Abstract
Objective: To determine the extent to which a combination of existing machine-readable health terminologies cover the concepts and terms needed for a comprehensive controlled vocabulary for health information systems by carrying out a distributed national experiment using the Internet and the UMLS Knowledge Sources, lexical programs, and server.
Methods: Using a specially designed Web-based interface to the UMLS Knowledge Source Server, participants searched the more than 30 vocabularies in the 1996 UMLS Metathesaurus and three planned additions to determine if concepts for which they desired controlled terminology were present or absent. For each term submitted, the interface presented a candidate exact match or a set of potential approximate matches from which the participant selected the most closely related concept. The interface captured a profile of the terms submitted by the participant and for each term searched, information about the concept (if any) selected by the participant. The term information was loaded into a database at NLM for review and analysis and was also available to be downloaded by the participant. A team of subject experts reviewed records to identify matches missed by participants and to correct any obvious errors in relationships. The editors of SNOMED International and the Read Codes were given a random sample of reviewed terms for which exact meaning matches were not found to identify exact matches that were missed or any valid combinations of concepts that were synonymous to input terms. The 1997 UMLS Metathesaurus was used in the semantic type and vocabulary source analysis because it included most of the three planned additions.
Results: Sixty-three participants submitted a total of 41,127 terms, which represented 32,679 normalized strings. More than 80% of the terms submitted were wanted for parts of the patient record related to the patient's condition. Following review, 58% of all submitted terms had exact meaning matches in the controlled vocabularies in the test, 41% had related concepts, and 1% were not found. Of the 28% of the terms which were narrower in meaning than a concept in the controlled vocabularies, 86% shared lexical items with the broader concept, but had additional modification. The percentage of exact meaning matches varied by specialty from 45% to 71%. Twenty-nine different vocabularies contained meanings for some of the 23,837 terms (a maximum of 12,707 discrete concepts) with exact meaning matches. Based on preliminary data and analysis, individual vocabularies contained <1% to 63% of the terms and <1% to 54% of the concepts. Only SNOMED International and the Read Codes had more than 60% of the terms and more than 50% of the concepts.
Conclusions: The combination of existing controlled vocabularies included in the test represents the meanings of the majority of the terminology needed to record patient conditions, providing substantially more exact matches than any individual vocabulary in the set. From a technical and organizational perspective, the test was successful and should serve as a useful model, both for distributed input to the enhancement of controlled vocabularies and for other kinds of collaborative informatics research.
Controlling the vocabulary used in electronic health records is one of the prerequisites for data that are unambiguous, sharable, and aggregatable. Whether interfaces guide health professionals to enter data in controlled terms or programs attempt to convert unconstrained clinical text into controlled terms or some combination of these techniques is employed, the use of controlled vocabulary in health care and public health systems is likely to increase the quality, effectiveness, and efficiency of health care and to facilitate clinical research, public health surveillance, and health services research.
Accordingly, “The medical informatics community—vendors and users—have been seeking a common, comprehensive clinical vocabulary for the past decade.”1 In the inaugural issue of JAMIA, the Board of Directors of the American Medical Informatics Association called for the use of a combination of existing codes and vocabularies as a practical approach to standardizing the content of patient data, e.g., one system for drugs, one for devices, one for observations.2 Comparative studies3,4 of the content coverage, structure, and other features of existing single systems (including SNOMED International5 and the Read Clinical Classification,6 two large and granular clinical terminologies) have provided evidence that much of the vocabulary needed for health data is already available and that a combination of existing systems may provide the best foundation for building a comprehensive controlled clinical vocabulary.
NLM and AHCPR began planning a large scale vocabulary test late in 1994,7 in conjunction with the award of eight cooperative agreements for research and development related to electronic medical records. The cooperative agreement partners* were seen as an appropriate core set of participants for a distributed national experiment to determine the extent to which a combination of existing health-related terminologies covers the vocabulary needed in information systems supporting health care, public health, and health services research.8
Background
In addition to a reasonable base of controlled terms, the development of a standard health data vocabulary will require an open and sustainable process for enhancing and updating the vocabulary as well as appropriate mappings from the vocabulary to administrative and statistical classifications. Efficient and low cost electronic distribution methods and mandates and incentives to use the vocabulary will also be needed. The need to establish and maintain this supporting infrastructure has been recognized by organizations interested in promoting robust computer-based patient record systems, including the Computer-Based Patient Record Institute, HL7,1 and AMIA. Progress toward the necessary infrastructure has been hampered by resource constraints, intellectual property issues, the absence of one or more de facto standards for granular clinical vocabulary, and the lack of a clear mandate for federal action on health data standards.
The passage of the Health Insurance Portability and Accountability Act of 19969 (HIPAA, also known as Kassebaum—Kennedy, Kennedy—Kassebaum, or K2) has addressed one of these barriers by assigning to the Secretary of Health and Human Services (HHS) a major role in the promulgation of health data standards for administrative transactions, including claims attachments that may have extensive clinical information. The Act also requires HHS and its external advisory committee, the National Committee on Vital and Health Statistics (NCVHS), to advise Congress on any legislative or regulatory actions needed to promote full electronic medical records. The HHS Data Council has been given the assignment to implement the provisions of the law, acting through its Health Data Standards Committee and Interagency Health Privacy Working Group.
The Health Data Standards Committee, which includes members from the National Library of Medicine (NLM) and the Agency for Health Care Policy and Research (AHCPR), has appointed six implementation teams to lead work on the different types of standards covered by the law. A Coding and Classification Implementation Team has responsibility for codes, classifications, and vocabulary for diseases, injuries, other health problems, and their manifestations; for procedures or other actions taken to prevent, diagnose, or treat individual health problems; and for any related drugs, equipment, and supplies. The implementation team's charge includes both a shortterm agenda to designate the codes and classifications that will be used for administrative transactions in the year 2000 and longer-term requirements to recommend the more granular vocabularies needed for full patient records, to ensure effective mapping between these and the administrative classifications designated as standards, and to ensure appropriate means for maintaining and distributing the classifications and vocabularies.
In this context, additional data about the extent to which existing terminologies provide the controlled vocabulary needed for granular health data may assist HHS and the NCVHS in formulating plans and resource estimates for making progress toward controlled vocabulary useful in clinical and public health information systems.
Although initial planning for the test preceded the establishment of the HHS Data Council in 1995 and the passage of the HIPAA in 1996, NLM and AHCPR expected the results of the test to provide useful input to the development of Federal policy on health data standardization. The results should also help clarify the nature and extent of the gaps in the combination of existing vocabularies and will help the National Library of Medicine in setting priorities for expansion of the Unified Medical Language System (UMLS) Metathesaurus.
The NLM/AHCPR Large Scale Vocabulary test had two principal hypotheses: (1) That existing machinereadable health terminologies cover the majority of concepts and terms needed in a comprehensive controlled vocabulary for health information systems; and (2) that together, the Internet and the UMLS Knowledge Sources, lexical programs, and the UMLS Knowledge Source Server provide an appropriate foundation for a distributed national experiment to assess the combined coverage of many health-related vocabularies.
The test differed from previous studies (e.g., Chute et al.3 and Campbell et al.4) of the clinical coverage of controlled vocabularies in several ways: (1) Its purpose was to assess the aggregate concept coverage of more than 30 terminology systems, rather than to evaluate or compare individual vocabularies; (2) the terms searched in the test represented concepts for which test participants desired controlled vocabulary or links to the UMLS Metathesaurus, rather than terms extracted from clinical free text; (3) participants determined the presence or absence of the exact meanings of their terms and, if possible, identified a closely related concept for those without exact meaning matches, but they did not. They were not required to construct matches from combinations of concepts present in the test vocabularies or assign values or scores to less than synonymous matches; (4) all the terminologies in the test were searched via the same interface and the same search algorithms, thus avoiding the potentially confounding effects of different browsers, different term indexing methods, etc.4,10; and (5) the test was a distributed experiment involving widely dispersed participants using a common Web-based system.
