Abstract
Generation of reactive oxygen species (ROS) has been shown to be important for many physiological processes, ranging from cell differentiation to apoptosis. With the development of the genetically encoded photosensitiser KillerRed (KR) it is now possible to efficiently produce ROS dose-dependently in a specific cell type upon green light illumination. Zebrafish are the ideal vertebrate animal model for these optogenetic methods because of their transparency and efficient transgenesis. Here we describe a zebrafish model that expresses membrane-targeted KR selectively in motor neurons. We show that KR-activated neurons in the spinal cord undergo stress and cell death after induction of ROS. Using single-cell resolution and time-lapse confocal imaging, we selectively induced neurodegeneration in KR-expressing neurons leading to characteristic signs of apoptosis and cell death. We furthermore illustrate a targeted microglia response to the induction site as part of a physiological response within the zebrafish spinal cord. Our data demonstrate the successful implementation of KR mediated ROS toxicity in motor neurons in vivo and has important implications for studying the effects of ROS in a variety of conditions within the central nervous system, including aging and age-related neurodegenerative diseases, such as Alzheimer's disease, Parkinson's disease and amyotrophic lateral sclerosis.
Keywords: Zebrafish, Optogenetics, Oxidative stress, Motor neurons, Microscopy
Graphical abstract
Highlights
-
•
Motor neurons can be targeted for oxidative stress using optogenetics in zebrafish.
-
•
KillerRed expressing neurons undergo characteristic sequence of neurodegeneration.
-
•
Targeted neurons show microglial activation as part of the physiological response.
-
•
ROS toxicity has important implications for mechanisms driving neurodegeneration.
1. Introduction
The generation of oxidative stress (OS) is an imbalance in the homeostasis of oxidation-reduction (redox) reactions and develops as a result of increased reactive oxygen species (ROS) in excess of available antioxidants. This disturbance of the normal redox state in favour of pro-oxidative factors is associated with numerous pathophysiological processes. While it is well-appreciated that ROS are physiologically important signalling molecules, excessive levels are associated with aging and the development of neurodegenerative diseases such as Parkinson's disease, Alzheimer's disease and amyotrophic lateral sclerosis (ALS) [1], [2], [3], [4], [5], [6]. Pathogenic disruptions in ALS have been linked to OS via redox dysregulation including protein aggregation, hyper-excitability, mitochondrial dysfunction, and impaired axonal transport [7], [8], [9], [10]. Motor neurons (MNs) seem particularly sensitive to these pathological effects and ROS have been demonstrated to result in DNA and tissue damage, inflammation and subsequent cellular apoptosis [11], [12], [13], [14], [15], [16]. Overall ROS-mediated OS appears to be an important factor in the progression of neurodegenerative diseases.
The standard approach for experimental exposure of cells or organisms to ROS has been through the global application of ROS or ROS-generating reagents. New approaches have become available to selectively target ROS to individual cells or particular cellular structures [17], [18], [19]. Optogenetic approaches use genetically encoded, light-inducible, ROS-generating photosensitizers (RGP) that can be expressed in specific tissues and cell types (e.g. intestine or neurons), subcellular compartments (e.g. nucleus, cellular membrane or lysosome), or even fused to individual proteins. This transgenic approach allows for advanced temporal and spatial regulation of (sub-) lethal ROS production [20], [21], [22] that has not been possible previously. One RGP that has been successfully used to specifically deliver ROS under the optogenetic control is the phototoxic fluorescent protein KillerRed (KR) [23], [24], [25]. Upon prolonged green light excitation (absorption spectrum 540–580 nm) KR generates high levels of ROS along with photo-bleaching of the fluorophore. While photoexcitation of KR generates both singlet oxygen and superoxide radicals, singlet oxygen is considered the primary damaging agent produced by this photosensitizer [26]. KR-mediated cell death has been shown to occur via apoptosis and its phototoxic activity is directly related to level of excitation (light intensity), duration of excitation (illumination time) and expression level [26].
Zebrafish embryos provide an excellent model system for these optogenetic studies, as they are transparent and allow long-term live-imaging studies. Furthermore, increased efficiency of transposon-mediated transgenesis now makes it possible to generate transgenic zebrafish models expressing tissue-specific RGPs targeting single cells or whole organs in vivo. As a vertebrate model, 70% of human genes have a zebrafish orthologue, and 82% of genes known to be associated with human disease have a zebrafish counterpart [27]. KR has been successfully applied previously in cell culture, worms [Caenorhabditis elegans], and zebrafish [Danio rerio]. Kobayashi et al. (2013) utilised membrane-tagged KillerRed (memKR) to target chemosensory neurons in C. elegans [20]. Korzh et al. (2011) generated transgenic zebrafish lines expressing memKR to generate ROS in the hindbrain, the habenular commissure and the optic tectum [28]. Teh et al. (2014) described a memKR zebrafish model of light-induced cardiac deficiency i.e. changes in heart rate and heart contractility [25]. Here we utilised the advantages of the zebrafish system to investigate ROS-mediated OS and neuronal degeneration of individual MNs in the vertebrate spinal cord. Using single-cell resolution, real-time confocal live imaging of zebrafish spinal MNs we visualise for the first time the vulnerability of MN to ROS-induced OS, leading to their apoptotic degeneration. This approach provides a novel and important platform to investigate OS and its role in spinal cord MN degeneration in vivo.
2. Results
2.1. Selective expression of KillerRed in the zebrafish spinal cord
To express KillerRed (KR) in zebrafish spinal motor neurons (MNs), we generated transgenic zebrafish expressing KR targeted to the inner cell membrane via a neuromodulin N-terminal membrane localization signal (MLS) under the control of the MN-specific promoter mnx1 (Fig. 1A). The mnx1 gene encodes a homeobox transcription factor that has conserved functions in vertebrate MN differentiation [29] and is expressed primarily in post-mitotic spinal cord MNs and a small set of interneurons [30], [31]. We used Tol2-mediated transgenesis as described previously [24] (Fig. 1B) to generate two stable transgenic zebrafish lines that either i) express KR in all spinal cord MNs (Tg[mnx1:MLS-KillerRed]) or ii) randomly integrate KR in a sub-set of spinal cord MNs (Tg[4xnrUAS:MLS-KillerRed, cryaa: EGFP]) . Germ line transmission was confirmed by PCR analysis of genomic DNA from F1 generation embryos for the genetically encoded photosensitizer KR (Fig. 1C). Tg[mnx1:MLS-KillerRed] expresses KR in selected primary MNs (Fig. 1D, E). Mosaic expression of KR-positive MNs (KR+ve) and/or EGFP+ve MNs was achieved by crossing of Tg[4xnrUAS:MLS-KillerRed, cryaa:EGFP] and Tg[met:GAL4,UAS:EGFP] (ed6) [32] (Fig. 2).
Fig. 1.
Motor neuron (MN) specific expression of KillerRed (KR). (A) A membrane localization signal (MLS) targets the photosensitizer protein KillerRed (KR) to the intracellular cell membrane of MNs (mnx1 promoter). Upon green light illumination (LI,), KR induces lipid oxidization generating reactive oxygen species (ROS) alongside photo-bleaching of KR. (B) Synthetic transposase mRNA and a Tol2 transposon plasmid DNA construct containing the Tol2 element, the mnx1 promoter and the sequence encoding MLS-KR were co-injected into one cell stage zebrafish eggs. The Tol2 construct is excised from the plasmid DNA and integrated into the genomic DNA. Tol2 insertions in germ cells are transmitted to the F1 generation (modified after Kawakami et al., 2007). (C) PCR analysis of genomic DNA extracted from 24hpf F1 generation zebrafish embryos, confirmed germ line transmission of KR. Expected product size for MLS-KR was 531 bp, b-actin served as a positive control (housekeeping gene). (D-E) MN specific MLS-KR expression (red) at 3 dpf (Tg[mnx1:MLS-KillerRed]). Images are lateral views, anterior to the left, dorsal to the top. Scale bar 25 µm.
