Abstract
Background
STARD13 has been revealed to suppress tumor progression. However, the roles in regulating the stemness of hepatocellular carcinoma (HCC) cells are unclear.
Methods
Quantitative real-time PCR (qRT-PCR) was used to detect STARD13 expression in HCC tissues and normal adjacent tissues. Kaplan Meier (KM)-plotter analysis was performed to analyze the correlation between STARD13 expression and overall survival of HCC patients. Cell spheroid formation and ALDH1 activity analysis were carried out to detect the effects of STARD13 on the stemness of HCC cells. Furthermore, immunofluorescent, luciferase reporter, RhoA GTPase and F-actin visualization assays were performed to explore the mechanisms contributing to STARD13-mediated effects.
Results
STARD13 expression was significantly downregulated in HCC tissues compared with normal adjacent tissues, and was positively correlated with the overall survival of HCC patients. Functionally, overexpression of STARD13 inhibited cells stemness and enhanced 5-FU sensitivity in HCC cells. Mechanistically, STRAD13 overexpression suppressed RhoGTPase signaling and thus inhibited transcriptional factor YAP translocation from nuclear to cytoplasm, leading to the downregulation of transcriptional activity of YAP. Notably, the inhibitory effects of STARD13 on HCC cells stemness and 5-FU sensitivity were rescued by RhoA or YAP-5SA overexpression.
Conclusion
Our results indicate that STARD13 could enhances 5-FU sensitivity by suppressing cancer stemness in hepatocellular carcinoma cells via attenuating YAP transcriptional activity.
Keywords: 5-FU, hepatocellular carcinoma, STARD13, stemness, YAP
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors worldwide.1 Although surgery, chemotherapy, and radiotherapy have largely improved the survival of HCC patients, it could not be cured after metastasis. Thus, it is still a major task to elucidate the pathogenesis, biological characteristics of HCC, in enforcing liver cancer prevention and control.
Cancer stem cells (CSCs) or cancer cell stemness has been suggested to contribute to cancer initiation and metastasis.2 Thus, treatments targeting CSCs were being developed to inhibit tumor progression.3 A previous study has shown that StAR-related lipid transfer domain 13 (STARD13) 3′-UTR suppresses breast cancer metastasis via inhibiting epithelial–mesenchymal transition (EMT) process.4 And it acts as a tumor suppressor in various cancers.5,6 However, it is still unclear whether STARD13 holds critical roles in HCC cell stemness.
Transcriptional factor YAP is a critical effector of Hippo signaling, in which YAP transcriptional activity is regulated by the upstream effector LATS1/2,7 and it has been shown that YAP could facilitate CSC formation or enhance tumor cell stemness.8 It is considered as “stemness factor” in several types of stem cells.9 Notably, YAP transcriptional activity could also be modulated in a LATS1/2-independent manner, such as mechanotransduction signaling in which YAP transcriptional activity was driven by Rho-GTPase/filamentous actin (F-actin) signaling. Importantly, previous studies have identified that STARD13 could block RhoA-ROCK signaling axis by acting as a Rho GTPase activating protein, thus disorganizing F-actin structures.10 Thus, we speculated that STARD13 could block YAP transcriptional activity through its Rho GTPase activity, suppressing RhoA-ROCK signaling axis, and finally attenuating HCC cell stemness.
In the present study, STARD13 expression was examined in HCC tissues and normal adjacent tissues. Kaplan–Meier (KM) plotter analysis was used to analyze the correlation between STARD13 expression and HCC patient’s survival. Further cell spheroid formation was used to examine the effects of STARD13 on HCC cell stemness. Additionally, 5-FU sensitivity of HCC cells with STARD13 over-expression or not was evaluated. Mechanistically, RhoA or YAP-5SA overexpression was found to rescue the decreased HCC cell stemness and increased 5-FU sensitivity induced by STARD13 overexpression. Thus, our results identified STARD13 as an inhibitor of HCC cells stemness and an enhancer of 5-FU sensitivity.
Materials and methods
Clinical tissues and cells culture
Human HCC cell lines HepG2, SMMC7721, Bel-7402, Hep3B, Huh7, and normal hepatic cell line L02 were purchased from the ATCC (American Type Culture Collection; Manassas, VA, USA). All HCC cell lines were cultured in DMEM medium (Gibco™, Grand Island, NY, USA) containing 10% FBS, penicillin, and streptomycin at 37°C under humidified air with 5% CO2. L02 cells were cultured in 1640 medium with the same condition of HCC cell lines. Thirty-six HCC tumors paired with adjacent normal tissues were obtained from patients who underwent surgery at The Third Hospital of Mianyang from February 2014 to June 2017. Approval from the institute research ethics committee of the Third Hospital Of Mianyang was obtained for the use of these clinical materials for research purposes, and patients whose tissues were used in this research have provided written informed consent. YAP-5SA (Cat # 33093) and 8xGTIIC-luciferase (Cat # 34615) plasmids were purchased from Addgene.
KM plotter analysis
KM plotter analysis (http://kmplot.com/) was performed to evaluate the correlation between STARD13 expression and the overall survival of HCC patients, where 364 HCC patients were included.11 The best cutoff was auto-selected.
