Abstract
DNA-encoded chemical libraries (DECLs) are collections of compounds, individually coupled to DNA tags serving as amplifiable identification barcodes. Since individual compounds can be identified by the associated DNA tag, they can be stored as a mixture, allowing the synthesis and screening of combinatorial libraries of unprecedented size, facilitated by the implementation of split&pool synthetic procedures or other experimental methodologies. In this review, we briefly present relevant concepts and technologies, which are required for the implementation and interpretation of screening procedures with DNA-encoded chemical libraries. Moreover, we illustrate some success stories, detailing how novel ligands were discovered from encoded libraries. Finally, we critically review what can realistically be achieved with the technology at the present time, highlighting challenges and opportunities for the future.
Keywords: DNA, DNA-encoded chemical libraries, drug discovery, high-throughput DNA sequencing, combinatorial chemistry
Introduction
The discovery of small organic ligands, capable of selective binding to protein targets of interest, represents a central problem in Chemistry, Biology and Pharmaceutical Sciences. Until recently, the main approach for the discovery of binding molecules (“hits”), that may subsequently be improved using Medicinal Chemistry techniques, has relied on the screening of large collections of compounds (“chemical libraries”), one molecule at a time. In general, the screening of chemical libraries is possible if a suitable bioassay is in place. For example, chemical compounds can be individually tested in microtiter plates using high-throughput screening (HTS) procedures for their ability to inhibit the biochemical activity of a target enzyme. This approach, which requires complex logistics, is typically limited to the screening of libraries, which in most cases are smaller than one million compounds. It has been estimated that the construction of a conventional library, containing 106 individual molecules in sufficient quantity and quality for pharmaceutical screening campaigns, may cost between 400 million and 2 billion U.S. dollars [1]. While certain academic efforts for the HTS of chemical libraries have been implemented, those activities can rarely compete with the ones performed by large pharmaceutical companies, in terms of hit productivity, as well as size and diversity. Importantly, conventional HTS approaches often fail to deliver ligands of sufficient affinity and specificity for pharmaceutical applications. This underlines the need for improved library synthesis and screening technologies.
DNA-encoded chemical libraries are large collections of organic molecules, individually coupled to DNA fragments, serving as amplifiable identification barcodes [2] As molecules are individually tagged with DNA barcodes, library members can be stored as a mixture and interrogated by affinity capture procedures. The use of DNA tags as barcodes allows the construction and screening of chemical libraries of unprecedented size, potentially exceeding one billion compounds. The identity and relative quantity of individual compounds can be determined by polymerase chain reaction (PCR) procedures, followed by high-throughput DNA sequencing. The libraries used for affinity-based screening can be very large, since even few copies of compounds (i.e., library members present at low concentration) can be reliably detected and quantified, thanks to the amplification power of PCR.
The advent of DNA-encoded chemical libraries was inspired by the successful introduction of technologies for the construction and screening of encoded combinatorial libraries of proteins and other bio-macromolecules. For example, the display on filamentous phage of antibody libraries allows the facile identification of human monoclonal antibodies, specific to target proteins of interest [3–6]. Similarly, large libraries of oligonucleotides (“aptamers”) can be interrogated by affinity capture procedures on target proteins of interest and “decoded” by DNA sequencing [7, 8]. While DNA acts as a barcode that drives the synthesis of the corresponding bio-macromolecule (e.g., DNA transcription and translation), in the case of chemical libraries the individual DNA fragments merely serve as identification barcodes, without a biosynthetic relation to the corresponding organic molecule.
The origin of DNA-encoded chemical library (DECL) technology dates back to the proposal of Brenner and Lerner to use beads for the alternate synthesis of polymers (e.g., peptides) and of cognate oligonucleotides serving as amplifiable barcodes [2, 9]. In 2004, three groups independently described that DNA-encoded chemical libraries can be synthesized and screened without beads [10–12]. Since then, in virtually all cases, DNA-encoded chemical libraries are synthesized and screened by direct coupling of organic molecules to the corresponding DNA tags [1, 13–20]. There are, however, multiple strategies for the synthesis of encoded libraries, as illustrated in a later section of this manuscript.
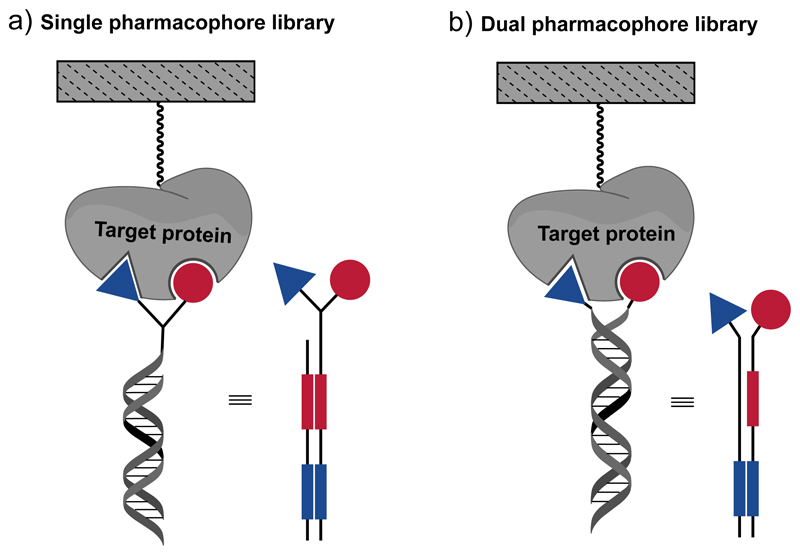
We normally distinguish between two main types of DNA-encoded chemical libraries: (i) “single-pharmacophore libraries”, in which individual compounds (no matter how complex) are attached to one DNA fragment; and (ii) “dual-pharmacophore libraries”, in which pairs of compounds are coupled to the extremity of the complementary strands of the DNA heteroduplex [Figure 1]. In this second example, binding fragments are identified, which need to be chemically linked at a later stage, in order to yield organic molecules which can be used in the absence of DNA. An appropriate linker optimization procedure may help further improve the affinity constants towards the protein target [21].
Figure 1.
Schematic representation of (a) a single-pharmacophore DNA-encoded chemical library and (b) of a dual pharmacophore DNA-encoded chemical library. In the scheme, the building blocks (triangles and circles) and the corresponding DNA-barcodes (rectangles) are depicted using the same color.
