neurites. CGN plated at a density of 3.105 cells/ml were labeled with the tubulin tracker green kit (300 nm, Thermo Fischer Scientific) into an incubation chamber (37 °C, 5% CO2 and 95% O2) for 30 min and 5 images per well were randomly acquired with an inverted microscope (Leica DMI 6000B). Analyses were performed by measuring for each cell, neurite lengths with the ImageJ software (version 1.46).
RNA Extraction
Cell RNA or total cerebella RNA from the six groups (P2, P4, P8, P16, P21, and P90) were extracted using Tri-Reagent (Sigma-Aldrich) and further purified with NucleoSpin RNA kit (Macherey Nagel, Duren, Germany). The concentration of RNA was measured with a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientfic) at an absorbance of 260 nm and 1 μg of total RNA from each sample was reverse transcribed with the ImProm II Reverse Transcription System (Promega).
Microdissection
Laser microdissection experiments were conducted as previously described (25). Briefly, cerebella from P8 and P21 rats were cut in the frontal-plane into 14 μm-thick sections. After cresyl violet coloration, sections were dehydrated in ethanol. For P8 cerebella, the 4 layers were sampled (i.e. IGL, PL, ML, and EGL), for P21 cerebella, the 3 remaining layers were cut (i.e. GL, PL and ML) with a laser microdissector (LMD 7000, Leica). For each cerebellar layer and each age, 0.4 mm2 of tissue was collected. RNA was extracted with the NucleoSpin RNA XS kit (Macherey Nagel), and then reverse transcribed and amplified with the SMARTer Pico PCR cDNA kit (Clontech, Kusatsu, Japan). To ensure that tissue from each cerebellar layer had been properly collected without any cross contamination, control real-time PCR experiments were performed with 8 different sets of primers, which amplify 2 genes specifically expressed in each of the 4 cerebellar cortex layers (Table I). These genes were chosen from the literature, the Brain Transcriptome Database (http://www.cdtdb.neuroinf.jp/CDT/Top.jsp) or the Allen Brain Atlas (http://mouse.brain-map.org/). Stathmin 1 (Stmn1; Allen Brain Atlas, experiment 68632984) and atonal bHLH transcription factor 1 (Atoh1; Brain Transcriptome Database, CD03233) genes were selected as markers of the EGL. Calcium-dependent secretion activator 2 (Cadps2; 23) and tenascin R (Tnr; Brain Transcriptome Database CD01342) genes were selected as markers of the ML. Cholecystokinin (CCK; 24) and SDF-1 receptor (CXCR4; 25) genes were selected as markers of PL. Homer scaffolding protein 1 (homer1; Brain Transcriptome Database CD03205) and neurogenic differentiation 1 (Neurod1; Allen Brain Atlas experiment 75650865) genes were selected as makers of the IGL at P8 and GL at P21.
Table I. Sequences of primers used for quantitative PCR experiments.
EGL: external granule cell layer. IGL: internal granule cell layer. GL: granule cell layer. ML: molecular layer. PL: Purkinje cell layer. Atoh1: atonal bHLH transcription factor 1. Cadps2: calcium-dependent secretion activator 2. CCK: cholecystokinin. CXCR4: SDF-1 receptor. GAPDH: glyceraldehyde-3-phosphate dehydrogenase, internal standard. Homer1: homer scaffolding protein 1. Neurod1: neurogenic differentiation 1. Oprl1: nociceptin receptor. Pnoc: nociceptin. Stmn1: stathmin 1. Tnr: tenascin R
| Cell layer | Gene name | Primer sequence |
|---|---|---|
| GAPDH | Reverse CAATGTCCACTTTGTCACAAGAGAA | |
| Forward CAGCCTCGTCTCATAGACAAGATG | ||
| Pnoc | Reverse GCGGACGTGAGCTCTGGAT | |
| Forward GCTCCGGGCAGCTTCAA | ||
| Oprl1 | Reverse CAATGTCCACTTTGTCACAAGAGAA | |
| Forward CAGCCTCGTCTCATAGACAAGATG | ||
| EGL | Stmn1 | Reverse CTTTCACCTGAATATCAGAAGATGC |
| Forward CTTTCCTTGCCAGTGGATTG | ||
| Atoh1 | Reverse CCGACAGAGCGTTGATGTAGATC | |
| Forward AACAACGACAAGAAGCTGTCCAA | ||
| ML | Cadps2 | Reverse CAGGTACCCACTATGCTTCATGTG |
| Forward AGGACTCGGACCTAAAGATCAAATT | ||
| Tnr | Reverse ACACCAATGAGCATGTTCTTCAA | |
| Forward AGGACGACATCACGAGATTCCTA | ||
| PL | CCK | Reverse CCAGGCTCTGCAGGTTCTTAA |
| Forward GCACTGCTAGCGCGATACATC | ||
| CXCR4 | Reverse GATGGTGGGCAGGAAGATCCTA | |
| Forward CATGGAACCGATCAGTGTGAGT | ||
| IGL/GL | Homer1 | Reverse GGGGTGATGGTGCTGTTTAT |
| Forward GGAACAGCTATCGGATCATCA | ||
| Neurod1 | Reverse GATGGTTCGTGTTTGAAAGAGAAGT | |
| Forward AAAGCCCCCTAACTGATTGCA | ||
As expected, Atoh1 and Stmn1 were significantly over expressed (+83.2% ± 9.6% and +84.7% ± 8.1%, respectively) in the EGL when compared with all other layers (supplemental Fig. S1); Cadps2 and Tnr were significantly over expressed in the ML at P8 (+255.5% ± 26.2% and +459.9% ± 36.8%, respectively), when compared with the EGL at P8 (supplemental Fig. S2) and in the ML at P21 (+294% ± 17.6% and +535.6% ± 35.9%, respectively), when compared with the EGL at P8 (supplemental Fig. S2); CCK and CXCR4 were significantly over expressed in the PL at P8 (+701.9% ± 48.6% and +322.1% ± 29.9%, respectively), when compared with the EGL at P8 (supplemental Fig. S3) and in the PL at P21 (+754.4% ± 53.2% and +258.7% ± 18.7%, respectively), when compared with the EGL at P8 (supplemental Fig. S3); Homer1 and Neurod1 were significantly over expressed in the IGL at P8 (+522.6% ± 43.6% and +329.8% ± 22.5%, respectively), when compared with the EGL at P8 (supplemental Fig. S4) and in the GL at P21 (+377.1% ± 30.2% and +459.1% ± 34.3%, respectively), when compared with the EGL at P8 (supplemental Fig. S4).
Quantitative PCR
Two ng complementary DNA (cDNA) was amplified in the presence of 2 X SYBR Green Mastermix (Applied Biosystems) containing preset concentrations of dNTPs, MgCl2 as well as forward and reverse primers, with a QuantStudio™ 12KFlex (Applied Biosystems™). Proper controls without reverse transcriptase or with H2O instead of cDNA were conducted. Primers targeting nociceptin (Pnoc) and its receptor (Oprl1) were designed using the Primer Express software (Applied Biosystems) and validated by real-time PCR (Table I). The relative amount of cDNA in each sample was determined using the comparative quantification cycle (Cq) method and expressed as 2−ΔΔCq using glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which was constant whatever the stage of cerebellar cortex development, as an internal standard for variations in amounts of input mRNA.
