Abstract
Cytological examination of urine is the most widely used noninvasive pathologic screen for bladder urothelial carcinoma (BLCA); however, inadequate diagnostic accuracy remains a major challenge. We performed mass spectrometry-based proteomic analysis of urine samples of ten patients with BLCA and ten paired patients with benign urothelial lesion (BUL) to identify ancillary proteomic markers for use in liquid-based cytology (LBC). A total of 4,839 proteins were identified and 112 proteins were confirmed as expressed at significantly different levels between the two groups. We also performed an independent proteomic profiling of tumor tissue samples where we identified 7,916 proteins of which 758 were differentially expressed. Cross-platform comparisons of these data with comparative mRNA expression profiles from The Cancer Genome Atlas identified four putative candidate proteins, AHNAK, EPPK1, MYH14 and OLFM4. To determine their immunocytochemical expression levels in LBC, we examined protein expression data from The Human Protein Atlas and in-house FFPE samples. We further investigated the expression of the four candidate proteins in urine cytology samples from two independent validation cohorts. These analyses revealed AHNAK as a unique intracellular protein differing in immunohistochemical expression and subcellular localization between tumor and non-tumor cells. In conclusion, this study identified a new biomarker, AHNAK, applicable to discrimination between BLCA and BUL by LBC. To our knowledge, the present study provides the first identification of a clinical biomarker for LBC based on in-depth proteomics.
Keywords: Clinical proteomics, Bladder cancer, Bioinformatics, Biomarker: Diagnostic, Immunohistochemistry, AHNAK, Biomarker, Bladder urothelial carcinoma, Cancer proteomics, Liquid-based cytology
Urothelial carcinoma of the bladder is a disease with high morbidity and mortality (1). Urinary cytology represents the 'gold standard' for surveillance of urothelial carcinoma diagnosis and recurrence; however, the diagnostic accuracy of the test has been doubted owing to considerable variability in its diagnostic positive prediction rate (31–72%) for identification of patients with bladder cancer (2, 3). In addition, inter-observer variability had a range of 38–65% across institutions (4). To overcome these drawbacks, several molecular tests have been developed; however, used alone, the overall specificity and sensitivity of these tests are like those of cytology, because of the lack of simultaneous assessment of cytological appearance (5).
Recent rapid advances in proteomic technologies, including computational algorithms and biochemical techniques, have enabled quantitative evaluation of novel diagnostic markers to determine their levels in tumor tissues (6, 7). In-depth proteomic analyses of clinically available urine specimens have been performed in several previous studies (8). Urine is a useful source of proteins for biomarker discovery and comprehensive assessment, because it is readily available, can be obtained by non-invasive collection methods, and enables disease monitoring; however, the proteins in urine can originate from various types of cells or secretions, such as blood cells, epithelial cells from the glomerulus or urinary tract, and can include a mixture of benign and malignant cells and excreted plasma, which can contribute to misinterpretation and misleading results. In contrast, cytological urine specimens almost exclusively contain epithelial cells from the lining of the urinary tract, which are selected under microscopic examination. Therefore, genomic or proteomic information obtained from cytological preparations is expected to exclusively reflect the molecular landscape in urothelial cells and be suitable for identification of novel biomarkers.
In this study, for the first time, we employed MS-based in-depth proteomics to identify novel biomarkers in voided urine cytology samples collected by the liquid-based method, which has technical advantages (9). To discover suitable biomarkers, we designed an integrative workflow, including comparative analyses of results from a cytological proteomic platform with those in a public transcriptomic database, and from an in-house generated formalin fixed paraffin embedded (FFPE)1-based proteomic experiment, followed by immunostaining validation in two independent liquid-based cytology cohorts.
EXPERIMENTAL PROCEDURES
Sample Selection
All pathologic specimens enrolled in this study were collected from the Seoul National University Hospital biorepository operated by the department of pathology. A discovery set consisting of a total of 20 voided urine cytology samples was collected from 10 patients with primary bladder urothelial carcinoma and 10 with benign urothelial lesion as a negative control. Separately, six FFPE urinary bladder tissue samples from three patients each with bladder urothelial carcinoma and benign urothelial lesion (previously diagnosed with cystitis cystica) were included for comparative proteomic analysis. For immunocytochemical validation of selected proteomic biomarkers, an independent cohort of 140 voided urine liquid-based cytology samples, containing both urothelial carcinoma and normal cells, were collected. All cases were histologically confirmed, using samples obtained within one month before corresponding surgical examination. All slides were reviewed by two experienced urologic pathologists and classified according to the WHO/ISUP system for surgical biopsy (10) and the Paris system for liquid-based cytology (11), respectively. The clinicopathologic features are presented in Table I.
Table I. Clinicopathologic characteristics of the patients in cohort of bladder urothelial carcinomas.
| Bladder urothelial carcinoma (BLCA) | Benign urothelial lesion (BUL) | |||
|---|---|---|---|---|
| Gender (%, n) | ||||
| Male | 100% | 10/10 | 100% | 10/10 |
| Age at diagnosis (years) | ||||
| 50–60 | 20% | 2/10 | 30% | 3/10 |
| 60–70 | 20% | 2/10 | 10% | 1/10 |
| 70 | 60% | 6/10 | 60% | 6/10 |
| Pathologic diagnosis | ||||
| Invasive urothelial carcinoma, high grade | 40% | 4/10 | ||
| Papillary urothelial carcinoma, high grade | 60% | 6/10 | ||
| Urothelial papilloma | 30% | 3/10 | ||
| Cystitis cystica | 70% | 7/10 | ||
| Stroma invasion | ||||
| Present | 90% | 9/10 | ||
| Pathologic stage | ||||
| pTis | 10% | 1/10 | ||
| pT1 | 60% | 6/10 | ||
| pT2 | 30% | 3/10 | ||
| AJCC stage | ||||
| 0is | 10% | 1/10 | ||
| I | 60% | 6/10 | ||
| II | 30% | 3/10 | ||
| Treatment | ||||
| Transurethral resection | 36% | 4/10 | 100% | 10/10 |
| Cystectomy | 9% | 1/10 | ||
| Nephroureterectomy | 9% | 1/10 | ||
| No treatment | 18% | 2/10 | ||
| Adjuvant chemotherapy+cystectomy | 9% | 1/10 | ||
| Cystectomy+ureterectomy | 9% | 1/10 | ||
| Status | ||||
| Follow up loss | 30% | 3/10 | ||
| No evidence of disease recurrence | 30% | 3/10 | 100% | 10/10 |
| Local recurrence | 30% | 3/10 | ||
| Dead of complication | 10% | 1/10 | ||
All liquid-based cytology slides were previously scanned with an Aperio AT2 Digital Whole Slide Scanner (Leica Biosystems, IL), and the number of cells initially screened by pathologists, and counted using an Aperio ImageScope (Leica Biosystems) with Aperio's nuclear algorithm (Leica Biosystems). The study protocol was approved by the Institutional Review Board at Seoul National University Hospital (IRB no. 1602-150-747).
Sample Preparation for Proteomics Analysis of Cytology Samples
All urine samples were fixed with BD CytoRichTM Clear Preservative Fluid (BD Diagnostics-TriPath Imaging, Burlington, NC) and prepared using the SurePath liquid-based preparation method according to the manufacturer's instructions (12). Briefly, the samples were collected in individual 20-ml specimen containers based on the midstream clean catch method, which is widely-used in daily practice. Each 12-ml sample was transferred to a 50 ml tube (BD Prepstain™ system). The supernatant was discarded and 10 ml of preservative fluid (BD CytoRich™ Clear) was added. After being vortexed for 15 ± 5 s and left static for a minimum of 30 min, the contents were transferred to a 12-ml tubes and centrifuged for 5 min at 600 g (3240 rpm). The urine samples were stored in a refrigerator (4 °C) without freezing for less than 5 min on average until further preparation. After samples were taken out from the refrigerator, the entire amount of each unfixed urine sample was transferred to a centrifuge tube and centrifuged for 5 min at 600 × g (3240 rpm). The supernatant fluid was decanted and vortexed for 15 ± 5 s at room temperature to homogenize the sample, followed by loading into a 12-ml centrifuge tube holder onto the BD Prepstain™ system for processing. BD SurePath™ PreCoat slides with settling chambers were placed on the slide rack in the same position as the tubes in the centrifuge tube holder. The NON-GYN program was run in the instrument. In the Prepstain™ system, 500 μl of buffered distilled water (Sigma-Aldrich, Cat #T6664; pH 8.0) was added to each 12-ml centrifuge tube. finally, 300 μl was aspirated from each sample and added to the corresponding slide (supplemental Fig. S1).