Quantitative real-time PCR (qRT-PCR) was used to examine the relative expression of mRNA. Briefly, total RNA was extracted using TRIzol® Reagent (Thermo Fisher Scientific, Waltham, MA, USA). Then cDNA was synthesized using TIANScript RT Kit (Cat # KR104-01; Tiangen, Beijing, China) following the standard protocols provided by the manufacturer. Afterward, mRNA expression was examined using Quant qRT-PCR kit (SYBR Green) (Cat # FP302-01; Tiangen) and determined on an ABI Prism 7500 Detection System (Applied Biosystems, Inc., Foster City, CA, USA). Relative mRNA expression was calculated with 2−∆∆ct method. GAPDH was served as an internal control for mRNA expression.
Western blot
Protein was extracted using protein extraction kit (KeyGEN BioTECH, Nanjing, China). Protein concentration was examined by Bradford assay. A total of 30 µg of protein was separated by SDS-PAGE, and transferred onto polyvinylidene fluoride (PVDF) membranes, which was followed by incubation with 5% nonfat milk for 1.5 hours at room temperature. Afterward, membranes were incubated with primary antibodies against STARD13 (ab126489), ALDH1 (ab52492), Nanog (ab21624), Ki67 (ab15580), cleaved caspase 3 (ab2302), Caspase-3 (ab13847), cleaved PARP (ab32064), PARP (ab74290), YAP (ab52771), RhoA (ab187027), and CTGF (ab6992), which were purchased from Abcam (Cambridge, UK). Primary antibodies against pMLC-S19 (Cat # 3675) and MLC (Cat # 3672) were purchased from Cell Signaling Technology (Danvers, MA, USA). Primary antibody against β-actin (Cat # AF0003) was purchased from Beyotime (Beijing, China). After incubating with primary antibodies, blots were washed and incubated with a peroxidase-conjugated antibody, and chemiluminescence was detected using an enhanced chemiluminescence kit (Thermo Fisher Scientific), followed by exposure in Tanon 5200 (Tanon, Shanghai, China).
Lentivirus plasmids and stable-infected cell lines construction
STARD13 coding sequences were inserted into pLVX-IRES-ZsGreen1, named as Lenti-STARD13. Lentiviral particles were packaged in HEK293T cells by co-transfecting Lenti-STARD13, pCMV-dR8.2, and pMD2.G constructs. Thereafter, 48 and 72 hours later, supernatants were collected and centrifuged to remove cell debris, which is referred to as virus. Virus supernatants were added to HCC cells with 2 µg/mL polybrene for 72 hours. The infected cells were selected with puromycin (2 µg/mL; Sigma-Aldrich Co., St Louis, MO, USA) for 2 weeks. qRT-PCR and Western blots were used to confirm the infection efficiency. Cells were named as HepG2-S and Huh7-S.
Cell spheroid formation assay
A total of 3,000 single cells were seeded into the 24-well ultra-low attachment plate (Corning Incorporated, Corning, NY, USA) in serum-free DMEM/F12 (Thermo Fisher Scientific), supplemented with B27 (1:50; Thermo Fisher Scientific), 20 ng/mL EGF (Peprotech, Rehovot, Israel), 20 ng/mL bFGF (Thermo Fisher Scientific), 100 U/mL penicillin, 100 mg/mL streptomycin, 4 mg/mL insulin (Sigma-Aldrich Co.), and 20% methylcellulose (Sigma-Aldrich Co.). Number of nonadherent spheres and size were detected and photographed using a microscope fitted with a ruler.
ALDH1 activity assay
ALDH1 activity was assayed by ALDH Activity Assay Kit (Colorimetric) (Cat # KA3742; Abnova, Taipei City, Taiwan) according to manufacturer’s protocols.
Cell viability assay
Cell viability assay was carried out to evaluate the cell viability of HCC cells with different treatments. Briefly, 5,000 cells/well were seeded into 96-well plates, and after 12 hours, 5-FU was added into the medium, which was followed by culturing for 24, 48, and 72 hours. Then cell viability was detected by Cell Counting Kit-8 (Cat # C0037; Beyotime) following the manufacturer’s recommendation. Cell viability was shown as relative value.
Cell apoptosis assay
Cell apoptotic rate was determined by Annexin V-FITC and propidium iodide (PI) kit (Cat # C1062; Beyotime) via flow cytometry (BD Biosciences, San Jose, CA, USA). Cells were stained with Annexin V-FITC and PI, followed by flow cytometry analysis according to the manufacturer’s recommendation.
Immunofluorescent assay
Immunofluorescent assay was used to examine YAP cytoplasm retention. Cells were cultured on glass bottom plates. After 24 hours, cells were washed with PBS, and then fixed with 4% paraformaldehyde for 15 minutes, followed by the permeabilization with 0.1% Triton X-100 for 15 minutes, and blocked with 5% BSA in PBS for 1 hour at room temperature. Then cells were incubated with antibody against YAP overnight at 4°C, and washed thrice with PBS for 5 minutes and incubated with FITC-conjugated secondary antibody (Cat # A0562; Beyotime) for 1 hour at room temperature, and washed with PBS for three times again and observed under the confocal microscopy.