Methods
Design and Implementation of the LSVT Application
The design of the Large Scale Vocabulary Test (LSVT) application was influenced by a number of factors. Since the primary hypothesis of the experiment was that a combination of existing terminologies will cover the majority of the concepts needed for a broad range of health information systems, and since many of these terminologies are already represented in the UMLS, the application was designed to allow participants to search local terms and concepts in the Metathesaurus. Local terminologies would be mapped to the UMLS Knowledge Sources, with the Internet-based UMLS Knowledge Source Server forming the foundation of the system.11,12 The extensive diffusion of World Wide Web technology, and particularly, the easy access to Web browsers influenced our decision to design a special Web application for use in conducting the large scale, distributed experiment that we envisioned. Our goal was to encourage participation by any interested persons with a real health-related task for which controlled vocabulary was desired, who were able to participate during the time frame of the experiment, and who had a good Internet connection. This latter was necessary, since users would be actively interacting with our system during relatively lengthy sessions, making multiple decisions about the data they had submitted.
The LSVT application was designed to query all constituent terminologies in the UMLS Metathesaurus simultaneously. The Metathesaurus contains all or part of some 30 vocabularies, including broad coverage terminologies such as SNOMED, ICD-9-CM, and MeSH, as well as terminologies in specialized domains, such as PDQ for oncology and DSM IV for psychiatry. Nursing vocabularies, e.g., the Nursing Interventions Classification and the Omaha system for community health nursing, an epidemiologic terminology developed at McMaster University, and several terminologies for adverse reactions are also included. The Metathesaurus constituent vocabularies formed the basis of the terminology searched by the LSVT application. In addition, and primarily as a result of the recommendations made at the December 1994 meeting on the vocabulary needs of computer-based patient record systems,7 we incorporated three planned additions to the Metathesaurus in the terminologies searched by the LSVT application. The planned additions were the rest of SNOMED International not already in the Methathesaurus, the Logical Observations Identifiers, Names, and Codes (LOINC) terminology, and the Read Clinical Classification system.†
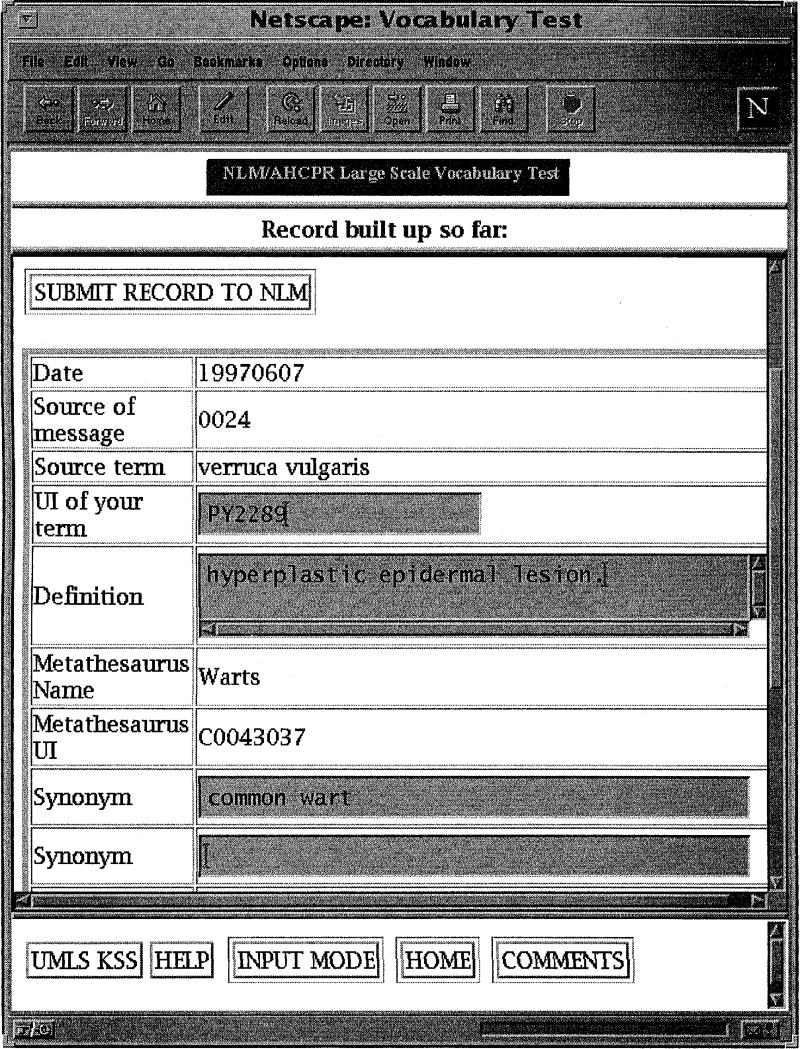
The LSVT application is a concept-based query system. This means that, as users submit their terms to the system, the system searches for the terms and maps them to concepts as these are represented in the Metathesaurus and its planned additions. Because it is well known that there is extensive variation in the way in which terminology is expressed, the LSVT search routines invoke the UMLS lexical programs.13 These programs were enhanced with an additional table to handle the spelling variation between American and British English, accounting for such variation as “esophagus,” “oesophagus,” and “hemophilia,” “haemophilia.” Figure 1 illustrates a sample interaction with the LSVT interface.
Figure 1.
The term “verruca vulgaris” has mapped to the concept “warts,” and the user is asked to decide whether this means the same thing as the submitted term.
The user is asked whether the concept “warts” is equivalent in meaning to the submitted term, “verruca vulgaris.” Basic information about the concept mapped to is presented on the right-hand side of the screen. The semantic types, definitions, synonyms, and source vocabulary hierarchies in which the concepts appear are all presented. Note that these are scrolling windows, indicating that there is more information available. For example, in this case, the concept appears in several vocabularies, including SNOMED as shown, and also MeSH, WHOART, and CRISP. Once the assessment of the correctness of the match is made, the next screen is presented (Fig. 2).
Figure 2.
The user is asked to verify the information captured by the LSVT application and may add information to the editable fields.
The system automatically captures the date and source of the message (the user's unique identifier), the user's source term, in this case “verruca vulgaris,” and the Metathesaurus default concept name and unique identifier. Several type-in windows are also presented, allowing the addition of certain information, such as a local unique identifier (“PY2289,” in this case), an additional definition if desired, and additional synonyms. Once the user is satisfied with the information presented in the completed record, clicking the button “submit record to NLM” sends the data back to the LSVT Web server, where it is stored in the system database.
If the system is not able to map the user's term directly into an existing concept, the approximate matching routines are invoked. These routines use the lexical programs, together with a lexical distance algorithm14 that computes a rank-ordered list of the ten most closely related concepts in the Metathesaurus and the ten most closely related terms in the planned additions (Fig. 3).
Figure 3.
A rank-ordered list is presented to the user who chooses the most closely related concept to the input term “achilles reflex” and assigns a relationship to it.
Here the user has previously submitted a file containing a list of terms through the batch processing input mode. The system searches for all the terms in the file and returns up to three output files: one for the exact matches found, one for the approximate matches found, and a third file containing those terms that matched nothing in either the Metathesaurus or the planned additions. The user is asked to make a decision about the single concept in the rank-ordered list of items that is most closely related to the input term. Once a choice is made, the user is asked to choose one of four possible relationships between the input term and the concept: synonymy, narrower in meaning, broader in meaning, or associated with. In this case, the user decides that the top-ranked item in the list, “ankle jerk,” is a synonym of “achilles reflex.”