Individual liquid-based cytology and FFPE tissue sections of 13 mm diameter and 10 μm thickness were scraped for each case to collect well-preserved populations of stained or unstained cells in individual Eppendorf tubes. Cell pellets were lysed with 100 μl of SDS extraction buffer (4% SDS; 100 mm Tris, pH 7.4; and 1 mm TCEP). Samples were lysed by sonication and boiling at 95 °C for 30 min. Proteins were digested using the filter-aided sample preparation procedure, as previously described (13). Briefly, 50 μl of samples were mixed with 0.2 ml 8 m urea in 0.1 m Tris/HCl, pH 8.5, loaded onto a 30 k spin filter (EMD Millipore, Billerica, MA). Buffer was exchanged with urea solution by centrifugation. Reduced cysteines were alkylated with iodoacetamide solution in darkness at room temperature for 30 min. An additional 50 mm ammonium bicarbonate was added to exchange the urea solution. Finally, proteins were digested at 37 °C overnight with trypsin at an enzyme to protein ratio of 1:100. After an overnight incubation, the filtration unit was transferred to new collection tubes, followed by centrifugation for 20 min. Peptides that were retained in the filtration units were eluted with 50 μl 0.5 m NaCl to enhance the yield of digested protein. The resultant supernatants were acidified with 1% TFA.
Sample Preparation for Proteomic Analysis of FFPE Samples
FFPE sections (10 μm) were incubated twice in xylene for 5 min, followed by 100% (v/v) ethanol twice for 3 min. Sections were then hydrated twice in 85% (v/v) ethanol for 1.5 min, and distilled water for 3 min. Tissue samples were then scraped off the slides into microfuge tubes, and extraction buffer (4% SDS; 1 mm TCEP; and 0.3 m Tris, pH 8.0) added. After sonication, samples were incubated at 95 °C for 2 h. Extracted proteins were precipitated by adding chilled acetone at a volume ratio of 1:5 buffer to acetone, followed by incubation at −20 °C for 16 h. After washing with 200 μl of chilled acetone, protein pellets were collected by centrifugation at 15,000 rpm for 10 min and air-dried. Protein concentrations were measured using a bicinchoninic acid reducing agent compatible kit (Thermo Fisher Scientific Inc., Rockford, IL). Protein (100 μg per sample) was digested using the filter-aided sample preparation procedure, as described above.
Desalting
Eluted peptides were desalted using C18 Stage Tips, as previously described (14). C18 Empore disk membranes (3 m, Bracknell, UK) were packed into the bottom of 200 μl yellow pipette tips. POROS 20 R2 reversed-phase media (Applied Biosystems, Foster City, CA) was dissolved in 1 ml MeOH and 100 μl of the mixture loaded separately into the tip for two rounds of filtration with MeOH. Packed microcolumns were washed three times with 100 μl of MeOH and 100% acetonitrile (ACN) consecutively and equilibrated three times with 100 μl 0.1% TFA, by applying air pressure from a syringe. After samples were loaded, microcolumns were washed three times with 100 μl 0.1% TFA, and peptides subsequently eluted with 100 μl of a series of elution buffers containing 40%, 60%, and 80% ACN in 0.1% formic acid. Finally, all eluates were dried in a vacuum centrifuge and stored at −80 °C until LC-MS/MS analysis.
LC-MS/MS Analysis
LC-MS/MS analysis was performed using a Q Exactive Plus Hybrid Quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific Inc.), coupled to an Ultimate 3000 RSLC system (Dionex, Sunnyvale, CA) via a nano electrospray source, as previously described (13, 15), with some modifications. Peptide samples were separated on a two-column system, consisting of a trap column and an analytic column (75 μm × 50 cm) with a 120 min gradient from 7% to 32% acetonitrile at 300 nl/min and analyzed by mass spectrometry. Column temperature was maintained at 60 °C using a column heater. Survey scans (350 to 1650 m/z) were acquired with a resolution of 70,000 at m/z 200. A top-20 method was used to select precursor ions with an isolation window of 1.2 m/z. MS/MS spectra were acquired at an HCD-normalized collision energy of 30, with a resolution of 17,500, at m/z 200. The maximum ion injection times for the full scan and MS/MS scan were 20 and 100 ms, respectively. The detailed data analysis process is described in supplemental Materials and Methods.
Data Analysis and Peptide Identification
Mass spectra were processed using MaxQuant version 1.5.3.1 (16). MS/MS spectra were searched against the Human Uniprot protein sequence database (December 2014, 88,657 entries) using the Andromeda search engine (17). Primary searches were performed using a 6-ppm precursor ion tolerance for total protein level analysis. The MS/MS ion tolerance was set to 20 ppm. Cysteine carbamidomethylation was set as a fixed modification. N-acetylation of protein and oxidation of methionine were set as variable modifications. Enzyme specificity was set to full tryptic digestion. Peptides with a minimum length of six amino-acids and up to two missed cleavages were considered. The required false discovery rate (FDR) was set to 1% at the peptide, protein, and modification level. To maximize the number of quantification events across samples, we enabled the 'Match between Runs' option on the MaxQuant platform.
Label Free Quantification and Statistical Analyses
For label-free quantification, the Intensity Based Absolute quantification (iBAQ) algorithm (18) was used as a part of the MaxQuant platform. Briefly, iBAQ values calculated by MaxQuant are raw intensities divided by the number of theoretical peptides. Thus, iBAQ values are proportional to the molar quantities of the proteins. All statistical analyses were performed using Perseus software (19). For quantitative analysis of iBAQ cytology data, we first filtered out proteins with at least 20 quantified values in each group. Missing values were imputed on the basis of a normal distribution (width = 0.15, down-shift = 1.8) to simulate signals of low abundance proteins. Finally, data were normalized using width adjustment, which subtracts the medians and scales all values in a sample to have equal interquartile ranges (20). For normalization, the first, second and third quartile (q1, q2, and q3) are calculated from the distribution of all iBAQ values. The second quartile that is the median is subtracted from each value to center the distribution. Then, we divide by the width in an asymmetric way. All values that are positive after subtraction of the median are divided by values that calculated from (q3–q2) while all negative values are divided by values that calculated from (q2–q1). For quantitative analysis of iBAQ bladder urothelial carcinoma FFPE data, we first filtered out proteins with at least 3 quantified values in each group. Missing values were imputed by normal distribution as described above. The iBAQ values of each protein were normalized against the sum of quantitative values in individual runs. For pairwise comparison of proteomes, two-sided t-tests were performed using permutation-based FDR and a significance level of 5%. In case of FFPE data, a protein was considered statistically significant if its fold change was ≥ 2 and if it had an FDR ≤ 0.05.
TCGA Data Process
For bladder urothelial carcinoma RNA sequencing data, we downloaded the level 3 RNA sequencing version 2 data set from TCGA with upper quartile normalized RSEM count estimates from Broad Institute GDAC FireBrowse (TCGA data version 20160128, http://firebrowse.org/). The RNA sequencing version 2 dataset were produced on the Illumina HiSeq 2000 platform and processed by the algorithms of MapSplice for aligning sequenced reads and RSEM for quantifying the gene expression levels. For the data normalization between samples, gene expression levels are scaled by upper-quartile normalization method. Among the data downloaded from TCGA, there are 408 bladder urothelial carcinoma samples which include 19 benign urothelial lesion-matched bladder urothelial carcinoma samples.
Integrated Analysis of Data from the Cancer Genome Atlas TCGA and Proteomic Platforms
Proteins identified in the comparative proteomic analysis as differentially abundant between benign urothelial lesion and bladder urothelial carcinoma in the liquid-based cytology cohort were aligned to transcripts expressed in the benign urothelial lesion and bladder urothelial carcinoma cohort data in FFPE-based quantitative proteomic analyses to compare the cytology proteome profiles of bladder urothelial carcinoma samples. Finally, the external public repository, TCGA data portal, was employed for comparative bioinformatics analyses. Bladder urothelial carcinoma RNA sequencing data were sourced independently for comparative analysis with protein expression data obtained using MS-based proteomic assays to evaluate reliable ancillary biomarkers for bladder urothelial carcinoma diagnosis using liquid-based cytology and FFPE samples (supplemental Fig. S2).
Immunostaining
To select immunoreactive markers, protein expression value data and criteria for antibodies listed from comparative analyses were obtained from The Human Protein Atlas data set, a public repository of immunohistochemistry data (6, 21). Immunohistochemical expression for each protein was evaluated using whole-slide histopathology images from The Human Protein Atlas. After exclusion of antibodies with indistinguishable expression between the benign urothelial lesion and bladder urothelial carcinoma groups based on micrographs from the data set, four candidate proteins were subsequently validated by immunohistochemistry of 25 FFPE cases and four immunocytochemistry of voided-urine liquid-based cytology samples (supplemental Table S1). Standard immunohistochemistry and immunocytochemical procedures for slides prepared by fixation in 10% neutral buffered formalin solution or 95% ethanol were performed using a Benchmark automatic immunostaining device (Ventana BenchMark XT Staining System, Tucson, AZ) without any antigen retrieval process. Tumor and normal cells (n = 100 each) were counted across representative areas of each slide under an Olympus BX51 microscope (Olympus, Tokyo, Japan) and the proportions of stained cells recorded separately.