In full analogy to antibody phage-display libraries or aptamer libraries, DNA-encoded chemical libraries are typically screened by performing affinity-capture procedures on a target protein of interest, immobilized on a solid support. For example, biotinylated proteins can be conveniently immobilized on streptavidin-coated magnetic beads [22] or on streptavidin-coated resin [17]. Molecules within the library, which are able to interact with the target protein, are preferentially enriched using this procedure, while non-binding compounds can be washed away. A comparative analysis of the relative abundance of the DNA tags in the library, before and after affinity capture, provides direct information about the identity of molecules, which are enriched on the target protein of interest. In general, it is good practice to confirm the enrichment results by re-synthesis of the hit compounds and by dedicated affinity measurements (“hit validation”).
The scope of DNA-encoded chemical library technology has been extended by exploiting the chemical opportunities offered by peptide-nucleic acids (PNAs) [23]. The chemical structure of PNAs allows the execution of broader set of chemical reactions, which may not always be possible with DNA [24]. However, since PNA molecules cannot be amplified by PCR, PNA-encoded libraries need to be analyzed by hybridization with complementary DNA molecules [23–25] rather than by PCR amplification and direct DNA sequencing.
Encoding Strategies
In theory, it would be conceivable to generate a DNA-encoded chemical library by coupling one by one individual compounds to distinctive oligonucleotides, serving as identification barcodes. Such an approach, however, would be expensive, unpractical and inefficient for many reasons. Firstly, it would be difficult to find millions of organic molecules, suitable for coupling to DNA. Secondly, it would be very expensive to synthesize or purchase millions of oligonucleotides. Thirdly, the freedom to generate and encode novel molecular designs would be severely limited.
Luckily, efficient strategies for the synthesis and encoding of chemical libraries have been developed, which are modular in nature and which require a limited number of oligonucleotides and of chemical building blocks. It is convenient to make a distinction between single- and dual-pharmacophore chemical libraries.
Single-pharmacophore libraries
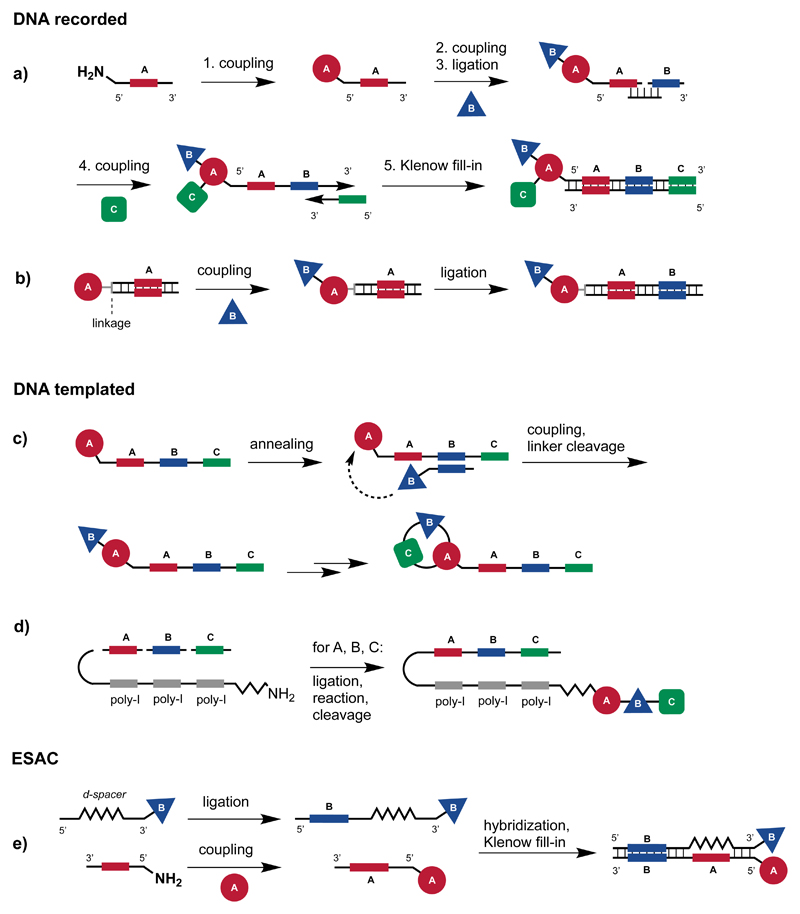
The most commonly used strategy for encoding single-pharmacophore libraries is called “DNA-recorded synthesis” and relies on the use of split-and-pool procedures [Figure 2a, b] [26, 27]. The basic concept behind this technology relates to the synthesis of chemical compounds using multiple steps, each of which is “recorded” by the addition of a DNA fragment that uniquely identifies a given chemical transformation. For illustrative purposes, we could consider the stepwise assembly of a library, generated by the reaction of two sets of building blocks (for example, 1’000 x 1’000 building blocks, leading to 106 chemical entities). In this case, it is convenient to couple the first 1’000 building blocks to 1’000 different oligonucleotides, which unambiguously identify the attached cognate chemical moiety. In most cases, the oligonucleotides that are used are similar in sequence, but differ in a central region (serving as “barcode”). The 1’000 encoded compounds can now be mixed, used as a pool and split into 1’000 reaction vessels for a subsequent chemical transformation (e.g., with 1’000 building blocks). At the end of the second set of reactions, a barcode could be added, providing an “identifier” for the chemical transformation which has been performed.
Figure 2.
(a) Schematic representation of strategies for DNA-recorded synthesis. In the first reaction cycle, chemical building blocks are encoded by direct coupling with 5' amino-modified oligonucleotides containing individual coding DNA-sequences (Code “A”). After a split & pool step which introduces the second set of building blocks B the DNA-code "B" is introduced by enzymatic DNA-ligation. In the last reaction cycle, the introduced building can be encoded by annealing with a partially-complementary oligonucleotide containing code "C", followed by the fill-in of the DNA-heteroduplex by Klenow polymerase. (b) In a variation of the encoding procedures described in (a), the organic moiety is connected by a linker (termed “headpiece”) to a double-strand DNA which is extended by subsequent ligation with coding DNA heteroduplexes, in parallel with the split-and-pool based synthesis. (c) In DNA-templated synthesis, pre-formed DNA-template molecules containing coding parts are annealed with code-specific reagent oligonucleotides, which mediate the transfer of the chemical moiety by its high effective molarity. After cleavage of the chemical moiety from the reagent oligonucleotide, it can be removed and the template undergo a new round of template-based synthesis.
(d) In a further implementation of this procedure, a template containing poly-inosine (poly-I) segments allows the annealing with various code-building block oligonucleotide conjugates. The building blocks are then transferred from the code-building block oligonucleotide conjugates to the main oligonucleotide template which, after hybridization, is encoded by ligation.