Luciferase reporter assay
Luciferase reporter assay was performed to measure YAP transcriptional activity via detecting the luciferase activity of 8xGTIIC-luciferase plasmid, which is a YAP-responsive synthetic promoter driving luciferase expression plasmid. Briefly, 8xGTIIC-luciferase plasmid was co-transfected into HCC cells with β-Galactosidase (Ambion, Thermo Fisher Scientific) plasmid using Lipofectamine™ 2000 followed for 72 hours. After that, the luciferase activity of 8xGTIIC-luciferase plasmid was measured using a Luciferase Reporter Assay Kit (Cat # K801-200; BioVision, Inc., Milpitas, CA, USA). β-gal activity was determined using a β-Galactosidase Enzyme Assay System with Reporter Lysis Buffer (Cat # E2000; Promega Corporation, WI, USA) following the manufacturer’s protocols, which was used as a normalization control for luciferase activity.
RhoA GTPase assay
RhoA GTPase activity was analyzed by RhoA G-LISA (Cat # BK132, Cytoskeleton, Inc., CO, USA) following the manufacturer’s protocol. Briefly, cells were lysed, and protein concentration was measured. Lysis buffer was added to equalize the cell extracts to give identical protein concentrations in each sample. Samples were handled following the technical guide. Finally, absorbance at 490 nm was measured using a microplate spectrophotometer to reflect the RhoA GTPase activity shown as relative value.
F-actin visualization
F-actin was stained with Rhodamine phalloidin (Cat # BK005; Cytoskeleton, Inc.) for 30 minutes at room temperature following the manufacturer’s protocol. The nuclei were stained by DAPI (Cat # C1002; Beyotime) for 10 minutes and observed with the confocal microscopy. Images were analyzed using Olympus fluoview version 3.0 viewer software.
Statistical analysis
All data were expressed as mean ± SD. Mean was compared using Student’s unpaired t-test. P<0.05 was considered statistically significant.
Results
STARD13 expression decreases in HCC cells and tissues, and correlates with poor prognosis
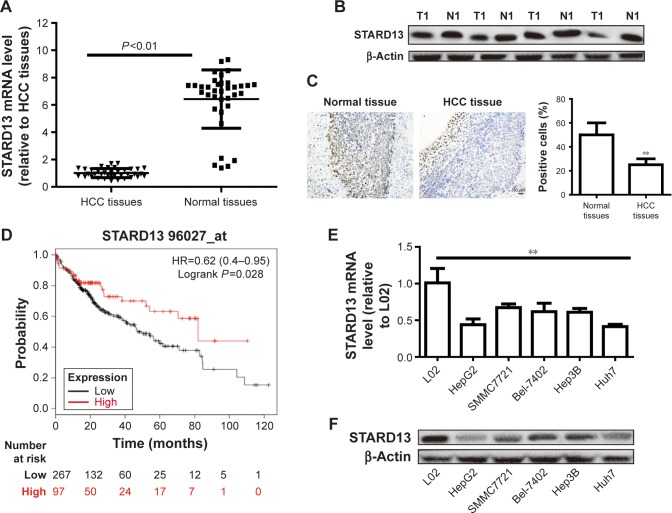
STARD13 expression was examined in HCC tissues and normal adjacent tissues, and showed a lower level in HCC tissues than that in normal adjacent tissues (Figure 1A and B). Consistent result was obtained by immunohistochemistry assay (Figure 1C). KM plotter assay indicated that patients with higher STARD13 expression held longer overall survival (Figure 1D). Additionally, STARD13 expression was decreased in HCC cells compared with that in normal hepatic cells (Figure 1E and F). These results demonstrate that STARD13 may suppress HCC progression.
Figure 1.
STARD13 expression is decreased in HCC cells and tissues and correlates with poor prognosis.
Notes: (A) STARD13 mRNA level was detected in HCC tissues and normal adjacent tissues. (B and C) STARD13 protein level was examined in HCC tissues and normal adjacent tissues. (D) KM plotter analysis was used to predict the correlation between STARD13 expression and overall survival of HCC patients. (D and E) STARD13 mRNA (D) and protein (E and F) expressions were measured in HCC cells and normal hepatic cells. Data were presented as mean ± SD; **P<0.01 vs HCC tissues or L02.
Abbreviations: HCC, hepatocellular carcinoma; KM plotter, Kaplan–Meier plotter; N, normal; STARD13, StAR-related lipid transfer domain 13; T, tumor.
