Abstract
We have used SWATH mass spectrometry to quantify 3648 proteins across 76 proteomes collected from genetically diverse BXD mouse strains in two fractions (mitochondria and total cell) from five tissues: liver, quadriceps, heart, brain, and brown adipose (BAT). Across tissues, expression covariation between genes' proteins and transcripts—measured in the same individuals—broadly aligned. Covariation was however far stronger in certain subsets than others: only 8% of transcripts in the lowest expression and variance quintile covaried with their protein, in contrast to 65% of transcripts in the highest quintiles. Key functional differences among the 3648 genes were also observed across tissues, with electron transport chain (ETC) genes particularly investigated. ETC complex proteins covary and form strong gene networks according to tissue, but their equivalent transcripts do not. Certain physiological consequences, such as the depletion of ATP synthase in BAT, are thus obscured in transcript data. Lastly, we compared the quantitative proteomic measurements between the total cell and mitochondrial fractions for the five tissues. The resulting enrichment score highlighted several hundred proteins which were strongly enriched in mitochondria, which included several dozen proteins were not reported in literature to be mitochondrially localized. Four of these candidates were selected for biochemical validation, where we found MTAP, SOAT2, and IMPDH2 to be localized inside the mitochondria, whereas ABCC6 was in the mitochondria-associated membrane. These findings demonstrate the synergies of a multi-omics approach to study complex metabolic processes, and this provides a resource for further discovery and analysis of proteoforms, modified proteins, and protein localization.
Keywords: Protein Translocation, Mitochondria function or biology, SWATH-MS, Tissues, Gene Expression, Mouse models, Subcellular Separation, Subcellular analysis, BXD mouse population, electron transport chain, multiomics, transcriptomics
Mitochondria are dynamic organelles essential to a range of metabolic processes, including the generation of ATP via oxidative phosphorylation (OXPHOS)1. Mitochondrial homeostasis is carefully maintained through many processes (1), including rapid modulation of mitochondrial protein expression (2). Protein quality control pathways in the mitochondria, such as the unfolded protein response, are essential for robust mitochondrial activity (2–4). Despite our increased knowledge of mitochondrial functions, we do not yet completely understand the mitochondria's variable proteomic composition and regulation—even for core functions such as OXPHOS. Several longstanding questions in mitochondria, such as the existence and makeup of OXPHOS supercomplexes (5), are now being addressed thanks to technical improvements in proteomics and protein imaging (6–8). Recent progress in further defining the composition of the mitochondrial proteome has been facilitated by improved organelle isolation procedures, such as by differential centrifugation (9), by mass spectrometry (MS)-based proteomics (10, 11), imaging of epitope-tagged proteins (12), and computational modeling (13). By combining many approaches, studies such as the human protein atlas (HPA) and MitoCarta have identified large parts of the mitochondrial proteome (14–17), e.g. MitoCarta 2.0 reports 1158 mitochondrial proteins (“mitoproteins”). These resources provide an excellent reference for subcellular protein localizations, yet it is also known that proteins are differentially localized in the cell depending on environmental conditions (18) and across tissue and cell types (15). Across all organelles it is estimated that less than half of the proteome can be assigned consistently to distinct, clear subcellular compartments (19). Consequently, it is of major interest to be able to identify and rapidly quantify proteins at large scale. In this study, we have applied SWATH-MS—which can consistently quantify up to ∼4000 unlabeled proteins in single injections of digested total cell extracts (8, 20, 21) with the aim of following proteins across widely varying cellular conditions and locations.
SWATH-MS, typically optimized for studying samples of similar overall protein composition, was adapted here to study the proteome across 5 distinct tissues and 2 fractions (total cell or isolated mitochondria) and 8 strains of the genetically diverse BXD mouse population (22). Across all samples, we quantified 3648 unique proteins. Among these 3648 included 774 of the 1158 proteins annotated as mitochondrial in MitoCarta (67%). Genes' tissue localizations likewise broadly aligned with the HPA database (72% clear alignment, 20% unclear, 8% clear disagreement). These data were aligned to transcriptional data from the same cohorts (23, 24) for multi-omic analysis, with just under half of variance at the mRNA level translating to variance at the protein level (∼42%), in line with previous estimates (25, 26). However, the likelihood of a relationship between a gene's transcript and protein is also a function of the gene's variance and expression, with ∼65% of highly variable and highly expressed transcripts covarying significantly with their protein, compared with only ∼8% of lowly variable and lowly expressed transcripts. This indicates that massive effects to transcript expression generally manifest at the protein level, as in gain- and loss-of-function studies, but subtler transcriptional effects, as seen across complex trait studies, are difficult to predict.
Next, we sought to examine the physiological relevance of proteomic expression variance, and how this may differ from conclusions drawn from transcriptome data. Among the 71 measured proteins from the OXPHOS pathway, clear differences were observed in protein abundances across tissue which hinted at functional differences in the mitochondria, e.g. that ATP synthase (Complex V, or CV) component proteins are depleted in BAT despite high mitochondrial content. This aligns to function, as BAT predominantly uses the ETC for mitochondrial uncoupling to generate heat (27), bypassing CV. Notably, this effect is not apparent for the same 71 OXPHOS genes' transcript levels measured at the same time in the same individuals. We last compared the abundances of proteins measured in the total cell and mitochondrial fractions to determine mitochondrial localization. This differential, or mitochondrial enrichment factor (EFmito), permitted empirical inference for proteins' probability to be mitochondrially localized. Most proteins with high EFmito are reported as mitoproteins in the literature, though not all. Four candidates were selected for validation—SOAT2, MTAP, IMPDH2, and ABCC6—by ultra-purified mitochondrial fractions and antibody-based approaches. The first three were observed inside the mitochondria, whereas ABCC6 was observed as part of the mitochondria-associated membrane (MAM). Together, both the novel and literature-supported findings indicate that fractionations measured by SWATH can be used to estimate proteins' localizations en masse, similar to other recent mass spectrometry technologies such as APEX (28) and hyperLOPIT (19).
EXPERIMENTAL PROCEDURES
Sample Selection
The 5 tissues studied—liver, BAT, quadriceps, brain, and heart—were collected from eight 29-week old male mice, one from each of 8 different BXD strains after overnight fasting and perfusion. All phenotyping and in vivo handling was approved by the Swiss cantonal veterinary of Vaud authority under licenses 2257 and 2257.1. Following the sacrifice, tissues were frozen and stored in liquid nitrogen. Later, tissues were broken by mortar and pestle and 30 to 100 mg were taken each for mRNA extraction, protein extraction, and for mitochondrial isolation followed by subsequent protein extraction. Our mitochondrial isolation technique has been recently described (29), and further details are available in supplemental methods. Whole cell samples and isolated mitochondrial samples were processed the same way for protein isolation as described (30), and further details are also available in supplemental. All MS runs were on a SCIEX (Framingham, MA) TripleTOF 5600 mass spectrometer. For transcriptomics, followed by microarray analysis on Affymetrix (Santa Clara, CA) Mouse Gene 1.0 ST (liver, quadriceps) or 2.0 ST arrays (heart, BAT).