Fig. 2.
Generation of mosaic KR expression utilising GAL4-UAS regulation. (A) The GAL4/upstream activating sequence (UAS) system is a powerful method for analysing cell function in vivo. The yeast GAL4 transcription factor activates the transcription of target genes by binding to UAS cis-regulatory sites. The Gal4/UAS system can be used as two-component gene expression system carried in separate lines. The driver line (Tg[met:Gal4; UAS:EGFP]) provides tissue-specific GAL4 expression and the responder line (Tg[4xnrUAS:MLS-KR, cryaa:EGFP]) carries the coding sequence for the gene of interest under the control of the UAS site. In the double transgenic F1 embryos, Gal4 expressing cells are visualized by fluorescent reporters, providing mosaic expression of the gene of interest (modified after Asakawa & Kawakami, 2008). (B) Zebrafish embryo at 3 dpf expressing both met:EGFP (green) and KR (red) (Tg[met:Gal4;UAS:EGFP;4xnrUAS:MLS-KillerRed, cryaa:EGFP]). (C-E) 5 dpf zebrafish larvae show mosaic expression for both KR-ve/EGFP+ve (C, E (green)), KR+ve/EGFP-ve (D, E (red)) and KR+ve/EGFP+ve (E (yellow)) motor neurons. All images are lateral views, anterior to the left, dorsal to the top. Scale bar 50 µm.
2.2. Quantification of KillerRed-induced ROS production upon light illumination
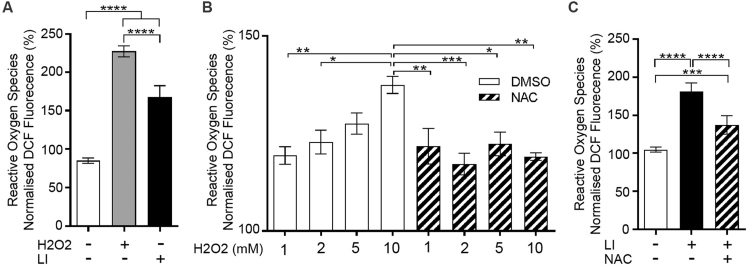
Overall intracellular oxidant activity was measured in an unbiased approach using the cell-permeable chemical reporter CM-H2DCFDA. This innately non-fluorescent reporter yields a highly fluorescent adduct following oxidation [33], thereby becoming a sensitive oxidant sensor. Microplate reader quantification of the oxidized fluorescent reporter served as a proxy for intracellular reactive oxygen species (ROS) concentration in vivo [34]. The oxidative stressor hydrogen peroxide (H2O2) was used as positive control and induced strong ROS-mediated fluorescence in KR+ve control fish (Fig. 3A). At 3 days post fertilisation (dpf) light-illuminated KR+ve zebrafish demonstrated significantly increased fluorescence levels compared to non-illuminated KR+ve clutch mates (Fig. 3A).
Fig. 3.
KR-activated ROS generation is rescued by the antioxidant N-acetyl Cysteine (NAC). The cell-permeable reporter CM-H2DCFDA was used to quantify ROS production 30 min after the OS event. (A) Intracellular ROS accumulation was quantified via microplate reader detection of the oxidized fluorescent reporter in control larvae (white bar, DMSO 0.1%, 1 h), H2O2 treated fish (grey bar, 5 mM, 1 h) and light-illuminated KR+ve zebrafish (black bar, LI, 2 h light-illumination). (B) The potent ROS scavenging compound Nacetylcysteine (NAC) rescued H2O2 induced ROS production. Zebrafish embryos (3 dpf) were pre-treated with DMSO or NAC before subsequent exposure to H2O2. (C) Light-illumination of KR+ve embryos resulted in an increase in ROS detection that was partially rescued by NAC pre-incubation. The arbitrary units of fluorescence measured in duplicate were normalized to blank readings. *P < 0.05, **P < 0.01; ***P < 0.001; ****P < 0.0001; one-way ANOVA and Tukey's post-hoc test (n = 3, SEM).
We next investigated the impact of antioxidant treatment upon KR-mediated ROS generation. The potent ROS scavenging compound Nacetylcysteine (NAC) has been shown to successfully rescue overall cellular and oxidative stress [35], [36], [37]. Application of increasing H2O2 concentrations (1 mM, 2 mM, 5 mM, and 10 mM) led to significant and dose-dependent increase in ROS-mediated fluorescence in 3 dpf zebrafish embryos (Fig. 3B; [38], [39], [40], [41]). While ROS accumulation and fluorescence in 1 and 2 mM treated fish was close to control levels and did not increase significantly, administration higher H2O2 concentrations led to a significant increase in ROS-mediated fluorescence (Fig. 3B). Embryos pre-treated with NAC showed no changes in their ROS levels with increasing H2O2 concentrations (Fig. 3B). To assess whether KR activated ROS generation can be prevented through NAC pre-treatment, both NAC-treated and non-NAC-treated KR+ve zebrafish embryos (3 dpf) were illuminated for 2 h on a stereo microscope (4x objective, 535–575 nm excitation filter, 80 mW/cm2). While green light illumination (LI) increased ROS-mediated fluorescence reliably and significantly, NAC treatment effectively decreased ROS accumulation and corresponding fluorescence levels compared to non-NAC-treated clutch mates (Fig. 3C).
2.3. Visualization of ROS-mediated cell death in selective motor neurons within the spinal cord
Apoptotic cells undergo a series of stereotypical morphological changes including progressive anterograde degeneration (blebbing), soma shrinkage, and nuclear condensation followed by nuclear fragmentation and chromosomal DNA degradation [42]. To visualise individual MN death in response to ROS exposure we generated transgenic zebrafish expressing mosaic KR and EGFP in spinal cord MNs (Tg[met:Gal4,UAS: EGFP] crossed to Tg[4xnrUAS:MLS-KillerRed, cryaa:EGFP]) (Fig. 4A). Light illumination (60 min initial continuous illumination plus ongoing scanning every 15 min; 40x objective, 535–575 nm excitation filter, 10 mW/cm2) of a subset of KR+ve/EGFP+ve MNs in 3 dpf zebrafish was sufficient to completely photo-bleach KR expression (Fig. 4B), concurrently inducing ROS production and initiating MN degeneration (Fig. 4C). The time-course of neuronal death was dependent upon illumination intensity and duration. In this example, the first visible signs of MN death presented 1.25 h post light-illumination in the form of axonal blebbing, followed by cell shrinkage 5 h post-illumination and complete cell loss around 8 h post-illumination (Fig. 4, Supplementary Video 1). Our data confirm that we can selectively target KR+ve MNs within the zebrafish spinal cord, leading to cell stress and death.
Fig. 4.
KR-mediated ROS production leads to MN degeneration. (A) A transgenic zebrafish (Tg[met:Gal4;UAS:EGFP];Tg[UAS:MLS-KR]) mosaically expressing EGFP (green; ai) and KR (red, aii) in individual neurons prior to light illumination and KR-activation (aiii). (B) Light illumination of fluorescent neurons leads to bleaching of KR fluorescence, evident 60 min post-illumination (bii), while EGFP fluorescence intensities remained unchanged (bi). (C) Live imaging of KR activated neurons for up to 10 h revealed a series of morphological changes including progressive anterograde degeneration (ci-ciii, arrowheads), soma shrinkage (arrows) and MN death (asterisks). Scale bar 25 µm.