STARD13 overexpression attenuates HCC cell stemness
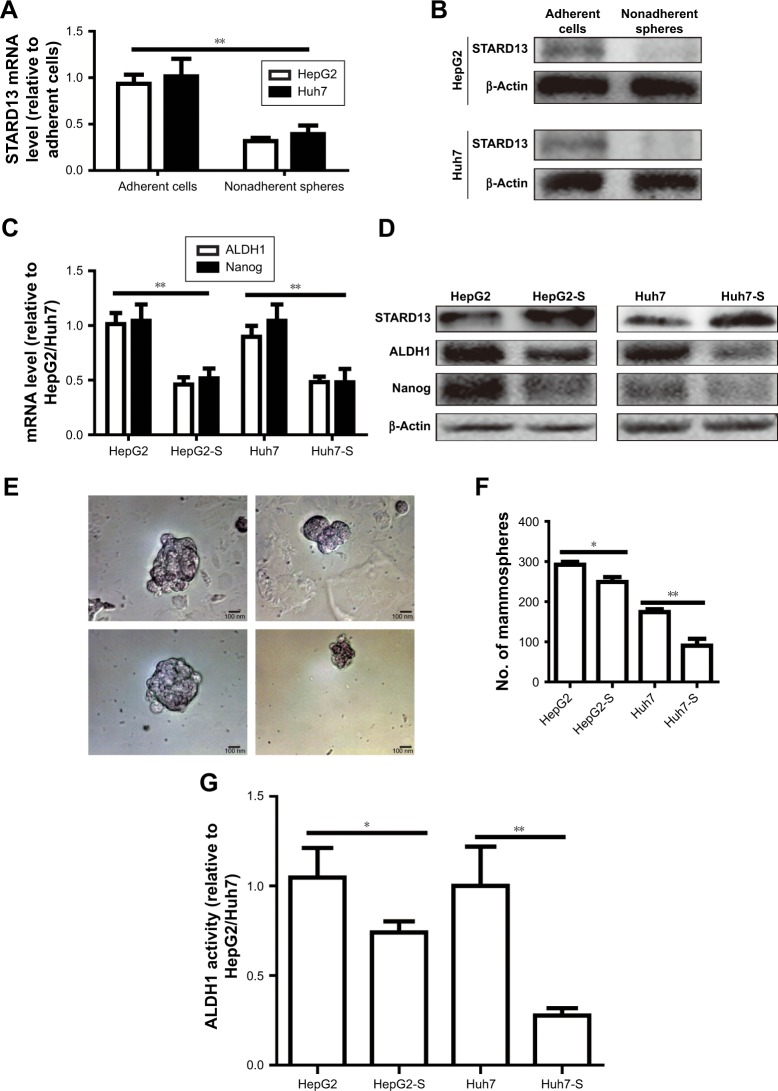
Since tumor cell stemness could lead to tumor recurrence and chemoresistance,12 we speculated that STARD13 over-expression could attenuate HCC cell stemness. Based on the fact that nonadherent spheres are highly enriched for CSC,13 and as STARD13 expression displayed relative lower level in HepG2 and Huh7 cells compared with other HCC cell lines (Figure 1D), HepG2 and Huh7 cells were chosen for the following experiments. STARD13 expression was firstly determined in nonadherent spheres and adherent cells. qRT-PCR and Western blot assays indicated that STARD13 expression was significantly decreased in nonadherent spheres compared with adherent cells (Figure 2A and B). Furthermore, the expression of stemness markers (ALDH1 and Nanog) was decreased in HCC cells with STARD13 overexpression (Figure 2C and D). The infection efficiency was confirmed by Western blot analysis (Figure 2D). Cell spheroid formation assay indicated that overexpression of STARD13 significantly decreased sphere’s size and number of HCC cells (Figure 2E and F). Importantly, ALDH1 activity was decreased by STARD13 overexpression (Figure 2G). Our results indicate that STARD13 could attenuate HCC cell stemness.
Figure 2.
STARD13 overexpression attenuates HCC cell stemness.
Notes: (A and B) STARD13 mRNA and protein expressions were examined in adherent HCC cells and nonadherent spheres. (C and D) The expression of stemness markers (ALDH1 and Nanog) was detected in HCC cells with STARD13 overexpression or not. (E and F) Cell spheroid potential (sphere’s size and number) was measured in cells depicted in (C). (G) ALDH1 activity was evaluated in cells depicted in (C). Data were presented as mean ± SD; *P<0.05; **P<0.01 vs adherent cells, HepG2, or Huh7.
Abbreviations: HCC, hepatocellular carcinoma; STARD13, StAR-related lipid transfer domain 13.