Experimental Design and Statistical Rationale
Peptide differential expression was investigated at 4 developmental stages (from P8 to P90). At each stage of the peptidomic analysis, 10 biological replicates were analyzed except for the targeted MS/MS where only 3 biological replicates were analyzed at P8 and P21. Expression levels of nociceptin and its receptor were measured at 6 different stages of development (from P2 to P90). For each stage, 4 biological replicates were performed with 3 technical replicates per biological sample (i.e. a total of 12 samples per group were analyzed). For laser microdissection experiments, the same experimental design was used with 4 biological replicates and 3 technical replicates (i.e. a total of 12 samples per condition) to analyze the 4 cell layers at 2 different developmental stages. To examine gene expression levels of nociceptin and its receptor in CGN and cultured astrocytes, 4 biological replicates and 3 technical replicates (i.e. a total of 12 samples per day of culture) were analyzed. To study the effect of nociceptin on cultured granule neurons, 4 parameters were analyzed, i.e. cellular caspase activity, survival, motility and differentiation. For cell survival quantification, 10 independent experiments were performed, each one being the mean of 4 technical replicates. Concerning caspase activity measurement, 8 independent experiments were conducted with 4 technical replicates each. Regarding cell motility, for each condition, 12 microexplants were examined and for each of them, at least 24 cells were averaged (i.e. over 288 cells analyzed per condition). Finally, for neuronal differentiation investigations, 5 images were randomly taken from 5 independent experiments for each condition. Results from each image were the average measurement from at least 28 cells, (i.e. over 700 cells were measured per condition). Statistical analysis was performed with GraphPad Prism® (version 6.04). Normality was tested with a Shapiro-Wilk test; when the data were parametric an ANOVA was used, when not a Kruskal-Wallis was used instead. Statistically significant differences were estimated by ANOVA followed by Bonferroni's tests for measurement of cell survival, motility and differentiation, and with Kruskal-Wallis followed by Dunn's tests for differentially expressed peptides, qPCR and caspase experiments and with a Mann-Whitney test for the targeted MS/MS. The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD006874.
RESULTS
Identification and Quantification of Peptides Within the Rat Cerebellum
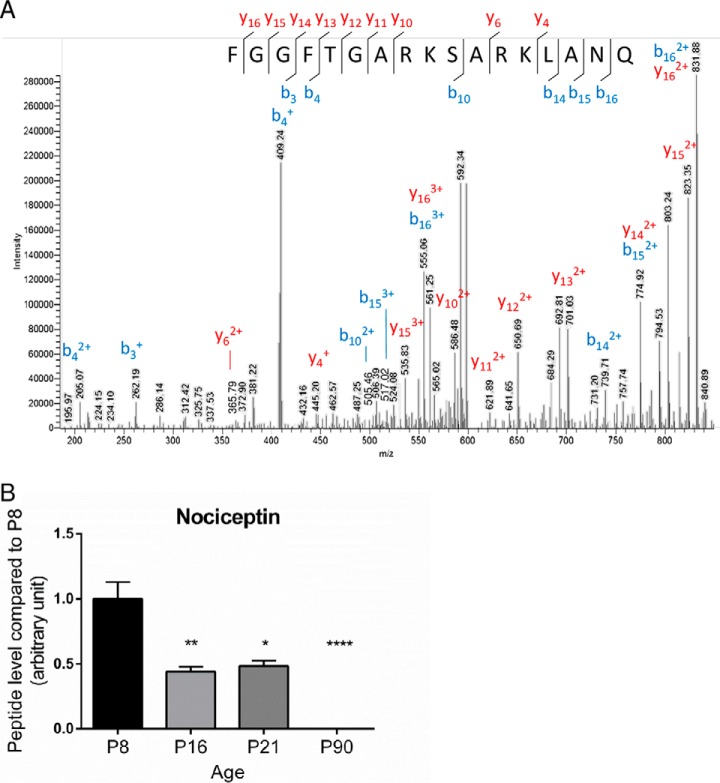
The peptides expressed in the cerebellum during development were characterized by mass spectrometry and identified by means of a database containing 184 rat neuropeptides or neuropeptide precursors (supplemental Table S1). To confirm the reproducibility of the extraction protocol and mass spectrometry analysis, HSA level in the four groups was measured as an external standard and no significant difference was found (supplemental Fig. S5). Analysis with SpectrumMill and Mascot software packages revealed 33 peptides expressed in at least 7 out of 10 samples, at one or several stages of development (supplemental Table S2). Quantification of peptide relative expression level with Progenesis software showed that 8 peptides were differentially expressed between 2 or more developmental stages (Table II). Among these 8 peptides, 4 had an expression level which increased gradually after the second week of life (Fig. 1A; p < 0.05) whereas the 4 others, i.e. nociceptin, VGF [353–372], cerebellin-2, and somatostatin exhibited a high expression level at birth which subsequently significantly declined until adulthood (Fig. 1B; p < 0.05). As illustrated with nociceptin (Fig. 2A), all mass spectra were manually verified. Nociceptin and VGF [353–372] levels of expression were confirmed by targeted MS/MS (supplemental Fig. S7). The data revealed that nociceptin expression was markedly reduced between P8 and P16 (Fig. 2B), remained stable between P16 and P21, and became undetectable at P90. Among the four peptides with this remarkable expression profile, somatostatin and cerebellin-2 distribution have already been reported in the cerebellum and VGF [353–372] is a putative new neuropeptide. Nociceptin has known functions in other brain structures, but has never been studied in the cerebellum, therefore it was selected as a putative new neurotrophic factor and considered for subsequent functional studies.
Table II. Proteomics study of the rat cerebellar peptidome with the Progenesis software. Eight peptides were found differentially expressed from birth to adulthood. Fold changes were calculated compared to P8. Score and ANOVA were calculated with Progenesis. *p < 0.05, **p < 0.01, ***p < 0.001. ND: not detected; P: postnatal day; VGF: non-acronymic; WE-14: first and last amino acid of a 14 amino acids long peptide.
| Cerebellin 1 | Octadeca-neuropeptide | Secretogranin 1 [304–321] | WE-14 | Cerebellin 2 | Nociceptin | Somatostatin 28 | VGF [353–372] | ||
|---|---|---|---|---|---|---|---|---|---|
| Sequence | SGSAKVAF | QATVGDVNT | HAAGESKDAN | WSRMDQLA | SGSAKVAF | FGGFTGARK | SANSNPAM | GLQETQQERE | |
| SAIRSTNH | DRPGLLDLK | VATANLGE | KELTAE | SATRSTNH | SARKLANQ | APRE | NEREEEAEQE | ||
| m/z | 1631.83797 | 1583.81551 | 1624.78315 | 1747.85633 | 1619.80159 | 1807.98055 | 1243.56154 | 2460.06369 | |
| P16 | Fold change | 1.2 | 1.5 | 1.9 | −1.4 | −2.1 | −2.3 | −1.3 | −2.5 |
| Score | 54 | 45 | 46 | 26 | 46 | 40 | 39 | 45 | |
| ANOVA | ns | * | * | * | ** | *** | * | ** | |
| P21 | Fold change | 8.2 | 2.3 | 2.8 | 4.6 | −2.8 | −2.1 | −2.8 | −2.6 |
| Score | 53 | 59 | 53 | 25 | 55 | 38 | 65 | 35 | |
| ANOVA | *** | ** | *** | *** | ** | *** | *** | *** | |
| P90 | Fold change | 14.2 | 4.5 | 4.9 | 5.8 | −3.4 | ND | −5 | −2.8 |
| Score | 59 | 54 | 69 | 43 | 49 | 60 | 46 | ||
| ANOVA | *** | *** | *** | *** | *** | *** | *** |
Fig. 1.
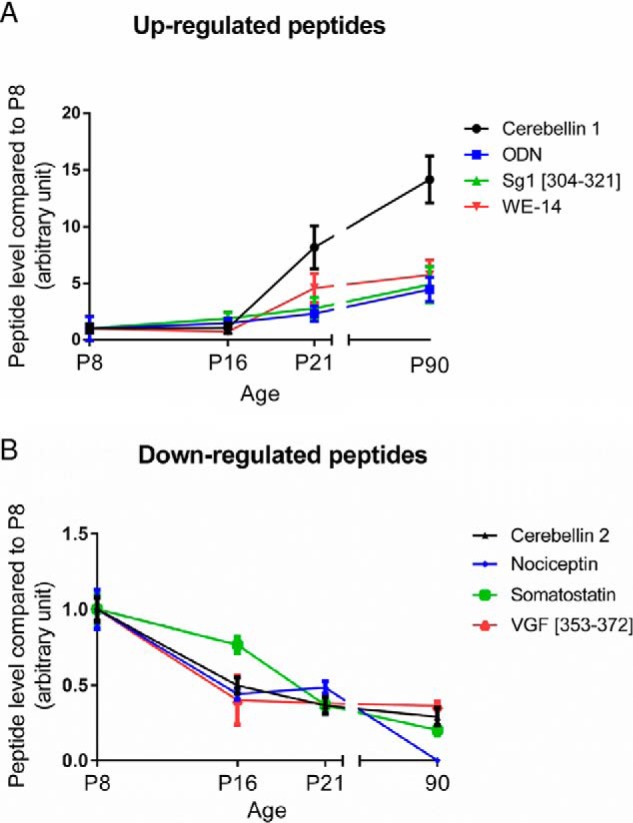
Identification of 8 neuropeptides differentially expressed during rat cerebellar development. nLC-MS/MS runs were analyzed with Progenesis to quantify their relative expression. A, Up-regulated peptides and (B) Down-regulated peptides from P8 (postnatal day 8) to P90. Each value is the mean (± S.E.) of 10 samples. ODN: octadecaneuropeptide; Sg1: secretogranin 1; SST: somatostatin; VGF: non-acronymic; WE-14: first and last amino acid of a 14-amino acids long peptide.