Supplementary material related to this article can be found online at doi:10.1016/j.redox.2018.08.011
The following is the Supplementary material related to this article Video 1.
Time-lapse video of MN degeneration illustrated in Fig. 4. KR activation in our transgenic zebrafish (Tg[met:Gal4;UAS:EGFP];Tg[UAS:MLS-KR]) resulted in axonal blebbing and MN degeneration within 10 h after light illumination.
2.4. KillerRed-activated motor neurons express apoptotic markers
A well-established method to detect apoptotic cells in vitro is based on loss of membrane asymmetry [43]. During apoptosis, the normal asymmetric distribution of phospholipids in the cell membrane is lost, and phosphatidylserine (PS) is exposed on the outer leaflet of the plasma membrane. The calcium-dependent protein ANNEXINV (A5) binds PS with high affinity [44], [45], [46]. While we have demonstrated that dying MNs present with blebbing and cell shrinkage upon prolonged ROS exposure, we additionally investigated if light-exposed KR+ve MNs developed similar loss of membrane asymmetry. To evaluate this dynamic response, we created a triple-fluorescent zebrafish, co-expressing KR and blue fluorescent protein (TagBFP) in MNs, as well as A5 ubiquitously (Fig. 5). Fluorescent A5 (mVenus) was driven by the ubiquitin promoter [47] and had a secretion signal attached that enabled the extracellular labelling of apoptotic cells [45], [48]. In fact, successful A5 injections resulted in the ubiquitous transgene expression and a weak yellow background fluorescence throughout the whole animal. 2 dpf triple-transgenic embryos positive for all three markers (KR+ve/BFP+ve/A5 +ve) (Fig. 5A) were subjected to selective light illumination of a small subset of MNs (40x objective, 535–575 nm excitation filter, 10 mW/cm2) (Fig. 5B). Two day old zebrafish were used in these experiments to ensure high A5 expression and least amount of pigmentation in these fish (allowing shorter illumination times). 75 min of light illumination led to complete photo-bleaching of the targeted KR fluorescent protein but had no bleaching effect on the TagBFP expression. In fact, the TagBFP signal revealed that MN integrity was unaffected immediately after light irradiation (Fig. 5C). 2.25 h post-illumination fluorescent A5 labelling was observed predominantly along illuminated axons and soma while neighbouring neurons seemed unaffected (Fig. 5D, Supplementary Video 2&3). Taken together this data confirmed that light-induced KR activation leads to selective death of these MNs accompanied by A5 accumulation.
Fig. 5.
KR activation lead to ANNEXINV (A5) accumulation and degeneration of MNs. (A) Triple fluorescent zebrafish (2 dpf) positive for KR (red) and TagBFP (blue) selectively in MNs, as well ubiquitous expression of A5 (yellow), were used to visualise apoptotic processes after oxidative stress induction through KR illumination. (B) Prior to light illumination within a restricted area (green dotted line) of the zebrafish spinal cord, MNs showed high intensities of KR and TagBFP expression (bi). (C) Following light illumination for 75 min the fluorescence intensity of KR (red) was markedly reduced in the light-exposed region while no changes in TagBFP-intensities could be observed. (D) Time-lapse imaging following KR activation revealed A5 accumulation (di, arrowheads) along the axon and cell soma selectively within the light-activated area (2 h post-illumination). Scale bars 25 µm.
Supplementary material related to this article can be found online at doi:10.1016/j.redox.2018.08.011
The following is the Supplementary material related to this article Video 2, Video 3.
Time-lapse demonstration of ANNEXINV activation within the light-targeted MNs illustrated in Fig. 5.
3D representation of Video 3 illustrating the selective ANNEXINV activation along the light-targeted soma and axon of the KR-activated neuron.
2.5. Directed response of microglia to the KillerRed-activation site
To examine whether KR-induced ROS generation induces phagocytosis of neuronal debris via microglia (resident macrophages of the CNS), we crossed our transgenic zebrafish lines to create triple-labelled fish that expressed KR and TagBFP throughout the MNs, as well as EGFP-labelled macrophages/microglia (Fig. 6A; Tg[mnx1:MLS-KillerRed; mnx1:mTagBFP] crossed to Tg[mpeg1:EGFP]). Using confocal microscopy we were able to clearly distinguish between macrophages and microglia and visualise the microglial interaction with motor neurons in the spinal cord. We recently reported that microglia in the mpeg1 line respond to UV-laser mediated MN death with characteristic morphological changes and phagocytosis of neuronal remnants [48], [49]. We also observed a microglia response in 3 out of 6 KR illumination experiments (n = 6 zebrafish). Following light illumination for 60 min the KR-fluorophore bleached at the illumination site (Fig. 6B) as expected, while TagBFP expression and MN morphology remained unaffected (Fig. 6B). As a result of the photo-bleaching, neighbouring microglia underwent characteristic morphological changes such as branched morphology and extension/retraction of processes towards the illumination site, seemingly inspecting light-targeted MN-bodies (Fig. 6C-D). After approximately 2 h the microglia moved away from the illumination site (Fig. 6E). Several hours after this initial ‘inspection’ a microglia returned to the illumination site selectively surrounding a subset of MN cell bodies and changing morphology to a round amoeboid body that is characteristic of a phagocytic and an ‘activated’ state (Fig. 6F–H) [48]. Additionally, near the site of microglia activity a TagBFP+ve MN soma disappeared, indicating its death due to KR activation (Fig. 6H, Supplementary Video 4). Taken together, KR activation in MNs in the spinal cord triggered a physiological microglia response towards the illumination site.
Fig. 6.
Microglia migrate towards the site of KR activation. (A-H) Time lapse imaging of a zebrafish expressing green fluorescent microglia (mpeg1:EGFP), and MNs labelled in blue (mnx1:mTagBFP) and expressing red KR (mnx1:MLS-KillerRed) throughout the spinal cord. Post-illumination (pI, B, white dotted line) KR fluorescence was significantly reduced while TagBFP fluorescence remained unaffected. Green fluorescent microglia within close proximity extended its processes towards to the KR activation site within the first two hours (C-D), seemingly inspecting light-targeted MN-bodies. Microglia subsequently moved away from the illumination site (E). Approximately 9 h post-illumination, microglia were again observed at the illumination site (F). These microglia underwent characteristic morphological changes (amoeboid body) and remained at the KR activation site for several hours (G-H). Notably, near the site of microglia activity a TagBFP+ve MN disappeared, conceivably indicating its death due to KR activation (Supplementary Video 4). Scale bar 50 µm.
Supplementary material related to this article can be found online at doi:10.1016/j.redox.2018.08.011
The following is the Supplementary material related to this article Video 4.
Microglia response (green) towards the KR activation site over a time course of 10 h. Stills are presented in Fig. 6.