STARD13 enhances 5-FU sensitivity in HCC cells
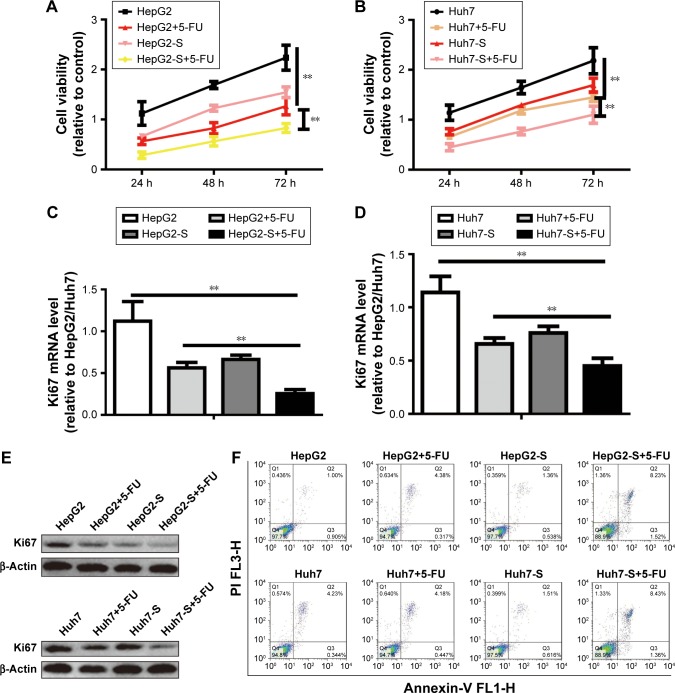
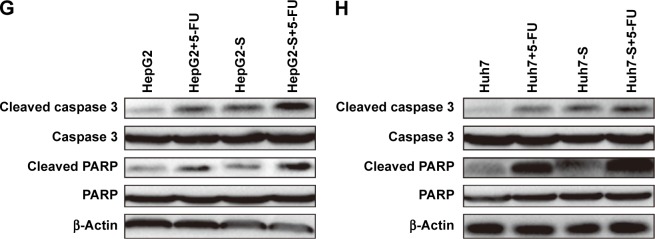
We further explored whether STARD13 could regulate chemotherapy sensitivity of HCC cells. HCC cells with STARD13 overexpression or not were treated with 5-FU, and subjected to cell viability assay, as Figure 3A and B results indicated that STARD13 overexpression enhanced 5-FU sensitivity in HCC cells. Moreover, qRT-PCR and Western blot analyses showed that overexpression of STARD13 promoted 5-FU-induced downregulation of proliferation marker Ki67 expression (Figure 3C–E). Cell apoptosis assay demonstrated that the increase of cell apoptosis mediated by 5-FU treatment was enhanced by STARD13 overexpression (Figure 3F). Additionally, 5-FU-induced increased expression of apoptosis executors (cleaved caspase 3 and cleaved PARP) was promoted by STARD13 overexpression (Figure 3G and H). Thus, these results suggest that STARD13 positively regulates chemotherapy sensitivity in HCC cells.
Figure 3.
STARD13 enhances 5-FU sensitivity in HCC cells.
Notes: (A and B) HCC cells with STARD13 overexpression or not were treated with 5-FU or not, and followed by examining the cell viability via CCK-8 analysis. (C–E) Ki67 mRNA and protein expressions were detected in cells depicted in (A). (F) Cell apoptotic rate was measured in cells depicted in (A). (G and H) The expression of apoptosis executors (cleaved caspase 3 and cleaved PARP) was determined in cells depicted in (A). Data were presented as mean ± SD; **P<0.01 vs HepG2 or Huh7.
Abbreviations: HCC, hepatocellular carcinoma; STARD13, StAR-related lipid transfer domain 13; CCK-8, Cell Counting Kit-8.
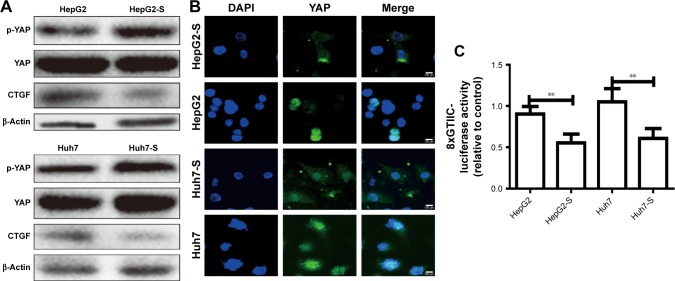
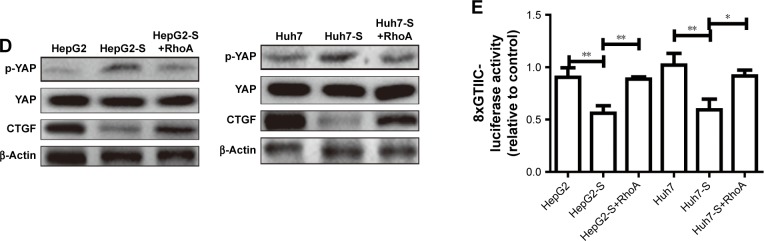
STARD13 suppresses YAP translocation from nuclear to cytoplasm via inhibiting RhoA activity
As YAP is considered as “stemness factor” in several types of stem cells,9 we detected whether STARD13 could regulate YAP activity in HCC cells. First, the phosphorylation level of YAP, which associated to YAP cytoplasm retention,14 was increased in STARD13 overexpressed cells (Figure 4A), and STARD13 inhibited the expression of CTGF, the target of YAP. Second, enforcing the expression of STARD13 in HCC cells increased the YAP cytoplasm retention (Figure 4B). Additionally, luciferase reporter assay showed that STARD13 overexpression decreased YAP transcriptional activity characterized as the decrease of 8xGTIIC-luciferase activity, a YAP-responsive synthetic promoter driving luciferase expression plasmid (Figure 4C). Indeed, G-LISA RhoA activation assay showed that STARD13 overexpressed cells showed a lower basal level of RhoA activity compared with that in control cells (Figure S1A), confirming STARD13 Rho GTPase activity. Immunofluorescence staining of F-actin with rhodamine-labeled phalloidin revealed that stress fiber formation was markedly inhibited in cells with STARD13 overexpression (Figure S1B). We further determined the effects of STARD13 on regulation of RhoA downstream effector MLC phosphorylation level, and found that pMLC-S19 level was significantly decreased in STARD13 overexpressed cells. Notably, STARD13 had no effect on expression of RhoA in HCC cells (Figure S1C). Then, we explored whether STARD13 regulated YAP cytoplasm retention dependent on its Rho GTPase activity. HCC cells with STARD13 overexpression was co-transfected with RhoA expression plasmid followed by detecting YAP phosphorylation level and CTGF expression (Figure 4D), and 8xGTIIC-luciferase activity (Figure 4E). As expected, the inhibition of STARD13 overexpression on YAP transcriptional activity was rescued by RhoA overexpression. In summary, these data indicate that STARD13 suppresses YAP translocation from nuclear to cytoplasm via inhibiting RhoA activity.
