Abstract
Interaction with the extracellular matrix (ECM) is one of the successful colonization strategies employed by nontypeable Haemophilus influenzae (NTHi). Here we identified Haemophilus lipoprotein e (P4) as a receptor for ECM proteins. Purified recombinant P4 displayed a high binding affinity for laminin (Kd = 9.26 nM) and fibronectin (Kd = 10.19 nM), but slightly less to vitronectin (Kd = 16.51 nM). A P4-deficient NTHi mutant showed a significantly decreased binding to these ECM components. Vitronectin acquisition conferred serum resistance to both P4-expressing NTHi and Escherichia coli transformants. P4-mediated bacterial adherence to pharynx, type II alveolar, and bronchial epithelial cells was mainly attributed to fibronectin. Importantly, a significantly reduced bacterial infection was observed in the middle ear of the Junbo mouse model when NTHi was devoid of P4. In conclusion, our data provide new insight into the role of P4 as an important factor for Haemophilus colonization and subsequent respiratory tract infection.
Keywords: adhesion, epithelial cells, extracellular matrix, fibronectin, Haemophilus influenzae, laminin, serum resistance, vitronectin
Nontypeable Haemophilus influenzae (NTHi) is one of the major causes of mucosal infections in the upper and lower respiratory tract. The pathogen is associated with acute otitis media in pre-school children and exacerbations in patients with chronic obstructive pulmonary disease, with recent reports also suggesting a role in invasive disease [1].
We and others have reported that NTHi interacts with extracellular matrix (ECM) proteins, including fibrinogen, collagens (I, III, IV and V), vitronectin (Vn), laminin (Ln), and fibronectin (Fn) [2–8]. Most ECM proteins are located underneath the epithelial cell barrier, and are thus unexposed to microbes. During viral infections and/or mechanical and chemical trauma, however, the epithelial cell barrier will be damaged and tight junctions break, exposing the ECM components. Interactions with the ECM thus promote the opportunistic pathogen NTHi to colonize and consequently infect the host [9–12]. These interactions require the expression of several NTHi surface molecules such as Haemophilus adhesion and penetration protein (Hap), protein E (PE), and protein F (PF) [4–7].
NTHi with deletion of single or multiple of the currently known ECM receptors still show residual ECM binding [4–6, 13]. This fact prompted us to speculate that there might be other yet unidentified ECM receptor(s) in NTHi. Here we identified Haemophilus outer membrane lipoprotein e (P4) as a novel multifunctional ECM receptor. Haemophilus P4 is a 28 kDa acid phosphatase recognized as a potential vaccine candidate due to its immunogenic properties [14, 15]. The lipoprotein is found in both NTHi and encapsulated H. influenzae, and is important for nicotinamide adenine dinucleotide (NAD) uptake and hemin utilization [16–19]. Importantly, we observed both an enhanced serum resistance and increased adherence with bacteria expressing P4 as a result of Vn acquisition and targeting of Ln/Fn, respectively. The P4-deficient NTHi mutant had a decreased survival in the middle ear of Junbo mice, suggesting an important role of P4 in acute otitis media caused by NTHi. Our work thus provides additional insight into the role of P4 for NTHi virulence.
Materials and Methods
Bacterial Strains and Eukaryotic Cells
NTHi 3655, isogenic mutants, and nasopharyngeal clinical isolates (KR164, KR314, KR315, and KR336) were cultured as indicated (Table 1). Escherichia coli DH5α (Stratagene, Santa Clara, California) and BL21 (DE3) (Novagen, Darmstadt, Germany) were cultured in Luria–Bertani (LB) broth or on LB agar. Antibiotics selection for each genetically modified bacterial strain is summarized in Table 1. The alveolar epithelial cell line A549 (ATCC CCL-185) was maintained in Ham’s Nutrient F12 medium (Gibco, Life Technologies, Carlsbad, California), the pharynx epithelial cell line Detroit 562 (ATCC CCL-138) in Minimum Essential Medium (Gibco), and the bronchial epithelial cell line NCI-H292 in Roswell Park Memorial Institute 1640 (Gibco). All media were supplemented with 10% fetal calf serum (FCS) (Gibco). Primary human bronchial epithelial (HBE) cells were obtained from healthy donors (Swedish Research Ethical Committee in Lund [FEK 413/2008]). HBE were maintained in bronchial epithelial cell growth medium (Clonetics, Basel, Switzerland).
Table 1. List of Bacterial Strains Used in This Study.
| Bacterial Straina,b,c | Description/Genotyped | Reference |
|---|---|---|
| NTHi 3655 | Clinical isolate from a 10-year-old child with acute otitis media with effusion. | [20] |
| NTHi 3655 (SmR) | SmR. Spontaneous streptomycin resistant isogenic mutant. The strain is used for mouse middle-ear infection. | This study |
| NTHi 3655Δhel | CmR. Isogenic hel deletion mutant of NTHi 3655 by cat replacement. The strain is devoid of P4 expression. | This study |
| NTHi 3655Δhel (SmR) | CmR and SmR. Spontaneous streptomycin-resistant isogenic mutant of NTHi 3655Δhel for mouse middle-ear infection. | This study |
| NTHi 3655 Δhel+hel | KanR and CmR. hel-transcomplemented isogenic mutant of NTHi 3655Δhel. The hel-bearing complementation construct amplified from pKR7.3-P4 (Supplementary Figure 1) was inserted into the CGSHi3655_06559 locus. The strain has a restored P4 expression. | This study |
| NTHi 3655 Δhel+hel (SmR) | CmR, KanR, and SmR. Spontaneous streptomycin-resistant isogenic mutant of NTHi 3655Δhel+hel for mouse middle-ear infection. | This study |
| NTHi 3655Δpe | ZeoR isogenic pe deletion mutant of NTHi 3655 by ble replacement. The strain is devoid of PE expression. | [21] |
| NTHi 3655Δhpf | CmR isogenic hpf deletion mutant of NTHi 3655 by cat replacement. The strain is devoid of PF expression. | [6] |
| NTHi 3655Δhap | CmR isogenic hap deletion mutant of NTHi 3655 by cat replacement. The strain is devoid of Hap expression. | [4] |
| NTHi 3655Δhel/pe | CmR and ZeoR isogenic pe and hel double-deletion mutant. The pe and hel were replaced with zeo and cat resistance genes, respectively. The strain is devoid of PE and P4 expression. | This study |
| NTHi 3655Δhel/hpf | CmR and KanR isogenic hpf and hel double-deletion mutant. The hpf and hel were replaced with cat and npt I resistance genes, respectively. The strain is devoid of PF and P4 expression. | This study |
| NTHi 3655Δhel/hap | CmR and KanR isogenic hpf and hel double-deletion mutant. The hpf and hel were replaced with cat and npt I resistance genes, respectively. The strain is devoid of PF and P4 expression. | This study |
| NTHi KR164 | NTHi blood isolate from 35-year-old patient with bacteremia. The wild-type strain binds ECM proteins. | [6] |
| NTHi KR164Δhel | CmR isogenic hel deletion mutant of NTHi KR164 by cat replacement. The strain is devoid of P4 expression. | This study |
| NTHi KR314 | NTHi nasopharyngeal isolate from 66-year-old patient with acute otitis media. The wild-type strain binds ECM proteins. | [6] |
| NTHi KR314Δhel | CmR isogenic hel deletion mutant of NTHi KR314 by cat replacement. The strain is devoid of P4 expression. | This study |
| NTHi KR315 | NTHi nasopharyngeal isolate from 79-year-old patient with bronchitis. The wild-type strain binds ECM proteins. | [6] |
| NTHi KR315Δhel | CmR isogenic hel deletion mutant of NTHi KR315 by cat replacement. The strain is devoid of P4 expression. | This study |
| NTHi KR336 | NTHi nasopharyngeal isolate from 32-year-old patient with sinusitis. The wild-type strain binds ECM proteins. | [6] |
| NTHi KR336Δhel | CmR isogenic hel deletion mutant of NTHi KR336 by cat replacement. The strain is devoid of P4 expression. | This study |
| E. coli–pET16b | AmpR E. coli BL21 (DE3) bearing the empty vector pET16b. | |
| E. coli–P4 | AmpR E. coli BL21 (DE3) bearing recombinant plasmid of pET16b with hel ORF insertion expressing P4 at surface. | This study |
Abbreviations: β-NMN, β-nicotinamide ribose monophosphate; Amp, ampicillin; BHI, brain heart infusion; Cm, chloramphenicol; ECM, extracellular matrix; Hap, Haemophilus adhesion and penetration protein; Kan, kanamycin; NAD, nicotinamide adenine dinucleotide; NTHi, nontypeable H. influenzae; ORF, open reading frame; P4, Haemophilus lipoprotein e; PE, protein E; PF, protein F; Sm, streptomycin; SmR, streptomycin-resistant; Zeo, zeocin.