(e) In the ESAC approach, two partially complementary sub-libraries A and B are combinatorially assembled. A Klenow fill-in reaction facilitates the transfer of code B onto the complementary strand, bearing code A. Following target-based selection, PCR amplification and DNA sequencing allows the identification of the preferentially enriched pairs of building blocks.
Figure 2a depicts a strategy for the DNA-recorded synthesis of a single-pharmacophore library, consisting of three sets of building blocks. Initially, a first set of building blocks (shown in red) is coupled to amino-tagged oligonucleotides, featuring a distinctive central sequence as barcode. After the coupling step, the conjugates can be pooled and split into several vessels, for the reaction with a second set of building blocks (shown in blue). After the reaction step, oligonucleotides can be extended by the use of a splint ligation procedure [28, 29], thus incorporating barcodes for the second reaction step (i.e., identifiers for the chemical nature of the individual building blocks used for the second reaction). After a further suitable pool and split procedure, a third set of chemical transformations can be performed (with building blocks depicted in green), thus generating the final compounds, consisting of three sets of building blocks. The chemical identity of the third reaction can be encoded by annealing with a set of partially complementary oligonucleotides, followed by a Klenow polymerization step [26, 29–31]. This procedure is efficient as it makes use of one oligonucleotide for each building block used for library construction. For example, in the case of a library consisting of 1’000 x 1’000 x 1’000 building blocks (109 compounds), only 3’000 oligonucleotides and chemical building blocks will be needed.
An alternative procedure, which is frequently used in the industrial environment, makes use of double-stranded DNA fragments, connected by a chemically-modifiable linker, serving as starting structure for the construction of encoded libraries [17] [Figure 2b]. Also in this case, sets of chemical transformations are performed using split and pool procedures, leading to large combinatorial diversity. As the nascent DNA structure is in double-stranded format, it is convenient to encode each chemical transformation by the ligation of an additional double-stranded DNA fragment. The procedure can be iterated. In this case, a library consisting of 1’000 x 1’000 x 1’000 building blocks will require 6’000 oligonucleotides (i.e., 3’000 double-stranded DNA fragments).
An alternative strategy for library synthesis and encoding, first developed by the group of David Liu [32, 33], relies on the use of long pre-synthesized oligonucleotides [bearing suitable coding sequences (here depicted as A, B and C)], serving as “templates” for the stepwise assembly of nascent organic molecules [Figure 2c]. According to this strategy, oligonucleotides carrying suitable reactive building blocks are hybridized to the cognate template sequences, thus facilitating a subsequent reaction step with a nascent molecule attached to the template. After a cleavage and oligonucleotide removal step, additional sets of reactions can be performed, thus leading to the sequential assembly of complex chemical structures. The transfer of building blocks may be facilitated by the high effective molarity associated with heteroduplex formation [11]. DNA-templated strategies have been particularly useful for the synthesis of encoded polypeptidic and of macrocyclic libraries [34]. A variation of the scheme, termed “Yoctoliter reactor™” has been proposed by Vipergen, featuring templated reactions which occur on DNA three-way junctions [18].
In order to avoid encoding errors and side reactions, the DNA-templated synthesis requires a high-fidelity annealing between complementary oligonucleotide sequences [Figure 2c]. This feature may represent a potential limitation of the technology. In order to solve this problem, Li and co-workers [35] have proposed a modified template architecture, featuring a “universal template” that contains poly-inosine segments, serving as promiscuous hybridization stretches [Figure 2d]. Short oligonucleotide derivatives, carrying suitable reactive building blocks, can be sequentially hybridized and ligated to the universal template, upon formation of a hairpin structure. The strategy allows the stepwise synthesis of chemical structures, accompanied by the incorporation of barcodes which can be PCR amplified and which unambiguously identify the chemical nature of the building blocks for all library members.
Dual-pharmacophore chemical libraries
Dual-pharmacophore libraries are often constructed on the basis of a synthetic strategy, leading to “encoded self-assembled chemical (ESAC) libraries”. This technology, initially proposed by our group in 2004 [10], makes use of two sets of partially complementary oligonucleotides, each containing a distinctive DNA barcode [Figure 2e]. The two sets of chemically modified oligonucleotides constitute “sub-libraries”, which can be mixed and hybridized to form stable DNA-heteroduplexes, which display pairs of building blocks. For example, the combinatorial assembly of two sub-libraries with 1’000 chemical moieties each leads to a 1-million membered ESAC library.
In order to generate ESAC libraries with DNA structures, which allow the identification of the chemical nature of the pairs of building blocks by DNA sequencing, an encoding strategy featuring the use of oligonucleotides with abasic sites has been described [28]. One sub-library (in this case, sub-library A, Figure 2e) is synthesized by coupling a set of building blocks to 5'-amino-modified oligonucleotides, which unambiguously code for the cognate compounds. A second set of building blocks is first coupled to a single oligonucleotide bearing a d-spacer region. The resulting conjugates can be subsequently encoded by splint ligation with distinct code-bearing oligonucleotides, yielding the sub-library B (Figure 2e). The d-spacer allows the hybridization of each member of sub-library B with each member of the complementary sub-library A, avoiding base pair mismatches and allowing the creation of very large combinatorial repertoires. A code-transfer strategy, which makes use of a Klenow-fill-in DNA polymerization step, allows the transfer of code B onto the complementary strand (sub-library A, Figure 2e). This feature allows the simultaneous determination of the binding fragments by PCR amplification and high-throughput DNA sequencing [28].
Dual-display libraries feature a higher degree of flexibility in the arrangement of the chemical building blocks compared to single-pharmacophore DECLs and may more easily reach adjacent binding sites on the surface of the cognate protein target. On the other hand, as soon as a combination of building blocks (A and B) has been identified as preferential binding pair, various linkers need to be tested, in order to yield binding molecules devoid of DNA, which can be used for practical applications. For that purpose, the use of a set of predefined bifunctional scaffolds and the implementation of affinity measurements on locked-nucleic acids (LNA) may facilitate the identification of the best linker [21].
Selection Strategies
Several strategies for affinity-based selections using DNA encoded chemical libraries have been proposed and implemented in practice [19, 36]. These methods can be grouped into two main categories: (i) solid-phase selections, in which the target proteins are anchored on a solid support, allowing the physical isolation of preferential binders; and (ii) solution phase selections, which do not require the need of immobilized protein for library screening.