3. Discussion
We have established and characterised transgenic zebrafish lines expressing the genetically encoded photosensitiser KillerRed (KR) selectively in spinal cord motor neurons (MNs). Our results confirm that light illumination of the KR fluorophore in MNs leads to the generation of reactive oxygen species (ROS) and subsequent MN degeneration. Furthermore single-cell visualization studies confirmed that KR-mediated oxidative stress in MNs triggers a degenerative process combined with A5 accumulation, a physiological marker of apoptosis. Microglial migration towards the illumination site was also observed after KR activation, indicating a recognition of the ‘disturbance’ as part of their physiological response. Overall, our data demonstrate the successful implementation of KR-mediated ROS toxicity in MNs in vivo. This approach provides a unique platform to investigate the progression of neurodegeneration in real-time and how ROS mediated imbalances in MNs may contribute to the selective loss of these cells in diseases such as ALS.
Previous studies have utilised the cytotoxic properties of KR for optogenetic manipulation of specific tissue, distinct cell populations and cellular activity. Del Bene et al. demonstrated that KR-mediated ablation of interneurons in the optic tectum of the zebrafish leads to impaired prey capture [50]. Both Teh et al. and Korzh et al. generated multiple enhancer trap zebrafish lines expressing KR in specific tissue e.g. the hypothalamus, the hindbrain, in the endocardium and the myocardium [18]. In 2014 Teh and Korzh described a KR-zebrafish model of light-induced cardiac deficiency, with a reduction in both heartbeat and contractility after KR illumination [25]. Our aim was to selectively express KR in MNs in the zebrafish spinal cord, allowing a highly targeted and precise spatial control over KR activation and ROS generation in these cells. Membrane-targeted motor neuronal KR enabled the activation of KR on a single cell level. We created two stable transgenic KR zebrafish lines: one expressing KR within spinal cord MNs (Fig. 1D), and a UAS-KR zebrafish line, utilising the GAL4/UAS reporter system, to obtain mosaic labelling of MNs (Fig. 2).
To verify the KR mediated production of ROS we applied the cell-permeable ROS reporter CM-H2DCFDA and measured fluorescence accumulation in the individual zebrafish embryo. Analysis of the fluorescent reporter revealed significant ROS production in KR+ve zebrafish upon experimental exposure to green light. Treatment with hydrogen peroxide served as a positive control and led to a dose-dependent increase of ROS that could be prevented by pre-incubation with the ROS scavenging compound Nacetylcysteine (NAC). NAC treatment of the KR+ve zebrafish embryos was sufficient to reduce ROS levels, demonstrating the specific production of ROS as a result of KR activation. While the exact mechanism of light-induced ROS production remains uncertain [51], it has become clear that ROS readily react with all types of biological molecules and high concentrations can cause damage to various cellular and extracellular structures, such as proteins, lipids, and nucleic acids, often inducing irreversible functional alterations or even complete destruction [52], [53]. Today ROS are understood to play a crucial role in human (patho-)physiology [54].
To untangle the KR-mediated cascade of MN degeneration we applied our established zebrafish imaging approach, allowing longer-term and high-resolution visualization of multiple fluorescent reporter at a single-cell level [48], [49], [55]. Through co-labelling of individual MNs with KR and EGFP (Fig. 2, Fig. 4) we were able to i) observe the photo-bleaching of the KR-fluorophore on a single cell level and in real-time, ii) confirm that prolonged light illumination had no immediate or unspecific effect on MNs not expressing KR, and iii) visualise the cellular effects of the ROS production on individual neurons. Time-lapse observation revealed the typical cascade of MN degeneration, characterised by axonal blebbing followed by soma shrinkage and complete cell loss. The time course of these morphological changes was highly dependent on parameters such as the light intensity, illumination duration, and KR expression levels [26]. ROS have been reported to have a very short half-life (nanoseconds) and diffuse only short distances within the cell (20 nm) [56]. These qualities make ROS’ signalling specificity highly reliant on the amount, type and subcellular localization and offer a high level of experimental control. In our settings, positive responses could be observed between 1 and 4 h post-illumination, ranging from MN stress to MN death.
In order to visualise potential apoptotic alterations following KR activation we utilised yellow fluorescent A5, a marker of phosphatidylserine-exposed apoptotic cells, by co-expressing A5 ubiquitously with KR and blue fluorescent protein (TagBFP) in MNs (Fig. 5). If any cell undergoes cell death accompanied by PS exposure to the outer membrane, mVenus fluorescence becomes visible within a short time frame, reflecting the extracellular accumulation of A5. In our experiments we observed A5 accumulation specifically along the soma and the axonal projections within 2 h after KR activation. This observation fits the overall understanding that the apoptotic time-course depends upon several factors such as cell type, tissue and apoptosis-inducing agent, and that A5 is generally considered to present during the earlier stages of apoptosis. Next, we investigated weather KR-mediated neuron stress/death could trigger a physiological response such as microglia engagement. We utilised our mpeg1 transgenic zebrafish that express a fluorescent reporter, labelling both macrophages and microglia alike [57]. Microglia are defined as the resident macrophages of the CNS [58] and can be clearly differentiated from peripheral macrophages using standard (confocal) microscopy approaches. We have previously demonstrated that UV-laser mediated neuron degeneration triggers microglial uptake of neuronal debris within the zebrafish spinal cord [48], [55] and that inhibition of this process can lead to the spread of disease associated protein aggregates [49]. KR-mediated OS also initiated a directed microglia response towards the illumination site, revealing typical features of these ‘activated’ cells (extensive branching, rapid movement, and a changing morphology to a round amoeboid body), leading to uptake of neuronal debris. This response highlights the physiological relevance of KR-induced ROS production in the zebrafish spinal cord and makes it a powerful model to study neuron stress/death and its corresponding glial responses in vivo. Overall, we demonstrate that the KR-mediated MN degeneration reveals characteristic features of apoptotic cell death and prompts the engagement of microglia.
Our neuronal KR zebrafish model allows for the first time to observe the cascade of OS induced neurodegeneration in vivo and in real-time. Compound transgenic zebrafish expressing fluorescent MNs, ROS and apoptotic markers are useful to detail the precise time course of MN degeneration, to study the molecular mechanisms underlying MN death and to compare the susceptibility of MNs towards ROS-mediated degeneration versus other neurons and surrounding glia. While ROS production and OS are only two potential triggers of neurodegeneration (in ALS and other diseases), many more factors (such as excitotoxicity, mitochondrial dysfunction, protein aggregation etc.) have been identified to play an important role [9], [59]. Zebrafish present an excellent platform to test for pharmacological interventions that may regulate these processes of MN degeneration. In summary, the single-cell resolution approach combined with real-time live imaging of the zebrafish spinal cord establishes the KR-mediated degeneration of MNs as a result of ROS production following light illumination. Importantly, we demonstrate that this model is scalable for high throughput screening in vivo, with the ability to quantitatively measure ROS generation using a plate-reader. In future this platform may allow researcher to study with high cellular specificity mechanisms underlying ROS mediated oxidative imbalances, which are associated with aging as well as age-related diseases, including Alzheimer's disease, Parkinson's disease and ALS [4], [5], [6], [60].
4. Methods
4.1. Zebrafish husbandry and transgenic lines
Zebrafish (Danio rerio) were maintained at 28 °C in a 13 h light and 11 h dark cycle. Embryos were collected by natural spawning and raised at 28.5 °C in E3 solution according to standard protocol [61].
Wildtype fish were also treated with the tyrosinase inhibitor 1-phenyl-2-thiourea (200 nM PTU; Sigma) to reduce pigment formation starting at 16–24 hpf onwards. All experimental procedures, i.e. light irradiation, ROS quantification and live-imaging, were carried out on zebrafish embryos anesthetized with 0.01% (w/v) tricaine methansulfonate (MS-222, Sigma). Experimental protocols were approved by Macquarie University Animal Ethics Committee (Zebrafish models of neural disorders; protocol no. 2012/050; Using zebrafish to understand how the central nervous system responds to neuronal stress and death caused by neurodegenerative diseases, 2015/033). Previously described transgenic zebrafish lines used in this study include: Tg(met: GAL4,UAS:EGFP) (ed6)[32] and Tg(mnx1:mTagBFP) (mq10) [62]. The Tg(met:GAL4,UAS:EGFP) line is partially silenced and fish show therefore mosaic expression of green neurons within the spinal cord.