Fig. 2.
Identification spectrum and quantification of nociceptin during rat cerebellar development. A, Mass spectrometry sequence determination of nociceptin after collision-induced dissociation of the 1807.98055 m/z z = 1 precursor fragment ion, exportation from Excalibur and manual annotation. B, Relative expression level of nociceptin during development between P8 (postnatal day 8) and P90. Each value is the mean (± S.E.) of 10 samples. Statistical analysis was performed with the Kruskal-Wallis test followed by Dunn's test: *p < 0.05, **p < 0.01, ****p < 0.0001 versus P8.
Expression of the mRNA of Nociceptin and Its Receptor in the Developing Rat Cerebellum
The evolution of the relative expression levels of nociceptin and its receptor (NOP: Nociceptin/Orphanin FQ receptor) in the developing rat cerebellum were investigated by real time PCR (Fig. 3). When compared with P2, the nociceptin mRNA expression level was stable until P8 but then dropped by P16 (Fig. 3A) and remained at a low level up to adulthood. The mRNA encoding NOP (Fig. 3B) was detected at a low level from P2 to P4 but then increased tremendously at P8 and remained high until P16. Nevertheless, this was only a transient increase as, by P21, NOP mRNA level subsequently decreased compared with P16 and remained low at adulthood.
Fig. 3.
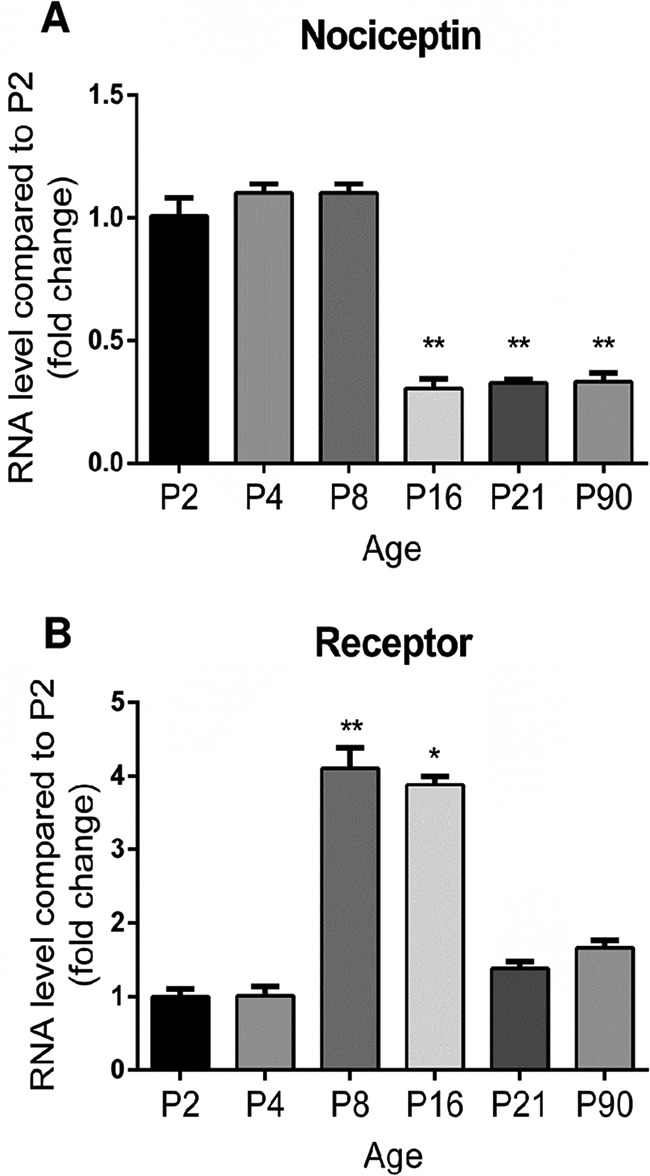
Transitory expression of nociceptin and its receptor mRNA during rat cerebellar development. The evolution of the relative expression level of (A) nociceptin and (B) its receptor in the cerebellum was measured by real-time PCR between P2 (postnatal day 2) and P90. Each value is the mean (± S.E.) of 6 cerebella. Statistical analysis was performed with the Kruskal-Wallis test followed by Dunn's tests: *p < 0.05, **p < 0.01 versus P2.
Laser microdissection of the different cerebellar cortex layers revealed that, at P8 (Fig. 4), nociceptin mRNA was mainly expressed in the IGL and appeared significantly less abundant in the EGL, ML, and PL. In the same experiments, NOP mRNA was only expressed at P8 in the IGL/GL with no detectable expression in the other layers (data not shown). Because the IGL is mainly composed of CGN and astrocytes, real time PCR was performed on these two types of cells in primary cultures to measure gene expression of the nociceptin and its receptor. Cultured CGN exhibited strong expression of nociceptin gene with a gradual decrease until culture day 5 (supplemental Fig. S6). Concurrently, NOP gene expression increased at culture day 1 (supplemental Fig. S6) and then decreased until day 5 (supplemental Fig. S6). On the contrary, in cultured astrocytes only low levels of expression of both genes were observed when compared with CGN (19.6% ± 1.9% for nociceptin mRNA and 1.5% ± 0.1% for NOP mRNA, data not shown).
Fig. 4.
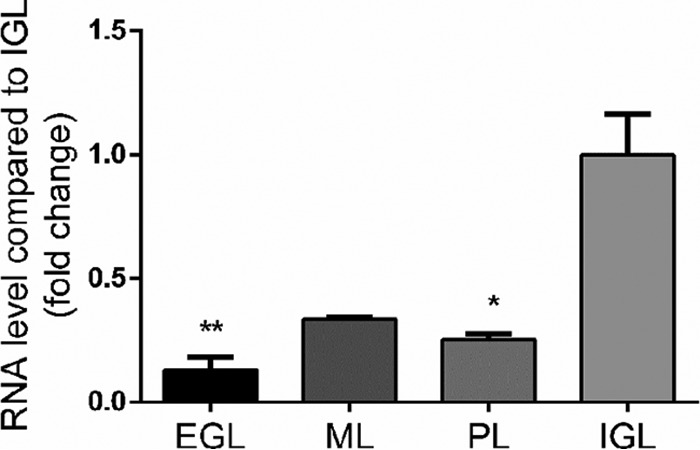
Nociceptin mRNA is mainly expressed in the internal granule cell layer during early phases of development. Gene expression of nociceptin was measured by real-time PCR at P8 (postnatal day 8). Each value is the mean (± S.E.) of 4 cerebella. Statistical analysis was performed with the Kruskal-Wallis test followed by Dunn's test: *p < 0.05, **p < 0.01 versus IGL. EGL: external granule cell layer; ML: molecular layer; PL: Purkinje cell layer; IGL: internal granule cell layer.