Conducting the Test
The test was conducted over a 5-month period, from August 1996 through January 1997. Participants included individuals from the sites that received grants from NLM and AHCPR to conduct research and development related to electronic medical records, a contract arrangement awarded specifically to obtain data for this test, several specialty groups coordinated by the Duke University Center for Outcomes Research, and from a number of self-selected organizations, companies, and universities. When registering for the test, participants provided their name, title, institution, and address, and in some cases a short narrative description of the terminology they planned to submit. During the test, each participant completed a term profile for the vocabulary submitted. The term profile information included the general data task for which the vocabulary was needed (e.g., record or display information about individuals), the general purpose of the task (e.g., direct patient care, clinical research, or public health surveillance), and, if applicable, the care setting or facility (e.g., ambulatory care, inpatient care, or clinical laboratory), the specific type of care (e.g., internal medicine, dentistry, or pediatrics), and the specific segment of the patient record for which controlled vocabulary was sought (e.g., chief complaint, patient history, or progress notes).
Participants registered throughout the entire test period. Ninety-five individuals registered for the test and 63 of these participated fully in the official test. The majority of the participants (73%) hold an MD, RN, DDS, or Pharmacy degree. The remainder are medical informaticians, medical librarians, or medical students. Participants contributed terminology from 21 states, the District of Columbia, and Ontario, Canada. Upon registering, each participant was asked to complete a short pre-test before beginning the official test. The LSVT team reviewed the pre-test data submitted, notified the participants of any problems or misunderstandings (these were quite rare), and told them that they were free to begin the official test, but only after reading the official test instructions that were available on the first screen. The official test instructions included a discussion of the term profile information as well as instructions for file preparation and saving completed records for possible incorporation in a local database. A full description of each of the potentially 16 data elements collected by the system and down-loadable by the participant was given. The 16 elements included the date of submission, the tester's unique identifier (assigned at the time of registration), the input term, the unique identifier of the input term, if any, the matched UMLS concept, the unique identifier of the UMLS concept, the type of match made by the LSVT system as it searched for the user's query term, and the nature of the relationship between the user's term and the matched concept in the case of approximate matching. Tester decisions about the matches made, and user supplied information such as a definition, synonyms, and comments, were also included in each record. All test data were collected using the LSVT application over the Internet, with the exception of the terms submitted by a single tester who was affected by the much-publicized December 1996—January 1997 America Online (AOL) access problems. Because the tester could not obtain reasonable access to the Internet during that time, she recorded her decisions on paper, and data entry was done by the LSVT team.
Data Review
After the data collection phase was complete in mid-January 1997, the content review phase of the project began. The purpose of the review phase was to search for additional matches—i.e., matches that are in the constituent vocabularies but were not found by the LSVT algorithms—and to correct any gross errors. Errors included misrepresentation of the direction of a relationship; e.g., if the local term was said to be broader in meaning than the concept found, and if, in fact, it is narrower in meaning, then content review revealed that the participant had simply reversed the direction of the relationship. Some misspellings were also caught in the content review phase, and the correctly spelled term was resubmitted to the LSVT, often resulting in a match. Five outside experts, holding either an MD, PhD, or RN degree, participated in the content review. The reviewers used their expert knowledge of medicine, their knowledge of the UMLS vocabularies, the UMLS Knowledge Source Server, and other reference sources to review the submitted records. All review was done over the Internet through another Web interface designed for this purpose. The information presented to the reviewer included the test terms with the participant decisions, the term information as seen by the participant, and the UMLS search tools. Several meetings of the five reviewers and the LSVT team were held at NLM, and weekly conference calls and e-mail discussions further facilitated the review process.
To assess the inter-rater reliability of the five reviewers, we conducted two tests in which the same records were placed on each reviewer's worklist. The first set of 15 records was placed on the reviewers' worklists early in the review process and became the basis for a conference call in which several additional guidelines for the review process were established. The second set of 40 records was placed on each content reviewer's worklist in the latter half of the review process. These 40 randomly selected records were used to do a formal assessment of the degree of inter-rater reliability among the reviewers. The inter-rater reliability was measured using an extension of Cohen's Kappa statistic (K) to the case of more than two raters.15 We computed the K values based on two methods: (1) categorizing the terms as not-matched, related, and matched; and (2) categorizing the terms as in (1) but with the related terms further subdivided into the broader than, narrower than, and associated with categories. The results for Kappa given the above assumptions are:
-
- for
1) K=0.83, s.e.(K)=0.07, z-statistic=11.82 (p value <0.0001),
-
- for
2) K=0.81, s.e.(K)=0.05, z-statistic=15.95 (p-value<0.0001)
The Kappa values for both methods are comparable and quite high. These Kappa values indicate a high degree of inter-rater agreement.16
A second level of review was done by the editors of two of the major clinical vocabularies, SNOMED and Read. Each was provided a list of 591 randomly selected terms for which exact matches were not found. The sample size was chosen by consulting statistical tables using p=0.5 (since we didn't know the true proportion of terms in the set which could be mapped to SNOMED or Read codes, we used the worst-case scenario of the most random situation), a cushion size =0.4, and a 95% confidence level. The groups were asked to identify if a synonym were available in their current version for any of the terms on the list, or if a combination of codes in their vocabulary would create a synonym. The results of this review were further studied by the LSVT team in consultation with two physicians. A small number of the single concepts that the Read and SNOMED reviewers had matched to text terms were not, in fact, synonyms according to the strict criteria used in the test. The analysis of the review included these corrections.
One of the authors (BLH), also in consultation with two physicians, reviewed a 21% (991 term) random sample of the terms assigned either an “associated with” relationship to a concept or “nothing found.” The terms in the sample were assigned semantic types (5% were too ambiguous to determine the correct semantic type), and the terms were further examined to determine other characteristics: e.g., presence of acronyms, abbreviations, or misspellings.
Results
Terms Submitted
The final test data included 41,127 terms. The average number of terms submitted by each tester was 653; the median was 504; and the range was 1 to 5,300. These figures do not include terms submitted in the pre-test conducted by each participant. Content review involved review of over 20,000 records. The records examined by the reviewers were the following: (1) all terms identified as synonyms by the tester that were not identified as exact matches by the interface; (2) all terms considered by the tester as related to a concept in the test vocabularies: (3) all terms for which the tester found no appropriate matching concept in the list of concepts presented by the interface; and (4) a small sample of the exact matches. The reviewers found additional synonyms that the testers had not identified and also judged some terms that had been classified as synonyms as actually not being equivalent in meaning. The median percentage of an individual tester's decisions changed was 16% (range 8-80%), excluding testers who submitted fewer than 100 terms.
Matching Results
Following content review, 58% of testers' terms were found as exact meanings, 41% were related in meaning, and another 1% were not found at all. Table 1 shows the decisions made by the testers, the final values as adjusted by reviewers, and the amount of change resulting from the review process in each category. Table 2 shows the variation in the final percentages of exact meanings, related concepts, and no related concepts for the sets of terminology submitted by individual test participants. Of the nine sets with 36% or fewer exact meaning matches, six were from a single expert diagnostic system. The two sets with more than 85% exact meaning matches came from large hospitals with substantial ambulatory care patient populations.
Table 1.
Summmary of LSVT Results
| Terms Matching As | Tester Data (%) | Reviewed Data (%) | Changed by Review (%) |
|---|---|---|---|
| Exact meaning | 22,674 (55) | 23,837 (58) | +1,163 (3) |
| Related concept | |||
| Broader than | 1,079 (3) | 1,162 (3) | +83 (<1) |
| Narrower than | 10,112 (25) | 11,387 (28) | +1,275 (3) |
| Associated with | 2,247 (5) | 4,150 (10) | +1,903 (5) |
| No related concept | 5,015 (12) | 591 (1) | -4,424 (11) |
| Total number of terms submitted = 41,127. | |||
Table 2.