4.2. Generation of transgenic KillerRed zebrafish lines
Stable transgenic zebrafish expressing KR were generated using the Tol2-based transposon system [63]. pME-MLS-KillerRed was generated by blunt cloning KR (pKillerRed-mem, Evrogen) into a modified pDONR221 vector [63] using EcoRI/BamHI restriction enzymes. p5E-4xnrUAS was generated by PCR amplifying the UAS-E1b cassette from the previously described 4xnrUAS plasmid [64] and recombining into pDONR P1-P4R (Invitrogen). The primers used were: forward 5’- ggggactgcttttttgtacaaacttgAATTTCGAGGTCGAGGGAAT -3’, reverse 5’- ggggacaactttgtatagaaaagttgCGGTGGCTTCTAATCCGTGAG -3’. Tg[mnx1:MLS-KillerRed] (mq12) and Tg[4xnrUAS:MLS-KillerRed, myl7:EGFP] (mq11) transgenic fish were made by using recombined p5E-mnx1 (-6 to −2869 bp) [65], pME-MLS-KillerRed, p3E-pA and pDest-Tol2-pA2 [63] on roya9; mitfaw2 strain background and p5E-4xnrUAS, pME-MLS-KillerRed, p3E-pA [63] and pDest-Tol2-pA2-acrys-EGFP [66] on AB/TU strain background respectively. The new constructs used in this study have been deposited with the Addgene Repository (IDs #115517, #115516, #115515). To facilitate Tol2-mediated random integration into the genome cDNA (25 ng/μl) was co-microinjected with Tol2 transposase mRNA (25 ng/μl) into zebrafish embryos at the one cell stage as described previously [67]. Positive transgenic embryos (mosaic KR expression) were raised to adulthood and screened for germ line incorporation. Successful integration was evident through KR expression and confirmed via PCR analysis in F1 generation embryos. For PCR analysis, genomic DNA was extracted from 24 hpf embryos following standard protocols [61] and PCR-screened for the presence of incorporated KR using the primers: forward 5’- TGTTCCAGAGCGACATGACC -3’, reverse 5’- TGAAGGTCATCTTGCTGTCG -3’ and b-actin primers as a positive control [68]. AnnexinV experiments were performed as follows: Tg[mnx1:MLS-KillerRed] was crossed to Tg[mnx1:mTagBFP](mq10) [62] and eggs injected with ubiquitous A5 fused to the yellow fluorescent protein mVenus (ubb:secHsa. ANNEXINV-mVenus) [45], [48]. Microglia experiments we performed by crossing Tg[mnx1:MLS-KillerRed]; Tg(mnx1:mTagBFP] to Tg[mpeg1:Gal,UAS-mCherry] [57] (gl22Tg).
4.3. Zebrafish Reactive Oxygen Species (ROS) assays
The compound CM-H2DCFDA (ThermoFisher), a chloromethyl derivative of 2',7'-dichlorodihydrofluorescein diacetate (H2DCFDA), was used to monitor accumulation of reactive oxygen species in embryonic zebrafish. Upon cleavage of the acetate groups by intracellular esterases and oxidation, the nonfluorescent CM-H2DCFDA is converted to the highly fluorescent 2',7'-dichlorofluorescein (DCF). To validate the specify of the ROS assay 6 hpf embryos were pre-treated with either dimethyl sulphoxide (DMSO, 0.1%) or with DMSO plus the potent ROS scavenging compound Nacetylcysteine (NAC, 200 ɥM) until 3 dpf. Solutions were changed daily. At 3 dpf embryos were incubated for 30 min in hydrogen peroxide (H2O2) at increasing concentrations (1 mM, 2 mM, 5 mM, and 10 mM). To assess ROS production through KR activation 3 dpf zebrafish embryos illuminated for 2 h on a Leica Stereo Microscope (Leica M165FC, 4x objective, 535–575 nm excitation filter, 80 mW/cm2). CM-H2DCFDA (50 ɥM) was added afterwards for 30 min to 1 h, and embryos transferred individually to a black 96-well microwell plate for post-treatment scan as described previously [34]. Fluorescence values were obtained using a PHERAstar plate reader (485 excitation, 520 emission), with well-scanning from the top in a 5 × 5 matrix. Samples were run in triplicate. Values were normalized to blank readings. Data are represented as mean ± S.E.M. Statistical significance was determined using one-way ANOVA followed by Tukey's multiple comparisons test. All statistical tests used GraphPad Prism.
4.4. Photo-activation of KillerRed
Whole animal KR activation for ROS quantification purposes was carried out on a stereo microscope (Leica M165FC) with a 4× objective, at 535–575 nm excitation (mercury arc lamp and respective filter) for 2 h. Light intensities were measured at an average of 80 mW/cm2 using a Power Meter (PM16–130 Compact USB Power Meter with Slim Photodiode Sensor, ThorLabs). KR activation in specific motor neurons within the spinal cord for live imaging purposes was performed on a confocal microscope (Leica SP5) using a 40× objective, a 535–575 nm excitation (mercury arc lamp and respective filter) for 60–75 min, resulting in a sample illumination intensity of 10 mW/cm2.
4.5. Time-lapse imaging
To visualise KR mediated cell death as well as A5 and microglia responses to KR activation we utilised a confocal imaging approach described previously [48], [55]. Briefly, transgenic fish (2–5 dpf) positive for the respective transgenes (Tg[met:Gal4,UAS: EGFP;4xnrUAS:MLS-KillerRed,cryaa:EGFP], Tg[mnx1:MLS-KillerRed; mnx1:mTagBFP] injected with ubb:secHsa. ANNEXINV-mVenus, Tg[mnx1:MLS-KillerRed; mnx1:mTagBFP; mpeg1:Gal,UAS-mCherry]) were anesthetized in 0.01% tricaine and embedded in 1.5% low-melting agarose. After initial illumination time-lapse imaging was carried out on an upright Leica SP5 confocal microscope (40x objective) and a tuneable white-light laser was used for excitation of the fluorophores. KR activation was performed by green light illumination using a mercury lamp and corresponding filter. The diaphragm of the SP5 confocal microscope was used to restrict light illumination to selected areas of the zebrafish spinal cord. Z-stacks spanning the depth of field (~20–40 µm) were imaged repetitively and final images were collapsed to maximum intensity projections using ImageJ or Fiji software (http://imagej.nih.gov/; http://fiji.sc/Fiji). Images were brightness and contrast adjusted for visualization and illustration. Videos were generated using the ImageJ and Imaris software.
Acknowledgements
This work was supported by Motor Neurone Research Institute of Australia (GIA 1838, GIA 1638 - Cure for MND Research Grant), Australian Research Council (Discovery Grants DP140103233 and DP150104472), and donations made towards MND research at Macquarie University. The authors would like to thank the Snow Foundation for their generous support towards establishing the transgenic zebrafish facility at Macquarie University. The authors thank Dr Tom Hall for the 4xnrUAS plasmid, and Jason Martin-Powell and his team for assistance in zebrafish care.
Acknowledgments
Competing interests
The authors declare that they have no conflicts of interest.