Molecular Validation
For molecular validation using high-purity fresh mitochondrial samples, tissues samples were collected from young male mice from C57BL/6J or DBA/2J backgrounds (see labeling in figures) as the ultra-purification protocol followed for the validations (31) cannot be performed on frozen tissue. The protocol is also described further in supplemental methods. For Western blotting of these samples, antibodies (TARGET: catalogue number) were purchased by Santa Cruz Biotechnology (Dallas, TX) (IMPDH: sc-166551; reacts with human IMPDH1 but only with mouse IMPDH2), (MTAP: sc-100782), (SOAT2: sc-69837), (Tubulin: sc-5286), (TOM20, sc-11415), (ABCC6: S-20: sc-5787), (CREBH: sc-377332), and (KDEL: sc-58774); by Sigma Aldrich (LONP1: HPA002192); by BD Transduction Laboratories (Nucleoporin p62: 610497); by Abcam (Cambridge, UK) (Calnexin: ab13504) and (UQCRC2 & ATP5A OXPHOS mixture: ab110413); and by Cell Signaling Technology (Danvers, MA) (Caspase-3, CST #9662). LONP1, NUP62, and tubulin are, respectively, used as the positive controls of mitochondrial, nuclear, and cytosolic fractions. Western blotting primary antibodies were all run at dilution of 1:1000. ICC used the same antibodies at a dilution of 1:200. TOM20 was also used as a control for immunocytochemistry (ICC) and for ABCC6, whereas DAPI (4′,6-diamidino-2-phenylindole) was used as the nuclear staining control in ICC. ICC was performed as described previously (32). Here, C2C12 myotube and AML12 hepatocytes were used cultured on Nunc™ Thermanox™ Coverslip. The fixation was done with Formal-Fixx (Thermo Fisher Scientific), followed by permeabilization with 0.4% Triton. Cells were stained with the same rabbit polyclonal TOM20 (1:100) primary antibody; and anti-rabbit Alexa Fluor 568 secondary antibody (1:500, A10042 Life Technologies). The same MTAP (1:100) and SOAT2 (1:100) antibodies as above were stained together with TOM20 and DAPI. All images were obtained were acquired using Zeiss AxioPlan 2 Microscope System (Carl Zeiss AG, Oberkochen, Germany) under non-saturating exposure conditions. Cell line provenance and other details are described further in supplemental methods.
Experimental Design and Statistical Rationale
Eight was selected as the n per tissue because it is the minimum number of strains required for performing quantitative trait locus (QTL) analysis in two-parental crosses like the BXDs (33). QTLs were used to perform quality control to ensure that data were broadly aligned with expectations, i.e. certain strong cis-pQTLs could be expected if the data are of high quality (detailed under the Sample Size section in supplemental methods). Use of eight strains also permits sufficient sample size for standard statistical tests (e.g. two-way ANOVA and gene set enrichment analyses (GSEA)). We have previously shown the variation between biological and technical replicates in SWATH is less than the difference between strains (technical error is ∼1/3 of biological error, and biological error is ∼1/2 strain differences (8)). The variance in protein expression linked to tissue and organelle was expected to significantly exceed differences across genotypes within a tissue, which was indeed observed in the final data set, where ∼50% of all protein variance is attributed to differences between tissue, compared with ∼20% being driven by differences across strain within tissue. All datapoints were maintained in the analyses and in the supplemental datasets 2 and 3 except for comparisons between HPA and SWATH data, where protein quantifications were suppressed if the proteins had non-significant m-scores for all 8 measurements for a given tissue or fraction. For instance, SNCB (Fig. 2A) is clearly detected in all 8 brain samples, but not in any sample from the other tissues. However, SWATH can search for specific peptides at a specific retention time and mass/charge ratio with the requantification feature even if they are not highly abundant (34). For quantitative comparisons (i.e. all comparisons except tissue localization alignments in Fig. 1B and Fig. 2A–2B), e.g. the comparison of mRNA to protein level (Figs. 2E–2G), we retain all values, as the mRNA data can be used as a secondary validation of the protein quantification.
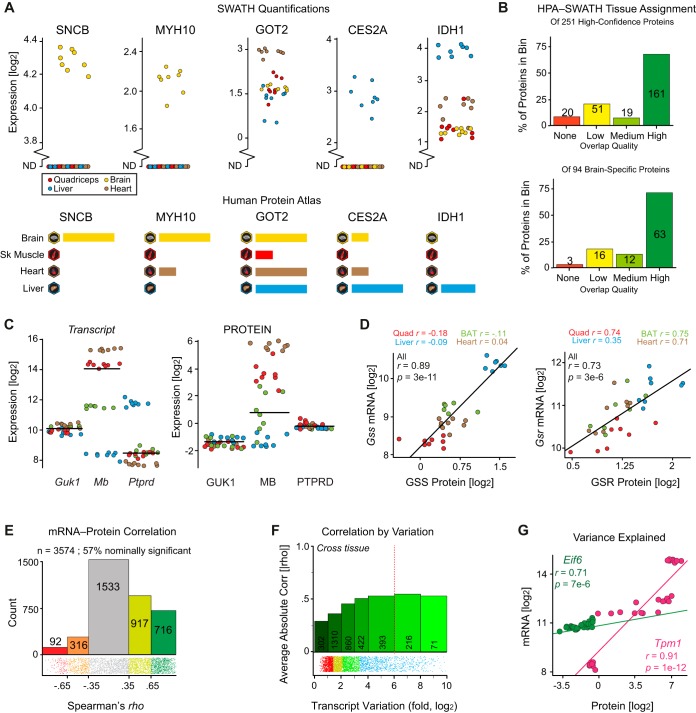
Fig. 2.
Variation and covariation across transcripts and proteins. A, Excerpt of the 251 comparisons between SWATH quantifications and HPA data for the four overlapping tissues (BAT is not in HPA). Each dot represents a different strain for the total proteome, with representative examples shown for exact matches (e.g. SNCB), close matches (MYH10, CES2A), moderate matches (GOT2), and discrepancies (IDH1). A broken axis with points at ND indicates “no data” (i.e. not quantified in the raw signal). B, Histogram of HPA-SWATH alignment (top) all tissues and (bottom) brain-only. HPA and SWATH broadly agree, with exact matches for ∼60%, moderate matches for ∼30%, and clear discrepancies for ∼10%. Further details can be found in the supplemental Table S1. C, Transcript and protein variation tend to be similar across layers (e.g. Guk1 and Mb), though exceptions are readily found (e.g. Ptprd). D, Although some gene product pairs covary only under situations of high variance, such as Gss—which correlates only when all tissues are considered—others are more consistently linear irrespective of variance, such as Gsr. E, Transcripts and proteins broadly covary. 57% of genes' proteins and transcripts correlate nominally (p < 0.05) across the data, at an average of rho = 0.42. F, More variable transcripts are more likely to correlate with their associated protein. Among highly variable transcripts (e.g. variance ≥ 26 fold, right of red dotted line), 72% correlate least nominally with their protein levels with an average correlation coefficient of rho = 0.53. Among the least variable transcripts (variance ≤ 21 fold), only 29% nominally correlate with an average correlation coefficient of rho = 0.29. G, Eif6 transcript and protein levels are strongly correlated despite relatively little variation across tissues. Tpm1 transcript and protein levels are highly correlated, but only in the context of massive cross-tissue variance (∼100-fold). Pearson correlation is used as the visual difference in expression variance is lost with Spearman correlations (which are rho = 0.69 and rho = 0.93 for Eif6 and Tpm1, respectively).
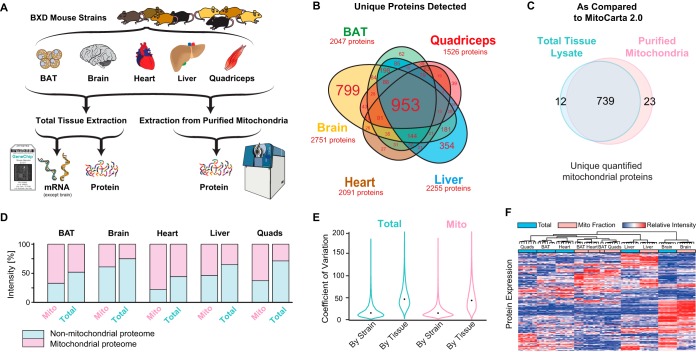
Fig. 1.