Effect of Nociceptin on Cultured CGN
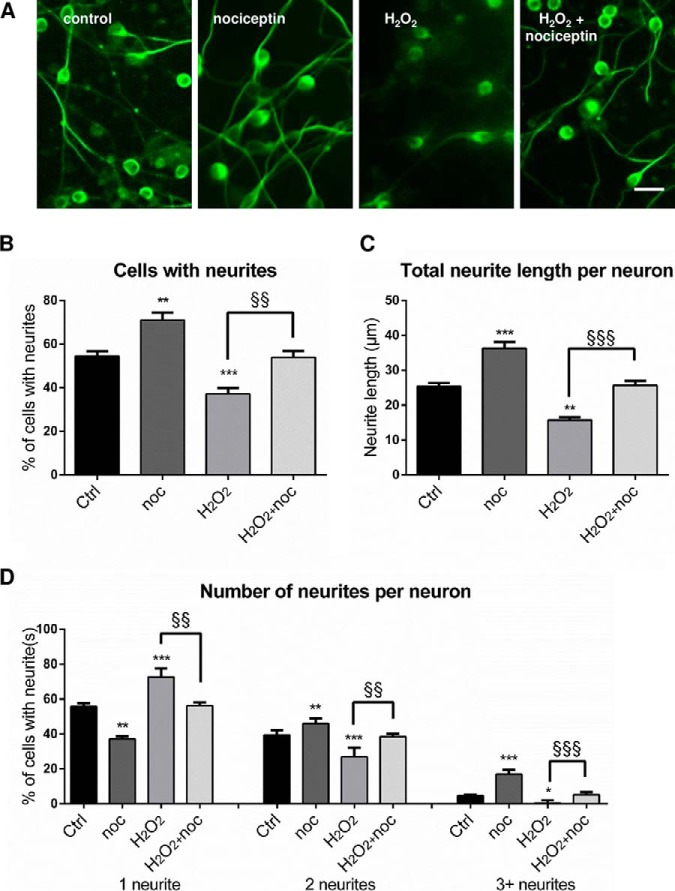
To determine whether nociceptin exerts neuroprotective activities during development, cultured CGN were treated with nociceptin 3 h after plating in the presence or absence of H2O2 (Fig. 5). After 24 h of culture, in control conditions, granule cells were alive and evenly distributed (Fig. 5A). The presence of nociceptin alone did not modify the aspect of the culture. Exposure to H2O2 (10 μm) decreased the number of living CGN. This deleterious effect of H2O2 was prevented by cotreatment of the cells with nociceptin (Fig. 5A). Measurements of cell survival and caspase activity confirmed these observations. H2O2 exposure induced a 25% reduction in the number of living cells (Fig. 5B) and a 49% increase in caspase-3/7 activity (Fig. 5C). Coincubation of CGN with H2O2 and graded concentrations of nociceptin (from 10−12 to 10−7 m; Fig. 5B) resulted in a dose-dependent increase in the number of surviving neurons with a half-maximum effect at a dose of 10 nm. Maximum prevention of cell death was observed at a concentration of 10−8 m nociceptin. Furthermore, if nociceptin had no effect per se on the basal level of caspase activity, 10−8 m nociceptin blocked caspase-3/7 activation induced by a 3-h exposure of CGN to H202 (Fig. 5C). Similar results were observed when cells were treated with nociceptin and/or H2O2, 24 h after plating (data not shown).
Fig. 5.
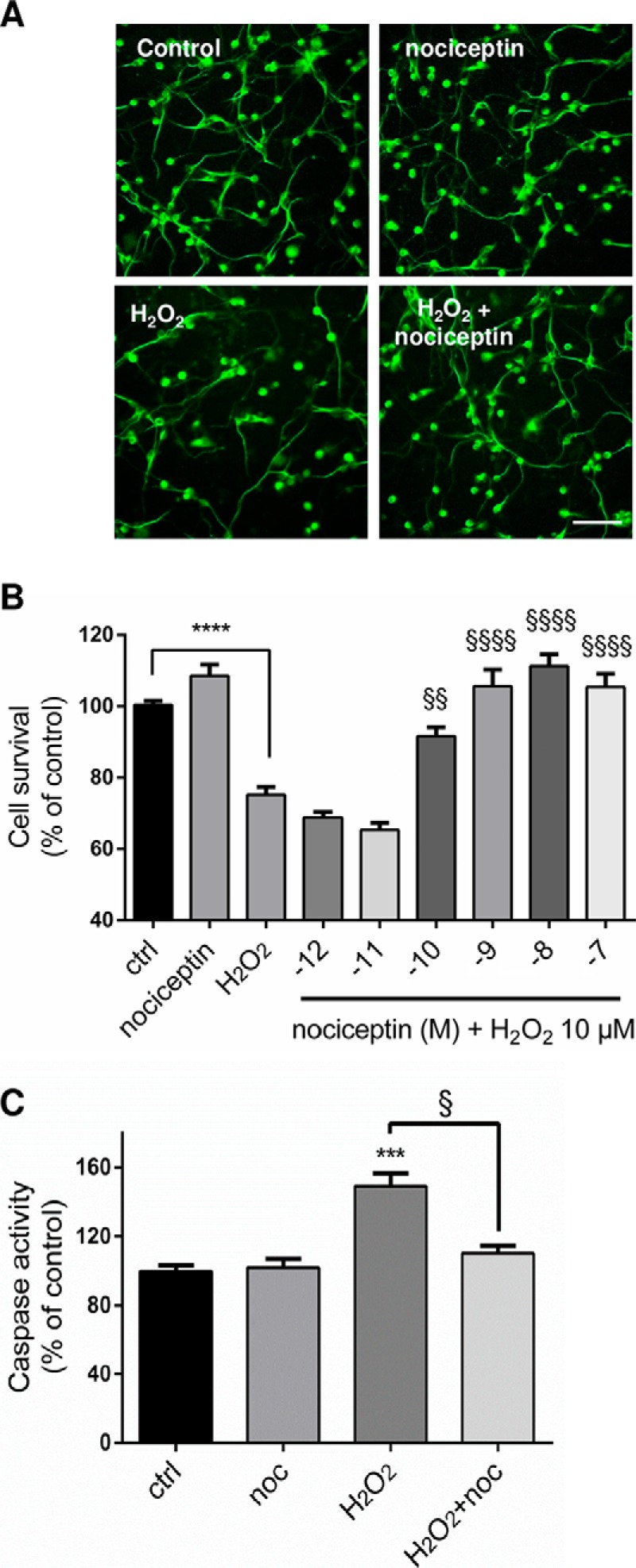
Nociceptin protects granule neurons from hydrogen peroxide-induced apoptosis. A, Typical images illustrating the protective effect of nociceptin against hydrogen peroxide (H2O2)-induced cerebellar granule cell death. Scale bar = 65 μm. Cells were treated for 24 h with 10−5 m H2O2 and in the presence or absence of 10−8 m nociceptin and labeled with tubulin tracker green. B, Effect of graded concentrations of nociceptin (noc; 10−12 to 10−7 m) on survival of cultured cerebellar granule cells exposed to 10−5 m of H2O2 after 24 h of treatment. Cell survival was quantified by measuring fluorescein diacetate intensity. Results are expressed as percentages of control. Each value is the mean (± S.E.) of 4 wells from at least 10 independent cultures. For the nociceptin alone condition, cells were treated with 10−8 m nociceptin. Each value is the mean (± S.E.) of 4 wells from at least 8 independent cultures. Statistical analysis was performed with ANOVA followed by Bonferroni's test: ****p < 0.001 versus control; §§p < 0.01, §§§§p < 0.0001 versus H2O2. (C) Caspase activity of cells treated with 10−5 m H2O2 in the presence or absence of 10−8 m nociceptin. Statistical analysis was performed with the Kruskal-Wallis test followed by Dunn's test: ***p < 0.001 versus control, §p < 0.05 versus H2O2.
To investigate the possible effect of nociceptin on granule neuron migration during development, cell motility was measured. Microexplants from P8 cerebella were cultured for 24 h and then treated with nociceptin. In control conditions, granule neurons moved regularly from their origin toward the outside of the microexplant during the whole recording period (Fig. 6A) at an average speed of 344 μm/h (Fig. 6B). Addition of nociceptin (10−8 m) after 45 min of recording did not affect CGN motility (Fig. 6B) while, as expected, addition of PACAP (10−7 m) as a positive control decreased CGN motility (Fig. 6B).
Fig. 6.