Contributor Information
Roger S. Chung, Email: roger.chung@mq.edu.au.
Marco Morsch, Email: marco.morsch@mq.edu.au.
References
- 1.Yoshikawa T. Free radicals and their scavengers in Parkinson's disease. Eur. Neurol. 1993;33(Suppl 1):S60–S68. doi: 10.1159/000118539. [DOI] [PubMed] [Google Scholar]
- 2.Perry G., Cash A.D., Smith M.A. Alzheimer disease and oxidative stress. J. Biomed. Biotechnol. 2002;2:120–123. doi: 10.1155/S1110724302203010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blasco H., Garcon G., Patin F., Veyrat-Durebex C., Boyer J., Devos D., Vourc'h P., Andres C.R., Corcia P. Panel of oxidative stress and inflammatory biomarkers in ALS: a pilot study. Can. J. Neurol. Sci. J. Can. Des. Sci. Neurol. 2016:1–6. doi: 10.1017/cjn.2016.284. [DOI] [PubMed] [Google Scholar]
- 4.Ferrante R.J., Browne S.E., Shinobu L.A., Bowling A.C., Baik M.J., MacGarvey U., Kowall N.W., Brown R.H., Jr., Beal M.F. Evidence of increased oxidative damage in both sporadic and familial amyotrophic lateral sclerosis. J. Neurochem. 1997;69:2064–2074. doi: 10.1046/j.1471-4159.1997.69052064.x. [DOI] [PubMed] [Google Scholar]
- 5.Qin B., Cartier L., Dubois-Dauphin M., Li B., Serrander L., Krause K.H. A key role for the microglial NADPH oxidase in APP-dependent killing of neurons. Neurobiol. Aging. 2006;27:1577–1587. doi: 10.1016/j.neurobiolaging.2005.09.036. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y., Dawson V.L., Dawson T.M. Oxidative stress and genetics in the pathogenesis of Parkinson's disease. Neurobiol. Dis. 2000;7:240–250. doi: 10.1006/nbdi.2000.0319. [DOI] [PubMed] [Google Scholar]
- 7.Chen S., Zhang X., Song L., Le W. Autophagy dysregulation in amyotrophic lateral sclerosis. Brain Pathol. 2012;22:110–116. doi: 10.1111/j.1750-3639.2011.00546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cleveland D.W., Rothstein J.D. From Charcot to Lou Gehrig: deciphering selective motor neuron death in ALS. Nat. Rev. Neurosci. 2001;2:806–819. doi: 10.1038/35097565. [DOI] [PubMed] [Google Scholar]
- 9.Cozzolino M., Pesaresi M.G., Gerbino V., Grosskreutz J., Carri M.T. Amyotrophic lateral sclerosis: new insights into underlying molecular mechanisms and opportunities for therapeutic intervention. Antioxid. Redox Signal. 2012;17:1277–1330. doi: 10.1089/ars.2011.4328. [DOI] [PubMed] [Google Scholar]
- 10.Rojas F., Gonzalez D., Cortes N., Ampuero E., Hernandez D.E., Fritz E., Abarzua S., Martinez A., Elorza A.A., Alvarez A., Court F., van Zundert B. Reactive oxygen species trigger motoneuron death in non-cell-autonomous models of ALS through activation of c-Abl signaling. Front. Cell. Neurosci. 2015;9:203. doi: 10.3389/fncel.2015.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilgun-Sherki Y., Melamed E., Offen D. Oxidative stress induced-neurodegenerative diseases: the need for antioxidants that penetrate the blood brain barrier. Neuropharmacology. 2001;40:959–975. doi: 10.1016/s0028-3908(01)00019-3. [DOI] [PubMed] [Google Scholar]
- 12.Soto C. Unfolding the role of protein misfolding in neurodegenerative diseases. Nat. Rev. Neurosci. 2003;4:49–60. doi: 10.1038/nrn1007. [DOI] [PubMed] [Google Scholar]
- 13.Shaw P.J., Eggett C.J. Molecular factors underlying selective vulnerability of motor neurons to neurodegeneration in amyotrophic lateral sclerosis. J. Neurol. 2000;247(Suppl 1):I17–I27. doi: 10.1007/BF03161151. [DOI] [PubMed] [Google Scholar]
- 14.Beal M.F. Oxidatively modified proteins in aging and disease. Free Radic. Biol. Med. 2002;32:797–803. doi: 10.1016/s0891-5849(02)00780-3. [DOI] [PubMed] [Google Scholar]
- 15.Genova M.L., Pich M.M., Bernacchia A., Bianchi C., Biondi A., Bovina C., Falasca A.I., Formiggini G., Castelli G.P., Lenaz G. The mitochondrial production of reactive oxygen species in relation to aging and pathology. Ann. N.Y. Acad. Sci. 2004;1011:86–100. doi: 10.1007/978-3-662-41088-2_10. [DOI] [PubMed] [Google Scholar]
- 16.Lenaz G., Bovina C., D'Aurelio M., Fato R., Formiggini G., Genova M.L., Giuliano G., Merlo Pich M., Paolucci U., Parenti Castelli G., Ventura B. Role of mitochondria in oxidative stress and aging. Ann. N.Y. Acad. Sci. 2002;959:199–213. doi: 10.1111/j.1749-6632.2002.tb02094.x. [DOI] [PubMed] [Google Scholar]
- 17.Baier H., Scott E.K. Genetic and optical targeting of neural circuits and behavior--zebrafish in the spotlight. Curr. Opin. Neurobiol. 2009;19:553–560. doi: 10.1016/j.conb.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teh C., Chudakov D.M., Poon K.L., Mamedov I.Z., Sek J.Y., Shidlovsky K., Lukyanov S., Korzh V. Optogenetic in vivo cell manipulation in KillerRed-expressing zebrafish transgenics. BMC Dev. Biol. 2010;10:110. doi: 10.1186/1471-213X-10-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wojtovich A.P., Foster T.H. Optogenetic control of ROS production. Redox Biol. 2014;2:368–376. doi: 10.1016/j.redox.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobayashi J., Shidara H., Morisawa Y., Kawakami M., Tanahashi Y., Hotta K., Oka K. A method for selective ablation of neurons in C. elegans using the phototoxic fluorescent protein, KillerRed. Neurosci. Lett. 2013;548:261–264. doi: 10.1016/j.neulet.2013.05.053. [DOI] [PubMed] [Google Scholar]
- 21.Sarkisyan K.S., Zlobovskaya O.A., Gorbachev D.A., Bozhanova N.G., Sharonov G.V., Staroverov D.B., Egorov E.S., Ryabova A.V., Solntsev K.M., Mishin A.S., Lukyanov K.A. KillerOrange, a genetically encoded photosensitizer activated by blue and green light. PLos One. 2015;10:e0145287. doi: 10.1371/journal.pone.0145287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu S., Chisholm A.D. Highly efficient optogenetic cell ablation in C. elegans using membrane-targeted miniSOG. Sci. Rep. 2016;6:21271. doi: 10.1038/srep21271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bulina M.E., Lukyanov K.A., Britanova O.V., Onichtchouk D., Lukyanov S., Chudakov D.M. Chromophore-assisted light inactivation (CALI) using the phototoxic fluorescent protein KillerRed. Nat. Protoc. 2006;1:947–953. doi: 10.1038/nprot.2006.89. [DOI] [PubMed] [Google Scholar]
- 24.Liao Z.X., Li Y.C., Lu H.M., Sung H.W. A genetically-encoded KillerRed protein as an intrinsically generated photosensitizer for photodynamic therapy. Biomaterials. 2014;35:500–508. doi: 10.1016/j.biomaterials.2013.09.075. [DOI] [PubMed] [Google Scholar]
- 25.Teh C., Korzh V. In vivo optogenetics for light-induced oxidative stress in transgenic zebrafish expressing the KillerRed photosensitizer protein. Methods Mol. Biol. 2014;1148:229–238. doi: 10.1007/978-1-4939-0470-9_15. [DOI] [PubMed] [Google Scholar]