Bioinformatic and Statistical Analyses
All proteomic datasets were submitted to the ProteomeXchange Consortium (http://proteomecentral.proteomechange.org) via the PRIDE partner repository (http://www.ebi.ac.uk/pride/archive/projects/PXD006805, project ID: PXD006805) (22). Annotated MS/MS spectra can be accessed through MS-Viewer (23) (http://msviewer.ucsf.edu/prospector/cgi-bin/mssearch.cgi?report_title=MS-viewer&search_key=d4qfuhxipu&search_name=msviewer) with the following search keys: d4qfuhxipu. Venny web server 2.0 (http://bioinfogp.cnb.csic.es/tools/venny/index.html) was used to generate genomic distributions across different groups (24).
The area under the curve (AUC) values for four individual transcripts were estimated from an entire data set including 427 observations (408 tumor and 19 normal samples) and a paired data set including 38 observations (19 tumor and 19 normal samples). We use the receiver operating characteristics (ROC) function of the R package, pROC, to obtain AUC values and thresholds producing maximum values for the sums of sensitivities and specificities (https://www.r-project.org/).
In addition to AUC value, we calculated five performance measures including Sensitivity, Specificity, Accuracy, Positive predictive value (PPV), and Negative predictive value (NPV). By using R package, we calculated the five performance measures for all possible threshold value of marker expression and determined the performance measures at the optimal threshold value, which maximizes sums of sensitivity, specificity and accuracy.
Experimental Design and Statistical Rational
We designed a comprehensive stepwise approach to discover novel biomarkers to detect urothelial carcinoma in liquid-based cytology. Total 20 biological replicates and technical duplicates were performed using liquid-based cytology samples. The discovery approach was performed with two-sided t-tests using permutation-based FDR to identify proteins altered between benign urothelial lesion and bladder urothelial carcinoma liquid-based cytology samples. A convergent filtering approach with additional proteomic and mRNA data set was adopted to prioritize the proteins quantified in the discovery step. Proteomic benign urothelial lesion and bladder urothelial carcinoma in-house FFPE samples and in-silico TCGA data repository were analyzed permutation-based FDR and Wilcoxon rank sum tests for genes with adjusted p values by using Benjamini-Hochberg method, respectively. Total six biological replicates and one technical replicates were analyzed in bladder urothelial carcinoma FFPE samples. Predictive ability of finally selected four candidate biomarkers was further evaluated to identify base on receiver operating characteristic curve. These were brought forward to the next validation step in which Students t test were applied to test for immunocytochemical diagnostic ability in independent liquid-based cytology cohorts being followed by the results of immunohistochemistry in FFPE samples and screened data from The Human Protein Atlas.
RESULTS
Quantitative Proteomic and Bioinformatic Analyses Workflow
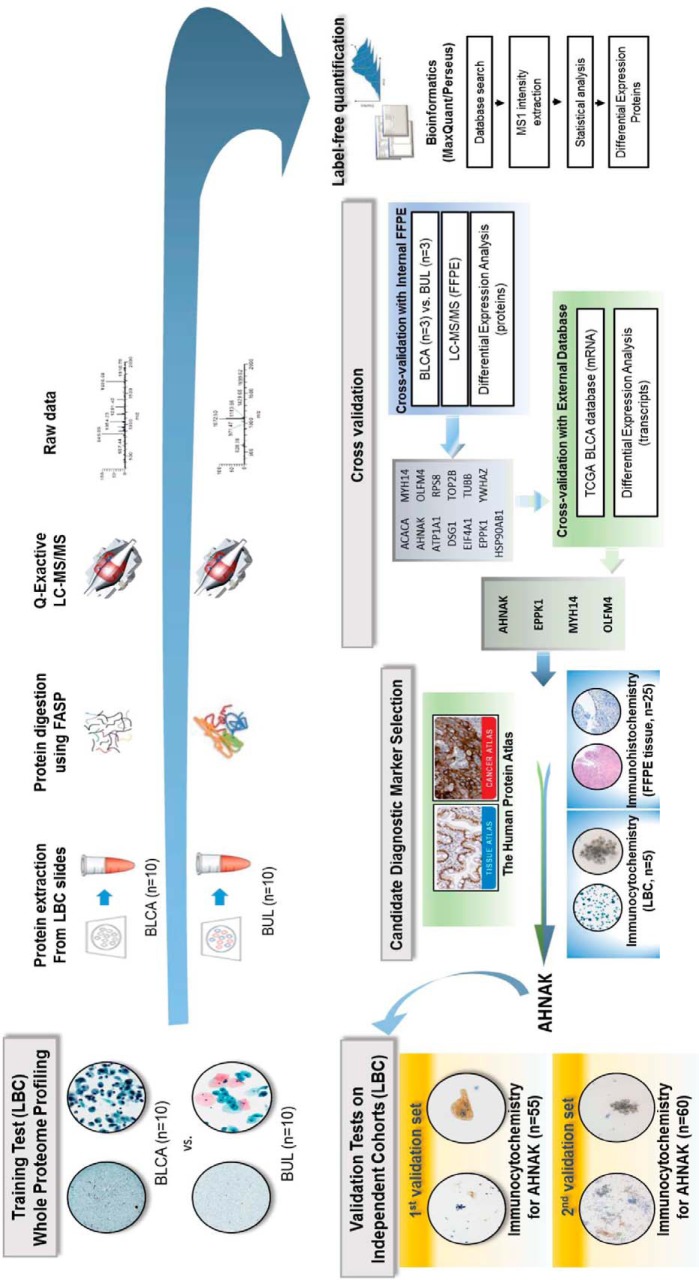
In this study, we developed a workflow to achieve identification of reproducible biomarkers with high ability to discriminate between benign urothelial lesion and bladder urothelial carcinoma in voided liquid-based cytology urine samples. A schematic summary of the overall multi-step workflow is presented in Fig. 1.
Fig. 1.
Overview of the study design. Proteomic analysis of urinary bladder urothelial carcinoma (BLCA) liquid-based cytology (LBC) samples.
Global Profiling of the Urothelial Carcinoma Proteome in Liquid-based Cytology
In liquid-based cytology, analysis of samples based on equal amounts of protein can cause serious problems, because even the normal range of total cell counts per urine sample volume is very wide. In this study, total cell counts of negative (benign urothelial lesion) samples ranged from 7 × 103 to 1 × 105, whereas those of positive (bladder urothelial carcinoma) samples ranged from 5 × 104 to 4 × 105. To address this problem, we analyzed all proteins obtained from one slide to ensure that the cells were derived from the same initial volume of urine. In addition, MS data were normalized based on total cell counts using a width adjustment algorithm to correct for systematic variations.
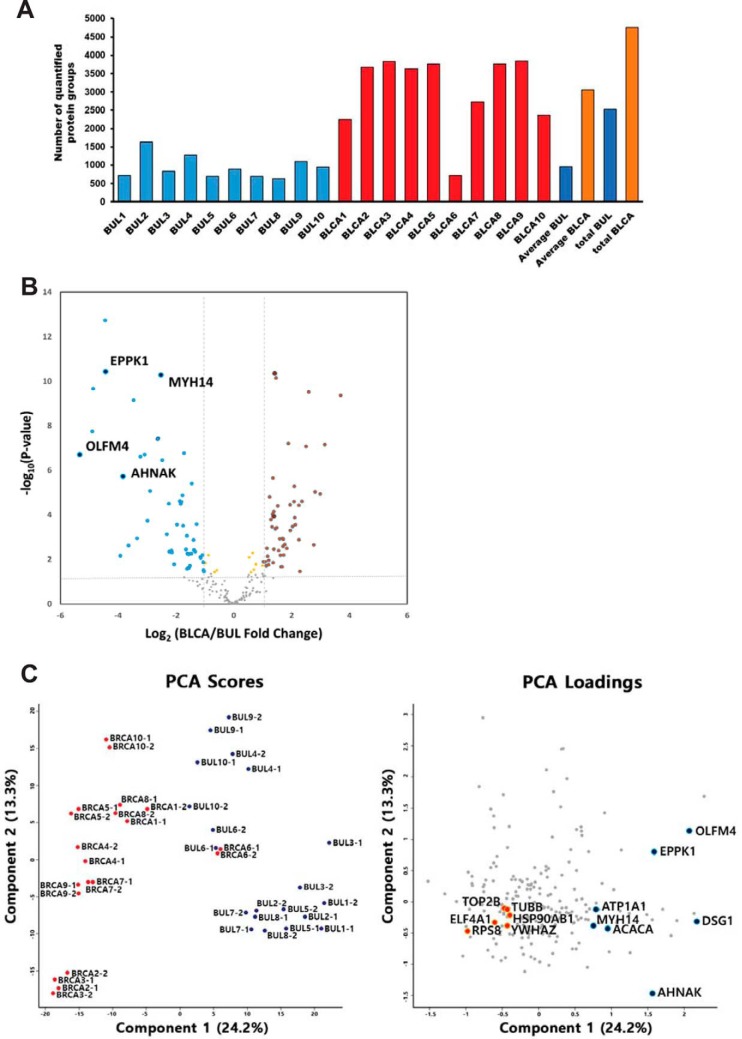
In total, 4839 proteins were identified at the 1% FDR level by single-shot proteomic analysis of the liquid-based cytology set. On average, we identified and quantified > 1000 and > 2500 proteins in benign urothelial lesion samples and bladder urothelial carcinoma samples, respectively (Fig. 2A and supplemental Table S2). To avoid endogenously biased comparisons, we only considered the subset of 214 proteins in our data for 100% valid values in all 20 single run analyses in each group (total 40 single-run analyses) for further analysis. We observed an excellent correlation between technical replicates (average R2 = 0.73–0.99) (supplemental Fig. S3). Interestingly, biological correlation indicated higher diversity among benign urothelial lesion samples (average R2 = 0.62) compared with bladder urothelial carcinoma samples (average R2 = 0.77). As expected, MS signals based on iBAQ value correlated well with total cell counts as overall correlation of 0.66 (supplemental Fig. S4). After implementation of a normalization process to correct for systematic bias across comparison groups (supplemental Fig. S5), label-free quantification and statistical analysis yielded 112 differentially expressed proteins from a total of 20 cases with an FDR-adjusted p value < 0.05 (supplemental Table S3). Hierarchical clustering and principal component analysis revealed tight clustering of two groups and their corresponding biological replicates, indicating distinct protein expression patterns within each group (bladder urothelial carcinoma and benign urothelial lesion; Fig. 2B and 2C and supplemental Fig. S6).