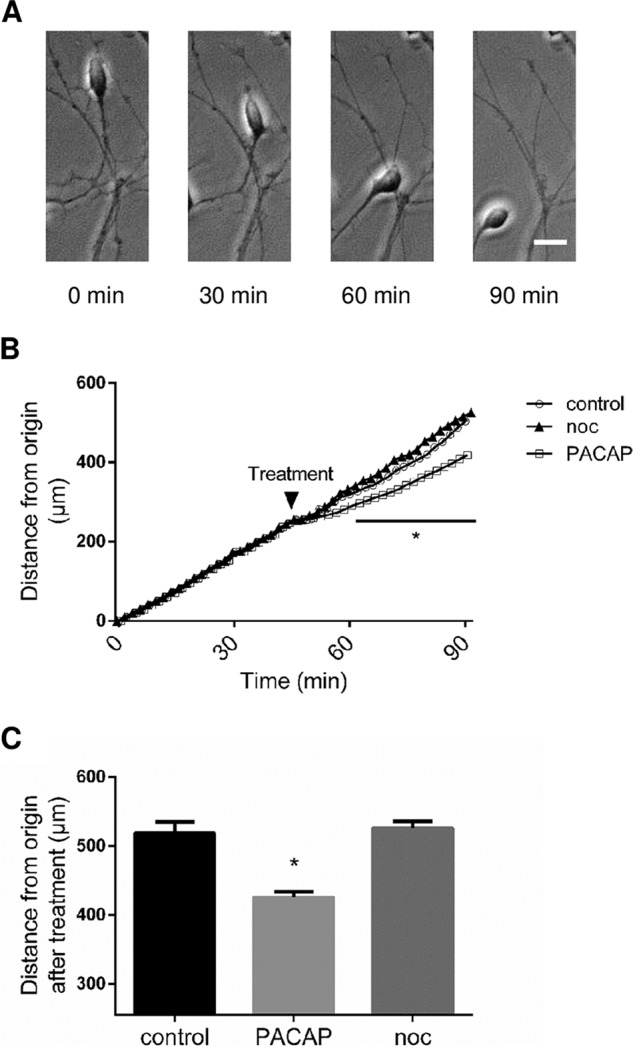
Nociceptin has no effect on cerebellar granule neuron motility. A, Typical images illustrating the migration of a granule neuron. Scale bar = 10 μm. B, Measurement of the distance from the origin. Cell movements were recorded for 45 min in control conditions prior to treatment with 10−8 m nociceptin (noc), 10−7 m PACAP or culture medium alone, and recorded for another 45 min. Each value is the mean (±S.E.) of 12 independent experiments. Statistical analysis was performed with ANOVA followed by Bonferroni's test: *p < 0.05 versus control.
To examine the possible effect of nociceptin on CGN differentiation, cultured cells were treated for 24 h with nociceptin in the presence or absence of H2O2 (Fig. 7). Addition of nociceptin increased the number and length of neurites per neuron and co-incubation of nociceptin with H2O2 prevented the deleterious effect of hydrogen peroxide on CGN differentiation (Fig. 7A). Quantification of neuritogenesis confirmed that nociceptin alone (10−8 m) caused an increase in the percentage of cells with neurites (Fig. 7B). Conversely, H2O2 (10 μm) exposure induced a marked reduction of the number of cells with neurites (Fig. 7B) and this effect was abolished by co-treatment with nociceptin (Fig. 7B). Nociceptin also increased the total neurite length per neuron (Fig. 7C) and reversed neurite shrinkage caused by H2O2. Finally, nociceptin by itself enhanced the number of neurites per neuron (Fig. 7D). Nociceptin also reversed the decrease of neurite per neuron provoked by H2O2 (Fig. 7D).
Fig. 7.
Nociceptin increases cerebellar granule neuron differentiation. Cells were treated for 24 h with 10−5 m hydrogen peroxide (H2O2) in the presence or absence of 10−8 m nociceptin (noc) and labeled with tubulin tracker green for microscopy imaging. A, Typical images illustrating the effect of nociceptin on cell differentiation. Scale bar = 20 μm. B, Percentage of granule neurons with neurites. C, Total neurite length per neuron. D, Number of neurites per neuron. Each value is the mean (± S.E.) of at least 5 images per well from 5 different experiments. Statistical analysis was performed with ANOVA followed by Bonferroni's test: *p < 0.05, **p < 0.01, ***p < 0.001 versus respective control; §§p < 0.01, §§§p < 0.001 versus H2O2.
DISCUSSION
The ontogeny of the brain requires several complementary processes including cell proliferation, elimination, migration and differentiation, to establish a proper architecture. In the postnatal developing cerebellar cortex, all these processes are under the signaling of various molecules, including growth factors, chemokines, neurotransmitters and/or neuropeptides, which are expressed in a specific spatial and temporal-dependent manner. Neuropeptides controlling cerebellar ontogenesis exhibit a high expression level during developmental stages and a lower expression level later at adulthood. In the present study we have searched for neuropeptides exhibiting such a specific expression pattern and we have investigated their possible implication in CGN development.
Rat Cerebellar Peptidome
Expression of neuropeptides during cerebellar development was studied by a peptidomic approach. Tissues were heat-denatured immediately after sampling to prevent degradation by proteases, and peptides were purified by filtering the tissue extract with a 10-kDa cutoff filter to eliminate heavy proteins. Analysis of the MS/MS data from all the samples with the SwissProt Rat database identified less than 100 proteins with only half of them reproducibly found in the cerebellum, showing the robustness of the peptide extraction procedure used. These proteins represent less than 1% of the entries of the database, indicating that the extraction protocol excluded virtually all proteins from the samples, and retained almost only peptides known to be expressed at low level compared with highly expressed proteins such as tubulin. Indeed, the present extraction protocol was chosen after optimization and comparison of several operating procedures. Nevertheless, the fact that a few proteins were also retained suggests that, despite all experimental precautions, some minor degradation still occurred in the time between tissue sampling and mass spectrometry analysis. It would thus be of interest to test additional extraction methods in which for instance tissues are immediately frozen in 2-methyl-butane (29) or peptides are directly extracted upon sampling (30). One of these approaches may reduce further sample contamination by protein fragments and improve the analysis of peptides present in the tissues. However, the protein fragments identified may also arise from the physiological turnover induced by enzymatic proteolysis through ubiquitin or proteasome pathways (31).
Peptide identification was conducted by means of a rat database which includes 184 sequences of known active peptides and peptide precursors. A size limit was set to 100 amino acids for peptides to be included in our database. Sequences encompassing over 100 amino acids, such as brain-derived neurotrophic factor or neurotrophin-3 (both 119 amino acid long) were excluded even though nLC-MS analysis revealed their presence in the cerebellum, in agreement with previous studies (32). From the 184 sequences present in the database, 70 peptides were identified in at least 1 of the 10 samples, at one or several stages. Nevertheless, to ascertain a reproducible presence, only 33 peptides found to be expressed in at least 7 out of 10 samples at one or several stages of development, were considered for further analysis. These 33 peptides represent almost 18% of all the entries of the database, indicating that a large diversity of peptides is expressed in the cerebellum. Furthermore, for all these identified neuropeptides, the MS/MS coverage rate was over 89%, showing the robustness of the identification approach.
Among these 33 peptides, 14 are already known peptides whereas 19 others represent putative new neuropeptides which have not yet been described but arise from established peptide precursors (i.e. chromogranin A, secretogranin 1, 2 and 3, and the neurosecretory protein VGF). All these putative new peptides share remarkable characteristics including: short sequences of less than 20 amino acids, presence of basic amino acid doublets at each extremity of the sequence as possible prohormone convertase cleavage sites, and localization in regions of their precursors already rich of known peptides (i.e. C-terminal or N-terminal part of the precursors which express WE-14, pancreastatin and catestatin for chromogranin and NERP-1, TLQP-21 for VGF). All these properties and the fact that the sequences were reproducibly detected, supports the hypothesis that they correspond to novel peptides (33) rather than to fragments of degradation, although further studies will be needed to identify their putative functions. Even though some peptides, such as cerebellin-1 and 2, or octadecaneuropeptide were already known to be expressed in the cerebellum (34), several others such as CNP-53, islet amyloid polypeptide, neuroendocrine regulatory peptide 1 and neuropeptide K were not, so that, their functions has never been investigated in this structure. It should be noted however that all the neuropeptides known to be expressed in the cerebellum were not detected. For instance, PACAP whose expression profile has already been extensively described in the literature (14) was not identified. As a matter of fact PACAP is only expressed by Purkinje cells in the developing cerebellum (35) and thus its abundance in the whole cerebellum remains very low. As whole cerebella were analyzed, despite our enrichment procedure, the samples remained very complex yielding to the collection of a huge number of peptide sequences (around 1400 per stage) and, consequently, low abundant peptides may have been hidden by other molecular species and not selected for fragmentation. This hypothesis is supported by the fact that although PACAP was not detected, the PACAP-related peptide, which originates from the same precursor, was found (36). It should also be pointed out that, as the analysis was conducted using a specific database containing only known neuropeptides and peptide precursors, we were unable to identify peptides coming from unknown precursors or from precursors not included in the database.