NTHi strains were grown on chocolate agar or cultured in BHI broth supplemented with NAD (Sigma-Aldrich) and hemin (Merck, Darmstadt, Germany) each at 10 µg/mL at 37°C in a humid atmosphere containing 5% CO2.
NTHi hel-deletion mutants with or without the other indicated genes were grown in medium supplemented with β-NMN (80 µM) (Sigma-Aldrich) instead of NAD.
SmR strains were generated as described [22].
Concentrations of antibiotics used: 100 µg/mL Amp; 10 µg/mL Cm; 15 µg/mL Kan; 300 µg/mL Sm; 4 µg/mL Zeo.
Construction of NTHi hel Deletion Mutants and Gene Transcomplementation
Deletion of the hel gene (GenBank Accession Number: EDJ92235; encodes P4) in single or double mutants of NTHi 3655 was performed as described [6] (Supplementary Figure 1). A linear hel-knockout vector containing cat (AY219687.1) or npt (HQ608521.1) inserted between upstream and downstream flanking regions of the hel gene (Supplementary Figure 1 and Table 1) was transformed into NTHi strains to yield Δhel mutant [23]. We used the pKR7.3 backbone plasmid to transcomplement hel in the NTHi 3655Δhel, yielding NTHi 3655Δhel+hel [24]. Spontaneous streptomycin-resistant (SmR) isogenic mutants used for animal experiments were also generated [22].
Recombinant Protein Expression and Antibody Production
Open reading frames of full-length and truncated variants of P4 were amplified from NTHi 3655 genomic DNA using restriction site-specific primers (Supplementary Table 1). Resulting DNA fragments were cloned into the expression vector pET26(b)+ for cytoplasmic recombinant protein production or pET16b for cell surface expression of P4. Clones were maintained in E. coli DH5α. His-tagged recombinant proteins were expressed in E. coli BL21 (DE3) induced with 1 mM isopropyl-1-thio-β-D-galactopyranoside followed by protein purification as described (Supplementary Figure 2) [6]. A rabbit anti-P4 polyclonal antibody (pAb) was prepared as described [25].
Enzyme-Linked Immunosorbent Assay
Purified proteins (50 nM) were immobilized on Polysorb microtitre plates (Nunc, Roskilde, Denmark) in 100 mM NaH2CO3 (pH 8.6). ECM proteins, human plasma Vn [26] and Fn (Sigma-Aldrich), and murine sarcoma Ln (Sigma-Aldrich), were added followed by detection with sheep anti-human Vn pAb (Abd Serotec, Kidlington, UK), rabbit anti-mouse Ln-111 pAb (Sigma-Aldrich), and anti-human Fn pAb (Dako, Glostrup, Denmark). In a separate experiment, we immobilized ECM proteins, incubated with recombinant P4 (rP4) and detected with horseradish peroxidase (HRP)–conjugated mouse anti-6 × His tag monoclonal antibody (mAb) (Abcam, Cambridge, UK). In a whole-cell enzyme-linked immunosorbent assay (ELISA), epithelial cells grown to confluency in 96-well plates were fixed with 4% formaldehyde. rP4 was loaded to the wells, washed, and detected with the anti-His mAb.
Bacterial Binding to ECM Protein-Coated Glass Slides
Bacterial adherence to immobilized ECM proteins was studied by adding bacterial suspensions to Vn-, Ln-, and Fn- (5 µg of each) coated glass slides (Menzel-Gläser, Thermo Scientific, Wilmington, Delaware) [6]. Slides were visualized by Gram-staining and analyzed in an Olympus IX73 microscope at 100× amplification.
Adherence Assay
NTHi adherence to mammalian cell lines was analyzed by flow cytometry as described [27]. For primary HBE cells, bacterial adherence was assayed by the counting of colony-forming units (CFUs) as described [28]. Epithelial cells grown to confluency in 24-well cell culture plates (non-polarized form) were incubated with fluorescein isothiocyanate (FITC)–labeled (for cell lines) or unlabeled NTHi (for HBE) (multiplicity of infection [MOI] = 100) at 37°C in medium without FCS for 1 hour. Unbound bacteria were removed by washing with phosphate-buffered saline (PBS); epithelial cells were detached and analyzed as described [27, 28].
Direct Protein Binding Assay
Purified ECM proteins were labeled with 0.05 M 125I (Amersham Biosciences, Buckinghamshire, UK) per mol protein using the chloramine-T method [29]. Iodine-labeled protein (300 kcpm) was added to a bacterial suspension (1 × 107 CFUs in 100 µL) in PBS-2.5% bovine serum albumin (BSA) and incubated for 1 hour at 37°C. Samples were washed, pelleted, and measured in a Tri-Carb B3110TR Liquid Scintillation Counter (Perkin Elmer, Waltham, Middlesex).
Serum Resistance Assay
Complement-mediated killing of NTHi and E. coli was assayed as described [6]. We used 1% and 5% normal human serum (NHS) for E. coli and NTHi, respectively. Vn-depleted serum (VDS) was prepared as described [25]. Bacterial suspensions (1.5 × 103 CFUs) in 150 µL of divalent-Veronal-buffered–BSA buffer [6] was added with NHS or VDS, and incubated at the indicated time points at 37°C. Percentage of bacterial killing was enumerated as described [6].
Mouse Infection Experiments
The heterozygote Junbo mice have a mutation of the Evi1 gene and display symptoms of chronic otitis media, including a thickened tympanic membrane and the presence of middle-ear fluid [30]. Eleven-week-old Junbo mice (n = 31) were inoculated intranasally with 106 CFUs of NTHi (SmR) strains. Animal were sampled terminally at day 7 postinoculation. Middle-ear fluids were collected and plated on brain heart infusion agar containing streptomycin. Bacterial colonies enumerated were used to calculate middle-ear infection rates. These studies were carried out under the authority of the appropriate UK Home Office Project License and were approved by the Medical Research Council Harwell ethical review committee.
Statistical Analyses
We used 2-tailed t test, and 1- or 2-way analysis of variance (ANOVA) tests as indicated. Differences were considered statistically significant at P ≤ .05. GraphPad Prism 5.0 (GraphPad Software, La Jolla, California) was used for statistical analyses.
Results
Identification of Haemophilus P4 as a Novel ECM-Binding Protein
NTHi has been shown to express multiple surface receptors, including PE, PF, and Hap to concurrently bind several ECM proteins [4–7]. When strains containing single or combinations of these genes mutated were tested, we observed that bacteria still bound some ECM proteins. This suggested that additional NTHi receptors may exist for certain ECM components. To determine which other NTHi protein(s) was involved in these interactions, we separated the NTHi 3655 outer membrane protein fraction on 2D–sodium dodecyl sulfate polyacrylamide gel electrophoresis, and probed with different ECM components. Interestingly, we found a protein spot (approximately 28 kDa) that was constantly reactive with Vn, Ln, and Fn (Supplementary Figure 3). Subsequent protein sequencing revealed that this putative ECM receptor corresponded to H. influenzae P4.