Solid-phase selections
Solid-phase affinity-based selections make use of target proteins, which are immobilized on a solid support and subsequently incubated with a DNA-encoded chemical library, allowing the physical separation of preferential binders from the other library members, which can be washed away. Convenient supports for library capture include magnetic beads [22, 26, 30] or resin-filled tips [17]. Washing conditions and the use of detergents may greatly impact on the selection results [22, 30]. After affinity capture, the barcodes of preferential binders are PCR amplified and submitted to a high-throughput sequencing procedure. A comparison of Illumina and 454 sequencing technologies has been reported [37]. In some cases, it may be preferable to incubate the library with a suitably tagged protein (e.g., a biotinylated protein) in solution, with a subsequent capture step on an affinity support (e.g., streptavidin-coated solid support) [17, 28, 30, 31, 38].
In full analogy to antibody-phage display technology [39], it is good practice to quantify the efficiency of the affinity capture. The use of quantitative PCR in model screening experiments (e.g., by spiking binders into large libraries) has recently been described [40].
Solution-phase selections
Solution-phase technologies allow the identification of hits without the use of a solid-phase affinity capture step. Various methodologies can be considered. The group of David Liu [41] proposed and successfully implemented the use of interaction-dependent PCR (IDPCR) [41] for the selective PCR amplification of barcodes associated with small molecules, capable of binding to the target protein of interest. The technology relies on the use of protein-oligonucleotide conjugates, capable of annealing to (and thus acting as primer for) the single-stranded DNA structures, attached to the chemical compounds in the library. The formation of a complex between a given small molecule and the target protein of interest stabilizes the DNA heteroduplex formation and facilitates the initiation of a PCR amplification step. This methodology has recently been extended, allowing the use of protein mixtures for the simultaneous identification of cognate ligands to multiple target proteins [42]. Instead of covalently attaching oligonucleotides to the protein of interest, DNA can be coupled to an antibody or to a SNAP-tagged protein [43].
Hansen et al. described Binder Trap Enrichment (BTE) as a screening methodology which takes place in water-in-oil emulsions [19]. The DNA-tagged protein target and the encoded library are encapsulated in water-in-oil droplets, which create physically isolated micro-environments, in which a splint ligation step can take place between DNA-tagged protein and cognate DNA-tagged ligands only if they are bound (i.e., found in the same water droplet).
A novel method, called “DNA-programmed photoaffinity labelling” (DPAL) [44, 45] features the use of small molecule-oligonucleotide conjugates, capable of forming a stable heteroduplex with a short complementary oligonucleotide, bearing a photoreactive group capable of covalent bond formation with protein residues [46]. When library members bind to the protein target in solution, an irradiation step ensures that preferential binders are “captured” on the target protein by photo-crosslinking.
Finally, members of DNA-encoded chemical libraries capable of protein binding can also be separated from non-binding compounds using capillary electrophoresis [47] or similar flow-based technologies.
Library Decoding
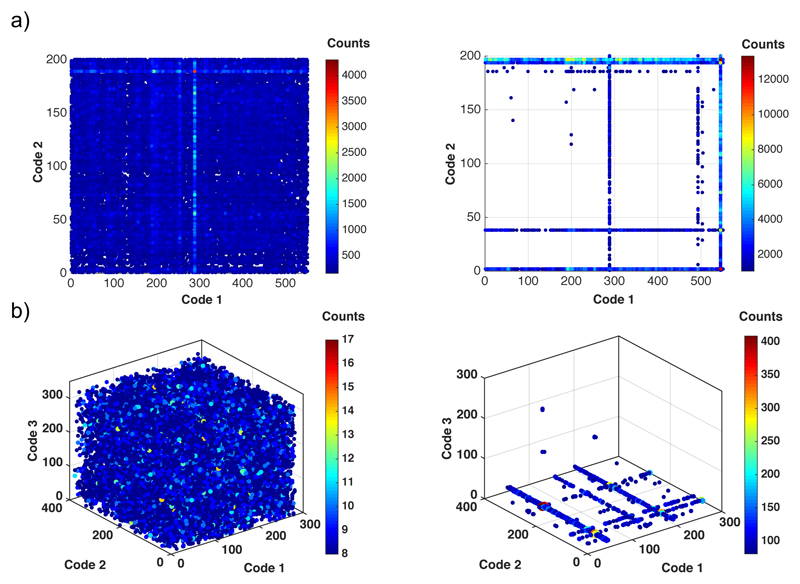
After the selection step (irrespective of whether this is performed with a solid-phase capture or in solution), the barcodes of preferential binders are amplified by PCR and read by high-throughput DNA sequencing. These results are deconvoluted by determining sequence counts for all library members, ideally comparing the frequency of barcodes before and after selection. We generally assume that the relative frequency, at which individually barcodes are found in the DNA sequencing experiment, corresponds to the relative abundance of the corresponding library members. The output data of library selections can be statistically evaluated [37] and represented by a suitable graphical display (Figure 3). The relative abundance for individual library members can be visualized on two- or three-dimensional plots (called “fingerprints”). For example, when considering a library consisting of two sets of building blocks (A and B respectively), the various AiBj combinations are displayed in the xy plane, while the corresponding numbers of counts (or the normalized enrichment factor) can be visualized in the z axis (not shown) or as spheres of different colour and/or size (Figure 3a). The display of selection results of a library based on three sets of building blocks requires a four-dimensional matrix. The three sets of building blocks (A, B and C respectively) are generally displayed in the xyz space, while enrichment factors can be represented as spheres of different colour and/or size (Figure 3b).
Figure 3.
Fingerprints of Naïve libraries (left panels) composed of (a) two sets of building blocks or (b) three sets of building blocks are compared with the same libraries selected against (a) horseradish peroxidase (HRP) or (b) carbonic anhydrase (CA) IX (right panels).
As shown in Figure 3, a successful selection experiment exhibits a clear difference in sequence counts for the various library members before and after the capture step. The enriched compounds are then resynthesized, in order to confirm the experimental findings and to measure affinity constants.
Ligands Isolated from DNA-Encoded Chemical Libraries
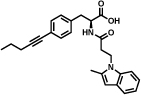
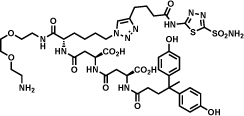
In this section, we report on some important hits that have been discovered by different implementations of DECL technology. The first example described in Table 1 (entry a) derives from a structurally-compact two-building block single-pharmacophore DNA-encoded chemical library [31]. The library contained 103`200 members formed from two sets of carboxylic acids (240x430). The hit inhibits tankyrase-1 with an IC50 value of 290 nM, as measured by an enzymatic PARylation assay [48].