Temporal Expression of Neuropeptides in the Rat Cerebellum During Development
The present study not only provides the first peptidome of the cerebellum but also investigates the relative expression level of the peptides identified during the developmental period. Among the 8 neuropeptides found to be differentially expressed during development, somatostatin which controls CGN migration (16, 15) and octadecaneuropeptide which promotes CGN survival (37), exhibited expression profiles that were consistent with previous reports from the literature, an observation which strengthens our results regarding the expression patterns of other peptides not yet described. Besides these already known peptides, a 20-amino acid long peptide derived from the neurosecretory protein VGF was found to be highly expressed from P8 to P16. Although this particular peptide has never been reported before, 3 other bioactive peptides originating from the same precursor protein have been described in the literature, which increase cellular calcium mobilization (38), and promote survival (39) and differentiation (40) of hippocampal neurons. The possible effect of this novel VGF [353–372] peptide on our CGN culture model thus deserves to be investigated.
Although, this study was initially conducted to identify peptides with a high expression level during development and a decline at adulthood, 4 peptides exhibited an opposite pattern of regulation. Among them, WE-14 is a 14-amino acid peptide resulting from post-translational processing of chromogranin A (41). Even though, the function of WE-14 remains unclear, chromogranin A is known to be involved in the biogenesis and exocytosis of secretory granules (42), two cellular mechanisms required for synaptic transmission. Because, during development, CGN and Purkinje cells create a functional network that relies on synaptic transmission, it is tempting to speculate that according to its expression profile, WE-14 may contribute to secretory mechanisms in the mature cerebellum.
Nociceptin as a Neurotrophic Factor
Among the 4 neuropeptides whose expression was down-regulated during cerebellar development, we have chosen to further investigate the expression profile and putative function of nociceptin because, despites its various functions in the brain, nothing is known regarding its involvement in the development and/or activity in the cerebellum. This 17 amino acid peptide was described for the first time for its pro-nociceptive effect during deorphanization of its receptor NOP (43). Since then, many functions of this peptide have been reported in the brain including impairment of learning abilities (44), participation in addictive behaviors (alcohol (45); food (46); cocaine and heroin (47)), and possible involvement in Parkinson disease (48). Nociceptin is also involved in cellular processes such as migration of tumor cells (49), differentiation of hippocampal neurons (50) and survival of dopaminergic neurons (51).
Expression of nociceptin and its receptor gene was transitory during development and the presence of these genes were also found to be particularly high in the IGL. Furthermore, cultured CGN, which are the most abundant cells in this cell layer, expressed these two genes. These observations led us to conclude that CGN were the cells expressing nociceptin and its receptor but further immunohistochemistry and in situ hybridization will confirm this assertion. During development of the cerebellar cortex, granule neuron precursors proliferate in the EGL, migrate through the ML and PL, to reach their destination in the IGL where they differentiate. This is accompanied by two main phases of apoptosis of CGN which do not properly complete these processes; a first one at P7 in the EGL and a second one in between P16 and P18 in the IGL. The presence of the NOP on cultured granule neurons and the specific expression of the genes encoding nociceptin and its receptor in the IGL suggest that the peptide may act on differentiating CGN rather than on the progenitors located in the EGL. Consistent with this hypothesis, functional studies showed that nociceptin increased the survival and differentiation of CGN but had no effect on their motility. This means that nociceptin does not act as a chemoattractive or chemorepulsive agent to modulate the migration of granule neurons as other neuropeptides do e.g. somatostatin (12) or PACAP (8). Because Lossi et al. (52) showed that the first phase of apoptosis in the EGL is synapse-independent whereas the second one in the IGL is synapse-dependent, by inhibiting apoptosis and promoting differentiation, nociceptin may contribute to the formation of networks between CGN and Purkinje cells and interneurons which, in the mature cerebellum, are crucial for proper motor and cognitive functions. Our data thus suggest that nociceptin, that displays strong expression during cerebellar development, protects CGN during this critical period of apoptosis but is no longer required at adulthood, as shown by its disappearance by P90. To confirm this hypothesis, future experiments should investigate the cerebellar architecture of nociceptin knockout mice to establish the role of the endogenous peptide and analyze the effect of nociceptin on the expression of contact molecules in between parallel fibers and Purkinje cells, such as NB-3 whose deficiency leads to CGN apoptosis within the IGL (53). Also, even if only one receptor for nociceptin has been reported in the literature so far, experiments using siRNA or knockout mice should be conducted both in vitro and in vivo to confirm that all the effects of nociceptin are mediated through NOP. But even if further investigations are needed to elucidate the exact contribution of nociceptin in brain development, the present results demonstrate the transient expression of the nociceptinergic system in the postnatal cerebellum IGL and provide first evidence that this peptide can, in this structure, promote CGN differentiation and survival.
DATA AVAILABILITY
Data were deposited in the ProteomeXchange consortium under the project number PXD006874. Data are available at https://www.ebi.ac.uk/pride/archive/projects/PXD006874.
Supplementary Material
Footnotes
* A.C. was the recipient of a doctoral fellowship from Normandy Region. This work was supported by INSERM (U1239), Rouen University, Normandy Region and the European Union (PeReNE and PACT-CBS projects). Europe gets involved in Normandy with European Regional Development Fund (ERDF).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- EGL
- external granule cell layer
- ACN
- acetonitrile
- ANOVA
- one-way analysis of variance
- Atoh1
- atonal bHLH transcription factor 1
- Cadps2
- calcium-dependent secretion activator 2
- CCK
- cholecystokinin
- cDNA
- complementary DNA
- CGN
- cerebellar granule neuron
- Cq
- quantification cycle
- CXCL14
- C-X-C motif chemokine ligand 14
- CXCR4
- SDF-1 receptor
- DMEM
- Dubecco's modified Eagle's medium
- FA
- formic acid
- FDA
- fluorescein diacetate
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- GL
- granule cell layer
- H2O2
- hydrogen peroxide
- Homer1
- homer scaffolding protein 1
- HAS
- human serum albumin
- IGL
- internal granule cell layer
- Neurod1
- neurogenic differentiation 1
- nLC-MS/MS
- nanoflow liquid chromatography-mass spectrometry
- ML
- molecular layer
- NOP
- nociceptin/orphanin FQ receptor
- Oprl1
- nociceptin/orphanin FQ receptor gene
- P
- postnatal day
- PACAP
- pituitary adenylate cyclase-activating polypeptide
- PL
- Purkinje cell layer
- Pnoc
- nociceptin gene
- Stmn1
- stathmin 1
- Tnr
- tenascin R.