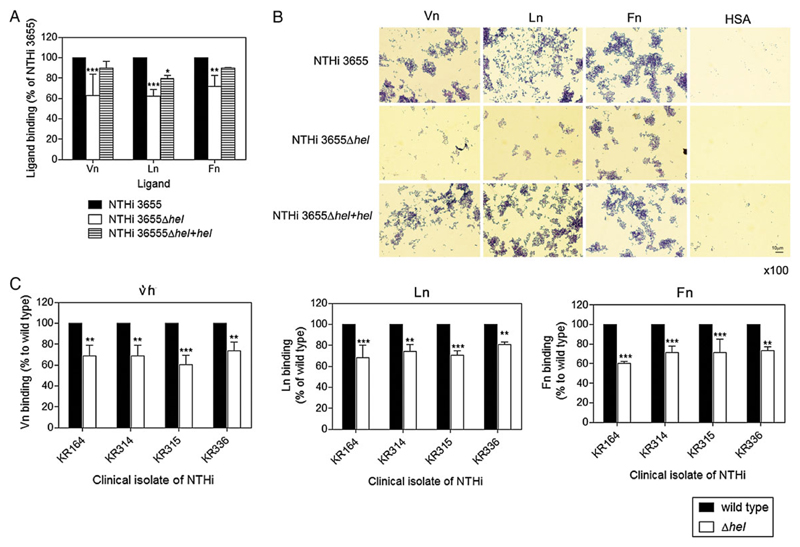
To determine whether expression of P4 may promote attachment of bacteria to purified ECM proteins, we compared binding of a P4-deleted mutant (NTHi 3655Δhel) with the NTHi wild-type (WT) strain. NTHi 3655Δhel bound significantly (P ≤ .01) less soluble [125I]–labeled ECM proteins as compared to the WT counterpart (Figure 1A). Importantly, the ECM-binding phenotype was restored in the transcomplemented NTHi 3655Δhel+hel. Because ECM proteins exist as tethered molecules at the basolateral surface of epithelial cells, bacterial adherence to immobilized ECM proteins was also evaluated. Assessment of bacterial adherence on Vn-, Ln-, and Fn-coated glass slides by the NTHi 3655 WT and transcomplemented mutant strains revealed numerous clustered bacteria associated with all three ECM proteins (Figure 1B). In contrast, the NTHi 3655Δhel mutant revealed only a few adhered bacteria on any of the glass slides. Importantly, a reduced deposition of ECM proteins was observed also with other clinical NTHi isolates when the hel gene was deleted (Figure 1C). Taken together, our results suggest that P4 interacts with Vn, Ln, and Fn to promote H. influenzae adherence.
Figure 1. Nontypeable NTHi devoid of P4 exhibits reduced binding capacities to ECM proteins.
A, Direct ligand binding assay of NTHi 3655 WT, hel-knockout mutant (NTHi 3655Δhel), and a hel-transcomplemented mutant (NTHi 3655 Δhel+hel), with soluble [125I]-labeled Vn, Ln, and Fn. B, Adherence of NTHi 3655 wild-type bacteria and P4 mutant to ECM-coated glass slides. HSA was included as a negative control. After a standard Gram staining, bacteria were visualized by light microscopy. Representative images of 2 independent experiments are shown. C, Binding of soluble ECM proteins to the clinical isolates NTHi KR164, KR314, KR315, and KR336, and corresponding hel-knockout mutants. Binding was assayed by a direct binding assay with [125I]-radiolabeled Vn (left panel), Ln (center panel), and Fn (right panel). In A and C, data are presented as percentage of bacterial binding to each ECM protein relatively to the NTHi 3655 WT strain that was set as 100%. Data represent mean values of 3 independent experiments, and error bars indicate standard deviations. The specificity of the P4-ECM protein interaction of NTHi 3655 was also confirmed by an inhibition assay with anti-P4 antibodies (Supplementary Figure 4). Differences between mutants and WTs in ECM protein binding were calculated by 2-way ANOVA. *P ≤ .05, **P ≤ .01 and ***P ≤ .001. Abbreviations: ANOVA, analysis of variance; ECM, extracellular matrix; Fn, fibronectin; HSA, human serum albumin; Ln, laminin; NTHi, nontypeable H. lipoprotein ; P4, Haemophilus lipoprotein e; Vn, vitronectin; WT, wild-type.
Interaction of P4 With ECM Proteins is Independent of the Core/α-Helix Domain
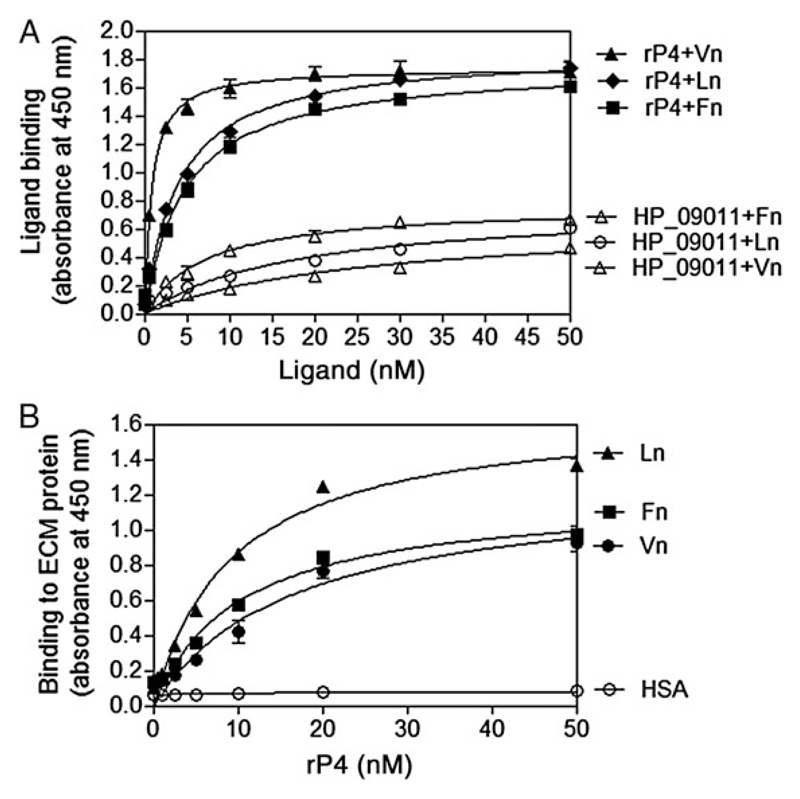
We continued to examine whether purified recombinant P4 (rP4) also interacted with ECM proteins. ELISA revealed that the immobilized full-length rP4 (rP4G22-K274) significantly bound soluble Vn, Ln, and Fn in a concentration-dependent manner (0.5–50 nM) (Figure 2A). We further quantitated the strength of the interactions between P4 and ECM proteins. A dose-dependent binding of rP4 (0.5–50 nM) to the wells coated with plasma Vn, Ln, and Fn was observed (Figure 2B). Based on a nonlinear regression analysis, the dissociation constant (Kd) of binding between P4 and Vn, Ln, and Fn was estimated to be 16.51 nM, 9.26 nM, and 10.19 nM, respectively. Our results suggested that binding of P4 to ECM proteins was a strong interaction and generally equivalent among the three ECM proteins studied.
Figure 2. Purified recombinant P4 binds soluble Vn, Ln, and Fn in a dose-dependent manner as measured by ELISA.
A, Binding of soluble ECM proteins to immobilized rP4. Full-length rP4 (rP4G22-K274) (50 nM) was coated on microtiter plates and incubated with increasing concentrations (0.5–50 nM) of Vn, Ln, or Fn. Bound Vn, Ln, and Fn was detected with specific Abs. A non-ECM-binding NTHi protein (HP_09011) (GI: 144986489) [6] was included as a negative control. B, Relative binding affinities of rP4G22-K274 to immobilized ECM proteins. Equal concentrations of Vn, Ln, and Fn were coated at 50 nM and incubated with increasing concentrations of rP4 (0.5–50 nM). Bound rP4 was detected with an anti-His mAb. rP4 did not bind to wells coated with HSA (negative control). The binding data were used to estimate the Kd value of P4 to Vn, Ln, and Fn. Data represent mean values of 3 independent experiments, and error bars indicate standard deviations. Abbreviations: Abs, antibodies; ECM, extracellular matrix; ELISA, enzyme-linked immunosorbent assay; Fn, fibronectin; HSA, human serum albumin; Ln, laminin; mAb, monoclonal antibody; NTHi, nontypeable H. influenzae; P4, Haemophilus lipoprotein e; rP4, recombinant P4; Vn, vitronectin.