Fig. 2.
Identification and label-free quantification of non-tumor benign urothelial lesion (BUL, control) and bladder urothelial carcinoma (BLCA, case) from urine liquid-based cytology (A) results of identified proteins in benign urothelial lesion and bladder urothelial carcinoma liquid-based cytology samples. Total and average number of protein groups identified in each case. B, Volcano plot depicting the variance in expression between benign urothelial lesion and bladder urothelial carcinoma samples. Highlighted data points indicate the four proteins selected using cross-platform based analysis (blue dots, decreased intensity in bladder urothelial carcinoma; red dots, increased intensity in bladder urothelial carcinoma). Dotted vertical lines indicate one standard deviation from the mean fold-change. C, Principal component analysis (PCA) of proteome data from 20 samples using the total set of proteins quantified with an expression value across all samples (left panel). Highlighted data points indicate the 13 proteins selected using cross-platform based analysis (blue dots, decreased intensity in bladder urothelial carcinoma; red dots, increased intensity in bladder urothelial carcinoma, right panel).
Comprehensive Identification of the Urothelial Carcinoma Proteome in FFPE Samples
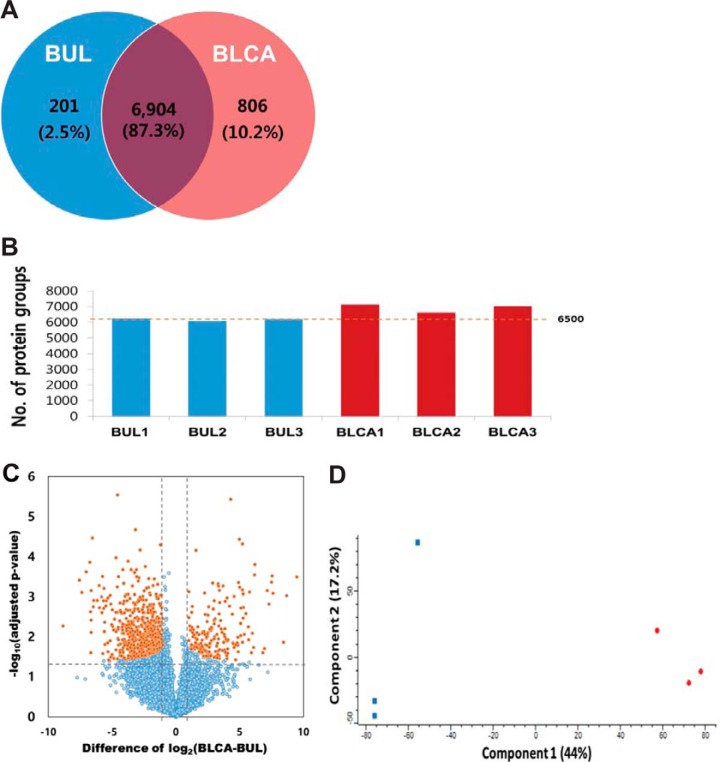
Next, we identified differentially expressed proteins between bladder urothelial carcinoma and cystitis cystica FFPE samples for cross-validation. MS analysis of the FFPE set yielded 7911 identified protein groups and 7870 quantified protein groups with high confidence at the 1% FDR level (supplemental Table S4 and Fig. 3A). On average, we identified > 6500 proteins in each sample (Fig. 3B), spanning seven orders of magnitude of signal intensity (supplemental Fig. S7). We observed a good correlation between biological replicates (Pearson correlation = 0.839–0.877 in cystitis cystica and 0.817–0.851 in urothelial carcinoma; supplemental Fig. S8). Analysis of spiked standard peptides for batch normalization revealed only small variations (coefficient of variation, CV < 6%) caused by processing, indicating that the quantified expression diversity stemmed from true biological difference between tumor types. Eventually, label free quantification, based on MS1 intensity, identified 758 differentially expressed proteins with an FDR-adjusted p value < 0.05 and > 2-fold-change in expression (Fig. 3C and supplemental Table S5). Principal component analysis revealed tight clustering between urothelial carcinoma and non-tumor cases, and their corresponding biological replicates, indicating distinct protein expression patterns within each sample (Fig. 3D).
Fig. 3.
Results of proteomics analysis in BUL (cystitis cystica) and BLCA (bladder urothelial carcinoma) FFPE samples. A, Number of proteins common to BUL and bladder urothelial carcinoma. B, Number of identified proteins in individual samples. C, Volcano plot depicting the variance in expression between bladder urothelial carcinoma and BUL samples. (Orange dot: p value <0.05, Blue dot: p value ≥ 0.05). D, Principal component analysis (PCA) of proteome data from six samples using the total set of proteins quantified with an expression value across all samples (Red dot: bladder urothelial carcinoma, Blue dot: benign urothelial lesion).
TCGA Analysis for External Validation
To validate the observations in our in-house data cohorts, we analyzed an external validation cohort of publicly available TCGA dataset which include 19 benign urothelial lesion and 408 bladder urothelial carcinoma mRNA sequencing samples. To identify differentially expressed genes between benign urothelial lesion and bladder urothelial carcinoma samples, we first applied a 2-fold cut-off criterion based on Fold Changes (FC). Subsequently, we performed Wilcoxon rank sum tests for the remaining 17,801 genes using the python module scipy.stats.ranksums. To correct for the occurrence of false-positives within the multiple statistical tests, we adjusted p values using the Benjamini-Hochberg method in Bioconductor q value package in R. After multiple testing corrections, we identified 4244 differentially expressed genes with Benjamini-Hochberg adjusted p values < 0.05. Among the selected differentially expressed genes, 1564 and 2680 genes were overexpressed and downregulated in bladder urothelial carcinoma, respectively (supplemental Table S6).
Protein Signature for Urothelial Carcinoma Diagnosis
To identify proteins that could serve as valuable tools for more accurate cytological diagnosis, we employed cross-platform comparisons and filtering procedures, using two quantitative proteomic data subsets derived from the bladder urothelial carcinoma and benign urothelial lesion liquid-based cytology samples and in silico analysis of publicly available TGCA data sets from the same groups.