REFERENCES
- 1. Hibi M., and Shimizu T. (2012) Development of the cerebellum and cerebellar neural circuits. Dev. Neurobiol. 72, 282–301 [DOI] [PubMed] [Google Scholar]
- 2. Ito M. (2006) Cerebellar circuitry as a neuronal machine. Prog. Neurobiol. 78, 272–303 [DOI] [PubMed] [Google Scholar]
- 3. Lewis P. M., Gritli-Linde A., Smeyne R., Kottmann A., and McMahon A. P. (2004) Sonic hedgehog signaling is required for expansion of granule neuron precursors and patterning of the mouse cerebellum. Dev. Biol. 270, 393–410 [DOI] [PubMed] [Google Scholar]
- 4. Kubo T., Nonomura T., Enokido Y., and Hatanaka H. (1995) Brain-derived neurotrophic factor (BDNF) can prevent apoptosis of rat cerebellar granule neurons in culture. Brain Res. Dev. Brain Res. 85, 249–258 [DOI] [PubMed] [Google Scholar]
- 5. Ruiz de Almodovar C., Coulon C., Salin P. A., Knevels E., Chounlamountri N., Poesen K., Hermans K., Lambrechts D., Van Geyte K., Dhondt J., Dresselaers T., Renaud J., Aragones J., Zacchigna S., Geudens I., Gall D., Stroobants S., Mutin M., Dassonville K., Storkebaum E., Jordan B. F., Eriksson U., Moons L., D'Hooge R., Haigh J. J., Belin M. F., Schiffmann S., Van Hecke P., Gallez B., Vinckier S., Chédotal A., Honnorat J., Thomasset N., Carmeliet P., and Meissirel C. (2010) Matrix-binding vascular endothelial growth factor (VEGF) isoforms guide granule cell migration in the cerebellum via VEGF receptor Flk1. J. Neurosci. Off. J. Soc. Neurosci. 30, 15052–15066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Komuro H., and Rakic P. (1993) Modulation of neuronal migration by NMDA receptors. Science 260, 95–97 [DOI] [PubMed] [Google Scholar]
- 7. Oostland M., Buijink M. R., Teunisse G. M., von Oerthel L., Smidt M. P., and van Hooft J. A. (2014) Distinct temporal expression of 5-HT(1A) and 5-HT(2A) receptors on cerebellar granule cells in mice. Cerebellum Lond. Engl. 13, 491–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cameron D. B., Galas L., Jiang Y., Raoult E., Vaudry D., and Komuro H. (2007) Cerebellar cortical-layer-specific control of neuronal migration by pituitary adenylate cyclase-activating polypeptide. Neuroscience 146, 697–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vaudry D., Gonzalez B. J., Basille M., Fournier A., and Vaudry H. (1999) Neurotrophic activity of pituitary adenylate cyclase-activating polypeptide on rat cerebellar cortex during development. Proc. Natl. Acad. Sci. 96, 9415–9420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vaudry D., Gonzalez B. J., Basille M., Pamantung T. F., Fournier A., and Vaudry H. (2000) PACAP acts as a neurotrophic factor during histogenesis of the rat cerebellar cortex. Ann. N.Y. Acad. Sci. 921, 293–299 [DOI] [PubMed] [Google Scholar]
- 11. Gonzalez B. J., Basille M., Vaudry D., Fournier A., and Vaudry H. (1997) Pituitary adenylate cyclase-activating polypeptide promotes cell survival and neurite outgrowth in rat cerebellar neuroblasts. Neuroscience 78, 419–430 [DOI] [PubMed] [Google Scholar]
- 12. Yacubova E., and Komuro H. (2002) Stage-specific control of neuronal migration by somatostatin. Nature 415, 77–81 [DOI] [PubMed] [Google Scholar]
- 13. Park C. R., Kim D.-K., Cho E. B., You D.-J., do Rego J. L., Vaudry D., Sun W., Kim H., Seong J. Y., and Hwang J. I. (2012) Spatiotemporal expression and functional implication of CXCL14 in the developing mice cerebellum. Mol. Cells. 34, 289–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tatsuno I., Somogyvari-Vigh A., and Arimura A. (1994) Developmental changes of pituitary adenylate cyclase activating polypeptide (PACAP) and its receptor in the rat brain. Peptides 15, 55–60 [DOI] [PubMed] [Google Scholar]
- 15. Inagaki S., Shiosaka S., Sekitani M., Noguchi K., Shimada S., and Takagi H. (1989) In situ hybridization analysis of the somatostatin-containing neuron system in developing cerebellum of rats. Brain Res. Mol. Brain Res. 6, 289–295 [DOI] [PubMed] [Google Scholar]
- 16. Villar M. J., Hökfelt T., and Brown J. C. (1989) Somatostatin expression in the cerebellar cortex during postnatal development. An immunohistochemical study in the rat. Anat. Embryol. 179, 257–267 [DOI] [PubMed] [Google Scholar]
- 17. Ellis J., Grimm R., Clark J. F., Pyne-Gaithman G., Wilbur S., and Caruso J. A. (2008) Studying protein phosphorylation in low MW CSF fractions with capLC-ICPMS and nanoLC-CHIP-ITMS for identification of phosphoproteins. J. Proteome Res. 7, 4736–4742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ellis J., Del Castillo E., Montes Bayon M., Grimm R., Clark J. F., Pyne-Geithman G., Wilbur S., and Caruso J. A. (2008) A preliminary study of metalloproteins in CSF by CapLC-ICPMS and NanoLC-CHIP/ITMS. J. Proteome Res. 7, 3747–3754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gonzalez B., Leroux P., Lamacz M., Bodenant C., Balazs R., and Vaudry H. (1992) Somatostatin receptors are expressed by immature cerebellar granule cells: evidence for a direct inhibitory effect of somatostatin on neuroblast activity. Proc. Natl. Acad. Sci. U.S.A. 89, 9627–9631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vaudry D., Pamantung T. F., Basille M., Rousselle C., Fournier A., Vaudry H., Beauvillain J. C., and Gonzalez B. J. (2002) PACAP protects cerebellar granule neurons against oxidative stress-induced apoptosis. Eur. J. Neurosci. 15, 1451–1460 [DOI] [PubMed] [Google Scholar]
- 21. Raoult E., Bénard M., Komuro H., Lebon A., Vivien D., Fournier A., Vaudry H., Vaudry D., and Galas L. (2014) Cortical-layer-specific effects of PACAP and tPA on interneuron migration during post-natal development of the cerebellum. J. Neurochem. 130, 241–254 [DOI] [PubMed] [Google Scholar]
- 22. Jolivel V., Basille M., Aubert N., de Jouffrey S., Ancian P., Le Bigot J. F., Noack P., Massonneau M., Fournier A., Vaudry H., Gonzalez B. J., and Vaudry D. (2009) Distribution and functional characterization of pituitary adenylate cyclase-activating polypeptide receptors in the brain of non-human primates. Neuroscience 160, 434–451 [DOI] [PubMed] [Google Scholar]
- 23. Vaudry D., Rousselle C., Basille M., Falluel-Morel A., Pamantung T. F., Fontaine M., Fournier A., Vaudry H., and Gonzalez B. J. (2002) Pituitary adenylate cyclase-activating polypeptide protects rat cerebellar granule neurons against ethanol-induced apoptotic cell death. Proc. Natl. Acad. Sci. U.S.A. 99, 6398–6403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Raoult E., Roussel B. D., Bénard M., Lefebvre T., Ravni A., Ali C., Vivien D., Komuro H., Fournier A., Vaudry H., Vaudry D., and Galas L. (2011) Pituitary adenylate cyclase-activating polypeptide (PACAP) stimulates the expression and the release of tissue plasminogen activator (tPA) in neuronal cells: involvement of tPA in the neuroprotective effect of PACAP: tPA mediates the neuroprotective effect of PACAP. J. Neurochem. 119, 920–931 [DOI] [PubMed] [Google Scholar]
- 25. Basille-Dugay M., Hamza M. M., Tassery C., Parent B., Raoult E., Bénard M., Raisman-Vozari R., Vaudry D., and Burel D. C. (2013) Spatio-temporal characterization of the pleiotrophinergic system in mouse cerebellum: evidence for its key role during ontogenesis. Exp. Neurol. 247, 537–551 [DOI] [PubMed] [Google Scholar]
- 26. Sadakata T., and Furuichi T. (2009) Developmentally regulated Ca2+-dependent activator protein for secretion 2 (CAPS2) is involved in BDNF secretion and is associated with autism susceptibility. Cerebellum Lond. Engl. 8, 312–322 [DOI] [PubMed] [Google Scholar]
- 27. Ingram M., Wozniak E. A. L., Duvick L., Yang R., Bergmann P., Carson R., O'Callaghan B., Zoghbi H. Y., Henzler C., and Orr H. T. (2016) Cerebellar Transcriptome Profiles of ATXN1 Transgenic Mice Reveal SCA1 Disease Progression and Protection Pathways. Neuron 89, 1194–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tissir F., Wang C.-E., and Goffinet A. M. (2004) Expression of the chemokine receptor Cxcr4 mRNA during mouse brain development. Brain Res. Dev. Brain Res. 149, 63–71 [DOI] [PubMed] [Google Scholar]