Cross-tissue mitochondrial proteomics using SWATH-MS. A, Project design. Eight genetically distinct BXD strains were selected for proteomic analysis in five tissues: BAT, brain, heart, liver, and quadriceps. Each tissue sample was measured for both whole-cell proteome and purified mitoproteome. mRNA was previously measured in the same individuals for all tissues except brain. B, Venn diagram showing the identification of proteins in each tissue. Brain has the most uniquely identified proteins. Font size corresponds to the number of proteins per category. C, Only a few additional mitoproteins are identified in purified mitochondria. This Venn diagram is collation of all tissues' data. D, Total proteome intensities summed and segregated into mitoproteins or non-mitoproteins based on MitoCarta. E, Coefficients of variation for protein intensities across strains and tissues. The median coefficient of variation within tissue across strains was ∼15%, compared with coefficients of variation of ∼45% within-strain across tissues. F, Hierarchical clustering of all protein expression levels across samples. Clustering (Euclidean distance, black lines at top) was performed using all samples proteins, although only 150 proteins are displayed below (y axis) for brevity and clarity in visualization.
RESULTS
Comprehensive Profiling of Mitochondrial Proteins Using SWATH-MS
In this study, we generated a multi-tissue and multi-omic dataset from 8 strains of the BXD mouse population, with the primary aim of discerning how the mitochondrial proteome varies across conditions. We have previously phenotyped these eight strains, and they are known to exhibit variation in mitochondrial phenotypes (8). mRNA data from the total tissue fraction from these individuals have also been generated (23, 24). Here, we took five tissues from these mice—BAT, brain, heart, liver, and quadriceps—and isolated protein from both the whole cellular fraction and from the purified mitochondrial fraction (Fig. 1A). To quantify the proteome, we first generated a spectral library from measuring the total tissue lysate and enriched mitochondrial samples for all tissues in data-dependent acquisition mode. The resulting consensus MS2 spectral library was merged with a prior library built from mouse liver lysate fractionated by off-gel electrophoresis (OGE) (8). This combined library contains 45,079 peptides (corresponding to 5152 distinct proteins). After the library was generated, all protein samples were then measured again on the mass spectrometer using data-independent acquisition mode with the aim of precisely and reproducibly quantifying as many mitochondrial proteins as possible in single MS measurements by SWATH-MS (20). The resulting data set quantified 3648 distinct proteins in at least one tissue. Of these proteins, 953 (26%) proteins were strongly identified in all five tissues, whereas 1281 (35%) proteins were tissue-specific (Fig. 1B). We next sought to determine our coverage of the mitoproteome by examining which of the 3648 quantified proteins are characterized in canonical mitoprotein databases, e.g. from MitoCarta 2.0 (17), AmiGO (35), and UniProt (36) (supplemental Table S1). Interestingly, mitoproteins are far more regularly detected across tissues than randomly selected proteins—only 87 of the 922 proteins which are reported as mitochondrial in at least one of the reference data sets are predominantly expressed in only a single tissue (9%), compared with 44% of nonmitochondrial proteins.
We next examined how mitoprotein detection varied between our total cell protein extraction and isolated mitochondria, using MitoCarta 2.0 as the primary reference. Among the 1158 mitoproteins listed in MitoCarta 2.0, we have quantified 774 (Fig. 1C). 756 mitoproteins were quantified in the purified mitochondrial fractions, whereas 726 were quantified in the total lysate. The majority (712 proteins, or 92%) were quantified in both extractions, whereas 12 MitoCarta proteins were detected only in whole cell fractions (e.g. Akr1b7, Arl2, As3mt, Cmc1, Sdsl), suggesting that these proteins may only be mitochondrially localized only under some conditions or tissues. In both fractions, differences in the total mitochondrial protein quantity across tissue were evident (Fig. 1D), even though the proteomic composition was consistent (i.e. 91% of mitoproteins were clearly quantified in all 5 tissues). In the total heart fraction, 55% of all protein signal stemmed from mitoproteins, in contrast to brain, where mitoproteins account for 25% of the total signal. These observations are consistent with the established literature for the mitochondrial density of different tissues (37) and may explain why the mitochondrial enrichment provided only a small increase in mitoprotein detections. We also observed that the variation in observed protein expression is driven primarily by the differences between tissues, with a secondary role played by differences between the 8 strains. (Fig. 1E). Given the magnitude of variation driven by tissue differences, hierarchical clustering of proteins across samples separated tissues completely by the whole cell fraction, and nearly completely separated mitochondrial fractions (Fig. 1F).
Genetic Variance and the Mitoproteome
As the pan-tissue protein data broadly aligned to expectations—e.g. heart and quadriceps have closer protein composition and expression than heart and brain—we next examined how tissue-specific proteins correspond to literature. To do so, we compared against the HPA which contains extensive, validated protein localization data for many tissues (although not BAT) and subcellular fractions for human samples. We selected the 764 proteins which were detected highly significantly in only one of these 4 tissues, or which were detected commonly to all (i.e. center and edges of Fig. 1B but using more stringent cutoffs for higher certainty; detailed in supplemental methods). Of these, 251 proteins have validated data from the HPA which also have clear and unique orthologs between the mouse and human gene. The putative localizations were compared between the two datasets (Fig. 2A), with most being in exact agreement (72%, e.g. SNCB, MYH10), whereas others were somewhat different (20%, e.g. GOT2), and a few in opposition (8%, e.g. CES2A, IDH1). Broadly, proteins' tissue localizations overlapped between SWATH and HPA (Fig. 2B), indicating that the clear distinction between tissue proteomes in the SWATH data are caused by underlying biological differences.
We next examined links between genes' transcript and protein expression for all 3574 genes with matching multi-omics measurements. Variances of transcript and protein expression were moderately covariable—genes with highly variable transcripts tend slightly to have more highly variable proteins and vice-versa (rho = 0.22 across all 3574 paired genes). However, exceptions to this trend are readily found, such as Ptprd (Fig. 2C). As for protein expression, tissue differences had a far larger impact on transcript expression than did strain differences. For instance, the median transcript variance is 4.4-fold across tissues for 8 strains, compared with a median variance of 1.8-fold across a single tissue (liver) for 40 strains (8). Many transcript-protein pairs covary only when data from all tissues are (e.g. GSS), although some genes have highly consistent covariance both within and across tissue (e.g. GSR) (Fig. 2D). Proteins and transcripts which have higher variation in their expression and with higher abundance are more likely to covary, and to covary more strongly. Across all samples, 57% of transcript-protein pairs covary with at least nominal significance (Fig. 2E; rho ≥ 0.35 corresponds to p < 0.05), with an average correlation of rho = 0.42. However, the expected covariance is higher for certain subsets of data. For instance, the 287 transcripts which vary by more than 64-fold (26) covary more strongly with their protein, with an average rho = 0.53 (Fig. 2F, right of dashed red line). In contrast, for the 302 transcripts which vary by less than 2-fold (21) the average correlation coefficient drops to r = 0.29 (Fig. 2F, leftmost group). A similar trend is observed for more abundant transcripts; the most abundant decile has an average rho of 0.52 versus 0.29 for the least-abundant decile of transcripts. Interestingly, expression level and expression variance do not covary (rho = 0.07 for transcript expression versus variance; rho = −0.005 for protein expression versus variance).
However, it is critical to note that the relationship between mRNA and protein cannot always be expected even for highly variable transcripts. 28% of transcripts which vary by ≥64-fold do not even nominally covary with their proteins (p > 0.05). Even in cases of significant correlation, one must also consider the utility of such findings. Eif6 varies across tissues by 1.8-fold and explains 44% of EIF6 protein levels (i.e. r2), whereas Tpm1 varies by 103-fold and explains 80% of TPM1 protein levels (Fig. 2G). Although the Eif6 connection is nominally weaker, it could provide more useful information for mechanistic examination: an increase in the Eif6 transcript of 1.5-fold can be expected to correspond to an increase in EIF6 protein levels, whereas Tpm1 would need to increase by >5-fold for one to confidently predict an increase in TPM1 levels. Fundamentally a gene's transcript and protein always covary—knocking out a transcript will knock out the corresponding protein. However, intermediate variances lead to highly variable relationships (e.g. linear models are only sometimes appropriate), which are not yet well understood. Although target genes in gain or loss-of-function models will have congruent effects on the mRNA and protein level, secondary or tertiary targets may not be reliable across omics layers. Similarly, population studies generally have low levels of gene expression variation (as compared with loss-of-function models) and are thus better served by a multi-omics approach. However, by incorporating information about transcripts' variance and expression levels, we may calculate the approximate probability for which we expect targets to validate at the protein level.