Figure 4.
STARD13 suppresses YAP translocation from nuclear to cytoplasm via inhibiting RhoA activity.
Notes: (A) The expression of p-YAP and its target CTGF were evaluated in HCC cells with STARD13 overexpression or not. (B) YAP translocation from nuclear to cytoplasm was detected in cells depicted in (A) via immunofluorescent assay. (C) The luciferase activity of 8xGTIIC-luciferase plasmid was examined in cells depicted in (A). (D) The expression of p-YAP and CTGF was measured in HCC cells with STARD13 overexpression as well as RhoA overexpression or not. (E) The luciferase activity of 8xGTIIC-luciferase plasmid was examined in cells depicted in (D). Data were presented as mean ± SD; *P<0.05; **P<0.01 vs HepG2 or Huh7.
Abbreviations: HCC, hepatocellular carcinoma; STARD13, StAR-related lipid transfer domain 13.
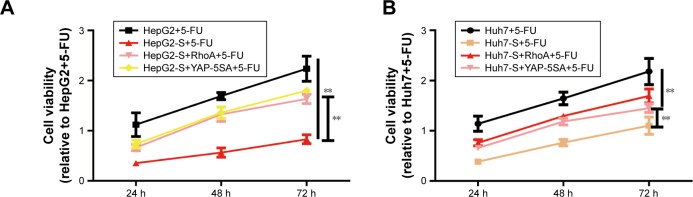
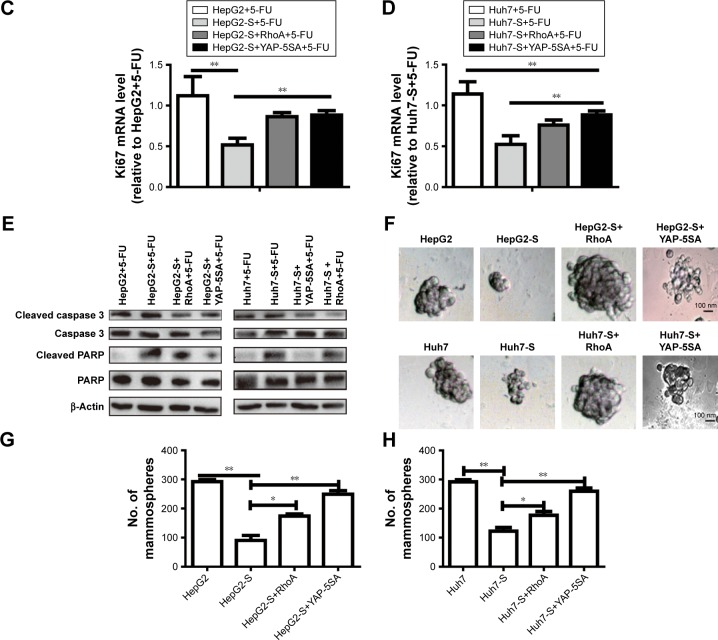
STARD13 enhances 5-FU sensitivity and cell stemness via inhibiting RhoA activity, thus enabling YAP translocation from nuclear to cytoplasm
Finally, we investigated whether STARD13 regulated 5-FU sensitivity and cell stemness through modulating RhoA activity and YAP cytoplasm retention. HCC cells with STARD13 overexpression was transfected with RhoA over-expression plasmid or YAP-5SA. Overexpression of RhoA or YAP-5SA attenuated the promotion of STARD13 on 5-FU sensitivity characterized as the decrease of cell viability (Figure 5A and B), Ki67 expression (Figure 5C and D), and cell apoptosis (Figure 5E). Additionally, STARD13 overexpression-induced downregulation of cell stemness was rescued or even reversed by RhoA or YAP-5SA overexpression characterized as the increase of cell spheroid formation ability (Figure 5F–H). Taken together, our results indicate that STARD13 could regulate 5-FU sensitivity via its Rho GTPase activity, thus suppressing RhoA/F-actin/YAP axis.
Figure 5.