Table 1. Examples of hit compounds identified from DNA-encoded chemical libraries.
The following abbreviations were used for the protein targets: TNKS1 = tankyrase-1, IL2 = interleukin-2, NK3 = Neurokinin 3, MAPK14 = Mitogen-activated protein kinase 14, sEH = soluble epoxide hydrolase, CAIX = carbonic anhydrase IX, AGP = alpha-1-acid glycoprotein.
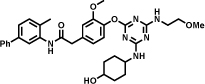
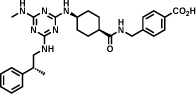
A ligand against interleukin-2 is reported in Table 1 (entry b). The library, constructed by Leimbacher et al. [30], was based on 100 different amino-acids, that had been coupled to 300 different carboxylic acids. The binder showed a dissociation constant (Kd) value of 2.5 µM, as measured by fluorescence polarization. The importance of the 2-methylindole moiety for interleukin-2 recognition was assessed by a comparative analysis of more than 100 different selections [30].
GlaxoSmith-Kline isolated a 2 nM antagonist for Neurokinin 3 (NK3) [49]. The hit compound, reported in Table 1 (entry c) was isolated from a 41-million member single-pharmacophore DECL using three sets of building blocks (64 hydroxy- or amino-acids, 854 and 758 amines respectively) [49]. The building blocks were connected to DNA through a central 1,3,5-triazine scaffold. In the validated hit (Table 1, entry c), the DNA moiety had been replaced by a 2-methoxyethan-1-amino linker [49].
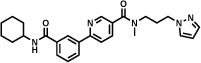
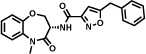
A highly specific 7 nM inhibitor of mitogen-activated protein kinase 14 (MAPK14, also called p38-α kinase) [50] was identified by Vipergen using a yoctoReactor™ DECL. The library consisted of three sets of building blocks (2 sets of aminoacids and 1 set of carboxylic acids) and had a size of 12.6 million members [18]. Affinity selections were performed in solution using Binder Trap Enrichment (BTE) technology [19]. Table 1 (entry d) reports the hit compound of the most enriched combination in which the DNA moiety had been replaced by an NH2 group.
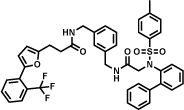
An extremely potent inhibitor of soluble epoxide hydrolase (sEH) was developed by X-Chem using an innovative DECL encoding strategy, featuring a DNA-ligation step that made use of Cu-catalyzed alkyne-azide cycloaddition (CuAAC) [51]. A 334-million member library was synthesized using 3 sets of building blocks (2'259 primary amines, 222 bromoaryl acids and 667 boronic acids). The hit compound Table 1 (entry e) in which the DNA portion was substituted by a methyl group, showed a 2 nM IC50 for sEH.
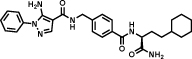
Dual-pharmacophore libraries based on ESAC technology [10] have also been successful [28]. A library containing more than 100,000 compounds allowed the isolation of an acetazolamide derivative (Table 1, entry f), that bound to carbonic anhydrase IX with a dissociation constant in the sub-nanomolar range [28]. The same ESAC library also produced a 190 nM binder against alpha-1-acid glycoprotein (AGP), thanks to the synergistic action of two building blocks that mediated a potent chelate binding to the target protein. Interestingly, the individual building blocks did not display any detectable protein binding when used on their own. The optimization of the linker connecting the two building blocks led to an improved binder Table 1 (entry g), with a dissociation constant of 9.9 nM [21].
The Road Ahead: Challenges and Opportunities
DECL technology works reliably for the isolation of hits against many target proteins of pharmaceutical interest. However, unlike human antibody technology [6], DECL sometimes fails to deliver hits to certain classes of protein targets, thus underlining the need for further technical improvements.
Thanks to advances in the quantification of library selection procedures through quantitative PCR methodologies, it is becoming increasingly clear that ligands with dissociation constants in the micromolar range are not efficiently recovered from encoded libraries in most experimental settings [54]. However, conventional technologies based on affinity capture methods are quite reliable for the isolation of nanomolar binders from encoded libraries, if and when they are present in the repertoire [40, 55].
Certain classes of proteins (e.g., integral membrane proteins) are not compatible with standard selection procedures, as they are difficult to keep in solution and to capture on solid supports (e.g., streptavidin-coated beads or resin) [22]. The use of stabilized GPCRs, selection on cells or on nanodisks may be considered [49, 56, 57].
DNA sequencing is an essential component of DECL technology and it would be desirable to further enhance the number of DNA tags that can be sequenced. At present, one lane of Illumina™ sequencing may allow the identification of up to 200 million sequence tags. As it is possible to synthesize libraries containing billions of members, it would be important to sequence billions of tags, in order to adequately characterize selection experiments with suitable statistical tools [37].
In principle, the creativity of Chemistry is unlimited and it is likely that future success in DECL technology will derive from the practical implementation of novel DNA-compatible reactions. The set of reactions which can be considered for library synthesis is constantly expanding [58]. However, it is important to remember that even simple transformations (such as the formation of amide bonds) do not proceed with complete yields on 100% of substrates. An extensive characterization and selection of building blocks, which give acceptable yields, represents an essential component for any new library synthesis effort. On the other hand, preserving the integrity of DNA is important [59].
Novel scaffolds and novel concepts in library design will likely contribute to the discovery of ligands to proteins of pharmaceutical interest. We have recently observed that the central chemical moiety, used for the display of two sets of building blocks, can greatly contribute to the affinity of the resulting hit, at least for a certain proportion of target proteins [60]. Macrocyclic scaffolds may facilitate the isolation of hits against "difficult" targets [29, 34, 38], but these larger chemical structures may not always be compatible with pharmaceutical applications and with oral administration procedures.
It has previously been noted that the number and quality of building blocks greatly contributes to library performance [13]. On the positive side, thousands of building blocks (e.g., carboxylic acids, amines, alkynes, azides, sulphonyl chlorides, boronates) are now commercially available at affordable prices, thus facilitating library construction activities even in the academic environment. On the other hand, large pharmaceutical industries may have access to proprietary building blocks and to privileged structures. Moreover, library size (especially for repertoires consisting of two sets of building blocks) depends on the financial resources that can be devoted to purchasing oligonucleotides and building blocks. For example, a library of 100 million compounds based on two sets of building blocks will require 20'000 oligonucleotides and 20'000 building blocks, while a three-building block library with 1 billion compounds can be created using 3'000 oligonucleotides and 3'000 building blocks.