Dynamic Composition of Mitochondrial Protein Modules
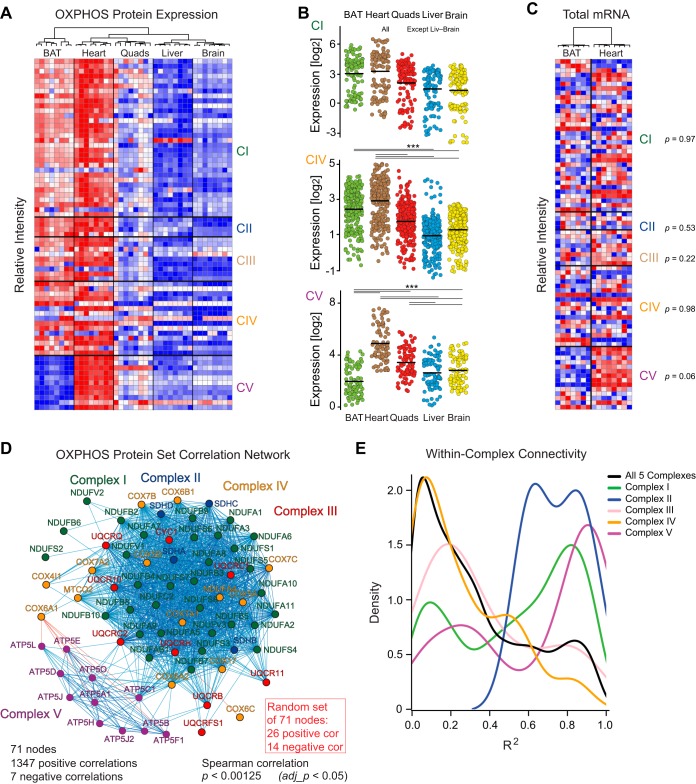
We hypothesized that the divergence in genes' mRNA and protein expression may result in distinct observations relating to mitochondria if separately examining the transcriptome or proteome data. Because of its core role in mitochondrial physiology, we elected to look at the 71 quantified proteins from the KEGG OXPHOS pathway (M19540). Distinct protein expression profiles were observed for each tissue, with average expression relatively the highest in the heart and relatively the lowest in liver and brain (Fig. 3A). Other patterns were visible as well, most noticeably that proteins from CV were expressed at low levels in BAT despite relatively high expression of proteins of other ETC complexes (Fig. 3A–3B). This relative down-regulation fits the tissue's functions, as BAT has high mitochondrial expression but relatively little ATP production because of its focus on mitochondrial uncoupling (38)—as the ETC is largely to produce heat (39). This is in clear contrast to the heart, where ATP regeneration is paramount (40). Interestingly, the tight coregulation of OXPHOS complex gene expression is weaker at the transcriptional level. Even the largest difference observed at the protein level—relative CV expression between BAT and heart—shows only a slight trend for the corresponding transcripts (Fig. 3C). Likewise, OXPHOS complex proteins are tightly coregulated both within and across complexes (Fig. 3D; CV is separate because of its disjunction in BAT). This observation agrees with previous findings that OXPHOS proteins tend to covary across complex, and they cluster particularly well within complex (8). The connectivity within complexes is highly variable which can be visualized through a density map of variance explained (Fig. 3E). Proteins in CI, CII, and CV all primarily correlated with proteins within the same complex, whereas proteins in CIII and CIV had equally strong and prevalent correlations across complex as within, mirroring the observations that CIII and CIV proteins are especially prone to dynamic interactions with other complexes (41, 42). Transcriptomic approaches are distinctly limited for the study of genes which form protein complexes (43), whereas antibody-based techniques for complex analysis can struggle with scaling up quantifying results across large numbers of samples or across multiple experiments. These data indicate that bottom-up MS proteomics measurements (i.e. fragmented peptide-based) can provide reliable overviews of expression of protein complexes, e.g. the OXPHOS complex.
Fig. 3.
Functional protein network based on large-scale proteomics. A, Relative protein intensities of the 71 measured OXPHOS proteins. BAT shows high expression of CI-IV, but the lowest expression of CV proteins (ANOVA post-hoc Dunnett test, p < 0.005 BAT versus all other tissues). B, Significant differences of CIV and CV protein expression across all five tissues (ANOVA with post-hoc Tukey tests, p < 0.005 indicated by ***). For CI, all differences are significant except for liver versus brain (p = 0.11). C, Transcript expression intensities in BAT and heart for the 66 OXPHOS transcripts that also have protein expression data (5 OXPHOS genes have protein measurements but no transcript measurements). Hierarchical clustering separates tissues, yet no complexes have different expression (paired Welch's t-tests, shown at side). D, Covariation network of all five tissues together shows major coexpression of CI-IV whereas CV is distinctly apart because of its contrary regulation in BAT. At adj_p < 0.05, there are 1347 positive correlations and 7 negative correlations between the 71 OXPHOS nodes (proteins). For a set of 71 proteins randomly selected from the same dataset, only 26 positive and 14 negative correlations are observed at the same cutoff. E, Covariation within complexes: Most correlations from CI, CII, and CV consistently correlate within their own complex, whereas CIII and CIV proteins are more variably distributed.