STARD13 enhances 5-FU sensitivity and cell stemness via inhibiting RhoA activity, and thus YAP translocate from nuclear to cytoplasm.
Notes: (A and B) HCC cells with STARD13 overexpression as well as RhoA or YAP-5SA overexpression were treated with 5-FU, followed by cell viability assay. (C and D) Ki67 mRNA level was detected in cells depicted in (A). (E) The expression of apoptosis executors (cleaved caspase 3 and cleaved PARP) was determined in cells depicted in (A). (F–H) Cell sphere’s size and number were measured and depicted in cells with STARD13 overexpression as well as RhoA or YAP-5SA overexpression. Data were presented as mean ± SD; *P<0.05, **P<0.01 vs HepG2 or Huh7.
Abbreviations: HCC, hepatocellular carcinoma; STARD13, StAR-related lipid transfer domain 13.
Discussion
The inhibitory roles of STARD13 have been established in various tumors, such as STARD13-mediated ceRNA network could inhibit EMT and metastasis of breast cancer,4 and STARD13 3′UTR could facilitate cells apoptosis in breast cancer15 and HCC cells.16 However, the roles and related mechanisms by which STARD13 regulates cancer cell stemness have never been found. To the best of our knowledge, this is the first study showing STARD13 roles in HCC cell stemness and 5-FU sensitivity.
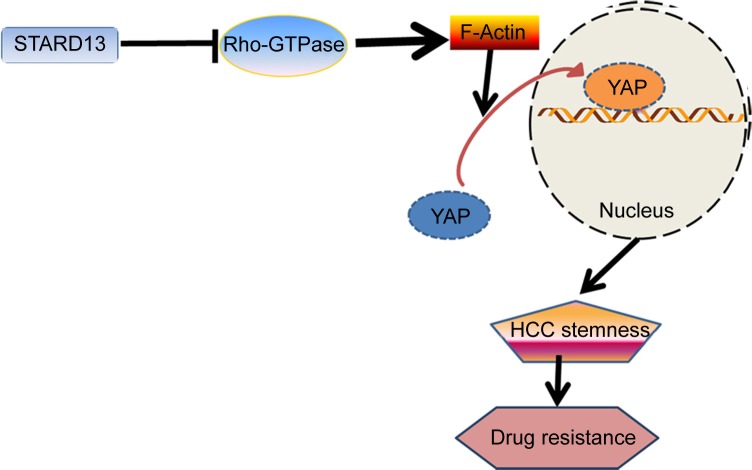
The carcinogenic roles of transcriptional factor YAP have been identified before, however, the methods targeting YAP were frustrating, indicating that there were other mechanisms contributing to regulate YAP transcriptional activity. In the present study, we confirmed that STARD13 indeed held Rho GTPase activity in HCC cells, which is consistent with the previous studies in other cancers.10,17 Additionally, YAP transcriptional activity was negatively regulated by STARD13 overexpression, which was rescued by restoring RhoA expression. These effects suggest that STARD13 could negatively regulate YAP transcriptional activity via its Rho GTPase activity. Importantly, overexpression of RhoA and YAP could attenuate the promotion of STARD13 overexpression on 5-FU sensitivity. Taken together, our results provided strong evidences confirming that STARD13 enhanced 5-FU sensitivity and cell stemness via its Rho GTPase activity, thus inhibiting RhoA and YAP activity. Our results provide novel clues that YAP transcriptional activity could be regulated in a classic Hippo pathway-independent manner (Figure 6). However, another downstream effector transcriptional co-activator with PDZ-binding motif (TAZ) transcriptional activity was not evaluated in this work, which could be examined in future work.
Figure 6.
Proposed model for STARD13 inhibiting HCC cell stemness, enhancing drug resistance.
Notes: STARD13 acts as a potential inhibitor of YAP transcriptional activity by blocking RhoA–F-actin signaling cascade through its Rho GTPase activity. This inhibition inhibits HCC stemness, thus enhancing drug sensitivity.
Abbreviations: F-actin, filamentous actin; HCC, hepatocellular carcinoma; STARD13, StAR-related lipid transfer domain 13.
Notably, we must admit that the effects identified here are just relied on in vitro experiments, further in vivo experiments should be performed to confirm the inhibitory roles of STARD13 in 5-FU sensitivity and cell stemness in HCC. Since we identified that STARD13 was significantly decreased in HCC tissues and cells, and positively correlated with the survival of STARD13 in HCC patients, we strongly believe that activation of STARD13 could be a potential novel therapeutic strategy to specifically target HCC stem cells and enhance chemotherapy sensitivity of HCC patients.
Supplementary material
Overexpression of STARD13 inhibits RhoA/F-actin axis via its Rho GTPase activity. (A) RhoA activity was determined in HCC cells with STARD13 overexpression or not via G-LISA analysis. (B) F-actin formation was evaluated in HCC cells with STARD13 overexpression or not. (C) The expression of p-MLC and RhoA was detected in HCC cells with STARD13 overexpression or not.
Notes: Data were presented as mean ± SD; **P< 0.01 vs HepG2 or Huh7.