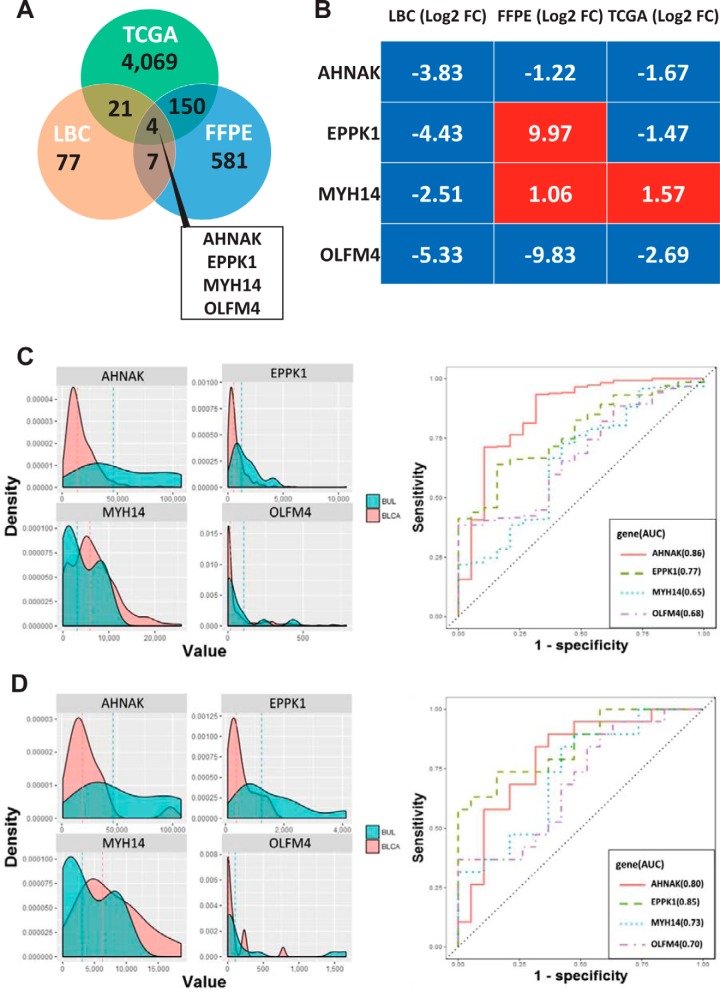
Co-expression analysis identified 11 shared proteins with FDR-adjusted p value < 0.05 from LBC and FFPE proteomics data (supplemental Table S7). Integrative analysis of these 11 proteins, along with 4244 genes identified as differentially expressed (q-value < 0.05) by analyzing TCGA-derived RNA sequencing expression data revealed a set of 4 candidate proteins including Neuroblast differentiation-associated protein AHNAK (AHNAK), Epiplakin (EPPK1), Myosin-14 (MYH14) and Olfactomedin-4 (OLFM4) which were able to discriminate between bladder urothelial carcinoma and benign urothelial lesion (Fig. 4A and supplemental Table S7). Among 4 candidates, EPPK1 and MYH14 were either elevated or decreased (LBC Log2FC range, −4.43 to −2.51; FFPE Log2FC range, 1.06 to 9.97; TCGA Log2FC range, −1.47 to 1.57) in their protein or mRNA expression in bladder urothelial carcinoma, respectively. AHNAK (LBC, Log2FC = −3.83; FFPE, Log2FC = −1.22; TCGA, Log2FC = −1.67) and OLFM4 (LBC, Log2FC = −5.33; FFPE, Log2FC = −9.83; TCGA, Log2FC = −2.69) were consistently downregulated in all data sets (Fig. 4B). To assess the discrimination power of the four transcripts, we compared the AUC values of each candidates (Fig. 4C and Fig. 4D and supplemental Table S8). The geom_density function computes and draws kernel density estimates, which are smoothed versions of the histogram. Through kernel density estimation, we found peaks for each transcript showing similar patterns in two separate sets including a paired and a total cohort in TCGA. The mRNA expression level of AHNAK and OLFM4 were significantly decreased in bladder urothelial carcinoma compared with normal control samples. EPPK1 and MYH14 were evenly distributed in both groups (Fig. 4C and Fig. 4D).
Fig. 4.
Distribution of differential mRNA (TCGA) and proteomic expression, and comparison of receiver operating characteristic (ROC) plots for the predictive power of four mRNA urinary biomarkers for distinguish bladder urothelial carcinoma (BLCA) from BUL. A, Venn diagram showing the genes differentially expressed between two proteomics and TCGA, and (B) heatmap of four common transcripts expression (Log2 fold change, FC) between benign urothelial lesion and bladder urothelial carcinoma. C, Total urothelial carcinoma cohort (n = 427) and (D) 19 paired cohort (n = 38) from TCGA. (left panel) Density distribution of correlation coefficients of mRNA expression between benign urothelial lesion (aqua) and bladder urothelial carcinoma (pink). Dashed lines indicate the mean correlation in each distribution (right panel). The area under the receiver operating characteristic (ROC) curve (AUC) of individual mRNA transcript to distinguish between benign urothelial lesion and bladder urothelial carcinoma.
AHNAK as a Single Immunocytochemical Biomarker for Urothelial Carcinoma Diagnosis
As a final step in the development of a reliable biomarker that can aid bladder urothelial carcinoma diagnosis from liquid-based cytology samples, four putative candidate proteins were queried to The Human Protein Atlas to annotate their comparative protein expression profiles between bladder urothelial carcinoma and normal urothelial samples, as determined by immunohistochemical staining (supplemental Table S9). Despite the limited information because of the lack of cases on the atlas, expression levels of EPPK1 and OLFM4 were not distinguishable between bladder urothelial carcinoma and normal urothelial samples as opposed to AHNAK and MYH14, which showed a tendency to be lower in bladder urothelial carcinoma compared with in non-tumor tissues (supplemental Table S9). Subsequently, we stained four immunostaining markers, AHNAK, EPPK1, MYH14, and OLFM4, to apply to the in-house 25 FFPE samples for further marker selection. Along with the immunohistochemical characteristics from The Human Protein Atlas, the in-house immunostaining results also indicated that, of the four candidate biomarkers, AHNAK was the only protein that could discriminate urothelial carcinoma from normal urothelial cells in preliminary immunocytochemical analysis of 25 FFPE tissue samples and five liquid-based cytology samples, separately.
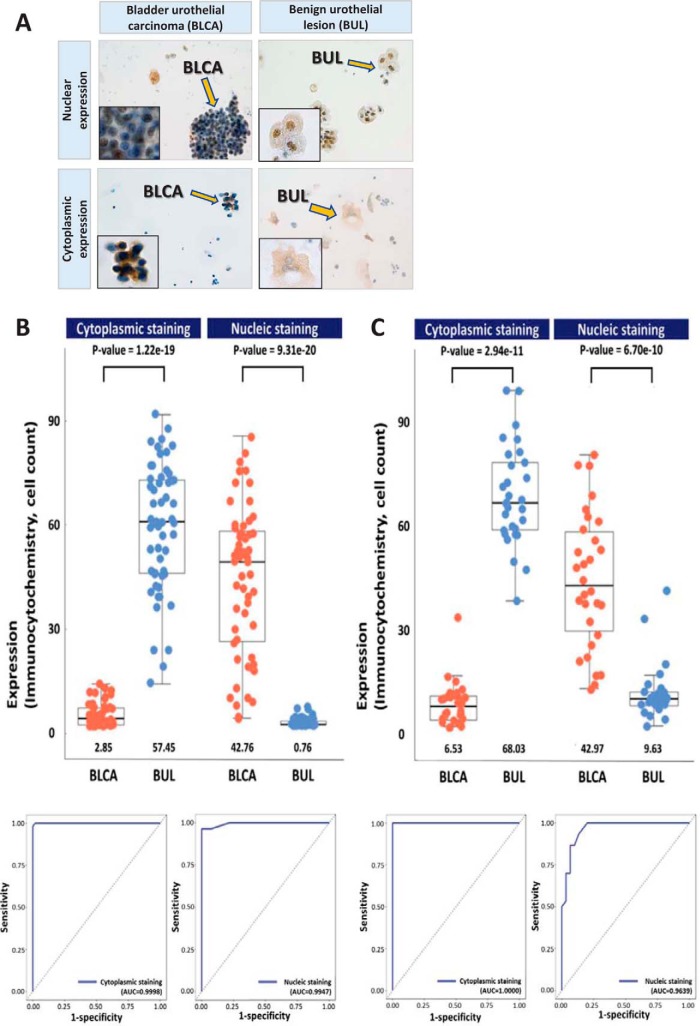
AHNAK was further investigated by immunocytochemistry in two independent validation cohorts of 55 and 60 voided-urine liquid-based cytology samples, which revealed that this biomarker demonstrated a cytological characteristic of cell-type specific subcellular localization. In urothelial carcinoma cells AHNAK immunocytochemistry was characterized by dominant nuclear staining, whereas in normal urothelial cells this protein was primarily localized to the cytoplasmic subcellular compartment (Fig. 5). The rate of positivity for nuclear AHNAK expression was significantly higher in carcinoma cells, compared with non-tumor urothelial cells (mean values, 42.7% and 42.9% versus 0.7% and 9.6%, p < 0.001, respectively for two independent validation tests; supplemental Table S10, supplemental Table S11 and Fig. 5), whereas notably lower cytoplasmic expression of AHNAK was observed in urothelial carcinoma cells compared with benign urothelial cells (2.8% and 6.5% versus 57.4% and 68.0%, p < 0.001, respectively for two independent validation tests; supplemental Table S10, supplemental Table S11 and Fig. 5). Diagnostic performances of AHNAK was summarized in Table II. In each of the two staining cases in Fig. 5, we calculated five performance measures (Sensitivity, Specificity, Accuracy, PPV, and NPV) using the AUC values for all possible threshold value of AHNAK expression and found the optimal threshold value which maximizes sums of sensitivity, specificity and accuracy. At the optimal threshold values for cytoplasmic and nucleic staining cases, the calculated performance measure values showed that AHNAK can be used to accurately discriminate between benign urothelial lesion and bladder urothelial carcinoma (Table II).
Fig. 5.