- 26.Bulina M.E., Chudakov D.M., Britanova O.V., Yanushevich Y.G., Staroverov D.B., Chepurnykh T.V., Merzlyak E.M., Shkrob M.A., Lukyanov S., Lukyanov K.A. A genetically encoded photosensitizer. Nat. Biotechnol. 2006;24:95–99. doi: 10.1038/nbt1175. [DOI] [PubMed] [Google Scholar]
- 27.Howe K., Clark M.D., Torroja C.F., Torrance J., Berthelot C., Muffato M., Collins J.E., Humphray S., McLaren K., Matthews L., McLaren S., Sealy I., Caccamo M., Churcher C., Scott C., Barrett J.C., Koch R., Rauch G.J., White S., Chow W., Kilian B., Quintais L.T., Guerra-Assuncao J.A., Zhou Y., Gu Y., Yen J., Vogel J.H., Eyre T., Redmond S., Banerjee R., Chi J., Fu B., Langley E., Maguire S.F., Laird G.K., Lloyd D., Kenyon E., Donaldson S., Sehra H., Almeida-King J., Loveland J., Trevanion S., Jones M., Quail M., Willey D., Hunt A., Burton J., Sims S., McLay K., Plumb B., Davis J., Clee C., Oliver K., Clark R., Riddle C., Elliot D., Threadgold G., Harden G., Ware D., Begum S., Mortimore B., Kerry G., Heath P., Phillimore B., Tracey A., Corby N., Dunn M., Johnson C., Wood J., Clark S., Pelan S., Griffiths G., Smith M., Glithero R., Howden P., Barker N., Lloyd C., Stevens C., Harley J., Holt K., Panagiotidis G., Lovell J., Beasley H., Henderson C., Gordon D., Auger K., Wright D., Collins J., Raisen C., Dyer L., Leung K., Robertson L., Ambridge K., Leongamornlert D., McGuire S., Gilderthorp R., Griffiths C., Manthravadi D., Nichol S., Barker G., Whitehead S., Kay M., Brown J., Murnane C., Gray E., Humphries M., Sycamore N., Barker D., Saunders D., Wallis J., Babbage A., Hammond S., Mashreghi-Mohammadi M., Barr L., Martin S., Wray P., Ellington A., Matthews N., Ellwood M., Woodmansey R., Clark G., Cooper J., Tromans A., Grafham D., Skuce C., Pandian R., Andrews R., Harrison E., Kimberley A., Garnett J., Fosker N., Hall R., Garner P., Kelly D., Bird C., Palmer S., Gehring I., Berger A., Dooley C.M., Ersan-Urun Z., Eser C., Geiger H., Geisler M., Karotki L., Kirn A., Konantz J., Konantz M., Oberlander M., Rudolph-Geiger S., Teucke M., Lanz C., Raddatz G., Osoegawa K., Zhu B., Rapp A., Widaa S., Langford C., Yang F., Schuster S.C., Carter N.P., Harrow J., Ning Z., Herrero J., Searle S.M., Enright A., Geisler R., Plasterk R.H., Lee C., Westerfield M., de Jong P.J., Zon L.I., Postlethwait J.H., Nusslein-Volhard C., Hubbard T.J., Crollius H. Roest, Rogers J., Stemple D.L. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496:498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korzh V., Teh C., Kondrychyn I., Chudakov D.M., Lukyanov S. Visualizing compound transgenic zebrafish in development: a tale of green fluorescent protein and KillerRed. Zebrafish. 2011;8:23–29. doi: 10.1089/zeb.2011.0689. [DOI] [PubMed] [Google Scholar]
- 29.Zelenchuk T.A., Bruses J.L. In vivo labeling of zebrafish motor neurons using an mnx1 enhancer and Gal4/UAS. Genesis. 2011;49:546–554. doi: 10.1002/dvg.20766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanabe Y., William C., Jessell T.M. Specification of motor neuron identity by the MNR2 homeodomain protein. Cell. 1998;95:67–80. doi: 10.1016/s0092-8674(00)81783-3. [DOI] [PubMed] [Google Scholar]
- 31.Seredick S.D., Van Ryswyk L., Hutchinson S.A., Eisen J.S. Zebrafish Mnx proteins specify one motoneuron subtype and suppress acquisition of interneuron characteristics. Neural Dev. 2012;7:35. doi: 10.1186/1749-8104-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hall T.E., Bryson-Richardson R.J., Berger S., Jacoby A.S., Cole N.J., Hollway G.E., Berger J., Currie P.D. The zebrafish candyfloss mutant implicates extracellular matrix adhesion failure in laminin alpha2-deficient congenital muscular dystrophy. Proc. Natl. Acad. Sci. USA. 2007;104:7092–7097. doi: 10.1073/pnas.0700942104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen X., Zhong Z., Xu Z., Chen L., Wang Y. 2',7'-Dichlorodihydrofluorescein as a fluorescent probe for reactive oxygen species measurement: forty years of application and controversy. Free Radic. Res. 2010;44:587–604. doi: 10.3109/10715761003709802. [DOI] [PubMed] [Google Scholar]
- 34.Walker S.L., Ariga J., Mathias J.R., Coothankandaswamy V., Xie X., Distel M., Koster R.W., Parsons M.J., Bhalla K.N., Saxena M.T., Mumm J.S. Automated reporter quantification in vivo: high-throughput screening method for reporter-based assays in zebrafish. PLos One. 2012;7:e29916. doi: 10.1371/journal.pone.0029916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dowling J.J., Arbogast S., Hur J., Nelson D.D., McEvoy A., Waugh T., Marty I., Lunardi J., Brooks S.V., Kuwada J.Y., Ferreiro A. Oxidative stress and successful antioxidant treatment in models of RYR1-related myopathy. Brain. 2012;135:1115–1127. doi: 10.1093/brain/aws036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu H., Gooneratne R., Huang X., Lai R., Wei J., Wang W. A rapid in vivo zebrafish model to elucidate oxidative stress-mediated PCB126-induced apoptosis and developmental toxicity. Free Radic. Biol. Med. 2015;84:91–102. doi: 10.1016/j.freeradbiomed.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 37.Wu C.Y., Lee H.J., Liu C.F., Korivi M., Chen H.H., Chan M.H. Protective role of L-ascorbic acid, N-acetylcysteine and apocynin on neomycin-induced hair cell loss in zebrafish. J. Appl. Toxicol. 2015;35:273–279. doi: 10.1002/jat.3043. [DOI] [PubMed] [Google Scholar]
- 38.Mugoni V., Camporeale A., Santoro M.M. Analysis of oxidative stress in zebrafish embryos. J. Vis. Exp.: JoVE. 2014 doi: 10.3791/51328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rieger S., Sagasti A. Hydrogen peroxide promotes injury-induced peripheral sensory axon regeneration in the zebrafish skin. PLoS Biol. 2011;9:e1000621. doi: 10.1371/journal.pbio.1000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seon-Heui Cha J.-H.L., Eun-Ah Kim, Chong Hyun Shin, Jun Hee-Sook, Jeon You-Jin. Phloroglucinol accelerates the regeneration of liver damaged by H2O2 or MNZ treatment in zebrafish. RSC Adv. 2017;7:46164–46170. [Google Scholar]
- 41.Da Costa M.M.J., Allen C.E., Higginbottom A., Ramesh T., Shaw P.J., McDermott C.J. A new zebrafish model produced by TILLING of SOD1-related amyotrophic lateral sclerosis replicates key features of the disease and represents a tool for in vivo therapeutic screening. Dis. Model Mech. 2014;7:73–81. doi: 10.1242/dmm.012013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kerr J.F., Wyllie A.H., Currie A.R. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fadok V.A., Voelker D.R., Campbell P.A., Cohen J.J., Bratton D.L., Henson P.M. Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J. Immunol. 1992;148:2207–2216. [PubMed] [Google Scholar]