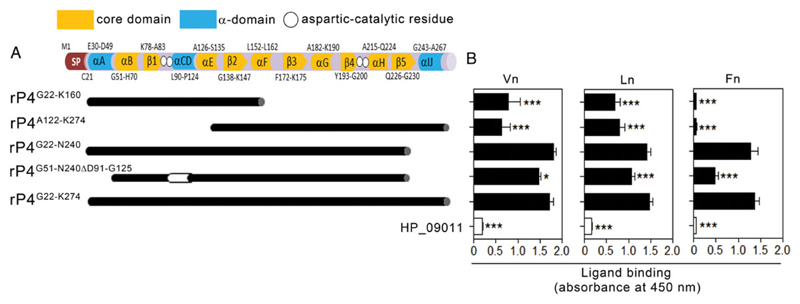
The crystal structure of P4 revealed that the protein consists of an α-helix domain and an α-helix/β-strand core domain (Figure 3A) [31]. The α/β-core domain is responsible for catalysis of phosphatase activity. Together with αB, αH, and β5 from the core domain, the α-helix domain is involved in the dimerization of rP4 that is crucial for dephosphorylation and hemin binding [14, 31, 32]. To pinpoint the relevance of the secondary structures of P4 in binding to ECM, we produced a series of rP4 in-frame deletion derivatives spanning different parts of the core and α-helix domains of the protein (Figure 3A). All ECM proteins bound efficiently to rP4G22-N240 but less to rP4G51-N240ΔD91-G125 that lacks the entire α-domain (Figure 3B). Further reduction in binding was observed with the N-terminal (rP4G22-K160) and C-terminal (rP4A122-K274) fragments that were devoid of the major parts of the core- and α-domain, respectively. Our results thus suggested that the interactions between P4 and the ECM proteins are independent of the core/α-helix domains.
Figure 3. Mapping of the ECM-binding sites on the P4 molecule.
A, Schematic representation of the identified secondary structures of P4. The α-domain (blue blocks) of P4 consists of 5 secondary structures (αA, αC, αD, αI, and αJ), with C-terminus 6-residues QAWDGK. The α/β core domain (yellow) is composed of αB, β1, αE, β2, αF, β3, αG, β4, αH, and β5. Location of “DDDD” aspartic-catalytic residues required for the P4 phosphomonoesterase activity (D64, D66, D181, and D185) are indicated with white circles. SP indicates the signal peptide. Truncated fragments spanning different parts of P4 are shown by black blocks and a deleted region in a mutated variant is indicated by a white block. B, Binding of ECM proteins to truncated P4 fragments as measured by ELISA. Equal molar concentrations (50 nM) of P4 variants were immobilized on microtiter plates and loaded with 10 nM Vn (left panel), Ln (center panel), and Fn (right panel). Bound ECM proteins were detected with specific antibodies. Negligible ECM binding was observed with the negative control HP_09011. In B, data represent mean values of 3 independent experiments, and error bars indicate standard deviations. Statistically significant differences between truncated fragments and full-length P4 were calculated by 2-way ANOVA. *P ≤ .05 and ***P ≤ .001. Abbreviations: ANOVA, analysis of variance; ECM, extracellular matrix; ELISA, enzyme-linked immunosorbent assay; Fn, fibronectin; Ln, laminin; P4, Haemophilus lipoprotein e; rP4, recombinant P4; Vn, vitronectin.
P4 Functions as an Additional Major Vitronectin- and Laminin-Binding Protein of NTHi, Whereas Hap Is the Major Fibronectin Receptor
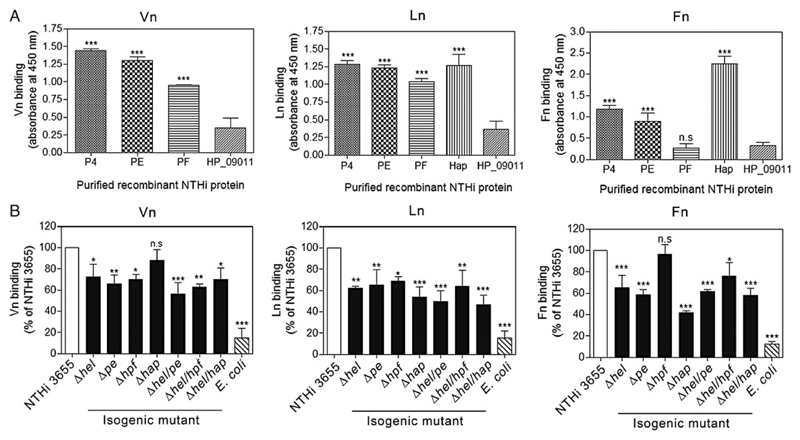
Because PE, PF, and Hap also promote H. influenzae–dependent adherence to ECM proteins, we aimed to evaluate the relative binding capacities of P4 in comparison with these receptors. As shown in Figure 4A, human Vn bound most efficiently to both immobilized rP4 and rPE, followed by rPF and minimally to rHapE523-L1036. Ln bound almost equally well to all immobilized proteins. Human Fn showed the highest interaction with Hap, followed by rP4 and rPE. In contrast, rPF and the negative control (HP_09011) did not bind Fn.
Figure 4. Relative importance of P4, PE, PF, and Hap in NTHi ECM binding.
A, Comparison of Vn (left panel), Ln (center panel), and Fn (right panel) binding to purified recombinant P4, PE, PF, and Hap as measured by ELISA. Recombinant PE, PF, and C-terminal Hap (E523-L1036) were produced as described [6, 13, 25]. Equal concentrations (50 nM) of NTHi recombinant proteins were coated on ELISA microtiter plates and loaded with 10 nM of Vn, Ln or Fn. Bound ECM proteins were detected with specific pAbs. The NTHi HP_09011 protein was included as a negative control. B, Direct ligand binding assay with NTHi 3655 strains and radiolabelled-ECM proteins. E. coli was included as a negative control. Binding of radiolabelled ECM proteins to single and double NTHi deletion mutants of pe, pf, hel, and hap were compared with the wild-type counterpart for Vn (left panel), Ln (center panel), and Fn (right panel) binding. Of note, we failed to generate triple or tetrad mutants of pe/pf/hap/hel due to antibiotic stress of NTHi 3655. Data represent mean values of 3 independent experiments and error bars indicate standard deviations. Statistically significant differences between ECM-binding to the negative control protein HP_09011 (A) or NTHi wild-type and mutants (B) in ECM protein binding were calculated by 2-way ANOVA. *P ≤ .05, **P ≤ .01 and ***P ≤ .001. Abbreviations: ANOVA, analysis of variance; ECM, extracellular matrix; ELISA, enzyme-linked immunosorbent assay; Fn, fibronectin; Hap, Haemophilus adhesion and penetration protein; Ln, laminin; n.s., not significant; NTHi, nontypeable H. influenzae; P4, Haemophilus lipoprotein e; PE, protein E; PF, protein F; Vn, vitronectin.
We further explored the relative contribution of each H. influenzae protein in binding to ECM using a direct binding assay. NTHi with single gene deletions of pe, hel, and hpf displayed significantly reduced binding (P ≤ .05); approximately 70% of the [125I]–Vn-binding capacity seen with NTHi 3655 (Figure 4B). When the bacteria were incubated with [125I]–Ln, all single-gene deletion strains showed significantly reduced binding (P ≤ .05). Deletion of Hap resulted in a major decrease in Fn binding (approximately 58%) (P ≤ .001) compared to NTHi WT, followed by a partially reduced binding (approximately 40%) with the in PE and P4 deletion mutant strains. In parallel to the results obtained by ELISA (Figure 4A), deletion of NTHi hap or hpf did not significantly affect Vn and Fn binding, respectively. For Vn and Ln, but not Fn, a slightly enhanced reduction in binding was observed in some double mutants compared to their Δhel counterpart, albeit this was not statistically significant. Taken together, these data indicate that in addition to PE and Hap, P4 serves as a major receptor for both Vn and Ln, whereas Hap remains the major Fn-binding protein in NTHi.