Abbreviations: F-actin, filamentous actin; HCC, hepatocellular carcinoma; STARD13, StAR-related lipid transfer domain 13.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391(10127):1301–1314. doi: 10.1016/S0140-6736(18)30010-2. [DOI] [PubMed] [Google Scholar]
- 2.Kim H, Lin Q, Glazer PM, Yun Z. The hypoxic tumor microenvironment in vivo selects the cancer stem cell fate of breast cancer cells. Breast Cancer Res. 2018;20(1):16. doi: 10.1186/s13058-018-0944-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ozsvari B, Bonuccelli G, Sanchez-Alvarez R, Foster R, Sotgia F, Lisanti MP. Targeting flavin-containing enzymes eliminates cancer stem cells (CSCs), by inhibiting mitochondrial respiration: Vitamin B2 (Riboflavin) in cancer therapy. Aging (Albany NY) 2017;9(12):2610–2628. doi: 10.18632/aging.101351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X, Zheng L, Zhang F, et al. STARD13-correlated ceRNA network inhibits EMT and metastasis of breast cancer. Oncotarget. 2016;7(17):23197–23211. doi: 10.18632/oncotarget.8099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nasrallah A, Saykali B, Al Dimassi S, Khoury N, Hanna S, El-Sibai M. Effect of StarD13 on colorectal cancer proliferation, motility and invasion. Oncol Rep. 2014;31(1):505–515. doi: 10.3892/or.2013.2861. [DOI] [PubMed] [Google Scholar]
- 6.Chang S, He S, Qiu G, et al. MicroRNA-125b promotes invasion and metastasis of gastric cancer by targeting STARD13 and NEU1. Tumour Biol. 2016;37(9):12141–12151. doi: 10.1007/s13277-016-5094-y. [DOI] [PubMed] [Google Scholar]
- 7.Ni W, Zhang Y, Zhan Z, et al. A novel lncRNA uc.134 represses hepatocellular carcinoma progression by inhibiting CUL4A-mediated ubiquitination of LATS1. J Hematol Oncol. 2017;10(1):91. doi: 10.1186/s13045-017-0449-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cordenonsi M, Zanconato F, Azzolin L, et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147(4):759–772. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- 9.Tremblay AM, Camargo FD. Hippo signaling in mammalian stem cells. Semin Cell Dev Biol. 2012;23(7):818–826. doi: 10.1016/j.semcdb.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 10.Tang F, Zhang R, He Y, Zou M, Guo L, Xi T. MicroRNA-125b induces metastasis by targeting STARD13 in MCF-7 and MDA-MB-231 breast cancer cells. PLoS One. 2012;7(5):e35435. doi: 10.1371/journal.pone.0035435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lanczky A, Nagy A, Bottai G, et al. miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2,178 breast cancer patients. Breast Cancer Res Treat. 2016;160(3):439–446. doi: 10.1007/s10549-016-4013-7. [DOI] [PubMed] [Google Scholar]
- 12.Song M, Lee H, Nam MH, et al. Loss-of-function screens of drug-gable targetome against cancer stem-like cells. FASEB J. 2017;31(2):625–635. doi: 10.1096/fj.201600953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dontu G, Abdallah WM, Foley JM, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17(10):1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo PD, Lu XX, Gan WJ, et al. RARγ downregulation contributes to colorectal tumorigenesis and metastasis by derepressing the Hippo-Yap pathway. Cancer Res. 2016;76(13):3813–3825. doi: 10.1158/0008-5472.CAN-15-2882. [DOI] [PubMed] [Google Scholar]
- 15.Guo X, Xiang C, Zhang Z, Zhang F, Xi T, Zheng L. Displacement of Bax by BMF mediates STARD13 3′UTR-induced breast cancer cells apoptosis in an miRNA-depedent manner. Mol Pharm. 2018;15(1):63–71. doi: 10.1021/acs.molpharmaceut.7b00727. [DOI] [PubMed] [Google Scholar]
- 16.Zhang H, Wang F, Hu Y. STARD13 promotes hepatocellular carcinoma apoptosis by acting as a ceRNA for Fas. Biotechnol Lett. 2017;39(2):207–217. doi: 10.1007/s10529-016-2253-6. [DOI] [PubMed] [Google Scholar]
- 17.Hu J, Li X, Guo X, et al. The CCR2 3′UTR functions as a competing endogenous RNA to inhibit breast cancer metastasis. J Cell Sci. 2017;130(19):3399–3413. doi: 10.1242/jcs.202127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Overexpression of STARD13 inhibits RhoA/F-actin axis via its Rho GTPase activity. (A) RhoA activity was determined in HCC cells with STARD13 overexpression or not via G-LISA analysis. (B) F-actin formation was evaluated in HCC cells with STARD13 overexpression or not. (C) The expression of p-MLC and RhoA was detected in HCC cells with STARD13 overexpression or not.
Notes: Data were presented as mean ± SD; **P< 0.01 vs HepG2 or Huh7.
Abbreviations: F-actin, filamentous actin; HCC, hepatocellular carcinoma; STARD13, StAR-related lipid transfer domain 13.