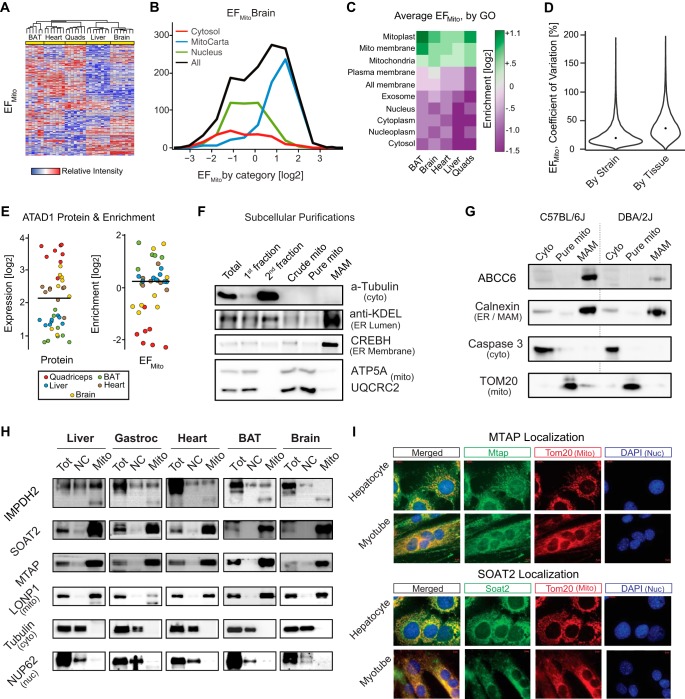
Mitochondrial Enrichment Factor as a Proxy of Mitochondrial Location
We next sought to examine how direct comparisons of the proteomic analysis of whole tissue extract and mitochondria-enriched fractions can provide complementary perspectives on the cellular state. First, we compared the protein quantifications of both fractions to calculate the mitochondrial enrichment factor (EFmito) for each detected protein (supplemental Table S1). The EFmito—the abundance ratio between the two fractions—quantitatively estimates to what extent a protein is relatively inside and outside the mitochondria. Proteins with high EFmito should be localized in the mitochondria, whereas proteins localized elsewhere should have low EFmito. As with the raw protein intensities, EFmito values cluster strongly by tissue (Fig. 4A, red = enriched in mitochondria; blue = diminished). We next collated the reported cytosolic and nuclear localization data for all proteins from UniProt (36) and the list of mitochondrial proteins from MitoCarta 2.0 (17) and examined their relative EFmito. As expected, protein sets reported as mitochondrial have the highest EFmito in all tissues, whereas proteins localized elsewhere, e.g. cytoplasm and nucleus, are always significantly depleted (Fig. 4B for brain; other tissues in supplemental Fig. S1A). On average across all five tissues, mitochondrial proteins were enriched by 4-fold compared with non-mitochondrial proteins (Fig. 4C; note that mitochondria are not removed from the whole cell fractions). As for protein intensities, the EFmito variation across tissues is larger than the variation caused by strain differences within any single tissue (coefficient of variation of 39% versus 22%, Fig. 4D). We next sought to identify how EFmito signatures compare with DNA-based prediction factors for mitochondrial localization: the mitochondrial-targeting sequence (MTS, using targetP 1.1 (44)). The MTS is a 20–40 amino acid sequence located in many precursor mitochondrial proteins and has been a good predictor for proteins destined to be imported to the mitochondria (Dinur-Mills et al., 2008). Among the 1158 genes reported as mitochondrially localized in MitoCarta, 729 are calculated to have likely MTS in mice (45). Among the 774 MitoCarta proteins quantified in this study, 486 have putative MTS—nearly two-thirds of the confirmed mitochondrial proteins. Conversely, among the 2808 non-MitoCarta proteins quantified, only 242 proteins have predicted MTS. Although proteins with predicted MTS tend to have higher EFmito, numerous exceptions are observed. Most noteworthy were two categories: (1) proteins reported in the literature as mitochondrial but which have low EFmito (i.e. similar to nuclear proteins), and (2) proteins reported as nuclear or cytoplasmic with high EFmito (i.e. similar to typical mitochondrial proteins) (Fig. 4B, tails). Furthermore, mitochondrial proteins in the outer membrane tend to not have MTS.
Fig. 4.
Using EFmito to identify mitochondrial localization. A, Cluster analysis of all 3648 proteins based on EFmito. Hierarchical clustering separates all tissues. B, Histogram of EFmito frequencies in brain for gene ontologies of nuclear, cytosolic, or mitochondrial localization. Mitochondrial proteins generally have high EFmito, whereas nuclear and cytosolic proteins generally have low EFmito, though significant overlap is observed. C, Heat map showing the average EFmito across all tissues in a selection of cell compartments, with consistent enrichment of mitoprotein sets. D, Coefficients of variation between strains and tissues for EFmito. Within strains, median is 22%, compared with 39% across tissues, i.e. tissue differences again have a larger impact than do strain differences. E, ATAD1 expression varied among tissues, with quadriceps having the highest expression yet lowest EFmito, suggesting tissue-dependent subcellular localization. F, Fractionations showing the ultracentrifugation purification of the mitochondria from the MAM. G, ABCC6 is consistently localized in the MAM across strains, though trace amounts appear in the cytosol. Abcc6 has a major sequence variant between C57BL/6J and DBA/2J (Williams et al., 2016), which substantially affects expression but not localization. H, Western blots indicate that MTAP, SOAT2, and IMPDH2 are localized in the mitochondria. LONP1, tubulin, and NUP62 were used as mitochondrial, cytosolic and nuclear markers, respectively. NC: Total cell minus the mitochondrial and MAM fractions. I, Mitochondrial and nuclear ICC staining of MTAP and SOAT2 shows the localization of both proteins with mitochondria in hepatocytes and myotubes. Tom20 and DAPI were used to stain mitochondria and nuclei, respectively.
All five tissues had similar overall localization and enrichment patterns for mitoproteins (e.g. Fig. 4C and supplemental Fig. S1A), though tissue-specific mitoproteins were also observed, such as UCP1 in BAT (supplemental Table S1). We first checked the 774 MitoCarta-reported mitochondrial proteins which have low EFmito scores in certain tissues where they are reliably detected (i.e. these proteins do not appear to be exclusively mitochondrial). This list includes ATPase family AAA-domain containing 1 (ATAD1), protein kinase A anchor protein 1 (AKAP1), and mitochondrial antiviral-signaling protein (MAVS) (Fig. 4E and supplemental Fig. S1B). In addition to the mitochondria, these three proteins have been reported in other cellular compartments: ATAD1 in the cytoplasm (46), MAVS in the peroxisome (47), and AKAP1 in the endoplasmic reticulum (48). This may be explained in part by differences between tissues and cell lines. For instance, ATAD1 is primarily mitochondrial in the BAT (EFmito = +1.0), but primarily non-mitochondrial in quadriceps (EFmito = −1.8) (Fig. 4E). Numerous other apparently tissue-specific localization differences are noted (supplemental Table S1, e.g. ADPRHL2 and ACBD3). The divergent organellar composition between tissues highlights the difficulty of maintaining a canonical database of protein localizations.
To validate our approach biochemically, we searched for novel potential mitochondrially localized proteins (i.e. those not reported in any of the databases examined in supplemental Table S1). Candidates were ranked based on their enrichment in each tissue, presence of MTS, and known literature such as antibody validations and reported subcellular localizations. The protein ATP-binding cassette, sub-family C (ABCC6; supplemental Fig. 1C) was the top candidate by these criteria and was selected first for validation. In the liver, ABCC6 has been controversially reported as a clear, unambiguous MAM protein (49), and as a clear, unambiguous plasma membrane protein (50), although it is not yet reported as mitochondrial in any standard database. Furthermore, ABCC6 has a major sequence variant between C57BL/6J and DBA/2J mice that leads to clear expression differences between the strains (51). We collected fresh livers for high purity mitochondrial isolations (31), which also allows for separation of the mitochondria-associated membrane by ultracentrifugation (MAM; Fig. 4F). In this validation experiment (Fig. 4G), ABCC6 was predominantly in the MAM fraction, with only trace signal coming from the cytosolic fraction (a longer exposure is shown in supplemental Fig. S1D). This confirmed our first hypothesis resulting from the EFmito calculations (MAM is enriched in the mitochondrial fractions for proteomics).
After ABCC6, the next strongest candidates are the genes 3110001D03Rik, Aqp4, Chmp6, Eml5, Efhc2, and Slc12a5. However, rather than go down the list linearly, we directly searched for novel candidate mitoproteins with closer to average enrichment to assess the general reliability of the approach. Several candidates were triaged which had signatures which would indicate potential mitoproteins and which also would indicate the general SWATH measurement is reliable. Sterol O-acyltransferase 2 (SOAT2) had reliable enrichment scores and substantially higher expression in liver than the other four tissues; inosine 5-phosphate dehydrogenase 2 (IMPDH2) had high enrichment in heart but low enrichment in other tissues, and methylthioadenosine phosphorylase (MTAP) was selected as a relatively ubiquitously expressed candidate. Several such candidates were considered for each category, with these three selected because of their antibody availability.
For these three candidates SOAT2, MTAP, and IMPDH2, we performed Western blots on three fractions: the total cell lysate (tot), the nucleus and cytoplasm-enriched fraction (NC), and the pure mitochondrial fraction (mito) across five fresh mouse tissues. SOAT2 and MTAP were detected consistently in the same fraction as the reference mitochondrial matrix protein LONP1—indicating these proteins are primarily mitochondrial under these experimental conditions (Fig. 4H). Conversely, differing isoforms of IMPDH2 appears to be localized either inside and/or outside the mitochondria. Finally, we elected to use immunocytochemistry (ICC) to visualize the two novel proteins which appear to be most distinctly located inside the mitochondria: SOAT2 and MTAP. Here, we observe substantial, though not complete, mitochondrial localization in both C2C12 myotubes and AML2 hepatocytes (Fig. 4I). As these proteins have been examined in other studies which have either not found them in the mitochondria, or which have been conflictingly reported in the mitochondria, it is likely that they are dynamically localized depending on the cell type and its environmental state. Together with the increasingly prevalent hypothesis that organellar protein composition is substantially dynamic (18), these findings emphasize the importance of implementing technologies and approaches which can perform rapid, comprehensive, and accurate assessments of proteins' locations.