Results of for immunocytochemical staining of bladder urothelial carcinoma cells (BLCA, A) and benign urothelial cells (BUL, A) for AHNAK, and boxplot and beeswarm plot with the area under the receiver operating characteristic (ROC) curve (AUC) of individual immunocytochemical staining in two validation cohorts (B&C). A, Positive immunocytochemical staining for AHNAK in bladder urothelial carcinoma cells (left upper: nuclear expression and left lower: cytoplasmic expression. ×40, inset ×1000). Positive immunocytochemical staining for AHNAK in benign urothelial cells (right upper: nuclear expression and right lower: cytoplasmic expression. ×40, inset ×1000). (B and C) Average protein expression values are indicated by lines, and the median and quartile values of cells with AHNAK immunocytochemical staining are represented by boxes. p values obtained by nonparametric Wilcoxon test are shown on top of each bar graph. The AUC of AHNAK immunocytochemistry is shown to distinguish between benign urothelial lesion and bladder urothelial carcinoma.
Table II. Differential diagnostic performance of AHNAK immunocytochemistry at the optimal threshold for two independent validation cohorts.
| Immunostaining characteristics | Sensitivity | Specificity | Accuracy | PPV | NPV |
|---|---|---|---|---|---|
| 1st validation cohort (n = 55) | |||||
| Positive cytoplasmic staining | 1 | 0.982 | 0.991 | 0.982 | 1 |
| Positive nucleic staining | 0.964 | 1 | 0.982 | 1 | 0.965 |
| 2nd validation cohort (n = 60) | |||||
| Positive cytoplasmic staining | 1 | 1 | 1 | 1 | 1 |
| Positive nucleic staining | 1 | 0.8 | 0.9 | 0.833 | 1 |
DISCUSSION
Liquid-based preparation has been used increasingly for various types of cytology samples (9) and has technical advantages over conventional smear tests (25, 26). In this study, all liquid-based cytology samples were prepared using the preservation solution, “CytoRich Clear Preservative Fluid,” which contains a mixture mainly composed of ethanol with small amounts of methanol and isopropanol, and has been confirmed to achieve superior DNA preservation for genomic analysis, compared with the conventional 95% ethanol-based fixation used in previous studies (27, 28). After scraping off all cytological material within an encircled spot, our single-shot MS analysis achieved identification of a total of 4839 proteins in a relatively short time without fractionation, which is a comparable depth of proteome coverage to that reported in a previous study (29). Carvalho et al. performed LC-MS of conventional bronchial cytology pellets obtained by centrifugation, unlike our harvesting of cellular material from one liquid-based cytology slide from each patient, which can maximize the application of ancillary tests, as well as preserve residual samples for the preparation of additional slides for further molecular analysis, unlike pellet-based testing (30). The harvest of cells directly from cytology slides is still challenging because of the unique characteristics of sample fixation and the limited number of cells from each cytology slide as opposed to FFPE- or FACs-based sorted samples, which are replicable for preparation in proteomic analysis. In our study, to minimize the technical challenge, we standardized the entire preparation process for mass spectrometry. All liquid-based cytology samples were obtained from the same volumes of urine for each case to minimize sample loading bias. Peptides were injected into the mass spectrometer after all cells were extracted from the liquid-based cytology slide to be more productive for protein identification. To remove any systemic bias inherent to our approach including the different amounts of starting protein materials, quantile normalization was performed. Missing values were imputed by normal distribution. In addition, all quantified proteins in each sample groups were only subjected to label-free quantification. Thus, we leveraged our proteomic strategy to generate the first and largest in-depth quantitative proteome analysis of bladder urothelial carcinoma using cytology specimens.
Further analysis used the Perseus software platform for pairwise comparisons of changes in expression, a Student t test applied with a permutation-based approach generated a filtered list of 112 significantly altered proteins. The following co-expression analysis subsequently identified 11 shared proteins with FDR-adjusted p value < 0.05 from cross-filtering between liquid-based cytology and FFPE proteomics data. Finally, TCGA RNA sequencing data set was adopted to filter out the candidate markers out of 11 proteins that had no discriminate power between benign urothelial lesion and bladder urothelial carcinoma, which has been the widely utilized data analyzing method in many previous researches for the selection or validation of candidate markers obtained from their in-house data (31–33).
Interestingly, we observed cell type-specific differences between urothelial carcinoma and normal urothelium cells in the subcellular translocation of the AHNAK protein. Immunoreaction was mainly detected in the nuclei of bladder urothelial carcinoma cells, compared with cytoplasmic localization in benign urothelial cells on liquid-based cytology slides. These are unique immunocytochemical features, compared with published findings of immunohistochemical staining of FFPE bladder urothelial carcinoma tissue samples (34). Okusa et al. demonstrated higher cytoplasmic membrane protein expression of AHNAK in urothelial carcinoma tissues compared with adjacent normal tissue and suggested that AHNAK may be a bladder urothelial carcinoma specific marker (34). Hence, the results of FFPE-based immunological analysis are partly consistent with our observation of AHNAK immunoreactivity in FFPE bladder urothelial carcinoma specimens (supplemental Fig. S9). Although there is no evidence to explain the disagreement in the findings generated by the two methods, our assumption is that variations in the extent of sample processing for immunohistochemistry and immunocytochemistry lead to differences in the results of antibody-mediated staining. No processes are required prior to antibody staining in immunocytochemistry, which may affect the ability of the antibodies to recognize target epitopes, unlike the upstream processes invariably required for immunohistochemical analyses.
Aside from the technical aspect, the biological functions of AHNAK are another ambiguous factor, and may be defined by the diversity of the subcellular localization of this protein (35–39). When AHNAK was initially identified in normal bovine muzzle epidermal cells, immunofluorescent microscopy revealed a patchy staining pattern along the cytoplasmic membrane (40); however, various other cell-type-specific intracellular locations have been described in further reports (38, 41). Absence of uniform expression of AHNAK has consistently been described in malignant tumors, with diverse intracellular localization depending on the type of tumor cell (38, 41–44). AHNAK was described as a nucleoprotein in neuroblastoma cell lines, where its expression was significantly suppressed (36), whereas in a study of melanoma, immunoreaction was observed mainly in the cytoplasm of normal cells, as oppose to loss of expression in melanoma cells (35). Based on these observations, the authors suggested that AHNAK may have a tumor suppressor role, which is in accord with our findings, as well as those of other studies (39, 43). In our study, the levels of AHNAK protein and transcript were both significantly decreased in bladder urothelial carcinoma compared with benign urothelial lesion samples, based on proteomics and TCGA data. Along with the results of the analyses, our immunocytochemistry data support significantly reduced AHNAK expression in bladder urothelial carcinoma relative to benign urothelial lesion samples (54.3% versus 37.5%) which is also consistent with the immunohistochemical characteristics provided from The Human Protein Atlas. In the data set, three different kinds of antibodies (HPA019010, HPA19070, and HPA026643; Sigma-Aldrich, Saint Louis, MI) were tested and demonstrated unique immunoreactivity depending on the type of antibody. HPA019010, which was the identical antibody employed in our study, also showed reduced immunoreactivity in bladder urothelial carcinoma compared with benign urothelial lesion (supplemental Table S12). According to the external data bases, including SurvExpress and The Human Protein Atlas, aside from a diagnostic role, AHNAK was revealed as the only prognostic markers out of 11 proteins which were selected as candidates in both datasets (supplemental Fig. S10, supplemental Fig. S11 and supplemental Table S13).
In conclusion, taking advantage of advanced proteomic techniques, the present study identified a novel promising diagnostic biomarker which can be applied as a new ancillary test for differentiation of bladder urothelial carcinoma from benign urothelial lesion using voided-urine liquid-based cytology samples, which are the most frequently used diagnostic sample in routine practice. We successfully demonstrated that the nano LC-MS/MS technique can be applied to in-depth -omics analysis for novel biomarker evaluation using cytology preparations with limited amounts of cellular content, which has been the main practical restriction to the application of high-throughput genomics to human cytological samples. Further investigations will be required in our future studies to understand the intracytoplasmic translocation of AHNAK, which is the limitation of our study.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (22) partner repository with the dataset identifier PXD006805 (http://www.ebi.ac.uk/pride/archive/projects/PXD006805). Annotated MS/MS spectra can be accessed through MS-Viewer (23) (http://msviewer.ucsf.edu/prospector/cgi-bin/mssearch.cgi?report_title=MS-Viewer&search_key=d4qfuhxipu&search_name=msviewer) with the following search keys: d4qfuhxipu. Project Name: In-depth protein profiling of cytology samples in urothelial carcinoma. Project ID : PXD006805.
Supplementary Material
Footnotes
* This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (NRF-2015R1C1A1A01055032) and a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number : HI17C0048). We declare no competing financial interests.
 This article contains supplemental Figures and Tables.
This article contains supplemental Figures and Tables.
1 The abbreviations used are:
- FFPE
- formalin fixed paraffin embedded
- ACN
- acetonitrile
- AHNAK
- neuroblast differentiation-associated protein AHNAK
- AUC
- the area under the curve
- BLCA
- bladder urothelial carcinoma
- BUL
- benign urothelial lesion
- CV
- coefficient of variation
- DEG
- differentially expressed gene
- DEP
- differentially expressed protein
- EPPK1
- Epiplakin
- FDR
- false discovery rate
- iBAQ
- intensity based absolute quantification
- LBC
- liquid-based cytology
- MYH14
- Myosin-14
- NPV
- Negative predictive value
- OLFM4
- Olfactomedin-4
- PPV
- Positive predictive value
- ROC
- the receiver operating characteristics
- TCGA
- the cancer genome atlas
- WHO/ISUP
- World Health Organization/the International Society of Urologic Pathology.