Variation in Matches Found in Terminology Submitted by Individual Test Participants (Data after Review—Excludes Sets of Fewer than 100 Terms)
| Terms | Median (%) | Range (%) |
|---|---|---|
| Exact meaning | 60 | 12-89 |
| Related concept | 37 | 11-81 |
| No related concept | 1 | 0-11 |
The 41,127 terms submitted by the testers represent 32,679 unique normalized strings. The normalization process described by McCray et al.13 ignores certain types of lexical variation between terms. For example, “adjustment disorder with anxiety and depression” was treated as the same normalized string as “adjustment disorder with depression and anxiety,” since they differ only in word order. Similarly, the submitted terms “Dehydration, with nausea & vomiting” and “Nausea & Vomiting, Dehydration” were treated as the same normalized string, differing in case, punctuation, word order, and the stopword “with.” “Corneal scar” and “corneal scarring” were normalized to the same string, as were all three submitted terms “WOUNDS,” “Wound,” and “wound,” which differ only in case and inflection.
The number of normalized strings that were found as exact meanings was 16,722. The number of normalized strings that were found as related concepts totaled 15,983, and in 579 cases no related concepts were found. The total of these three categories is slightly higher than the total number of unique normalized strings given above, since in some cases testers made different decisions about the concepts they found. For example, the term “atypical chest discomfort” was submitted twice. One tester considered it to be broader in meaning than the concept “atypical chest pain,” while another considered it to be a synonym of that concept.
Once normalized, tester terms were analyzed to see how many times a particular term appeared in the entire set of submitted terms. Of the 32,679 strings submitted, 28,049 appeared only once in the test set. In the remaining set of 4,630, almost 3,000 appeared twice, some 900 appeared 3 times, and 11 terms were submitted more than 10 times. Among the terms that were submitted most frequently were “diabetes mellitus” (14 instances), “hypertension” (13 instances), and “urinary tract infection” (12 instances). The greatest majority of the terms submitted more than once were either disorders or findings.
In those cases where tester submitted terms were narrower in meaning than the concepts they mapped to, 86% shared lexical items with the broader concept.
The term and the concept mapped to differed either by just a premodifier in the narrower term, just a postmodifier in the narrower term, or the combination of both a premodifier and a postmodifier. The 39 most frequent cases (appearing 10 or more times) where the narrower and broader terms differed solely by a premodifier included primarily simple adjectives, with the 5 most frequent being “mild” (134 instances), “left” (119 instances), “chronic” (118 instances), “right” (110 instances), and “bilateral” (105 instances). Other premodifiers included participal adjectives, such as “decreased,” “delayed,” and “improved.” Only 2 items on this list of 39 were not adjectives: “skin” (11 instances), and “grade” (10 instances). Overall, 75% of the single word premodifiers that appeared more than once were adjectives, and the rest were noun premodifiers. Multi-word premodifiers included multiple adjectives and nouns as premodifiers: e.g., “left hand” from “left hand surgery,” and “drug induced” from “drug induced gingival hyperplasia,” as well as phrases such as “edge to edge” from “edge to edge occlusion,” and “mild to moderate” from, for example, “mild to moderate aortic stenosis.” Other common premodifiers included such phrases as “history of,” “episodes of,” and “s/p” (status post). Postmodifiers included prepositional phrases, such as “in the colon,” “of the eyelid,” and “of recent onset.” Many narrower terms differed from the broader terms mapped to be post-modificational structures that were other types of phrases: e.g., “aggravated by exercise,” as in “lower extremity pain aggravated by exercise” and “sudden onset,” as in “low back pain sudden onset.” A wide range of phenomena can be observed in those cases in which the narrower and broader terms differed by both pre- and post-modifiers. For example, “caries” was chosen as broader in meaning than the tester's term, “salivary dysfunction caries secondary to medication.” Here “caries” is premodified by “salivary dysfunction” and postmodified by “secondary to medication.” Other examples involved pre- and post-modifiers expressed as abbreviations: e.g., “Ac Chest Pain R/o Mi” as narrower in meaning than “chest pain.” The example “chronic migraine headaches,” submitted by a tester whose chose “migraine” as the broader concept, illustrates cases in which the terms with and without the postmodifier, in this case “headaches,” actually mean the same thing.
In 14% of the narrower terms there was no lexical overlap with the broader concept mapped to. For example, the tester's term “allergic to iodine” and the broader term “hypersensitivity” share no lexical items, yet the first is narrower in meaning than the second. Similarly “arterial bleeding” is narrower in meaning than “hemorrhage,” but they do not share any lexical items. In some cases, the lexical dissimilarity was occasioned by the presence of an acronym or abbreviation—e.g., “ALL, new onset”—where the broader term was the fully expanded form of the acronym ALL—i.e., “acute lymphocytic leukemia.”
A total of 17,024 of the 23,837 exact meanings found in the test were also found directly as exact matches by the LSVT application (based on a normalized lexical match to a default preferred term, synonym, or abbreviation). Testers were able to identify 5,650 additional exact meanings from the ranked lists of approximate matches, and reviewers identified another 1,163 exact meaning matches as part of the content review process.
The second level review conducted by representatives of Read and SNOMED resulted in the identification of 94 exact meanings (16%) in the 591 term sample of terms for which an exact meaning had not been found by test participants or reviewers. The sample size is large enough, so that it is reasonable to project these results onto the full data set. The total number of nonexact matches is 17,290, and if 16% of these (2,766) are actually additional exact matches, then this could result in as high as a total of 65% (26,603) exact meanings found. An additional 100 synonyms were found in the sample by combining two or more concepts in either Read or SNOMED. Adding these to the exact meanings found and projecting this total onto the full data set would result in 79% exact meanings found. Since this method was used for only two of the vocabularies in the Metathesaurus, and since the versions of the two vocabularies were updated versions of these terminologies, these results are simply suggestive of the results of further, more detailed analysis methods.
The review of the 991-term sample of “associated with” and “not found” terms identified synonyms for 3% of the terms. Closely related broader concepts were found for an additional 6%, and closely related narrower concepts for 1%. When projected to the universe of associated with and not found terms, these increases in the number of synonyms, narrower than, and broader than terms would have a minor effect (<1% change) in the overall numbers of submitted terms that were synonyms or broader or narrower concepts in the controlled vocabularies. Analysis of the terms revealed that 11% were adjectives or adjective phrases: e.g., “ciliary,” “critical,” “platelike.” In other respects the terms were not different in kind from terms identified as narrower than concepts in the test vocabularies. Some of the terms contained abbreviations, acronyms, or mispellings—for example, “Concussion/MVA,” “A.M. HTN,” and “Reemer.” Some combined concepts, such as symptoms and their causes—e.g., “Abd pain/esrd”—and drugs and devices used to administer them were also found. A small number of terms included the dates of specific laboratory tests. More included a test name and its result. The majority of the terms were nouns or noun phrases of varying lengths: e.g., “posters,” “viral resistance,” “four chamber view,” “difficulty working with arms in a raised position.” Only one term appeared more than once in the sample, and very few terms were synonyms of each other. Acronyms for several of the common qualifiers described by Chute16 (e.g., “HX,” “S/P,” and “R/O”) appeared multiple times both in the set of terms assigned a narrower than relationship and those considered only related or not found.
Testers' Categorization of Terms Submitted
Appendix 2 shows the number and percentage of terms assigned to each element of the term profile, and the percentages of terms assigned that profile element for which exact meanings were found, related concepts were found, and no related concepts were found. Testers were allowed to select multiple elements in each section of the profile, and most did. The percentages in each section therefore add up to more than 100%. The selection of multiple categories was expected, given the intentional overlap between some categories, the multiple purposes for which a single set of terms can be used, and the extra effort involved in splitting input terms into sets used for discrete purposes. Due to the large numbers of terms assigned to individual elements of the profile, differences in percentages of exact meanings found have statistical significance, although some have little practical significance.
Data Tasks
The largest percentage (56%) of terms was identified as needed for recording or displaying individual patient data, and the smallest (12%) for retrieving information from knowledge bases. The percentage of exact meanings found ranged from a low of 51% (for building multi-purpose vocabulary databases or tools) to a high of 61% (for extracting or summarizing data about groups of patients).