DISCUSSION
In this study, we have generated a proteomics dataset across two cellular fractions and five tissues for eight inbred strains of the BXD mouse population in order to develop an overview into mitochondrial variation. The variation in expression across different tissues substantially outweighs the variation caused by differences caused by genetic factors and permits us to examine hypotheses that require multiple tissues to test and assists in examining hypotheses which benefit from highly variable data. First, SWATH is highly accurate and specific at determining the presence of proteins both across tissues and within cellular compartments and may thus be used as a parallel technology for localization experiments. The high variation in the expression of gene products across tissues also provides a resource for examining multi-omic relationships between mRNA and protein.
Multi-omic studies always observe positive average correlation between genes' transcript and protein levels, but with highly variable estimates—anywhere from around 0.25 to 0.85 (52–54). In this study, we were able to identify several variables which influence the average transcript-protein covariance. Particularly, transcripts which have more variable expression tend to correlate better with their corresponding protein (and vice-versa, as more variable proteins tend to correlate better with their transcript). Similarly, more abundant transcripts and proteins also tended to correlate better with one-another, and no association was observed between a gene product's abundance and its expression variance. Consequently, by subsetting our data on these characteristics, we were able to observe average correlation coefficients as high as 0.74—for genes with highly variable and highly expressed proteins and transcripts—and as low as 0.19—for gene products with low variation and low expression. Although expression level and variation alone are not likely the sole variables explaining the discrepancy in published studies on mRNA-protein relationships, they are a contributing factor.
These observed major variations in the expression of gene products within and across tissues was also linked to functional differences. For the mitochondria, we observed that OXPHOS proteins are co-expressed with approximate stoichiometry (55) and their absolute levels represent tissue mitochondrial density and OXPHOS state. However, the equivalent transcripts display far weaker functional connections, highlighting the importance of protein quantifications for mitochondrial analysis. Moreover, proteomics permits identification of which proteins are transiently localized to the mitochondria. Across the 14 different tissues of MitoCarta, only 1/3rd of the proteins are confidently detected in every tissue (17). Furthermore, recent meta-analyses have noted hundreds of discrepancies in the literature for organellar composition, with up to half of the proteome thought to be fluidly localized (19, 56). Standardized databases of protein localizations have been built up over decades of careful and painstaking research yet contain discrepancies even for well-studied proteins such as GSR—a high confidence mitochondrial protein according to MitoCarta, but a nuclear and cytoplasmic protein in HPA. Both sources may be correct, and rather the circumstances that differentially localize GSR are simply not understood. Here, we have identified several dozen potential novel mitoproteins, of which four were selected for validation: ABCC6, IMPDH2, SOAT2, and MTAP. Because of these validations, we can show at least three possible reasons for discrepancies between resources (1). The tissues and cell lines used for localization studies may not be generally extrapolated; whereas IMPDH2 is expressed in all five tissues, it is only localized in the mitochondria in four of them (2). Divergent localization of protein isoforms may be difficult to detect; in cases of mass spectrometry, when there are no available proteotypic peptides for each isoform (e.g. IMPDH2), and in cases of antibody-based approaches when two isoforms are of similar molecular weight (3). Organellar isolations are not always perfect, and enrichments may include off-target parts of other organelles.
Improvements in MS proteomics techniques, such as hyperLOPIT or SWATH-MS, are increasingly permitting researchers to comprehensively and precisely quantify the proteome at relatively modest investments of cost and time. Together, these results indicate that SWATH-MS can provide a rapid and comprehensive method to detect and quantify different protein expression patterns across tissues, genotype, and subcellular fractions. These technological advances can permit the generation and analysis of hypotheses on mitochondrial composition and function that could not be generated with transcriptome data alone. Consequently, it is now feasible to examine protein localization en masse as a function of several experimental conditions—a necessary development given the apparently dynamic state of much of the proteome. Genes which do not fit uniformly into reported locations of expression patterns can be picked up by multi-omic analyses, and subsequent experiments can then identify when, why, and how they move within the cell. Resources such as this study begin to provide a background for which later meta-analyses can mine to address observed differences in protein localization between databases or to address discrepancies between gene isoforms predicted from mRNA data, and those borne out in observation by proteomics. Future meta-analyses may then be able to observe additional patterns in localization.
DATA AVAILABILITY
ProteomeXchange Consortium via the PRIDE partner repository (57), under the identifier PXD005044 (www.ebi.ac.uk/pride/archive/projects/PXD005044). All MS runs were on a SCIEX TripleTOF 5600 mass spectrometer. The processed proteomic data are available in supplemental Tables 2 (peptide-level) and 3 (protein-level). Please note that protein level data is log2-scaled and centered at zero whereas peptide-level data are intensities and linear scale. Transcriptomics are available on GEO (58) for liver, BAT, and quadriceps under identifiers GSE60149, GSE60150, GSE60151 respectively (23). Heart transcriptomics data can be found on GEO under identifier GSE60489 (24). The transcriptome measurements are for the same individuals despite the non-continuous GEO numbers. All transcriptome data are also available in the supplemental data (supplemental Table 4), normalized in log2 scale and centered at 8.0.
Supplementary Material
Footnotes
* Research was further supported by the EPFL, ETHZ, ERC (Proteomics4D grant AdvG-670821 and Proteomics v3.0), the AgingX program of the Swiss Initiative for Systems Biology (51RTP0-151019, 2013/134, and 2013/153 to JA), SNSF (31003A-140780 and CSRII3-136201 to JA and 31003A_166435 to RA), and the NIH (R01AG043930 to JA). EGW was supported by an NIH F32 Ruth Kirschstein Fellowship (F32GM119190). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
 This article contains supplemental Figures and Tables.
This article contains supplemental Figures and Tables.
1 The abbreviations used are:
- OXPHOS
- oxidative phosphorylation (i.e. complex 1–4 + ATP synthase)
- SWATH
- sequential window acquisition of all theoretical mass spectra
- LOPIT
- localization of organelle proteins by isotope tagging
- BXD
- a recombinant inbred mouse cross between C57BL/6J and DBA/2J
- ETC
- electron transport chain (i.e. complex 1–4)
- CI/II/III/IV
- Complex I, II, III, IV of the electron transport chain
- CV
- ATP synthase (complex V of the electron transport chain)
- BAT
- brown adipose tissue
- MTS
- mitochondrial targeting sequence
- OGE
- off-gel electrophoresis
- MAM
- mitochondria-associated membrane
- NC
- nuclear and cytosolic (fraction)
- QTL
- quantitative trait locus
- GSEA
- Gene set enrichment analysis
- KEGG
- Kyoto Encyclopedia of Genes and Genomes
- HPA
- Human Protein Atlas
- GEO
- Gene Expression Omnibus
- ICC
- immunocytochemistry
- MS
- mass spectrometry.