Expression of P4 Enables NTHi to Subvert Serum Killing Through the Recruitment of Vitronectin
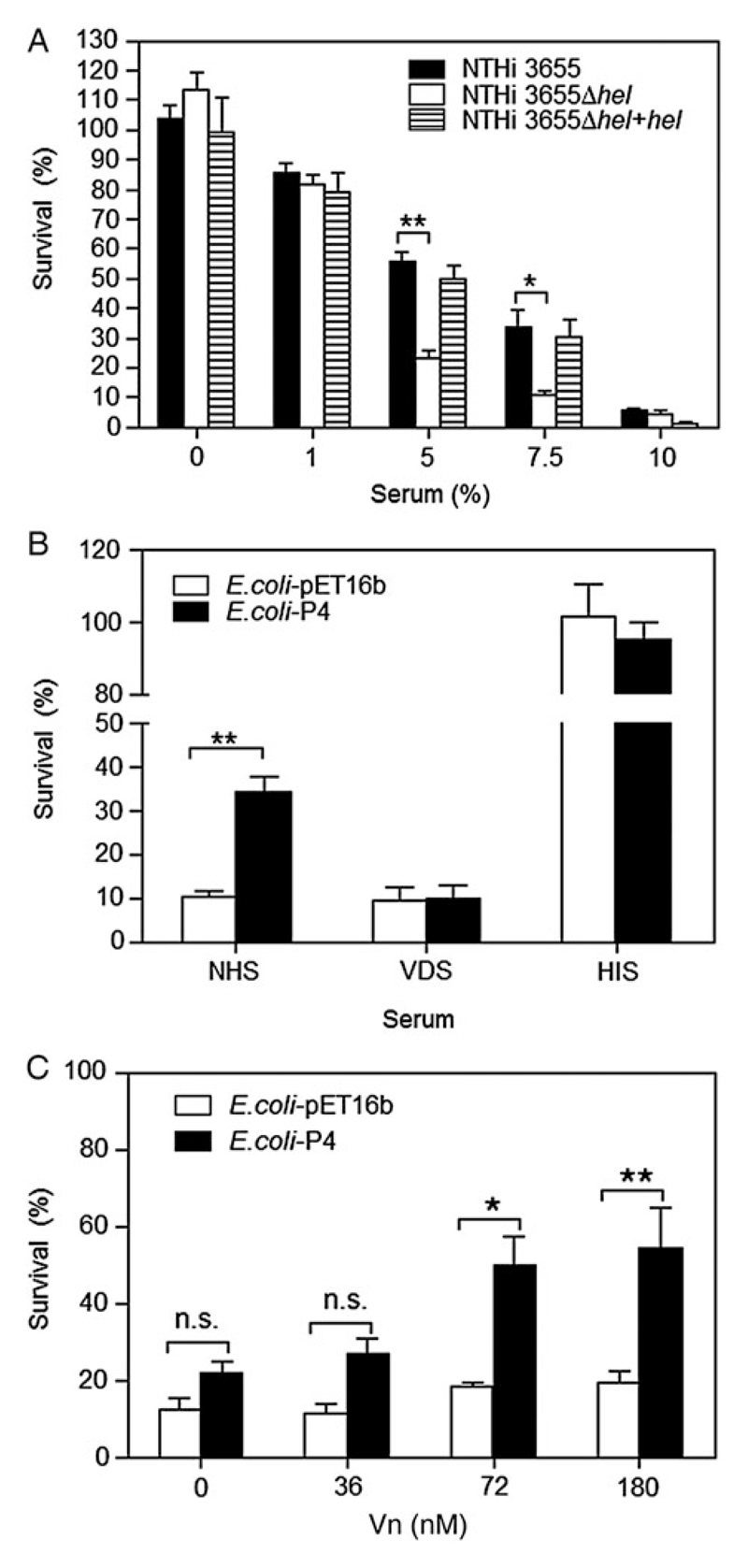
Vn functions as a complement regulator, preventing the host from self-damage, and inhibits the formation of the membrane attack complex at the terminal lytic step of the complement system [11]. Deposition of Vn at the bacterial surface thus confers resistance against complement-mediated killing, as exemplified by Vn-dependent PE- and PF-mediated serum resistance in NTHi [6, 25]. Here, we evaluated the potential role of Vn recruitment by P4 in NTHi serum resistance. Deletion of hel from NTHi 3655 significantly (P ≤ .05) rendered the P4-deficient knock-out mutant more serum sensitive when exposed to higher percentages of NHS compared to the WT (Figure 5A). However, this bactericidal effect of NHS was reversed in the NTHi 3655Δhel+hel–complemented strain.
Figure 5. Bacteria expressing P4 have enhanced serum resistance.
A, Survival of NTHi 3655 wild type, hel-deleted and transcomplemented mutants at various percentages (0%–10%) of NHS. B, Survival of E. coli with surface expression of P4 in 1% NHS and 1% VDS. A control experiment with 1% HIS was included. C, Serum resistance of E. coli strains in 1% VDS supplemented with various concentrations of Vn (36–180 nM). Addition of Vn (greater than 72 nM) significantly enhanced the serum resistance of P4-expressing E. coli. Vn at 72 nM corresponded to approximately 1% NHS (Vn concentration in NHS is approximately 400 µg/mL; 5.3 µM) [6, 35]. For A–C, samples (10 µL) at 0 minutes (T0) and indicated time points (Tt) were plated overnight at 37°C on chocolate agar for NTHi (A) and LB agar containing ampicillin for E. coli (B and C), and incubated at 37°C overnight. Percentage of bacterial killing was expressed as ((T0 CFUs−Tt CFUs)/T0 CFUs) × 100. Bacterial survival was determined at 15 minutes (A), and 10 minutes (B and C). Data represent mean values of 3 independent experiments, and error bars indicate standard deviations. Differences in serum resistance between wild-type and mutants were calculated by 2-way ANOVA. *P ≤ .05; **P ≤ .01. Abbreviations: ANOVA, analysis of variance; CFUs, colony-forming units; HIS, heat-inactivated serum; LB, Luria–Bertani; n.s., not significant; NHS, normal human serum; NTHi, nontypeable H. influenzae; P4, Haemophilus lipoprotein e; VDS, Vn-depleted serum; Vn, vitronectin.
To extend these results, we expressed P4 at the surface of the serum-sensitive and non-Vn-binding E. coli [6, 25]. When exposed to 1% NHS, P4-expressing E. coli survived significantly (P ≤ .01) better than control E. coli (Figure 5B). Interestingly, depletion of Vn from NHS (VDS) resulted in a similar susceptibility to serum killing for both P4-expressing and control E. coli (Figure 5B). Similar results were obtained with NTHi 3655 and its corresponding Δhel mutant (data not shown). However, supplementing VDS with Vn (≥72 nM) significantly rescued (P ≤ .05) the P4-expressing E. coli from serum-mediated killing (Figure 5C). In conclusion, P4-dependent Vn binding contributes to NTHi serum resistance.
NTHi P4 Mediates Bacterial Adhesion to Pulmonary Epithelial Cells Through ECM Proteins
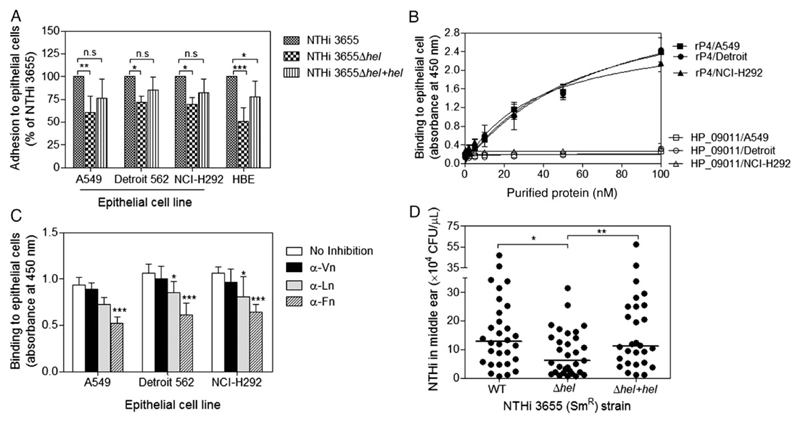
In the basal membrane (BM), Vn, Ln, and Fn are tethered with distinct integrin receptors anchored at the basolateral surface of epithelial cells [33–36]. Bacterial binding to these ECM proteins might thus promote adhesion to epithelial cells [9–12, 37, 38]. Accordingly, we further explored the involvement of P4 in H. influenzae adherence to cultured pulmonary epithelial cells. First, we found that bacterial adherence to cell lines A549, Detroit and NCI-H292, and primary HBE cells was significantly (P ≤ .05) reduced when P4 was deleted (NTHi 3655Δhel) (Figure 6A). Thereafter, the direct protein–cell interaction between rP4 and epithelial cells by a whole-cell ELISA was quantified. rP4 bound to all cell lines in a dose-dependent manner as compared to the negative control protein (HP_09011) (Figure 6B).
Figure 6. P4 promotes NTHi adhesion to pulmonary epithelial cells and infection in the middle ears of Junbo mice.