- 29. Van Dijck A., Hayakawa E., Landuyt B., Baggerman G., Van Dam D., Luyten W., Schoofs L., and De Deyn P. P. (2011) Comparison of extraction methods for peptidomics analysis of mouse brain tissue. J. Neurosci. Methods. 197, 231–237 [DOI] [PubMed] [Google Scholar]
- 30. Fälth M., Sköld K., Svensson M., Nilsson A., Fenyö D., and Andren P. E. (2007) Neuropeptidomics strategies for specific and sensitive identification of endogenous peptides. Mol. Cell. Proteomics 6, 1188–1197 [DOI] [PubMed] [Google Scholar]
- 31. Glickman M. H., and Ciechanover A. (2002) The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82, 373–428 [DOI] [PubMed] [Google Scholar]
- 32. Zhou X. F., and Rush R. A. (1994) Localization of neurotrophin-3-like immunoreactivity in the rat central nervous system. Brain Res. 643, 162–172 [DOI] [PubMed] [Google Scholar]
- 33. Hook V., Funkelstein L., Lu D., Bark S., Wegrzyn J., and Hwang S.-R. (2008) Proteases for processing proneuropeptides into peptide neurotransmitters and hormones. Annu. Rev. Pharmacol. Toxicol. 48, 393–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Malagon M., Vaudry H., Van Strien F., Pelletier G., Gracia-Navarro F., and Tonon M. C. (1993) Ontogeny of diazepam-binding inhibitor-related peptides (endozepines) in the rat brain. Neuroscience 57, 777–786 [DOI] [PubMed] [Google Scholar]
- 35. Nielsen H. S., Hannibal J., and Fahrenkrug J. (1998) Expression of pituitary adenylate cyclase activating polypeptide (PACAP) in the postnatal and adult rat cerebellar cortex. Neuroreport 9, 2639–2642 [DOI] [PubMed] [Google Scholar]
- 36. Vaudry D., Falluel-Morel A., Bourgault S., Basille M., Burel D., Wurtz O., Fournier A., Chow B. K., Hashimoto H., Galas L., and Vaudry H. (2009) Pituitary Adenylate Cyclase-Activating Polypeptide and Its Receptors: 20 Years after the Discovery. Pharmacol. Rev. 61, 283–357 [DOI] [PubMed] [Google Scholar]
- 37. Kaddour H., Hamdi Y., Vaudry D., Basille M., Desrues L., Leprince J., Castel H., Vaudry H., Tonon M. C., Amri M., and Masmoudi-Kouki O. (2013) The octadecaneuropeptide ODN prevents 6-hydroxydopamine-induced apoptosis of cerebellar granule neurons through a PKC-MAPK-dependent pathway. J. Neurochem. 125, 620–633 [DOI] [PubMed] [Google Scholar]
- 38. Sasaki K., Takahashi N., Satoh M., Yamasaki M., and Minamino N. (2010) A peptidomics strategy for discovering endogenous bioactive peptides. J. Proteome Res. 9, 5047–5052 [DOI] [PubMed] [Google Scholar]
- 39. Noda Y., Shimazawa M., Tanaka H., Tamura S., Inoue T., Tsuruma K., and Hara H. (2015) VGF and striatal cell damage in in vitro and in vivo models of Huntington's disease. Pharmacol. Res. Perspect. 3, e00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Thakker-Varia S., Behnke J., Doobin D., Dalal V., Thakkar K., Khadim F., Wilson E., Palmieri A., Antila H., Rantamaki T., and Alder J. (2014) VGF (TLQP-62)-induced neurogenesis targets early phase neural progenitor cells in the adult hippocampus and requires glutamate and BDNF signaling. Stem Cell Res. 12, 762–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Curry W. J., Barkatullah S. C., Johansson A. N., Quinn J. G., Norlen P., Connolly C. K., McCollum A. P., and McVicar C. M. (2002) WE-14, a chromogranin a-derived neuropeptide. Ann. N.Y. Acad. Sci. 971, 311–316 [DOI] [PubMed] [Google Scholar]
- 42. Elias S., Delestre C., Ory S., Marais S., Courel M., Vazquez-Martinez R., Bernard S., Coquet L., Malagon M. M., Driouich A., Chan P., Gasman S., Anouar Y., and Montero-Hadjadje M. (2012) Chromogranin A induces the biogenesis of granules with calcium- and actin-dependent dynamics and exocytosis in constitutively secreting cells. Endocrinology 153, 4444–4456 [DOI] [PubMed] [Google Scholar]
- 43. Meunier J. C., Mollereau C., Toll L., Suaudeau C., Moisand C., Alvinerie P., Butour J.-L., Guillemot J.-L., Ferrara P., Monsarrat B., Mazarguil H., Vassart G., Parmentier M., and Costentin J. (1995) Isolation and structure of the endogenous agonist of opioid receptor-like ORL1 receptor. Nature. 377, 532–535 [DOI] [PubMed] [Google Scholar]
- 44. Redrobe J. P., Calo G., Guerrini R., Regoli D., and Quirion R. (2000) [Nphe(1)]-Nociceptin (1–13)-NH(2), a nociceptin receptor antagonist, reverses nociceptin-induced spatial memory impairments in the Morris water maze task in rats. Br. J. Pharmacol. 131, 1379–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rorick-Kehn L. M., Ciccocioppo R., Wong C. J., Witkin J. M., Martinez-Grau M. A., Stopponi S., Adams B. L., Katner J. S., Perry K. W., Toledo M. A., Diaz N., Lafuente C., Jiménez A., Benito A., Pedregal C., Weiss F., and Statnick M. A. (2016) A Novel, Orally Bioavailable Nociceptin Receptor Antagonist, LY2940094, Reduces Ethanol Self-Administration and Ethanol Seeking in Animal Models. Alcohol. Clin. Exp. Res. 40, 945–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Statnick M. A., Chen Y., Ansonoff M., Witkin J. M., Rorick-Kehn L., Suter T. M., Song M., Hu C., Lafuente C., Jiménez A., Benito A., Diaz N., Martínez-Grau M. A., Toledo M. A., and Pintar J. E. (2016) A Novel Nociceptin Receptor Antagonist LY2940094 Inhibits Excessive Feeding Behavior in Rodents: A Possible Mechanism for the Treatment of Binge Eating Disorder. J. Pharmacol. Exp. Ther. 356, 493–502 [DOI] [PubMed] [Google Scholar]
- 47. Kallupi M., Scuppa G., de Guglielmo G., Calò G., Weiss F., Statnick M. A., Rorick-Kehn L. M., and Ciccocioppo R. (2017) Genetic Deletion of the Nociceptin/Orphanin FQ Receptor in the Rat Confers Resilience to the Development of Drug Addiction. Neuropsychopharmacol. Off. Publ. Am. Coll. Neuropsychopharmacol. 42, 695–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Marti M. (2005) Blockade of Nociceptin/Orphanin FQ Transmission Attenuates Symptoms and Neurodegeneration Associated with Parkinson's Disease. J. Neurosci. 25, 9591–9601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bedini A., Baiula M., Vincelli G., Formaggio F., Lombardi S., Caprini M., and Spampinato S. (2017) Nociceptin/orphanin FQ antagonizes lipopolysaccharide-stimulated proliferation, migration and inflammatory signaling in human glioblastoma U87 cells. Biochem. Pharmacol. 140, 89–104 [DOI] [PubMed] [Google Scholar]
- 50. Ring R. H., Alder J., Fennell M., Kouranova E., Black I. B., and Thakker-Varia S. (2006) Transcriptional profiling of brain-derived-neurotrophic factor-induced neuronal plasticity: A novel role for nociceptin in hippocampal neurite outgrowth. J. Neurobiol. 66, 361–377 [DOI] [PubMed] [Google Scholar]
- 51. Arcuri L., Viaro R., Bido S., Longo F., Calcagno M., Fernagut P. O., Zaveri N. T., Calò G., Bezard E., and Morari M. (2016) Genetic and pharmacological evidence that endogenous nociceptin/orphanin FQ contributes to dopamine cell loss in Parkinson's disease. Neurobiol. Dis. 89, 55–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lossi L., Mioletti S., and Merighi A. (2002) Synapse-independent and synapse-dependent apoptosis of cerebellar granule cells in postnatal rabbits occur at two subsequent but partly overlapping developmental stages. Neuroscience 112, 509–523 [DOI] [PubMed] [Google Scholar]
- 53. Sakurai K., Toyoshima M., Ueda H., Matsubara K., Takeda Y., Karagogeos D., Shimoda Y., and Watanabe K. (2009) Contribution of the neural cell recognition molecule NB-3 to synapse formation between parallel fibers and Purkinje cells in mouse. Dev. Neurobiol. 69, 811–824 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data were deposited in the ProteomeXchange consortium under the project number PXD006874. Data are available at https://www.ebi.ac.uk/pride/archive/projects/PXD006874.