REFERENCES
- 1. Nunnari J., and Suomalainen A. (2012) Mitochondria: in sickness and in health. Cell 148, 1145–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jovaisaite V., Mouchiroud L., and Auwerx J. (2014) The mitochondrial unfolded protein response, a conserved stress response pathway with implications in health and disease. J. Exp. Biol. 217, 137–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fox T. D. (2012) Mitochondrial protein synthesis, import, and assembly. Genetics 192, 1203–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Neupert W., and Herrmann J. M. (2007) Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76, 723–749 [DOI] [PubMed] [Google Scholar]
- 5. Schagger H., and Pfeiffer K. (2000) Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 19, 1777–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Letts J. A., Fiedorczuk K., and Sazanov L. A. (2016) The architecture of respiratory supercomplexes. Nature 537, 644–648 [DOI] [PubMed] [Google Scholar]
- 7. Gu J., Wu M., Guo R., Yan K., Lei J., Gao N., and Yang M. (2016) The architecture of the mammalian respirasome. Nature 537, 639–643 [DOI] [PubMed] [Google Scholar]
- 8. Williams E. G., Wu Y., Jha P., Dubuis S., Blattmann P., Argmann C. A., Houten S. M., Amariuta T., Wolski W., Zamboni N., Aebersold R., and Auwerx J. (2016) Systems proteomics of liver mitochondria function. Science 352, aad0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Frezza C., Cipolat S., and Scorrano L. (2007) Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat. Protoc. 2, 287–295 [DOI] [PubMed] [Google Scholar]
- 10. Foster L. J., de Hoog C. L., Zhang Y., Zhang Y., Xie X., Mootha V. K., and Mann M. (2006) A mammalian organelle map by protein correlation profiling. Cell 125, 187–199 [DOI] [PubMed] [Google Scholar]
- 11. Lu P., Vogel C., Wang R., Yao X., and Marcotte E. M. (2007) Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat. Biotechnol. 25, 117–124 [DOI] [PubMed] [Google Scholar]
- 12. Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., and O'Shea E. K. (2003) Global analysis of protein localization in budding yeast. Nature 425, 686–691 [DOI] [PubMed] [Google Scholar]
- 13. Calvo S., Jain M., Xie X., Sheth S. A., Chang B., Goldberger O. A., Spinazzola A., Zeviani M., Carr S. A., and Mootha V. K. (2006) Systematic identification of human mitochondrial disease genes through integrative genomics. Nat. Genet. 38, 576–582 [DOI] [PubMed] [Google Scholar]
- 14. Reinders J., Zahedi R. P., Pfanner N., Meisinger C., and Sickmann A. (2006) Toward the complete yeast mitochondrial proteome: multidimensional separation techniques for mitochondrial proteomics. J. Proteome Res. 5, 1543–1554 [DOI] [PubMed] [Google Scholar]
- 15. Uhlen M., Fagerberg L., Hallström B. M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., Olsson I., Edlund K., Lundberg E., Navani S., Szigyarto C. A., Odeberg J., Djureinovic D., Takanen J. O., Hober S., Alm T., Edqvist P. H., Berling H., Tegel H., Mulder J., Rockberg J., Nilsson P., Schwenk J. M., Hamsten M., von Feilitzen K., Forsberg M., Persson L., Johansson F., Zwahlen M., von Heijne G., Nielsen J., and Pontén F. (2015) Proteomics Tissue-based map of the human proteome. Science 347, 1260419. [DOI] [PubMed] [Google Scholar]
- 16. Ponten F., Jirstrom K., and Uhlen M. (2008) The Human Protein Atlas–a tool for pathology. J. Pathol. 216, 387–393 [DOI] [PubMed] [Google Scholar]
- 17. Calvo S. E., Clauser K. R., and Mootha V. K. (2016) MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 44, D1251–D1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kustatscher G., and Rappsilber J. (2016) Compositional dynamics: Defining the fuzzy cell. Trends Cell. Biol. 26, 800–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Christoforou A., Mulvey C. M., Breckels L. M., Geladaki A., Hurrell T., Hayward P. C., Naake T., Gatto L., Viner R., Martinez Arias A., and Lilley K. S. (2016) A draft map of the mouse pluripotent stem cell spatial proteome. Nat. Commun. 7, 8992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gillet L. C., Navarro P., Tate S., Röst H., Selevsek N., Reiter L., Bonner R., and Aebersold R. (2012) Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics 11, O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang H., Liu T., Zhang Z., Payne S. H., Zhang B., McDermott J. E., Zhou J. Y., Petyuk V. A., Chen L., Ray D., Sun S., Yang F., Chen L., Wang J., Shah P., Cha S. W., Aiyetan P., Woo S., Tian Y., Gritsenko M. A., Clauss T. R., Choi C., Monroe M. E., Thomas S., Nie S., Wu C., Moore R. J., Yu K. H., Tabb D. L., Fenyö D., Bafna V., Wang Y., Rodriguez H., Boja E. S., Hiltke T., Rivers R. C., Sokoll L., Zhu H., Shih I. M., Cope L., Pandey A., Zhang B., Snyder M. P., Levine D. A., Smith R. D., Chan D. W., Rodland K. D., CPTAC Investigators (2016) Integrated proteogenomic characterization of human high-grade serous ovarian cancer. Cell, 166, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang X., Pandey A. K., Mulligan M. K., Williams E. G., Mozhui K., Li Z., Jovaisaite V., Quarles L. D., Xiao Z., Huang J., Capra J. A., Chen Z., Taylor W. L., Bastarache L., Niu X., Pollard K. S., Ciobanu D. C., Reznik A. O., Tishkov A. V., Zhulin I. B., Peng J., Nelson S. F., Denny J. C., Auwerx J., Lu L., and Williams R. W. (2016) Joint mouse-human phenome-wide association to test gene function and disease risk. Nat. Commun. 7, 10464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Williams E. G., Mouchiroud L., Frochaux M., Pandey A., Andreux P. A., Deplancke B., and Auwerx J. (2014) An evolutionarily conserved role for the aryl hydrocarbon receptor in the regulation of movement. PLoS Genet. 10, e1004673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gu Q., Mendsaikhan U., Khuchua Z., Jones B. C., Lu L., Towbin J. A., Xu B., and Purevjav E. (2017) Dissection of Z-disc myopalladin gene network involved in the development of restrictive cardiomyopathy using system genetics approach. World J. Cardiol. 9, 320–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maier T., Guell M., and Serrano L. (2009) Correlation of mRNA and protein in complex biological samples. FEBS Lett. 583, 3966–3973 [DOI] [PubMed] [Google Scholar]
- 26. Edfors F., Danielsson F., Hallström B. M., Käll L., Lundberg E., Pontén F., Forsström B., and Uhlén M. (2016) Gene-specific correlation of RNA and protein levels in human cells and tissues. Mol. Syst. Biol. 12, 883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nicholls D. G., and Locke R. M. (1984) Thermogenic mechanisms in brown fat. Physiol. Rev., 64, 1–64 [DOI] [PubMed] [Google Scholar]
- 28. Lee S. Y., Kang M. G., Park J. S., Lee G., Ting A. Y., and Rhee H. W. (2016) APEX Fingerprinting Reveals the Subcellular Localization of Proteins of Interest. Cell Rep. 15, 1837–1847 [DOI] [PubMed] [Google Scholar]
- 29. Jha P., Wang X., and Auwerx J. (2016) Analysis of mitochondrial respiratory chain supercomplexes using blue native polyacrylamide gel electrophoresis (BN-PAGE). Curr. Protroc. Mouse Biol. 6, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu Y., Williams E. G., and Aebersold R. (2017) Application of SWATH Proteomics to Mouse Biology. Curr. Protoc. Mouse Biol. 7, 130–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wieckowski M. R., Giorgi C., Lebiedzinska M., Duszynski J., and Pinton P. (2009) Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nat. Protoc. 4, 1582–1590 [DOI] [PubMed] [Google Scholar]