A, Adhesion of NTHi 3655 to different pulmonary cell lines and primary HBE cells. For cell lines A549, Detroit 562, and NCI-H292, bacterial adherence was measured by flow cytometry as mean fluorescence intensity of the FITC-labeled bacteria adhered to epithelial cells. Bacteria (1 × 109 CFUs) were labeled with FITC (10 µg/mL) for 45 minutes and washed to remove unbound FITC. Epithelial cells were incubated with FITC-labeled NTHi (MOI 100) at 37°C in medium without FCS for 1 hour. Unbound bacteria were removed by washing with PBS before epithelial cells were detached and subjected to analysis by flow cytometry. In this assay, epithelial cells were weakly autofluorescent, whereas all FITC-labeled bacteria strains had equal fluorescence intensity (Supplementary Figure 6). The flow cytometer was set to exclude debris and bacterial clumps. In a separate control experiment, the incubation of cells with unlabeled bacteria in the absence of labeled bacteria did not affect the fluorescence of the cells (data not shown). For primary HBE cells, cells were incubated with unlabeled bacteria (MOI 100) for 1 hour. HBE cells were washed, detached, and lysed, followed by plating of cell lysates on chocolate agar at different dilutions. Plates were incubated overnight at 37°C in a humid atmosphere containing 5% CO2. Colonies of bacteria bound to cells were enumerated. Data are presented in percentage of bacterial adherence relative to the NTHi 3655 wild-type (WT) strain that was set as 100%. Deletion of hel resulted in reduced NTHi adhesion to pulmonary cell lines: alveolar (A549), pharyngeal (Detroit 562), and bronchial (NCI-H292) epithelial cells, and primary HBE cells. B, Direct interaction of rP4 to fixed and immobilized A549, Detroit 562, and NCI-H292 as measured by cell ELISA. Purified rP4 at increasing concentrations (0.5–50 nM) was loaded to each cell line. rP4 bound each cell line in a dose-dependent manner, whereas the negative control protein HP_09011 did not. C, Inhibition of rP4 binding to cell lines by specific anti-ECM antibodies. Fixed cells were preincubated with anti-human Vn, a cocktail of anti-Ln pAb (1:1 mixture of anti-Ln-111 (Sigma-Aldrich) and anti-Ln-332 mAb (AbCam); Supplementary Figure 5) or finally, anti-Fn pAb (1:10 dilution) followed by incubation with 10 nM rP4. Binding of rP4 was significantly inhibited by anti-Ln cocktail pAb and anti-Fn pAb. In our nonpolarized cell models, P4 might have targeted cellular Ln and Fn expressed at the cell surface or cell–cell contacts. In B and C, bound rP4 was detected with an anti-His mAb. Data represent mean values of 3 independent experiments, and error bars indicate standard deviations. D, P4-bearing NTHi is more virulent than the P4-deficient mutant in a mouse middle-ear infection model. Streptomycin-resistant NTHi strains (NTHi 3655 (SmR), NTHi 3655 Δhel (SmR), and NTHi 3655 Δhel+hel (SmR)) were used to infect Junbo mice. Mice were experimentally infected with 106 CFUs of bacteria via the intranasal route. Shown are the middle-ear colonization rates (recovered CFUs/µL of middle-ear bulla fluids) for mice (n = 30, 31, and 28) infected with NTHi 3655 (WT), hel-deleted mutant (Δhel), or hel-transcomplemented mutant (Δhel+hel) at day 7 postinfection, respectively. Of note, these strains all grow remarkably well in the inflamed mouse middle ear during early infection (data not shown). Each filled circle represents the recovered CFUs/µL middle-ear fluid from one mouse. The horizontal bars indicate the media of each data set. Differences in adherence between WT and mutants (A), and between conditions with and without antibody blocking (B) were determined by 2-way ANOVA. Differences in middle-ear colonization rates between 2 groups among WT and mutants (D) were analyzed by a 2-tailed t test. *P ≤ .05; **P ≤ .01; ***P ≤ .001. Abbreviations: ANOVA, analysis of variance; CFUs, colony-forming units; ECM, extracellular matrix; ELISA, enzyme-linked immunosorbent assay; FCS, fetal calf serum; FITC, fluorescein isothiocyanate; Fn, fibronectin; HBE, human bronchial epithelial; HIS, heat-inactivated serum; Ln, laminin; mAB, mouse anti-6 × His tag monoclonal antibody; MOI, multiplicity of infection; n.s., not significant; NTHi, nontypeable H. influenzae; P4, Haemophilus lipoprotein e; pAb, polyclonal antibody; PBS, phosphate-buffered saline; rP4, recombinant P4; SmR, streptomycin-resistant; Vn, vitronectin.
We speculated that binding to ECM proteins might be one of the factors that contribute to the rP4-dependent cell interaction. Epithelial cell–derived Vn, Ln, and Fn was first confirmed with anti-ECM pAbs (Supplementary Figure 5). Thereafter, we blocked rP4 binding by anti-ECM pAbs in the whole-cell ELISA (Figure 6C). Inhibition of the rP4–cell interaction was most striking in the experiment with anti-Fn pAb (P ≤ .001) followed by anti-Ln pAb (P ≤ .05). In contrast, preincubation with the anti-Vn pAb caused only a slight reduction in rP4 binding.
Our data suggest that the receptors for P4 on the nonpolarized epithelial cells include the cellular Ln and Fn that are probably expressed at the cell surface or cell–cell contacts, promoting adherence of rP4 to epithelial cells.
Haemophilus P4 Has an Important Role in Middle Ear Infection
Because NTHi also causes middle-ear infection, we further addressed the importance of P4 in NTHi-dependent acute otitis media. To this end, we examined the ability of NTHi 3655 and corresponding hel mutants to survive in the middle ear of the heterozygote Junbo mice. These genetically modified mice are defective in nuclear factor (NF)–κB signaling, have inflamed middle ears, and thus are highly susceptible to acute otitis media caused by NTHi [39]. At day 7 postinfection, we observed a significantly reduced colonization of the hel-deleted NTHi mutant (median 6.04 × 104 CFUs/µL of middle-ear fluid) in the middle ear, compared to the P4-expressing WT (12.72 × 104 CFUs/µL) or transcomplemented hel mutant (11.15 × 104 CFUs/µL) (Figure 6D). Our data thus indicate the significance of P4 in experimental acute otitis media caused by NTHi.
Discussion
In this study, we presented several lines of evidence that the NTHi lipoprotein P4, which is crucial for NAD uptake, also serves as an ECM receptor for Vn, Ln, and Fn. First, NTHi expressing P4 bound to Vn, Ln, and Fn more efficiently when compared to an isogenic strain without the hel gene (Figure 1). Second, deletion of the highly conserved P4 resulted in a reduced ECM-binding phenotype of several clinical NTHi isolates. Third, the strong interaction of rP4 with Vn, Ln, and Fn (low affinity constants; Kd) verified that the identification of P4 as a multiple ECM protein receptor was not an artefact due to non-native conformations of proteins seen in the initial 2D-gel electrophoresis (Supplementary Figure 3).
The ECM-binding sites of PE, PF, and Hap have been defined at the central, N- and C-terminal regions, respectively [4–6, 13, 25]. These binding sites do not, however, have consensus sequences for any specific ECM protein [6] (sequence alignment not shown). In contrast to Fn, no specific Vn- or Ln-binding domains of bacterial proteins have been reported. Most bacterial Fn-binding proteins comprise consensus repeats (ED(T/S)(X) nGG(X)n(I/V)DF) in the fibronectin-binding domains [10, 40]. However, this motif is not present in P4. The interaction of P4 with Vn, Ln, and Fn is unlikely to be site-specific as neither depends on the α/β-core- or α-helix-domains (Figure 3). P4 requires residues located between Gly22 and Asn240 for optimal ligand binding. It would be possible that rP4 does not contain a single binding site but is conformational-dependent or alternatively may employ multiple binding regions. In parallel, detailed analysis of Staphylococcus SdrI, and a site-directed mutagenesis study of Haemophilus Hap, revealed that both proteins bind Fn independently of the motif typical for Fn-binding proteins but involve multiple binding regions [41, 42]. Among the NTHi ECM receptors, we showed that PE and P4 are the major Vn receptors in NTHi. Despite PE, PF, P4, and Hap bound Ln equally well, Hap preferentially bound Fn, followed by P4 and PE (Figure 4).