General Purpose
Most terms were searched to find controlled vocabulary for individual health care and related decision support and research. Only 3,239 or 8% of the terms were searched for public health purposes. The percentage of exact meanings found was lowest for public health (50%) and highest for clinical research (62%).
Care Setting or Facility
More than half of the terms submitted were identified as needed for ambulatory care facilities, and an essentially equal number were labelled as needed for inpatient care facilities. About one fifth were needed for long-term care facilities and about one-tenth each for home care and free-standing clinical laboratories. The smallest percentage (7%) was associated with free-standing pharmacies. Free-standing pharmacies also had the smallest exact match percentage (41%), and ambulatory care facilities had the largest (60%).
Type of Care or Speciality
The percentage of terms submitted for different types of care or specialties ranged from a low of <1% (Anesthesiology, Pharmacy) to a high of 42% (Internal Medicine). Other categories assigned to more than 15% of the terms submitted were Surgery, Diagnostic Imaging, Emergency Medicine, Intensive Care/Critical Care, Family Practice, and Nursing. For types of care and specialities with more than 1,000 terms submitted, the percentages of exact meanings found ranged from a low of 45% for Veterinary Medicine to a high of 71% for Ophthalmology. Other categories with more than 1,000 terms and exact meaning percentages higher than 60% were Emergency Medicine, Family Practice, Internal Medicine, Intensive Care/Critical Care, Neurology, Nursing, Pediatrics, and Psychiatry/Clinical Psychology.
Segment of the Patient Record
Testers were primarily seeking terminology for the parts of the patient record that describe the patient's current or past condition. More than 80% of the terms were tagged with one or more of the segments of the patient record dealing principally with diseases, problems, and various kinds of findings. In contrast, 37% of the terms were tagged with segments of the patient record dealing with procedures and other interventions.
Concepts Selected
Of the concepts and terms presented to the testers as planned additions to the UMLS Metathesaurus, the remainder of SNOMED International, all of LOINC, and about one-third of the Read Codes have been integrated into the 1997 edition. Final counts of the number of unique concepts selected by test participants will not be possible until the rest of the Read system has been integrated into the Metathesaurus, but many of the terms selected by testers from the planned additions have now been linked as identical in meaning to concepts already in the Metathesaurus at the time of the test. For example, test participants identified 27 terms as narrower than “Hypersensitivity” in the Metathesaurus and an additional 22 terms as narrower than “Allergy” in the planned additions. Use of the table that maps the temporary identifiers assigned to the planned additions to the Metathesaurus concept identifiers (distributed with the 1997 UMLS Knowledge Sources) makes it clear that testers actually selected the same concept 49 times under 2 different names. The preliminary number of unique concepts selected during the test has therefore been generated using this table, and the subsequent analyses are based on this set.
Testers found matching or related concepts for 40,536 of the 41,127 terms submitted. These represent 14,311 unique concepts in the current Metathesaurus and an additional 2,868 terms in the planned additions, for a total unique number of 17,179. After all the planned additions are incorporated in the Metathesaurus, it is likely that the total number of unique concepts mapped to will be somewhat lower than 17,179, since some percentage of the terms in the planned additions will be synonyms of existing Metathesaurus concepts.
The concepts “diabetes mellitus, non-insulin-dependent” and “hypertension” were the most frequent exact matches (37 occurrences each). Other frequent exact matches included “atrial fibrillation,” “dyspnea,” “obesity,” “congestive heart failure,” and “osteoarthritis.” The actual terms submitted by the user may, of course, have differed from each other and from the default-preferred name of the concept. For example, when the exact meaning was found for “osteoarthritis,” tester submitted terms included, in addition to “osteoarthritis,” “degenerative joint disease,” “degenerative arthritis,” “DJD,” and even the misspelling “dejenerative joint disease.” The most frequently occurring concepts that were broader in meaning than the tester's term were “mass,” “ulcer,” “lesion,” “pain,” and “surgery.” The most frequently occurring concepts that were related in some other way to the tester's term included body parts (e.g., “eye,” “prostate,” and “colon”) and disorders and findings (e.g., “coronary disease,” and “pain”).
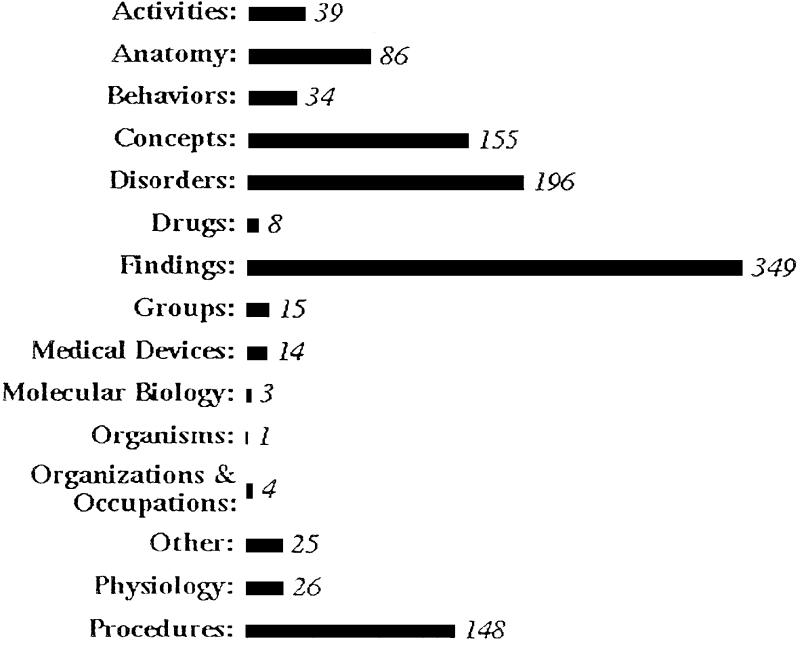
Each concept in the Metathesaurus is assigned to one or more semantic types. Grouping these semantic types into 15 major categories—including, for example, activities, anatomical concepts, disorders, drugs and findings—shows the categories of meaning that the submitted terminology was mapped to. Figure 4 shows the number of unique concepts that exactly matched the meaning of testers' terms in each of the 15 semantic type categories.
Figure 4.
Number of unique concepts that exactly matched the meaning of testers' terms in each of 15 semantic type categories.
The largest category, by far, is disorders. Next are findings, procedures, and anatomy. This may be contrasted with the semantic type distribution for the sample of 991 terms for which no exact or closely related meanings were found (Fig. 5). The largest number of the terms in this sample that were either simply “associated with” a concept or not found at all were findings.
Figure 5.
Semantic type distribution for sample of 991 terms for which no exact or closely related meanings were found.
Vocabulary Sources of Exact Meanings Found
The terms with exact meaning matches were mapped to single concepts in one or more of 29 different vocabularies.‡ The 23,837 exact meaning matches involve a maximum of 12,707 discrete concepts. Many concepts were present in multiple vocabularies. For example, “Hypertension” (one of the most frequently submitted concepts for which an exact meaning was found) appears in 12 of the test vocabularies. Other frequently submitted concepts appear in only one vocabulary.
Individual vocabularies contained single concepts for the exact meaning of from <1% to at least 63% of the 23,837 terms and from <1% to at least 54% of the 12,707 unique concepts. In the data currently available, only SNOMED International and the Read Codes have single concepts for more than 60% of the terms and more than 50% of the unique concepts. The percentages for all individual vocabularies are preliminary figures pending the full integration of the Read Codes into the Metathesaurus.§
Some vocabularies had matching meanings for much higher percentages of terms than of concepts. The ratios of terms to concepts matched in individual vocabularies ranged from 1:1 to 4.7:1. Vocabularies focused on procedures and equipment had relatively low ratios, reflecting the smaller number of terms submitted for these categories of concepts. The highest ratios occurred in vocabularies focused on common problems, conditions, and findings, which reflected the large numbers of such terms submitted by test participants. For example, in the current data, the COSTAR vocabulary present in the Metathesaurus contained the meanings of 43% of the terms for which exact meanings were found, but only 18% of the unique concepts. The ratios of terms to concepts for the larger, more comprehensive vocabularies (e.g., SNOMED International, Read Codes, ICD-9-CM, MeSH) were 2-3:1.