- 32. Ryu D., Mouchiroud L., Andreux P. A., Katsyuba E., Moullan N., Nicolet-dit-Félix A. A., Williams E. G., Jha P., Lo Sasso G., Huzard D., Aebischer P., Sandi C., Rinsch C., and Auwerx J. (2016) Urolithin A induces mitophagy and prolongs lifespan in C. elegans and increases muscle function in rodents. Nat. Med. 22, 879–888 [DOI] [PubMed] [Google Scholar]
- 33. Sen S., Satagopan J. M., Broman K. W., and Churchill G. A. (2007) R/qtlDesign: inbred line cross experimental design. Mamm. Genome 18, 87–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Röst H., Liu Y., D'Agostino G., Zanella M., Navarro P., Rosenberger G., Collins B. C., Gillet L., Testa G., Malström L., and Aebersold R. (2016) TRIC: an automated alignment strategy for reproducible protein quantification in targeted proteomics. Nat. Methods 13, 777–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Carbon S., Ireland A., Mungall C. J., Shu S., Marshall B., Lewis S., AmiGO Hub; Web Presence Working Group (2009) AmiGO: online access to ontology and annotation data. Bioinformatics 25, 288–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Magrane M., and UniProt Consortium. (2011) UniProt Knowledgebase: a hub of integrated protein data. Database 2011, bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Else P. L., and Hulbert A. J. (1985) An allometric comparison of the mitochondria of mammalian and reptilian tissues: the implications for the evolution of endothermy. J. Comp. Physiol. B. 156, 3–11 [DOI] [PubMed] [Google Scholar]
- 38. Kramarova T. V., Shabalina I. G., Andersson U., Westerberg R., Carlberg I., Houstek J., Nedergaard J., and Cannon B. (2008) Mitochondrial ATP synthase levels in brown adipose tissue are governed by the c-Fo subunit P1 isoform. FASEB J. 22, 55–63 [DOI] [PubMed] [Google Scholar]
- 39. Forner F., Kumar C., Luber C. A., Fromme T., Klingenspor M., and Mann M. (2009) Proteome differences between brown and white fat mitochondria reveal specialized metabolic functions. Cell Metab. 10, 324–335 [DOI] [PubMed] [Google Scholar]
- 40. Dorn G. W. 2nd, Vega R. B., and Kelly D. P. (2015) Mitochondrial biogenesis and dynamics in the developing and diseased heart. Genes Dev. 29, 1981–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cogliati S., Calvo E., Loureiro M., Guaras A. M., Nieto-Arellano R., Garcia-Poyatos C., Ezkurdia I., Mercader N., Vázquez J., and Enriquez J. A. (2016) Mechanism of super-assembly of respiratory complexes III and IV. Nature 539, 579–582 [DOI] [PubMed] [Google Scholar]
- 42. Greggio C., Jha P., Kulkarni S. S., Lagarrigue S., Broskey N. T., Boutant M., Wang X., Conde Alonso S., Ofori E., Auwerx J., Cantó C., and Amati F. (2017) Enhanced respiratory chain supercomplex formation in response to exercise in human skeletal muscle. Cell Metab. 25, 301–311 [DOI] [PubMed] [Google Scholar]
- 43. Liu Y., Borel C., Li L., Müller T., Williams E. G., Germain P. L., Buljan M., Sajic T., Boersema P. J., Shao W., Faini M., Testa G., Beyer A., Antonarakis S. E., and Aebersold R. (2017) Systematic proteome and proteostasis profiling in human Trisomy 21 fibroblast cells. Nat. Commun. 8, 1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Emanuelsson O., Nielsen H., Brunak S., and von Heijne G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016 [DOI] [PubMed] [Google Scholar]
- 45. Emanuelsson O., Brunak S., von Heijne G., and Nielsen H. (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protocols 2, 953–971 [DOI] [PubMed] [Google Scholar]
- 46. Chen Y. C., Umanah G. K., Dephoure N., Andrabi S. A., Gygi S. P., Dawson T. M., Dawson V. L., and Rutter J. (2014) Msp1/ATAD1 maintains mitochondrial function by facilitating the degradation of mislocalized tail-anchored proteins. EMBO J. 33, 1548–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dixit E., Boulant S., Zhang Y., Lee A. S., Odendall C., Shum B., Hacohen N., Chen Z. J., Whelan S. P., Fransen M., Nibert M. L., Superti-Furga G., and Kagan J. C. (2010) Peroxisomes are signaling platforms for antiviral innate immunity. Cell 141, 668–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ma Y., and Taylor S. (2002) A 15-residue bifunctional element in D-AKAP1 is required for both endoplasmic reticulum and mitochondrial targeting. J. Biol. Chem. 277, 27328–27336 [DOI] [PubMed] [Google Scholar]
- 49. Martin L. J., Lau E., Singh H., Vergnes L., Tarling E. J., Mehrabian M., Mungrue I., Xiao S., Shih D., Castellani L., Ping P., Reue K., Stefani E., Drake T. A., Bostrom K., and Lusis A. J. (2012) ABCC6 localizes to the mitochondria-associated membrane. Circ. Res. 111, 516–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pomozi V., Le Saux O., Brampton C., Apana A., Iliás A., Szeri F., Martin L., Monostory K., Paku S., Sarkadi B., Szakács G., and Váradi A. (2013) ABCC6 is a basolateral plasma membrane protein. Circ. Res. 112, e148–e151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Meng H. J., Vera I., Che N., Wang X., Wang S. S., Ingram-Drake L., Schadt E. E., Drake T. A., and Lusis A. J. (2007) Identification of Abcc6 as the major causal gene for dystrophic cardiac calcification in mice through integrative genomics. Proc. Natl. Acad. Sci. U.S.A. 104, 4530–4535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liu Y., Beyer A., and Aebersold R. (2016) On the dependency of cellular protein levels on mRNA abundance. Cell 165, 535–550 [DOI] [PubMed] [Google Scholar]
- 53. Jovanovic M., Rooney M. S., Mertins P., Przybylski D., Chevrier N., Satija R., Rodriguez E. H., Fields A. P., Schwartz S., Raychowdhury R., Mumbach M. R., Eisenhaure T., Rabani M., Gennert D., Lu D., Delorey T., Weissman J. S., Carr S. A., Hacohen N., and Regev A. (2015) Immunogenetics Dynamic profiling of the protein life cycle in response to pathogens. Science 347, 1259038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. de Sousa Abreu R., Penalva L. O., Marcotte E. M., and Vogel C. (2009) Global signatures of protein and mRNA expression levels. Mol. Biosyst. 5, 1512–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Houtkooper R. H., Mouchiroud L., Ryu D., Moullan N., Katsyuba E., Knott G., Williams R. W., and Auwerx J. (2013) Mitonuclear protein imbalance as a conserved longevity mechanism. Nature 497, 451–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lund-Johansen F., de la Rosa Carrillo D., Mehta A., Sikorski K., Inngjerdingen M., Kalina T., Røysland K., de Souza G. A., Bradbury A. R., Lecrevisse Q., and Stuchly J. (2016) MetaMass, a tool for meta-analysis of subcellular proteomics data. Nat Methods, 13, 837–840 [DOI] [PubMed] [Google Scholar]
- 57. Vizcaino J.A., Csordas A., Del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q. W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Edgar R., Domrachev M., and Lash A. E. (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ProteomeXchange Consortium via the PRIDE partner repository (57), under the identifier PXD005044 (www.ebi.ac.uk/pride/archive/projects/PXD005044). All MS runs were on a SCIEX TripleTOF 5600 mass spectrometer. The processed proteomic data are available in supplemental Tables 2 (peptide-level) and 3 (protein-level). Please note that protein level data is log2-scaled and centered at zero whereas peptide-level data are intensities and linear scale. Transcriptomics are available on GEO (58) for liver, BAT, and quadriceps under identifiers GSE60149, GSE60150, GSE60151 respectively (23). Heart transcriptomics data can be found on GEO under identifier GSE60489 (24). The transcriptome measurements are for the same individuals despite the non-continuous GEO numbers. All transcriptome data are also available in the supplemental data (supplemental Table 4), normalized in log2 scale and centered at 8.0.