- 44.van Ham T.J., Kokel D., Peterson R.T. Apoptotic cells are cleared by directional migration and elmo1- dependent macrophage engulfment. Curr. Biol.: CB. 2012;22:830–836. doi: 10.1016/j.cub.2012.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Ham T.J., Mapes J., Kokel D., Peterson R.T. Live imaging of apoptotic cells in zebrafish. FASEB J. 2010;24:4336–4342. doi: 10.1096/fj.10-161018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vermes I., Haanen C., Steffens-Nakken H., Reutelingsperger C. A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled Annexin V. J. Immunol. Methods. 1995;184:39–51. doi: 10.1016/0022-1759(95)00072-i. [DOI] [PubMed] [Google Scholar]
- 47.Mosimann C., Kaufman C.K., Li P., Pugach E.K., Tamplin O.J., Zon L.I. Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development. 2011;138:169–177. doi: 10.1242/dev.059345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morsch M., Radford R., Lee A., Don E., Badrock A., Hall T., Cole N., Chung R. In vivo characterization of microglial engulfment of dying neurons in the zebrafish spinal cord. Front. Cell. Neurosci. 2015;9:321. doi: 10.3389/fncel.2015.00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Svahn A.J., Don E.K., Badrock A.P., Cole N.J., Graeber M.B., Yerbury J.J., Chung R., Morsch M. Nucleo-cytoplasmic transport of TDP-43 studied in real time: impaired microglia function leads to axonal spreading of TDP-43 in degenerating motor neurons. Acta Neuropathol. 2018 doi: 10.1007/s00401-018-1875-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Del Bene F., Wyart C., Robles E., Tran A., Looger L., Scott E.K., Isacoff E.Y., Baier H. Filtering of visual information in the tectum by an identified neural circuit. Science. 2010;330:669–673. doi: 10.1126/science.1192949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shibuya T., Tsujimoto Y. Deleterious effects of mitochondrial ROS generated by KillerRed photodynamic action in human cell lines and C. elegans. J. Photochem. Photobiol. B. 2012;117:1–12. doi: 10.1016/j.jphotobiol.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 52.Droge W. Free radicals in the physiological control of cell function. Physiol. Rev. 2002;82:47–95. doi: 10.1152/physrev.00018.2001. [DOI] [PubMed] [Google Scholar]
- 53.Wiseman H., Halliwell B. Damage to DNA by reactive oxygen and nitrogen species: role in inflammatory disease and progression to cancer. Biochem. J. 1996;313(Pt 1):17–29. doi: 10.1042/bj3130017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Holmstrom K.M., Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat. Rev. Mol. Cell Biol. 2014;15:411–421. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- 55.Morsch M., Radford R.A., Don E.K., Lee A., Hortle E., Cole N.J., Chung R.S. Triggering cell stress and death using conventional UV laser confocal microscopy. J. Vis. Exp.: JoVE. 2017 doi: 10.3791/54983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nowis D., Makowski M., Stoklosa T., Legat M., Issat T., Golab J. Direct tumor damage mechanisms of photodynamic therapy. Acta Biochim. Pol. 2005;52:339–352. [PubMed] [Google Scholar]
- 57.Ellett F., Pase L., Hayman J.W., Andrianopoulos A., Lieschke G.J. Mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood. 2011;117:e49–56. doi: 10.1182/blood-2010-10-314120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Graeber M.B. Changing face of microglia. Science. 2010;330:783–788. doi: 10.1126/science.1190929. [DOI] [PubMed] [Google Scholar]
- 59.Cluskey S., Ramsden D.B. Mechanisms of neurodegeneration in amyotrophic lateral sclerosis. Mol. Pathol. 2001;54:386–392. [PMC free article] [PubMed] [Google Scholar]
- 60.Shaw P.J., Ince P.G., Falkous G., Mantle D. Oxidative damage to protein in sporadic motor neuron disease spinal cord. Ann. Neurol. 1995;38:691–695. doi: 10.1002/ana.410380424. [DOI] [PubMed] [Google Scholar]
- 61.Westerfield M. Univ. of Oregon Press; Eugene: 2000. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio) [Google Scholar]
- 62.Don E.K., Formella I., Badrock A.P., Hall T.E., Morsch M., Hortle E., Hogan A., Chow S., Gwee S.S., Stoddart J.J., Nicholson G., Chung R., Cole N.J. A Tol2 gateway-compatible toolbox for the study of the nervous system and neurodegenerative disease. Zebrafish. 2016 doi: 10.1089/zeb.2016.1321. [DOI] [PubMed] [Google Scholar]
- 63.Kwan K.M., Fujimoto E., Grabher C., Mangum B.D., Hardy M.E., Campbell D.S., Parant J.M., Yost H.J., Kanki J.P., Chien C.B. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn.: Off. Publ. Am. Assoc. Anat. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- 64.Akitake C.M., Macurak M., Halpern M.E., Goll M.G. Transgenerational analysis of transcriptional silencing in zebrafish. Dev. Biol. 2011;352:191–201. doi: 10.1016/j.ydbio.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Arkhipova V., Wendik B., Devos N., Ek O., Peers B., Meyer D. Characterization and regulation of the hb9/mnx1 beta-cell progenitor specific enhancer in zebrafish. Dev. Biol. 2012;365:290–302. doi: 10.1016/j.ydbio.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Berger J., Currie P.D. 503unc, a small and muscle-specific zebrafish promoter. Genesis. 2013;51:443–447. doi: 10.1002/dvg.22385. [DOI] [PubMed] [Google Scholar]
- 67.Clark K.J., Urban M.D., Skuster K.J., Ekker S.C. Transgenic zebrafish using transposable elements. Methods Cell Biol. 2011;104:137–149. doi: 10.1016/B978-0-12-374814-0.00008-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tang R., Dodd A., Lai D., McNabb W.C., Love D.R. Validation of zebrafish (Danio rerio) reference genes for quantitative real-time RT-PCR normalization. Acta Biochim. Biophys. Sin. 2007;39:384–390. doi: 10.1111/j.1745-7270.2007.00283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Time-lapse video of MN degeneration illustrated in Fig. 4. KR activation in our transgenic zebrafish (Tg[met:Gal4;UAS:EGFP];Tg[UAS:MLS-KR]) resulted in axonal blebbing and MN degeneration within 10 h after light illumination.
Time-lapse demonstration of ANNEXINV activation within the light-targeted MNs illustrated in Fig. 5.
3D representation of Video 3 illustrating the selective ANNEXINV activation along the light-targeted soma and axon of the KR-activated neuron.
Microglia response (green) towards the KR activation site over a time course of 10 h. Stills are presented in Fig. 6.