Discussion
Overall, test participants were able to detect whether the exact meanings of their terms were present in the vocabularies in the UMLS Metathesaurus and its planned additions with a high degree of accuracy. The credit for this must be shared by the testers, the content of the vocabularies included in the test, the additional synonymy and definitional information provided by the UMLS Metathesaurus, the use of UMLS lexical methods both to generate normalized indexes for all the test vocabularies and to produce normalized forms of the terms submitted by testers, and the approximate matching programs used in the LSVT application. The testers' ability to determine the presence or absence of specific concepts bodes well for a distributed Internet-based approach to the identification of synonyms and new concepts for addition to clinical vocabularies, provided that similarly robust tools are used.
The participation of people who desired controlled vocabulary for real tasks was a key feature of the test. The nature of the terms submitted was therefore determined to a large extent by the priorities of the participants. As is clear from the number of terms submitted, their distribution across care sites and specialties, and their breakdown by segment of the patient record and semantic type, the majority of participants were seeking controlled terminology for clinical diagnoses, problems, and findings. Some were working on problem list vocabularies for patient record systems, and others were attempting to link diseases and findings in expert diagnostic systems to the UMLS Metathesaurus. Given the very large number of test terms in these categories, the results are highly likely to present a valid picture of the coverage of disorders and findings in the existing controlled vocabularies included in the test. Although the numbers of terms for procedures, drugs, and medical devices are not small compared with numbers in previous vocabulary studies, in total they represent only 13% of the terms submitted. For this reason, our results may not provide a representative view of the controlled terminology desired to describe clinical interventions and their attendant drugs and devices. A number of potential explanations for the low percentage of terms in these categories come to mind, including the centrality of the description of the patient's condition to a clinical information system, the availability of several functional commercial systems for dealing with drug codes and related information, and the use in many settings of the American Medical Association's Current Procedural Terminology (CPT) to code physician procedures.
The large majority of test participants were clearly focused on obtaining terminology for clinical information systems. Only 8% of submitted terms were specifically identified as needed for population-based public health. Both anecdotal evidence and recent survey data from the National Association of County and City Health Officials support the hypothesis that the requirement for a high-speed Internet connection and a Web-capable workstation was a deterrent to participation by public health professionals.
The combination of controlled vocabularies included in the test contained single concepts for the exact meanings of 58% of the terms submitted. The percentage of exact matches found was considerably higher for terms needed for some specialties or types of care. A relatively high percentage of terms submitted (28%) was narrower in meaning than concepts found in the test vocabularies. After normalization, 86% of the narrower terms contained lexical items in common with the broader concept to which they were linked, plus pre-modification, post-modification, or both. Many of the frequently occurring modifiers are also present in some controlled vocabularies included in the test. Some of the narrower terms included quantitative measures (e.g., “3 cm”), which were stripped from test data in previous vocabulary studies.3 This quantitative information represents one of several categories of qualification that may be more appropriately represented as an attribute in a specific patient record, rather than included in a controlled vocabulary. The templates in the Read system reflect this approach. Other qualifiers that appeared in terms submitted in the test might not be needed in patient record systems that stored and displayed data in discretely labeled sections.
The data indicate that the combination of controlled vocabularies contained single concepts for the exact meanings of many more terms than were present in any individual vocabulary in the test set. This result is compatible with data from the study conducted by Campbell et al.,4 which found a higher percentage of exact matches in the UMLS Metathesaurus than in the individual vocabularies examined. The two largest clinical vocabularies in this test (SNOMED International and the Read Codes) contained the highest percentages (>60%) of exact meaning matches, but many exact meaning matches appeared only in smaller, more focused vocabularies that contain common clinical terminology. As previously explained, the percentages of exact meanings found in individual vocabularies are preliminary figures, which are likely to be revised upward once the Read codes are fully integrated into the Metathesaurus. This process could affect the percentage of exact matches for any vocabulary in the test set. (To illustrate: If one tester selected a Read term as an exact match and that term is later determined to be a synonym of a Metathesaurus concept found in 5 other vocabularies, the numbers of exact meaning matches for each of those 5 vocabularies will be increased by 1. If a Read term is found to be synonym of a Methathesaurus concept selected by one tester as an exact match, then the total of exact matches for the Read system will increase by 1.) As in other vocabulary tests, the percentage of exact meaning matches for some vocabularies may be lower because the test participants did not have access to the most current version of those sources.
Several of the largest test vocabularies (e.g., SNOMED International, the Read codes, MeSH) are combinatorial systems in which many additional meanings can be represented by combinations of concepts, by adding modifiers of various sorts to basic concepts, or by both of these methods. The differences in the current SNOMED and Read compositional approaches are discussed by Campbell et al.4 Given their clinical focus and the data from past vocabulary tests, it is virtually certain that synonymous or closely related combinations of SNOMED codes and of Read codes can be constructed for many of the single concepts that were found in other test set vocabularies, but not in these two large systems. Data from the second-level review of test data by the SNOMED and Read editors corroborate this view.
Based on a preliminary sample analysis, the 11% of the terms that had no closely related concept in the controlled vocabulary do not cluster in a small number of specialties or types of care or substantially differ in form from the narrower terms. As might be expected, they include a higher percentage of findings than was found in more closely related or synonymous concepts, but there were substantial percentages of findings in those other categories as well.
Conclusions
Due to the size of the test and the nature of the terms submitted, the data provide an excellent picture of the terminology desired to record patient conditions in a range of individual health care settings and in diagnostic decision-support systems. The combination of existing controlled vocabularies included in the test represents the exact meanings of the majority of this terminology as single concepts, providing substantially more exact matches than any individual vocabulary in the set. Primarily due to the presence of SNOMED International and the Read Codes, the set of existing vocabularies also includes the necessary constituent parts to form combinations of concepts that are synonymous with some of the narrower or otherwise related terms submitted during the test, although the exact percentage has yet to be determined. The test results indicate that most of the concepts and qualifiers needed to record data about patient conditions are already included in one or more of the existing controlled vocabularies. This has significant implications for the strategy for establishing, maintaining, and enhancing a comprehensive national health vocabulary.
From a technical and organizational perspective, the test was successful and should serve as a useful model, both for distributed input to the enhancement of controlled vocabularies and for other kinds of collaborative informatics research.
The data collected in the test represent a rich resource for exploring many questions related to clinical language, controlled vocabulary development, the design of efficient clinical data entry systems, and other important informatics concerns. We have reported the results of an initial analysis that addresses some of the key questions that motivated the test. Additional studies that make use of the data will be forthcoming from NLM and other test participants. To the extent approved by the test participants, NLM intends to make the test data generally available as a research data set.
Acknowledgments
Many people contributed to the work reported in this paper. We wish to thank all of the test participants, without whom the experiment could not have been conducted. We thank the content reviewers for the many long and arduous hours they spent in reviewing the data: Carol Bean, Suzanne Henry, Flora Hsieh, Patricia Michael, and Keyvan Rafei, who also developed the reviewer interface. We are grateful to Roger Cote (for SNOMED) and Philip Brown (for Read) for their rapid and thorough second review of a sample of terms. Thanks are due to Carolyn Lichtenstein, who provided statistics expertise to the project. Anantha Bangalore implemented the LSVT application and provided extensive reports for the data analysis phase of the project. Guy Divita, Amir Razi, and Zoe Stavri also contributed to the design and implementation of the LSVT application. Lexical Technology, Inc., provided the planned additions data in standard Metathesaurus format for use by the Knowledge Source Server. William Hole and Stuart Nelson assisted in the review of data samples and provided helpful comments on the manuscript. The careful review and comments by two anonymous JAMIA reviewers resulted in many improvements to the manuscript.