REFERENCES
- 1. Fitzmaurice C., Allen C., Barber R. M., Barregard L., Bhutta Z. A., Brenner H., Dicker D. J., Chimed-Orchir O., Dandona R., Dandona L., Fleming T., Forouzanfar M. H., Hancock J., Hay R. J., Hunter-Merrill R., Huynh C., Hosgood H. D., Johnson C. O., Jonas J. B., Khubchandani J., Kumar G. A., Kutz M., Lan Q., Larson H. J., Liang X., Lim S. S., Lopez A. D., MacIntyre M. F., Marczak L., Marquez N., Mokdad A. H., Pinho C., Pourmalek F., Salomon J. A., Sanabria J. R., Sandar L., Sartorius B., Schwartz S. M., Shackelford K. A., Shibuya K., Stanaway J., Steiner C., Sun J., Takahashi K., Vollset S. E., Vos T., Wagner J. A., Wang H., Westerman R., Zeeb H., Zoeckler L., Abd-Allah F., Ahmed M. B., Alabed S., Alam N. K., Aldhahri S. F., Alem G., Alemayohu M. A., Ali R., Al-Raddadi R., Amare A., Amoako Y., Artaman A., Asayesh H., Atnafu N., Awasthi A., Saleem H. B., Barac A., Bedi N., Bensenor I., Berhane A., Bernabe E., Betsu B., Binagwaho A., Boneya D., Campos-Nonato I., Castaneda-Orjuela C., Catala-Lopez F., Chiang P., Chibueze C., Chitheer A., Choi J. Y., Cowie B., Damtew S., das Neves J., Dey S., Dharmaratne S., Dhillon P., Ding E., Driscoll T., Ekwueme D., Endries A. Y., Farvid M., Farzadfar F., Fernandes J., Fischer F., G/Hiwot TT., Gebru A, Gopalani S., Hailu A., Horino M., Horita N., Husseini A., Huybrechts I., Inoue M., Islami F., Jakovljevic M., James S., Javanbakht M., Jee S. H., Kasaeian A., Kedir M. S., Khader Y. S., Khang Y. H., Kim D., Leigh J., Linn S., Lunevicius R., El Razek H. M., Malekzadeh R., Malta D. C., Marcenes W., Markos D., Melaku Y. A., Meles K. G., Mendoza W., Mengiste D. T., Meretoja T. J., Miller T. R., Mohammad K. A., Mohammadi A., Mohammed S., Moradi-Lakeh M., Nagel G., Nand D., Le Nguyen Q., Nolte S., Ogbo F. A., Oladimeji K. E., Oren E., Pa M., Park E. K., Pereira D. M., Plass D., Qorbani M., Radfar A., Rafay A., Rahman M., Rana S. M., Soreide K., Satpathy M., Sawhney M., Sepanlou S. G., Shaikh M. A., She J., Shiue I., Shore H. R., Shrime M. G., So S., Soneji S., Stathopoulou V., Stroumpoulis K., Sufiyan M. B., Sykes B. L., Tabares-Seisdedos R., Tadese F., Tedla B. A., Tessema G. A., Thakur J. S., Tran B. X., Ukwaja K. N., Uzochukwu B. S., Vlassov V. V., Weiderpass E., Wubshet Terefe M., Yebyo H. G., Yimam H. H., Yonemoto N., Younis M. Z., Yu C., Zaidi Z., Zaki M. E., Zenebe Z. M., Murray C. J., and Naghavi M. (2016) Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-years for 32 Cancer Groups, 1990 to 2015: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 3, 524–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wiener H. G., Vooijs G. P., and van't Hof-Grootenboer B. (1993) Accuracy of urinary cytology in the diagnosis of primary and recurrent bladder cancer. Acta Cytologica 37, 163–169 [PubMed] [Google Scholar]
- 3. Raitanen M. P., Aine R., Rintala E., Kallio J., Rajala P., Juusela H., and Tammela T. L. (2002) Differences between local and review urinary cytology in diagnosis of bladder cancer. An interobserver multicenter analysis. Eur. Urol. 41, 284–289 [DOI] [PubMed] [Google Scholar]
- 4. Karakiewicz P. I., Benayoun S., Zippe C., Ludecke G., Boman H., Sanchez-Carbayo M., Casella R., Mian C., Friedrich M. G., Eissa S., Akaza H., Huland H., Hedelin H., Rupesh R., Miyanaga N., Sagalowsky A. I., Marberger M. J., and Shariat S. F. (2006) Institutional variability in the accuracy of urinary cytology for predicting recurrence of transitional cell carcinoma of the bladder. BJU Int. 97, 997–1001 [DOI] [PubMed] [Google Scholar]
- 5. Moonen P. M., Merkx G. F., Peelen P., Karthaus H. F., Smeets D. F., and Witjes J. A. (2007) UroVysion compared with cytology and quantitative cytology in the surveillance of non-muscle-invasive bladder cancer. Eur. Urol. 51, 1275–1280; discussion 1280 [DOI] [PubMed] [Google Scholar]
- 6. Hughes C. S., McConechy M. K., Cochrane D. R., Nazeran T., Karnezis A. N., Huntsman D. G., and Morin G. B. (2016) Quantitative Profiling of Single Formalin Fixed Tumour Sections: proteomics for translational research. Sci. Reports 6, 34949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tyanova S., Albrechtsen R., Kronqvist P., Cox J., Mann M., and Geiger T. (2016) Proteomic maps of breast cancer subtypes. Nat. Commun. 7, 10259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Frantzi M., Latosinska A., Fluhe L., Hupe M. C., Critselis E., Kramer M. W., Merseburger A. S., Mischak H., and Vlahou A. (2015) Developing proteomic biomarkers for bladder cancer: towards clinical application. Nat. Rev. Urol. 12, 317–330 [DOI] [PubMed] [Google Scholar]
- 9. Hoda R. S. (2007) Non-gynecologic cytology on liquid-based preparations: A morphologic review of facts and artifacts. Diagnostic Cytopathol. 35, 621–634 [DOI] [PubMed] [Google Scholar]
- 10. Epstein J. I., Amin M. B., Reuter V. R., and Mostofi F. K. (1998) The World Health Organization/International Society of Urological Pathology consensus classification of urothelial (transitional cell) neoplasms of the urinary bladder. Bladder Consensus Conference Committee. Am. J. Surg. Pathol. 22, 1435–1448 [DOI] [PubMed] [Google Scholar]
- 11. Barkan G. A., Wojcik E. M., Nayar R., Savic-Prince S., Quek M. L., Kurtycz D. F., and Rosenthal D. L. (2016) The Paris System for Reporting Urinary Cytology: The Quest to Develop a Standardized Terminology. Acta Cytologica 60, 185–197 [DOI] [PubMed] [Google Scholar]
- 12. Lee C. H., Chung S. Y., Moon K. C., Park I. A., Chung Y. R., and Ryu H. S. (2015) A Pilot Study Evaluating Fine-Needle Aspiration Cytology of Clear-Cell Renal Cell Carcinoma: Comparison of Ancillary Immunocytochemistry and Cytomorphological Characteristics of SurePath Liquid-Based Preparations with Conventional Smears. Acta Cytologica 59, 239–247 [DOI] [PubMed] [Google Scholar]
- 13. Han D., Moon S., Kim Y., Kim J., Jin J., and Kim Y. (2013) In-depth proteomic analysis of mouse microglia using a combination of FASP and StageTip-based, high pH, reversed-phase fractionation. Proteomics 13, 2984–2988 [DOI] [PubMed] [Google Scholar]
- 14. Rappsilber J., Mann M., and Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nature Protocols 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 15. Han D., Jin J., Woo J., Min H., and Kim Y. (2014) Proteomic analysis of mouse astrocytes and their secretome by a combination of FASP and StageTip-based, high pH, reversed-phase fractionation. Proteomics 14, 1604–1609 [DOI] [PubMed] [Google Scholar]
- 16. Tyanova S., Temu T., and Cox J. (2016) The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nature Protocols 11, 2301–2319 [DOI] [PubMed] [Google Scholar]
- 17. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 18. Schwanhausser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., and Selbach M. (2011) Global quantification of mammalian gene expression control. Nature 473, 337–342 [DOI] [PubMed] [Google Scholar]
- 19. Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M. Y., Geiger T., Mann M., and Cox J. (2016) The Perseus computational platform for comprehensive analysis of (prote)omics data. Nature Methods 13, 731–740 [DOI] [PubMed] [Google Scholar]
- 20. Deeb S. J., Tyanova S., Hummel M., Schmidt-Supprian M., Cox J., and Mann M. (2015) Machine Learning-based Classification of Diffuse Large B-cell Lymphoma Patients by Their Protein Expression Profiles. Mol. Cell. Proteomics 14, 2947–2960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Uhlen M., Fagerberg L., Hallstrom B. M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjostedt E., Asplund A., Olsson I., Edlund K., Lundberg E., Navani S., Szigyarto C. A., Odeberg J., Djureinovic D., Takanen J. O., Hober S., Alm T., Edqvist P. H., Berling H., Tegel H., Mulder J., Rockberg J., Nilsson P., Schwenk J. M., Hamsten M., von Feilitzen K., Forsberg M., Persson L., Johansson F., Zwahlen M., von Heijne G., Nielsen J., and Ponten F. (2015) Proteomics. Tissue-based map of the human proteome. Science 347, 1260419. [DOI] [PubMed] [Google Scholar]