ESAC technology appears to be ideally suited for the discovery of fragments, which bind to adjacent sites on the target protein of interest. At present, the optimization of linkers connecting synergistic building blocks is still empirical, even though "on-DNA" procedures may facilitate this experimental task [21]. It remains to be seen whether linker optimization can be embedded in library construction and selection procedures. ESAC technology relies on pure components and facilitates the "affinity maturation" of existing hits, which can be paired to existing libraries in a facile manner.
In summary, DECL technology has reached a good level of maturity. Both pharmaceutical companies and academic laboratories use encoded libraries for the discovery of hits. Experimental procedures have been extensively validated and are well documented in the literature. Nonetheless, we are confident that improvements in library design, chemical procedures, selections and DNA sequencing methodologies will continue to broaden the scope of the methodology and to make it broadly accessible to the research community.
Acknowledgments
Financial support from ETH Zürich, the Swiss National Science Foundation (310030B_163479/1 Grant and CRSII2_160699/1 Sinergia Grant), the ERC Advanced Grant “Zauberkugel” and Philochem AG is gratefully acknowledged.
References
- 1.Goodnow RA, Jr, Dumelin CE, Keefe AD. DNA-encoded chemistry: enabling the deeper sampling of chemical space. Nat Rev Drug Discov. 2017;16:131–147. doi: 10.1038/nrd.2016.213. [DOI] [PubMed] [Google Scholar]
- 2.Brenner S, Lerner RA. Encoded combinatorial chemistry. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:5381–5383. doi: 10.1073/pnas.89.12.5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kang AS, Barbas CF, Janda KD, Benkovic SJ, Lerner RA. Linkage of recognition and replication functions by assembling combinatorial antibody Fab libraries along phage surfaces. PNAS. 1991;88:4. doi: 10.1073/pnas.88.10.4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCafferty J, Griffiths AG, W G, Chiswell D. Phage antibodies: filamentous phage displaying antibody variable domains. Nature. 1990;348:3. doi: 10.1038/348552a0. [DOI] [PubMed] [Google Scholar]
- 5.Winter G, Milstein C. Man-made antibodies. Nature. 1991;349:7. doi: 10.1038/349293a0. [DOI] [PubMed] [Google Scholar]
- 6.Winter G, Griffiths AD, Hawkins RE, Hoogenboom HR. MAKING ANTIBODIES BY PHAGE DISPLAY TECHNOLOGY. Annual Review of Immunology. 1994;12:23. doi: 10.1146/annurev.iy.12.040194.002245. [DOI] [PubMed] [Google Scholar]
- 7.Roberts RW, Szostak JW. RNA–peptide fusions for the in vitro selection of peptides and proteins. Proc Natl Acad Sci USA. 1997;94:5. doi: 10.1073/pnas.94.23.12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ochsner UA, Green LS, Gold L, Janjic N. Systematic selection of modified aptamer pairs for diagnostic sandwich assays. Biotechniques. 2014;56:125–8. doi: 10.2144/000114134. 130, 132-3. [DOI] [PubMed] [Google Scholar]
- 9.Lerner RA, Brenner S. DNA-Encoded Compound Libraries as Open Source: A Powerful Pathway to New Drugs. Angew Chem Int Ed Engl. 2017;56:1164–1165. doi: 10.1002/anie.201612143. [DOI] [PubMed] [Google Scholar]
- 10.Melkko S, Scheuermann J, Dumelin CE, Neri D. Encoded self-assembling chemical libraries. Nat Biotechnol. 2004;22:568–74. doi: 10.1038/nbt961. [DOI] [PubMed] [Google Scholar]
- 11.Gartner ZJ, Tse BN, Grubina R, Doyon JB, Snyder TM, Liu DR. DNA-Templated Organic Synthesis and Selection of a Library of Macrocycles. Science. 2004;305:1601. doi: 10.1126/science.1102629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halpin DR, Harbury PB. DNA display II. Genetic manipulation of combinatorial chemistry libraries for small-molecule evolution. PLoS Biol. 2004;2:E174. doi: 10.1371/journal.pbio.0020174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franzini RM, Neri D, Scheuermann J. DNA-encoded chemical libraries: advancing beyond conventional small-molecule libraries. Acc Chem Res. 2014;47:1247–55. doi: 10.1021/ar400284t. [DOI] [PubMed] [Google Scholar]
- 14.Zimmermann G, Neri D. DNA-encoded chemical libraries: foundations and applications in lead discovery. Drug Discov Today. 2016;21:1828–1834. doi: 10.1016/j.drudis.2016.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neri D, Lerner R. DNA-Encoded Chemical Libraries: A Selection System Based On Endowing Organic Compounds with Amplifiable Information. Annual Review of Biochemistry. 2018;87:24. doi: 10.1146/annurev-biochem-062917-012550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu Z, Shaginian A, Grady LC, O'Keeffe T, Shi XE, Davie CP, Simpson GL, Messer JA, Evindar G, Bream RN, Thansandote PP, et al. Design and Application of a DNA-Encoded Macrocyclic Peptide Library. ACS Chem Biol. 2018;13:53–59. doi: 10.1021/acschembio.7b00852. [DOI] [PubMed] [Google Scholar]
- 17.Clark MA, Acharya RA, Arico-Muendel CC, Belyanskaya SL, Benjamin DR, Carlson NR, Centrella PA, Chiu CH, Creaser SP, Cuozzo JW, Davie CP, et al. Design, synthesis and selection of DNA-encoded small-molecule libraries. Nat Chem Biol. 2009;5:647–54. doi: 10.1038/nchembio.211. [DOI] [PubMed] [Google Scholar]
- 18.Hansen MH, Blakskjær P, Petersen LK, Hansen TH, Højfeldt JW, Gothelf KV, Hansen NJV. A Yoctoliter-Scale DNA Reactor for Small-Molecule Evolution. J AM CHEM SOC. 2009;131:6. doi: 10.1021/ja808558a. [DOI] [PubMed] [Google Scholar]
- 19.Blakskjaer P, Heitner T, Hansen NJ. Fidelity by design: Yoctoreactor and binder trap enrichment for small-molecule DNA-encoded libraries and drug discovery. Curr Opin Chem Biol. 2015;26:62–71. doi: 10.1016/j.cbpa.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 20.Buller F, Mannocci L, Scheuermann J, Neri D. Drug Discovery with DNA-Encoded Chemical Libraries. Bioconjugate Chem. 2010;21:10. doi: 10.1021/bc1001483. [DOI] [PubMed] [Google Scholar]