Several studies have reported the potential role of P4 in NTHi infection. P4 expression was enhanced in NTHi during interaction with NCI-H292 [43]. The lipoprotein was also required for long-term survival of NTHi in the murine lung [44]. In yet another study, P4 was needed for aggressive infection of NTHi in influenza A virus–coinfected mice [45]. Except for its physiological role in acquisition of NAD and heme, the exact role for P4 in NTHi pathogenesis is less well understood. Not all bacterial receptors can bind efficiently to both soluble and cellular ECM proteins [9]. Interestingly, P4 significantly binds soluble (plasma) and insoluble (cellular) Vn, Ln, and Fn (Figures 2 and 6), which have distinct functions as immunomodulators during inflammation and for cell adherence [10, 33–35, 46, 47]. Our work thus reveals that P4-dependent recruitment of Vn and Ln/Fn is associated with NTHi serum resistance and adherence to pulmonary epithelial cells, respectively (Figures 5 and 6). Importantly, serum resistance and bacterial adherence were restored after re-introduction of the hel gene in the P4-deletion mutant. Moreover, P4 expression confers Vn-binding to E. coli, and thus promoted serum survival in 1% NHS and after the addition of exogenous Vn to VDS. We suggest that P4-mediated NTHi adherence involves at least cellular Fn and Ln, but less likely Vn (Figure 6). Interestingly, this is in sharp contrast to Hsf expressed by H. influenzae type b (Hib) that targets cellular Vn for optimal adherence [28]. At the BM, cellular Ln and Fn bind epithelial integrin receptors via the C-terminal LG1–3 domains of Ln α-chain and Arg-Gly-Asp-Ser motif of Fn type III domain, respectively [33, 34]. It is thus plausible that during NTHi colonization, P4 might target the cellular ECM proteins to adhere to epithelial cells by binding to non-integrin binding sites of cellular Ln and Fn. Hence, the role of P4 in ECM binding, and perhaps nutrient acquisition, might partly explain the decreased NTHi persistence in the highly inflamed middle ears of Junbo mice when P4 was deleted (Figure 6D). The importance of P4 for Hib causing invasive disease has also been proven in an adult rat bacteremia model [18].
In conclusion, we have described a novel role for P4 as a multifunctional ECM receptor that is important for serum resistance and adherence of NTHi in addition to bacterial survival in an otitis media mouse model. Identification of alternative ECM receptors in NTHi sheds light on the multifaceted strategies of the pathogen to interact with the host and evade its immune response, and this consequently impacts on the propensity of the bacteria to cause disease.
Supplementary Material
Supplementary materials are available at http://jid.oxfordjournals.org. Consisting of data provided by the author to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the author, so questions or comments should be addressed to the author.
Acknowledgments
We thank the Clinical Microbiology Laboratory at Labmedicin Skåne (Malmö, Sweden) for providing clinical isolates.
Financial support. This work was supported by grants from the Alfred Österlund, the Anna and Edwin Berger, Greta and Johan Kock, the Swedish Medical Research Council (grant number K2015-57X-03163-43-4, www.vr.se), the Physiographical Society (Forssman’s Foundation), and Skåne County Council’s research and development foundation.
Footnotes
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Van Eldere J, Slack MP, Ladhani S, Cripps AW. Non-typeable Haemophilus influenzae, an under-recognised pathogen. Lancet Infect Dis. 2014;14:1281–92. doi: 10.1016/S1473-3099(14)70734-0. [DOI] [PubMed] [Google Scholar]
- 2.Virkola R, Lahteenmaki K, Eberhard T, et al. Interaction of Haemophilus influenzae with the mammalian extracellular matrix. J Infect Dis. 1996;173:1137–47. doi: 10.1093/infdis/173.5.1137. [DOI] [PubMed] [Google Scholar]
- 3.Bresser P, Virkola R, Jonsson-Vihanne M, Jansen HM, Korhonen TK, van Alphen L. Interaction of clinical isolates of nonencapsulated Haemophilus influenzae with mammalian extracellular matrix proteins. FEMS Immunol Med Microbiol. 2000;28:129–32. doi: 10.1111/j.1574-695X.2000.tb01466.x. [DOI] [PubMed] [Google Scholar]
- 4.Hallstrom T, Singh B, Resman F, Blom AM, Morgelin M, Riesbeck K. Haemophilus influenzae protein E binds to the extracellular matrix by concurrently interacting with laminin and vitronectin. J Infect Dis. 2011;204:1065–74. doi: 10.1093/infdis/jir459. [DOI] [PubMed] [Google Scholar]
- 5.Jalalvand F, Su YC, Morgelin M, et al. Haemophilus influenzae protein F mediates binding to laminin and human pulmonary epithelial cells. J Infect Dis. 2013;207:803–13. doi: 10.1093/infdis/jis754. [DOI] [PubMed] [Google Scholar]
- 6.Su YC, Jalalvand F, Morgelin M, Blom AM, Singh B, Riesbeck K. Haemophilus influenzae acquires vitronectin via the ubiquitous Protein F to subvert host innate immunity. Mol Microbiol. 2013;87:1245–66. doi: 10.1111/mmi.12164. [DOI] [PubMed] [Google Scholar]
- 7.Fink DL, Green BA, St Geme JW., III The Haemophilus influenzae Hap autotransporter binds to fibronectin, laminin, and collagen IV. Infect Immun. 2002;70:4902–7. doi: 10.1128/IAI.70.9.4902-4907.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eberhard T, Ullberg M. Interaction of vitronectin with Haemophilus influenzae. FEMS Immunol Med Microbiol. 2002;34:215–9. doi: 10.1111/j.1574-695X.2002.tb00627.x. [DOI] [PubMed] [Google Scholar]
- 9.Chagnot C, Listrat A, Astruc T, Desvaux M. Bacterial adhesion to animal tissues: protein determinants for recognition of extracellular matrix components. Cell Microbiol. 2012;14:1687–96. doi: 10.1111/cmi.12002. [DOI] [PubMed] [Google Scholar]
- 10.Henderson B, Nair S, Pallas J, Williams MA. Fibronectin: a multidomain host adhesin targeted by bacterial fibronectin-binding proteins. FEMS Microbiol Rev. 2011;35:147–200. doi: 10.1111/j.1574-6976.2010.00243.x. [DOI] [PubMed] [Google Scholar]
- 11.Singh B, Su YC, Riesbeck K. Vitronectin in bacterial pathogenesis: a host protein used in complement escape and cellular invasion. Mol Microbiol. 2010;78:545–60. doi: 10.1111/j.1365-2958.2010.07373.x. [DOI] [PubMed] [Google Scholar]
- 12.Singh B, Fleury C, Jalalvand F, Riesbeck K. Human pathogens utilize host extracellular matrix proteins laminin and collagen for adhesion and invasion of the host. FEMS Microbiol Rev. 2012;36:1122–80. doi: 10.1111/j.1574-6976.2012.00340.x. [DOI] [PubMed] [Google Scholar]
- 13.Fink DL, Buscher AZ, Green B, Fernsten P, St Geme JW., III The Haemophilus influenzae Hap autotransporter mediates microcolony formation and adherence to epithelial cells and extracellular matrix via binding regions in the C-terminal end of the passenger domain. Cell Microbiol. 2003;5:175–86. doi: 10.1046/j.1462-5822.2003.00266.x. [DOI] [PubMed] [Google Scholar]
- 14.Reilly TJ, Chance DL, Smith AL. Outer membrane lipoprotein e (P4) of Haemophilus influenzae is a novel phosphomonoesterase. J Bacteriol. 1999;181:6797–805. doi: 10.1128/jb.181.21.6797-6805.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hotomi M, Ikeda Y, Suzumoto M, et al. A recombinant P4 protein of Haemophilus influenzae induces specific immune responses biologically active against nasopharyngeal colonization in mice after intranasal immunization. Vaccine. 2005;23:1294–300. doi: 10.1016/j.vaccine.2004.08.042. [DOI] [PubMed] [Google Scholar]
- 16.Reidl J, Mekalanos JJ. Lipoprotein e(P4) is essential for hemin uptake by Haemophilus influenzae. J Exp Med. 1996;183:621–9. doi: 10.1084/jem.183.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reidl J, Schlor S, Kraiss A, Schmidt-Brauns J, Kemmer G, Soleva E. NADP and NAD utilization in Haemophilus influenzae. Mol Microbiol. 2000;35:1573–81. doi: 10.1046/j.1365-2958.2000.01829.x. [DOI] [PubMed] [Google Scholar]