Appendix I
Source Vocabularies Included in the 1997 UMLS Metathesaurus
ACR92 Index for radiological diagnoses: including diagnostic ultrasound. Rev. 3rd ed. Reston (VA): American College of Radiology; 1986.
AIR93 AI/RHEUM. Bethesda, MD: National Library of Medicine; 1993.
BRMP97 Descritores em Ciencias de Saude. [Portuguese translation of MeSH] Latin American and Carribean Center on Health Sciences Information. BIREME/PAHO/WHO, Sao Paulo, Brazil; 1997.
BRMS97 Descriptores en Ciencias de la Salud. [Spanish translation of MeSH}] Latin American and Carribean Center on Health Sciences Information. BIREME/PAHO/WHO, Sao Paulo, Brazil; 1997.
COS89 COSTAR (ComputerStored Ambulatory Records) of Massachusetts General Hospital, 1989. (List of terms that occur frequently at 3 COSTAR sites, supplied by Massachusetts General Hospital)
COS92 COSTAR (ComputerStored Ambulatory Records) of Massachusetts General Hospital, 1992. (List of terms that occur frequently at 3 COSTAR sites, supplied by Massachusetts General Hospital)
COS93 COSTAR (ComputerStored Ambulatory Records) of Massachusetts General Hospital, 1993. (List of terms that occur frequently at 3 COSTAR sites, supplied by Massachusetts General Hospital)
COS95 COSTAR (ComputerStored Ambulatory Records) of Massachusetts General Hospital, 1995. (List of terms that occur frequently at 3 COSTAR sites, supplied by Massachusetts General Hospital)
CPM93 Columbia Presbyterian Medical Center Medical Entities Dictionary. New York, NY: Columbia Presbyterian Medical Center; 1993.
CPT96 Physicians' current procedural terminology: CPT. 4th ed. Chicago, IL: American Medical Association; 1996.
CSP94 CRISP thesaurus. Bethesda, MD: National Institutes of Health. Division of Research Grants, Research Documentation Section; 1994.
CST93 COSTART: coding symbols for thesaurus of adverse reaction terms. Rockville, MD: Food and Drug Administration, Center for Drug Evaluation and Research; 1993.
DMD97 German translation of the MeSH. Cologne, Germany: Deutsches Institut fuer Medizinische Dokumentation und Information; 1997.
DOR27 Dorland's Illustrated Medical Dictionary, 27th. ed. Philadelphia, PA: Saunders; 1988.
DSM3R Diagnostic and statistical manual of mental disorders: DSMIIIR. 3rd ed. rev. Washington, DC: American Psychiatric Association; 1987.
DSM4 Diagnostic and statistical manual of mental disorders: DSMIV. Washington, DC: American Psychiatric Association; 1994.
DXP94 DXplain, an expert diagnosis program, developed by Massachusetts General Hospital.
HHC93 Saba, Virginia. Home Health Care Classification of Nursing Diagnoses and Interventions. Washington, DC: Georgetown University; 1993.
ICD89 The International Classification of Diseases: 9th revision, Clinical Modifications: 3rd ed. Washington, DC: Health Care Financing Administration; 1989.
ICD91 The International Classification of Diseases: 9th revision, Clinical Modification: 4th ed. Washington, DC: Health Care Financing Administration; 1991.
ICD96 The International Classification of Diseases: 9th revision, Clinical Modification: 6th ed. Washington, DC: Health Care Financing Administration; 1996.
INS97 Thesaurus Biomedical Francais/Anglais. [French translation of MeSH]. Paris: Institut National de la Sante et Recherche Medicale; 1997.
LCH90 Library of Congress subject headings. 12th ed. Washington, DC: Library of Congress; 1989.
LNC10F Logical Observations Identifiers, Names, and Codes. LOINC. version 1.0f. Indianapolis, IN: The Regenstrief Institute; 1996.
MCM92 Haynes, Brian. Glossary of methodologic terms for clinical epidemiologic studies of human disorders. Hamilton, Ontario, Canada: McMaster University; 1992.
MIM93 Online Mendelian Inheritance in Man
MSH97 Medical subject headings, Bethesda, MD: National Library of Medicine; 1997.
MTH UMLS Metathesaurus.
NAN94 Carroll-Johnson, Rose Mary, editor. Classification of nursing diagnoses: proceedings of the 10th conference, North American Diagnosis Association. Philadelphia, PA: Lippincott; 1994.
NEU95 Bowden, Douglas M., Martin, Richard F., Dubach, Joev G. Neuronames Brain Hierarchy. Seattle, WA: University of Washington, Primate Information Center; 1995.
NIC94 McCloskey, Joanne C.; Bulechek, Gloria M., editors. Nursing interventions classification (NIC): Iowa intervention project. St. Louis, MO: Mosby Year Book; 1994.
OMS94 Martin, Karen S., Scheet, Nancy J. The Omaha System: Applications for Community Health Nursing. Philadelphia, PA: Saunders; 1992. (with 1994 corrections).
PDQ96 Physician Data Query Online System. National Cancer Institute; 1996.
PSY94 Thesaurus of Psychological Index Terms. Washington DC: American Psychological Association; 1994.
RCD95 The Read Thesaurus, version 3.1 October, 1995; National Health Service National Coding and Classification Centre; 1995.
SNM2 Cote, Roger A., editor. Systematized nomenclature of medicine. 2nd ed. Skokie, IL: College of American Pathologists; 1979. SNOMED update, 1982. Skokie, IL: College of American Pathologists; 1982.
SNMI95 Cote, Roger A., editor. Systemized Nomenclature of Human and Veterinary Medicine: SNOMED International. Northfield, IL: College of American Pathologists; Schaumburg, IL: American Veterinary Medical Association; 1995.
ULT93 Bell, Douglas. Ultrasound Structured Attribute Reporting. UltraSTAR. Boston, MA: Brigham & Womens Hospital; 1993.
UMD97 Universal medical device nomenclature system: product category thesaurus. Plymouth Meeting, PA: ECRI; 1997.
WHO93 WHO Adverse Drug Reaction Terminology. WHOART. Uppsala, Sweden: WHO Collaborating Centre for International Drug Monitoring; 1993.
Appendix II
Testers' Categorization of Terms Submitted
Table 3.