- 21.Bigatti M, Dal Corso A, Vanetti S, Cazzamalli S, Rieder U, Scheuermann J, Neri D, Sladojevich F. Impact of a Central Scaffold on the Binding Affinity of Fragment Pairs Isolated from DNA-Encoded Self-Assembling Chemical Libraries. ChemMedChem. 2017;12:1748–1752. doi: 10.1002/cmdc.201700569. [DOI] [PubMed] [Google Scholar]
- 22.Decurtins W, Wichert M, Franzini RM, Buller F, Stravs MA, Zhang Y, Neri D, Scheuermann J. Automated screening for small organic ligands using DNA-encoded chemical libraries. Nat Protoc. 2016;11:764–80. doi: 10.1038/nprot.2016.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Svensen N, Diaz-Mochon JJ, Bradley M. Decoding a PNA encoded peptide library by PCR: the discovery of new cell surface receptor ligands. Chem Biol. 2011;18:1284–9. doi: 10.1016/j.chembiol.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 24.Zambaldo C, Barluenga S, Winssinger N. PNA-encoded chemical libraries. Curr Opin Chem Biol. 2015;26:8–15. doi: 10.1016/j.cbpa.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 25.Gorska K, Huang KT, Chaloin O, Winssinger N. DNA-templated homo- and heterodimerization of peptide nucleic acid encoded oligosaccharides that mimick the carbohydrate epitope of HIV. Angew Chem Int Ed Engl. 2009;48:7695–700. doi: 10.1002/anie.200903328. [DOI] [PubMed] [Google Scholar]
- 26.Mannocci L, Zhang Y, Scheuermann J, Leimbacher M, De Bellis G, Rizzi E, Dumelin C, Melkko S, Neri D. High-throughput sequencing allows the identification of binding molecules isolated from DNA-encoded chemical libraries. Proceedings of the National Academy of Sciences. 2008;105:17670–17675. doi: 10.1073/pnas.0805130105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu R, Li X, Lam KS. Combinatorial chemistry in drug discovery. Curr Opin Chem Biol. 2017;38:117–126. doi: 10.1016/j.cbpa.2017.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wichert M, Krall N, Decurtins W, Franzini RM, Pretto F, Schneider P, Neri D, Scheuermann J. Dual-display of small molecules enables the discovery of ligand pairs and facilitates affinity maturation. Nat Chem. 2015;7:241–9. doi: 10.1038/nchem.2158. [DOI] [PubMed] [Google Scholar]
- 29.Li Y, De Luca R, Cazzamalli S, Pretto F, Bajic D, Scheuermann J, Neri D. Versatile protein recognition by the encoded display of multiple chemical elements on a constant macrocyclic scaffold. Nature Chemistry. 2018 doi: 10.1038/s41557-018-0017-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leimbacher M, Zhang Y, Mannocci L, Stravs M, Geppert T, Scheuermann J, Schneider G, Neri D. Discovery of small-molecule interleukin-2 inhibitors from a DNA-encoded chemical library. Chemistry. 2012;18:7729–37. doi: 10.1002/chem.201200952. [DOI] [PubMed] [Google Scholar]
- 31.Franzini RM, Ekblad T, Zhong N, Wichert M, Decurtins W, Nauer A, Zimmermann M, Samain F, Scheuermann J, Brown PJ, Hall J, et al. Identification of structure-activity relationships from screening a structurally compact DNA-encoded chemical library. Angew Chem Int Ed Engl. 2015;54:3927–31. doi: 10.1002/anie.201410736. [DOI] [PubMed] [Google Scholar]
- 32.Kanan MW, Rozenman MM, Sakurai K, Snyder TM, Liu DR. Reaction discovery enabled by DNA-templated synthesis and in vitro selection. nature. 2004;431:5. doi: 10.1038/nature02920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X, Liu DR. DNA-templated organic synthesis: nature's strategy for controlling chemical reactivity applied to synthetic molecules. Angew Chem Int Ed Engl. 2004;43:4848–70. doi: 10.1002/anie.200400656. [DOI] [PubMed] [Google Scholar]
- 34.Kleiner RE, Dumelin CE, Tiu GC, Sakurai K, Liu DR. In Vitro Selection of a DNA-Templated Small-Molecule Library Reveals a Class of Macrocyclic Kinase Inhibitors. J Am Chem Soc. 2010;132:13. doi: 10.1021/ja104903x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y, Zhao P, Zhang M, Zhao X, Li X. Multistep DNA-templated synthesis using a universal template. J Am Chem Soc. 2013;135:17727–30. doi: 10.1021/ja409936r. [DOI] [PubMed] [Google Scholar]
- 36.Chan AI, McGregor LM, Liu DR. Novel selection methods for DNA-encoded chemical libraries. Curr Opin Chem Biol. 2015;26:55–61. doi: 10.1016/j.cbpa.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buller F, Steiner M, Scheuermann J, Mannocci L, Nissen I, Kohler M, Beisel C, Neri D. High-throughput sequencing for the identification of binding molecules from DNA-encoded chemical libraries. Bioorg Med Chem Lett. 2010;20:4188–92. doi: 10.1016/j.bmcl.2010.05.053. [DOI] [PubMed] [Google Scholar]
- 38.Maianti JP, McFedries A, Foda ZH, Kleiner RE, Du XQ, Leissring MA, Tang WJ, Charron MJ, Seeliger MA, Saghatelian A, Liu DR. Anti-diabetic activity of insulin-degrading enzyme inhibitors mediated by multiple hormones. Nature. 2014;511:94–8. doi: 10.1038/nature13297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mutuberria R, Hoogenboom HR, Linden Evd, Bruine APd, Roovers RC. Model systems to study the parameters determining the success of phage antibody selections on complex antigens. Journal of Immunological Methods. 1999;231:17. doi: 10.1016/s0022-1759(99)00141-6. [DOI] [PubMed] [Google Scholar]