- 22. Vizcaino J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Rios D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolome S., Apweiler R., Omenn G. S., Martens L., Jones A. R., and Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Baker P. R., and Chalkley R. J. (2014) MS-viewer: a web-based spectral viewer for proteomics results. Mol. Cell. Proteomics 13, 1392–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Reza A. M., Choi Y. J., Yasuda H., and Kim J. H. (2016) Human adipose mesenchymal stem cell-derived exosomal-miRNAs are critical factors for inducing anti-proliferation signalling to A2780 and SKOV-3 ovarian cancer cells. Sci. Reports 6, 38498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Son S. M., Koo J. H., Choi S. Y., Lee H. C., Lee Y. M., Song H. G., Hwang H. K., Han H. S., Yun S. J., Kim W. J., Kim E. J., and Lee O. J. (2012) Evaluation of Urine Cytology in Urothelial Carcinoma Patients: A Comparison of CellprepPlus(R) Liquid-Based Cytology and Conventional Smear. Korean J. Pathol. 46, 68–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Norimatsu Y., Kawanishi N., Shigematsu Y., Kawabe T., Ohsaki H., and Kobayashi T. K. (2010) Use of liquid-based preparations in urine cytology: An evaluation of Liqui-PREP and BD SurePath. Diagnostic Cytopathol. 38, 702–704 [DOI] [PubMed] [Google Scholar]
- 27. Kawahara A., Taira T., Abe H., Watari K., Murakami Y., Fukumitsu C., Takase Y., Yamaguchi T., Azuma K., Akiba J., Ono M., and Kage M. (2014) Fixation effect of SurePath preservative fluids using epidermal growth factor receptor mutation-specific antibodies for immunocytochemistry. Cancer Cytopathol. 122, 145–152 [DOI] [PubMed] [Google Scholar]
- 28. Krevolin M. D., Hardy D., Pane J., Aslam S., and Behrens C. M. (2017) Development and validation of a preanalytic procedure for performing the cobas HPV test in SurePath preservative fluid. J. Mol. Diagnostics 19, 288–294 [DOI] [PubMed] [Google Scholar]
- 29. Carvalho A. S., Cuco C. M., Lavareda C., Miguel F., Ventura M., Almeida S., Pinto P., de Abreu T. T., Rodrigues L. V., Seixas S., Barbara C., Azkargorta M., Elortza F., Semedo J., Field J. K., Mota L., and Matthiesen R. (2017) Bronchoalveolar lavage proteomics in patients with suspected lung cancer. Sci. Reports 7, 42190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rossi E., Bizzarro T., Martini M., Longatto-Filho A., Schmitt F., Fagotti A., Scambia G., and Zannoni G. F. (2017) The role of liquid based cytology and ancillary techniques in the peritoneal washing analysis: our institutional experience. PloS One 12, e0168625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Worst T. S., von Hardenberg J., Gross J. C., Erben P., Schnolzer M., Hausser I., Bugert P., Michel M. S., and Boutros M. (2017) Database-augmented mass spectrometry analysis of exosomes identifies claudin 3 as a putative prostate cancer biomarker. Mol. cellular Proteomics 16, 998–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Michaut M., Chin S. F., Majewski I., Severson T. M., Bismeijer T., de Koning L., Peeters J. K., Schouten P. C., Rueda O. M., Bosma A. J., Tarrant F., Fan Y., He B., Xue Z., Mittempergher L., Kluin R. J., Heijmans J., Snel M., Pereira B., Schlicker A., Provenzano E., Ali H. R., Gaber A., O'Hurley G., Lehn S., Muris J. J., Wesseling J., Kay E., Sammut S. J., Bardwell H. A., Barbet A. S., Bard F., Lecerf C., O'Connor D. P., Vis D. J., Benes C. H., McDermott U., Garnett M. J., Simon I. M., Jirstrom K., Dubois T., Linn S. C., Gallagher W. M., Wessels L. F., Caldas C., and Bernards R. (2016) Integration of genomic, transcriptomic and proteomic data identifies two biologically distinct subtypes of invasive lobular breast cancer. Sci. Reports 6, 18517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Surinova S., Radova L., Choi M., Srovnal J., Brenner H., Vitek O., Hajduch M., and Aebersold R. (2015) Non-invasive prognostic protein biomarker signatures associated with colorectal cancer. EMBO Mol. Med. 7, 1153–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hiroshi Okusa., Yoshio Kodera., Masamichi Oh-Ishi, Yutaka Minamida, Mayumi Tsuchida, Nicholas Kavoussi, Kazumasa Matsumoto, Takefumi Sato, Masatsugu Iwamura, Tadakazu Maedaand Baba S. (2008) Searching for new biomarkers of bladder cancer based on proteomic analysis. J. Electrophoresis 52, 19–24 [Google Scholar]
- 35. Sheppard H. M., Feisst V., Chen J., Print C., and Dunbar P. R. (2016) AHNAK is downregulated in melanoma, predicts poor outcome, and may be required for the expression of functional cadherin-1. Melanoma Res. 26, 108–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shtivelman E., Cohen F. E., and Bishop J. M. (1992) A human gene (AHNAK) encoding an unusually large protein with a 1.2-microns polyionic rod structure. Proc. Natl. Acad. Sci. U.S.A. 89, 5472–5476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shankar J., Messenberg A., Chan J., Underhill T. M., Foster L. J., and Nabi I. R. (2010) Pseudopodial actin dynamics control epithelial-mesenchymal transition in metastatic cancer cells. Cancer Res. 70, 3780–3790 [DOI] [PubMed] [Google Scholar]
- 38. Huang Y., Laval S. H., van Remoortere A., Baudier J., Benaud C., Anderson L. V., Straub V., Deelder A., Frants R. R., den Dunnen J. T., Bushby K., and van der Maarel S. M. (2007) AHNAK, a novel component of the dysferlin protein complex, redistributes to the cytoplasm with dysferlin during skeletal muscle regeneration. FASEB J. 21, 732–742 [DOI] [PubMed] [Google Scholar]
- 39. Lee I. H., Sohn M., Lim H. J., Yoon S., Oh H., Shin S., Shin J. H., Oh S. H., Kim J., Lee D. K., Noh D. Y., Bae D. S., Seong J. K., and Bae Y. S. (2014) Ahnak functions as a tumor suppressor via modulation of TGFbeta/Smad signaling pathway. Oncogene 33, 4675–4684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hieda Y., Tsukita S., and Tsukita S. (1989) A new high molecular mass protein showing unique localization in desmosomal plaque. The Journal of cell biology 109, 1511–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hashimoto T., Gamou S., Shimizu N., Kitajima Y., and Nishikawa T. (1995) Regulation of translocation of the desmoyokin/AHNAK protein to the plasma membrane in keratinocytes by protein kinase C. Exp. Cell Res. 217, 258–266 [DOI] [PubMed] [Google Scholar]
- 42. Silva T. A., Smuczek B., Valadao I. C., Dzik L. M., Iglesia R. P., Cruz M. C., Zelanis A., de Siqueira A. S., Serrano S. M., Goldberg G. S., Jaeger R. G., and Freitas V. M. (2016) AHNAK enables mammary carcinoma cells to produce extracellular vesicles that increase neighboring fibroblast cell motility. Oncotarget 7, 49998–50016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen B., Wang J., Dai D., Zhou Q., Guo X., Tian Z., Huang X., Yang L., Tang H., and Xie X. (2017) AHNAK suppresses tumour proliferation and invasion by targeting multiple pathways in triple-negative breast cancer. J. Exp. Clin. Cancer Res. 36, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Weller P., Bankfalvi A., Gu X., Dominas N., Lehnerdt G. F., Zeidler R., Lang S., Brandau S., and Dumitru C. A. (2014) The role of tumour FoxP3 as prognostic marker in different subtypes of head and neck cancer. Eur. J. Cancer 50, 1291–1300 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (22) partner repository with the dataset identifier PXD006805 (http://www.ebi.ac.uk/pride/archive/projects/PXD006805). Annotated MS/MS spectra can be accessed through MS-Viewer (23) (http://msviewer.ucsf.edu/prospector/cgi-bin/mssearch.cgi?report_title=MS-Viewer&search_key=d4qfuhxipu&search_name=msviewer) with the following search keys: d4qfuhxipu. Project Name: In-depth protein profiling of cytology samples in urothelial carcinoma. Project ID : PXD006805.