- 18.Morton DJ, Smith A, VanWagoner TM, Seale TW, Whitby PW, Stull TL. Lipoprotein e (P4) of Haemophilus influenzae: role in heme utilization and pathogenesis. Microbes Infect. 2007;9:932–9. doi: 10.1016/j.micinf.2007.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kemmer G, Reilly TJ, Schmidt-Brauns J, et al. NadN and e (P4) are essential for utilization of NAD and nicotinamide mononucleotide but not nicotinamide riboside in Haemophilus influenzae. J Bacteriol. 2001;183:3974–81. doi: 10.1128/JB.183.13.3974-3981.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Musser JM, Barenkamp SJ, Granoff DM, Selander RK. Genetic relationships of serologically nontypable and serotype b strains of Haemophilus influenzae. Infect Immun. 1986;52:183–91. doi: 10.1128/iai.52.1.183-191.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ronander E, Brant M, Janson H, Sheldon J, Forsgren A, Riesbeck K. Identification of a novel Haemophilus influenzae protein important for adhesion to epithelial cells. Microbes Infect. 2008;10:87–96. doi: 10.1016/j.micinf.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 22.Roier S, Leitner DR, Iwashkiw J, et al. Intranasal immunization with nontypeable Haemophilus influenzae outer membrane vesicles induces cross-protective immunity in mice. PLOS One. 2012;7:e42664. doi: 10.1371/journal.pone.0042664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poje G, Redfield RJ. Transformation of Haemophilus influenzae. Methods Mol Med. 2003;71:57–70. doi: 10.1385/1-59259-321-6:57. [DOI] [PubMed] [Google Scholar]
- 24.Fleury C, Su YC, Hallstrom T, Sandblad L, Zipfel PF, Riesbeck K. Identification of a Haemophilus influenzae factor H-Binding lipoprotein involved in serum resistance. J Immunol. 2014;192:5913–23. doi: 10.4049/jimmunol.1303449. [DOI] [PubMed] [Google Scholar]
- 25.Hallstrom T, Blom AM, Zipfel PF, Riesbeck K. Nontypeable Haemophilus influenzae protein E binds vitronectin and is important for serum resistance. J Immunol. 2009;183:2593–601. doi: 10.4049/jimmunol.0803226. [DOI] [PubMed] [Google Scholar]
- 26.Akiyama SK. Purification of vitronectin. Curr Protoc Cell Biol. 2001 doi: 10.1002/0471143030.cb1006s02. [DOI] [PubMed] [Google Scholar]
- 27.Hytonen J, Haataja S, Finne J. Use of flow cytometry for the adhesion analysis of Streptococcus pyogenes mutant strains to epithelial cells: investigation of the possible role of surface pullulanase and cysteine protease, and the transcriptional regulator Rgg. BMC Microbiol. 2006;6:18. doi: 10.1186/1471-2180-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh B, Su YC, Al-Jubair T, et al. A fine-tuned interaction between trimeric autotransporter haemophilus surface fibrils and vitronectin leads to serum resistance and adherence to respiratory epithelial cells. Infect Immun. 2014;82:2378–89. doi: 10.1128/IAI.01636-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nordstrom T, Blom AM, Forsgren A, Riesbeck K. The emerging pathogen Moraxella catarrhalis interacts with complement inhibitor C4b binding protein through ubiquitous surface proteins A1 and A2. J Immunol. 2004;173:4598–606. doi: 10.4049/jimmunol.173.7.4598. [DOI] [PubMed] [Google Scholar]
- 30.Cheeseman MT, Tyrer HE, Williams D, et al. HIF-VEGF pathways are critical for chronic otitis media in Junbo and Jeff mouse mutants. PLOS Genet. 2011;7:e1002336. doi: 10.1371/journal.pgen.1002336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Felts RL, Ou Z, Reilly TJ, Tanner JJ. Structure of recombinant Haemophilus influenzae e (P4) acid phosphatase reveals a new member of the haloacid dehalogenase superfamily. Biochemistry. 2007;46:11110–9. doi: 10.1021/bi701016m. [DOI] [PubMed] [Google Scholar]
- 32.Reilly TJ, Green BA, Zlotnick GW, Smith AL. Contribution of the DDDD motif of H. influenzae e (P4) to phosphomonoesterase activity and heme transport. FEBS Lett. 2001;494:19–23. doi: 10.1016/s0014-5793(01)02294-3. [DOI] [PubMed] [Google Scholar]
- 33.Domogatskaya A, Rodin S, Tryggvason K. Functional diversity of laminins. Annu Rev Cell Dev Biol. 2012;28:523–53. doi: 10.1146/annurev-cellbio-101011-155750. [DOI] [PubMed] [Google Scholar]
- 34.Singh P, Carraher C, Schwarzbauer JE. Assembly of fibronectin extracellular matrix. Annu Rev Cell Dev Biol. 2010;26:397–419. doi: 10.1146/annurev-cellbio-100109-104020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leavesley DI, Kashyap AS, Croll T, et al. Vitronectin—master controller or micromanager? IUBMB Life. 2013;65:807–18. doi: 10.1002/iub.1203. [DOI] [PubMed] [Google Scholar]
- 36.Madsen CD, Sidenius N. The interaction between urokinase receptor and vitronectin in cell adhesion and signalling. Eur J Cell Biol. 2008;87:617–29. doi: 10.1016/j.ejcb.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 37.Tsang TM, Annis DS, Kronshage M, et al. Ail protein binds ninth type III fibronectin repeat (9FNIII) within central 120-kDa region of fibronectin to facilitate cell binding by Yersinia pestis. J Biol Chem. 2012;287:16759–67. doi: 10.1074/jbc.M112.358978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Agarwal V, Kuchipudi A, Fulde M, Riesbeck K, Bergmann S, Blom AM. Streptococcus pneumoniae endopeptidase O (PepO) is a multifunctional plasminogen- and fibronectin-binding protein, facilitating evasion of innate immunity and invasion of host cells. J Biol Chem. 2013;288:6849–63. doi: 10.1074/jbc.M112.405530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu X, Woo CH, Steere RR, et al. EVI1 acts as an inducible negative-feedback regulator of NF-kappaB by inhibiting p65 acetylation. J Immunol. 2012;188:6371–80. doi: 10.4049/jimmunol.1103527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwarz-Linek U, Werner JM, Pickford AR, et al. Pathogenic bacteria attach to human fibronectin through a tandem beta-zipper. Nature. 2003;423:177–81. doi: 10.1038/nature01589. [DOI] [PubMed] [Google Scholar]
- 41.Spahich NA, Kenjale R, McCann J, et al. Structural determinants of the interaction between the Haemophilus influenzae Hap autotransporter and fibronectin. Microbiology. 2014;160:1182–90. doi: 10.1099/mic.0.077784-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakinc T, Kleine B, Michalski N, Kaase M, Gatermann SG. SdrI of Staphylococcus saprophyticus is a multifunctional protein: localization of the fibronectin-binding site. FEMS Microbiol Lett. 2009;301:28–34. doi: 10.1111/j.1574-6968.2009.01798.x. [DOI] [PubMed] [Google Scholar]
- 43.van Ulsen P, van Schilfgaarde M, Dankert J, Jansen H, van Alphen L. Genes of non-typeable Haemophilus influenzae expressed during interaction with human epithelial cell lines. Mol Microbiol. 2002;45:485–500. doi: 10.1046/j.1365-2958.2002.03025.x. [DOI] [PubMed] [Google Scholar]
- 44.Gawronski JD, Wong SM, Giannoukos G, Ward DV, Akerley BJ. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc Natl Acad Sci USA. 2009;106:16422–7. doi: 10.1073/pnas.0906627106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wong SM, Bernui M, Shen H, Akerley BJ. Genome-wide fitness profiling reveals adaptations required by Haemophilus in coinfection with influenza A virus in the murine lung. Proc Natl Acad Sci USA. 2013;110:15413–8. doi: 10.1073/pnas.1311217110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.To WS, Midwood KS. Plasma and cellular fibronectin: distinct and independent functions during tissue repair. Fibrog Tissue Repair. 2011;4:21. doi: 10.1186/1755-1536-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Senkovich OA, Yin J, Ekshyyan V, et al. Helicobacter pylori AlpA and AlpB bind host laminin and influence gastric inflammation in gerbils. Infect Immun. 2011;79:3106–16. doi: 10.1128/IAI.01275-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.