- 40.Li Y, Zimmermann G, Scheuermann J, Neri D. Quantitative PCR is a Valuable Tool to Monitor the Performance of DNA-Encoded Chemical Library Selections. Chembiochem. 2017;18:848–852. doi: 10.1002/cbic.201600626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McGregor LM, Gorin DJ, Dumelin CE, Liu DR. Interaction-Dependent PCR: Identification of Ligand−Target Pairs from Libraries of Ligands and Libraries of Targets in a Single Solution-Phase Experiment. J Am Chem Soc. 2010;132:3. doi: 10.1021/ja107677q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McGregor LM, Jain T, Liu DR. Identification of ligand-target pairs from combined libraries of small molecules and unpurified protein targets in cell lysates. J Am Chem Soc. 2014;136:3264–70. doi: 10.1021/ja412934t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat Biotechnol. 2003;21:86–9. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- 44.Li G, Liu Y, Liu Y, Chen L, Wu S, Liu Y, Li X. Photoaffinity labeling of small-molecule-binding proteins by DNA-templated chemistry. Angew Chem Int Ed Engl. 2013;52:9544–9. doi: 10.1002/anie.201302161. [DOI] [PubMed] [Google Scholar]
- 45.Wang D-Y, Cao Y, Zheng L-Y, Chen L-D, Chen X-F, Hong Z-Y, Zhu Z-Y, Li X, Chai Y-F. Target Identification of Kinase Inhibitor Alisertib (MLN8237) by Using DNA-Programmed Affinity Labeling. Chemistry a European Journal. 2017;23:9. doi: 10.1002/chem.201702033. [DOI] [PubMed] [Google Scholar]
- 46.Zhao P, Chen Z, Li Y, Sun D, Gao Y, Huang Y, Li X. Selection of DNA-encoded small molecule libraries against unmodified and non-immobilized protein targets. Angew Chem Int Ed Engl. 2014;53:10056–9. doi: 10.1002/anie.201404830. [DOI] [PubMed] [Google Scholar]
- 47.Bao J, Krylova SM, Cherney LT, Hale RL, Belyanskaya SL, Chiu CH, Shaginian A, Arico-Muendel CC, Krylov SN. Predicting Electrophoretic Mobility of Protein-Ligand Complexes for Ligands from DNA-Encoded Libraries of Small Molecules. Anal Chem. 2016;88:5498–506. doi: 10.1021/acs.analchem.6b00980. [DOI] [PubMed] [Google Scholar]
- 48.Wahlberg E, Karlberg T, Kouznetsova E, Markova N, Macchiarulo A, Thorsell AG, Pol E, Frostell A, Ekblad T, Oncu D, Kull B, et al. Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors. Nat Biotechnol. 2012;30:283–8. doi: 10.1038/nbt.2121. [DOI] [PubMed] [Google Scholar]
- 49.Wu Z, Graybill TL, Zeng X, Platchek M, Zhang J, Bodmer VQ, Wisnoski DD, Deng J, Coppo FT, Yao G, Tamburino A, et al. Cell-Based Selection Expands the Utility of DNA-Encoded Small-Molecule Library Technology to Cell Surface Drug Targets: Identification of Novel Antagonists of the NK3 Tachykinin Receptor. ACS Comb Sci. 2015;17:722–31. doi: 10.1021/acscombsci.5b00124. [DOI] [PubMed] [Google Scholar]
- 50.Petersen LK, Blakskjær P, Chaikuad A, Christensen AB, Dietvorst J, Holmkvist J, Knapp S, Kořínek M, Larsen LK, Pedersen AE, Röhm S, et al. Novel p38α MAP kinase inhibitors identified from yoctoReactor DNA-encoded small molecule library. MedChemComm. 2016;7:1332–1339. [Google Scholar]
- 51.Litovchick A, Dumelin CE, Habeshian S, Gikunju D, Guie MA, Centrella P, Zhang Y, Sigel EA, Cuozzo JW, Keefe AD, Clark MA. Encoded Library Synthesis Using Chemical Ligation and the Discovery of sEH Inhibitors from a 334-Million Member Library. Sci Rep. 2015;5 doi: 10.1038/srep10916. 10916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harris PA, Berger SB, Jeong JU, Nagilla R, Bandyopadhyay D, Campobasso N, Capriotti CA, Cox JA, Dare L, Dong X, Eidam PM, et al. Discovery of a First-in-Class Receptor Interacting Protein 1 (RIP1) Kinase Specific Clinical Candidate (GSK2982772) for the Treatment of Inflammatory Diseases. J Med Chem. 2017;60:1247–1261. doi: 10.1021/acs.jmedchem.6b01751. [DOI] [PubMed] [Google Scholar]
- 53.Belyanskaya SL, Ding Y, Callahan JF, Lazaar AL, Israel DI. Discovering Drugs with DNA-Encoded Library Technology: From Concept to Clinic with an Inhibitor of Soluble Epoxide Hydrolase. Chembiochem. 2017;18:837–842. doi: 10.1002/cbic.201700014. [DOI] [PubMed] [Google Scholar]
- 54.Denton KE, Wang S, Gignac MC, Milosevich N, Hof F, Dykhuizen EC, Krusemark CJ. Robustness of In Vitro Selection Assays of DNA-Encoded Peptidomimetic Ligands to CBX7 and CBX8. SLAS Discovery. 2017:12. doi: 10.1177/2472555217750871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cuozzo JW, Centrella PA, Gikunju D, Habeshian S, Hupp CD, Keefe AD, Sigel EA, Soutter HH, Thomson HA, Zhang Y, Clark MA. Discovery of a Potent BTK Inhibitor with a Novel Binding Mode by Using Parallel Selections with a DNA-Encoded Chemical Library. Chembiochem. 2017;18:864–871. doi: 10.1002/cbic.201600573. [DOI] [PubMed] [Google Scholar]
- 56.Bayburt TH, Sligar SG. Membrane protein assembly into Nanodiscs. FEBS Lett. 2010;584:1721–7. doi: 10.1016/j.febslet.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Klenk C, Ehrenmann J, Schutz M, Pluckthun A. A generic selection system for improved expression and thermostability of G protein-coupled receptors by directed evolution. Sci Rep. 2016;6 doi: 10.1038/srep21294. 21294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Satz AL, Cai J, Chen Y, Goodnow R, Gruber F, Kowalczyk A, Petersen A, Naderi-Oboodi G, Orzechowski L, Strebel Q. DNA Compatible Multistep Synthesis and Applications to DNA Encoded Libraries. Bioconjug Chem. 2015;26:1623–32. doi: 10.1021/acs.bioconjchem.5b00239. [DOI] [PubMed] [Google Scholar]
- 59.Malone ML, Paegel BM. What is a "DNA-Compatible" Reaction? ACS Comb Sci. 2016;18:182–7. doi: 10.1021/acscombsci.5b00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Favalli N, Biendl S, Hartmann M, Piazzi J, Sladojevich F, Gräslund S, Brown PJ, Näreoja K, Schüler H, Scheuermann J, Franzini R, et al. A DNA-encoded library of chemical compounds based on common scaffolding structures reveals the impact of ligand geometry on protein recognition. 2018 doi: 10.1002/cmdc.201800193. in press. in press, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]