| No. of Terms Categ. Chosen | % of Terms Categ. Chosen | Exact Meaning Found (%) | Related Concept Found (%) | No Related Concept Found (%) | |
|---|---|---|---|---|---|
| A. DATA TASK (required) | |||||
| A1. Record or display data abt indivs | 22879 | 56 | 59 | 39 | 2 |
| A2. Extract or summarize data abt grps | 1436 | 35 | 61 | 37 | 1 |
| A3. Retrieve info from knowledge bases | 4861 | 12 | 56 | 43 | 1 |
| A4. Build multi-purpose vocabulary dbs | 15163 | 37 | 51 | 48 | 2 |
| A5. Link natural lang to controlled vocab | 19062 | 46 | 57 | 42 | 1 |
| A6. Other | 2 | 0 | 50 | 50 | 0 |
| B. GENERAL PURPOSE OF TASK (required) | |||||
| B1. Direct patient care | 30487 | 74 | 59 | 40 | 1 |
| B2. Decision support | 16080 | 39 | 56 | 43 | 1 |
| B3. Clinical research | 15875 | 39 | 62 | 37 | 1 |
| B4. Public health surveill or intervent | 3239 | 8 | 50 | 49 | 2 |
| B5. Outcomes, health service research | 11266 | 27 | 59 | 39 | 1 |
| B6. Development of guidelines, pathways | 695 | 17 | 57 | 41 | 2 |
| B7. Enhancement of health database | 11371 | 28 | 57 | 40 | 2 |
| B8. Other | 124 | 0 | 61 | 36 | 2 |
| C. CARE SETTING OR FACILITY (required if applicable) | |||||
| C1. Ambulatory care office/clinic | 23181 | 56 | 60 | 39 | 1 |
| C2. Inpatient care facility | 23401 | 57 | 58 | 40 | 2 |
| C3. Long term care facility | 7857 | 19 | 53 | 46 | 1 |
| C4. Home care | 3829 | 9 | 46 | 52 | 2 |
| C5. Free-standing clinical laboratory | 4151 | 10 | 48 | 51 | 1 |
| C6. Free-standing pharmacy | 2720 | 7 | 41 | 57 | 1 |
| C7. Other | 3168 | 8 | 30 | 69 | 1 |
| D. SPECIFIC TYPE OF CARE OR SPECIALITY (required, if applicable) | |||||
| D1. Anesthesiology | 163 | 0 | 54 | 45 | 1 |
| D2. Dentistry | 1347 | 3 | 59 | 40 | 2 |
| D3. Diagnostic Imaging | 7898 | 19 | 55 | 44 | 1 |
| D4. Emergency Medicine | 8007 | 19 | 61 | 38 | 1 |
| D5. Family Practice | 6827 | 17 | 62 | 38 | 1 |
| D6. Internal Medicine | 17252 | 42 | 65 | 34 | 1 |
| D7. Intensive Care/Critical Care | 7820 | 19 | 63 | 36 | 0 |
| D8. Neurology | 1096 | 3 | 65 | 34 | 1 |
| D9. Nursing | 6745 | 16 | 63 | 35 | 3 |
| D10. Obstetrics/Gynecology | 2187 | 5 | 58 | 41 | 1 |
| D11. Opthalmology | 3618 | 9 | 71 | 28 | 1 |
| D12. Orthopedics | 460 | 1 | 59 | 40 | 2 |
| D13. Pathology | 1261 | 3 | 55 | 45 | 1 |
| D14. Pediatrics | 1544 | 4 | 64 | 34 | 2 |
| D15. Pharmacology | 1601 | 4 | 49 | 51 | 0 |
| D16. Pharmacy | 190 | 8 | 55 | 45 | 0 |
| D17. Psychiatry/Clinical Psychology | 3680 | 9 | 66 | 33 | 1 |
| D18. Social Work | 868 | 2 | 68 | 31 | 1 |
| D19. Surgery | 10731 | 26 | 57 | 42 | 1 |
| D20. Urology | 604 | 1 | 59 | 38 | 3 |
| D21. Veterinary Medicine | 2075 | 5 | 45 | 53 | 1 |
| D22. Other | 3115 | 8 | 37 | 62 | 1 |
| E. SPECIFIC SEGMENT OF PATIENT RECORD FOR WHICH CONTROLLED TERMINOLOGY IS SOUGHT (required, if applicable) | |||||
| E1. Chief Complaint | 9792 | 24 | 55 | 44 | 1 |
| E2. Problem list | 19065 | 46 | 64 | 35 | 1 |
| E3. Discharge summary | 9880 | 24 | 60 | 39 | 1 |
| E4. Medications | 671 | 2 | 30 | 66 | 4 |
| E5. Diagnoses | 15392 | 37 | 60 | 39 | 1 |
| E6. Patient history | 8982 | 22 | 46 | 53 | 1 |
| E7. Physical Examination | 7023 | 17 | 55 | 44 | 1 |
| E8. Review of systems | 2299 | 6 | 47 | 52 | 1 |
| E9. Laboratory tests | 3267 | 8 | 59 | 40 | 2 |
| E10. Procedures | 12409 | 30 | 56 | 43 | 1 |
| E11. Progress notes | 5666 | 14 | 51 | 46 | 2 |
| E12. Immunizations | 6 | 0 | 50 | 50 | 0 |
| E13. Family history | 455 | 1 | 64 | 31 | 5 |
| E14. Assessment | 2927 | 7 | 63 | 33 | 4 |
| E15. Flowsheet | 885 | 2 | 59 | 40 | 1 |
| E16. Plan | 3928 | 10 | 52 | 46 | 2 |
| E17. Intake and output | 675 | 2 | 66 | 34 | 0 |
| E18. Environmental exposures | 118 | 0 | 34 | 47 | 19 |
| E19. Demographic data | 768 | 2 | 29 | 67 | 3 |
| E20. Functional status | 185 | 0 | 44 | 44 | 12 |
| E21. Consult/referral | 1904 | 5 | 45 | 54 | 1 |
| E22. Patient education/teaching record | 571 | 1 | 20 | 75 | 5 |
| E23. Other | 3 | 0 | 100 | 0 | 0 |
Footnotes
The cooperative agreement partners are Beth Israel Hospital, Boston; Children's Hospital, Boston; Columbia University; Indiana University; Massachusetts General Hospital; Mayo Foundation and Kaiser Permanente; Oregon Health Sciences University; and Washington University, St. Louis.
The versions of these terminologies used in the test are as follows: Systemized Nomenclature of Medicine, Version 3.1, 1995; The Read Thesaurus, National Health Service National Coding and Classification Centre, Version 3.1, 1995; Logical Observations Identifiers, Names, and Codes, Version 1.0f, the Regenstrief Institute, 1996.
This number excludes the non-English versions of the Medical Subject Headings (MeSH) and treats nonoverlapping sets of terms from single sources added into the Metathesaurus in different years as single sources. SNOMED International and SNOMED II are counted as discrete sources. The third edition, revised, and the fourth edition of the Diagnostic and Statistical Manual of Mental Disorders are also treated as different sources.
It is possible that the high end of these ranges will be adjusted upward when the Read Codes have been fully integrated into the Metathesaurus. In the current data, unlinked Read synonyms could artificially lower the percentage of exact matches found in any vocabulary in the test set.
References
- 1.Hammond WE. Call for a standard clinical vocabulary. J Am Med Inform Assoc. 1997;4: 454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Board of Directors of the American Medical Informatics Association. Standards for medical identifiers, and messages needed to create an efficient computer-stored medical record. J Am Med Inform Assoc. 1994;1: 1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chute CG, Cohn SP, Campbell KE, Oliver DE, Campbell JR for Computer-Based Patient Record Institute's Work Group on Codes & Structures. The content of clinical classifications. J Am Med Inform Assoc. 1996;3: 224-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell JR, Carpenter P, Sneiderman C, Cohn S, Chute CG, Warren J for CPRI Work Group on Codes and Structures. Phase II evaluation of clinical coding schemes: completeness, taxonomy, mapping, definitions, and clarity. J Am Med Inform Assoc. 1997;4: 238-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cote RA, Rothwell DJ, Palotay JL, Beckett RS, Brochu L (eds). The Systematized Nomenclature of Human and Veterinary Medicine: SNOMED International. Northfield, IL: College of American Pathologists, 1993.
- 6.O'Neil MJ, Payne C, Read JD. Read Codes Version 3: a user lead terminology. Meth Inform Med. 1995;34: 187-92. [PubMed] [Google Scholar]
- 7.Vocabularies for computer-based patient records: identifying candidates for large scale testing. Minutes of a December 5-6 meeting sponsored by the National Library of Medicine and the Agency for Health Care Policy and Research. Bethesda, MD: National Library of Medicine, 1994.
- 8.Humphreys BL, Hole WT, McCray AT, Fitzmaurice JM. Planned NLM/AHCPR large-scale vocabulary test: using UMLS technology to determine the extent to which controlled vocabularies cover terminology needed for health care and public health. J Am Med Inform Assoc. 1996;3: 281-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Health Insurance Portability and Accountability Act of 1996, Pub. L. 104-191, Sections 261, 262.
- 10.Mullins HC, Scanland PM, Collins D, et al. The efficacy of SNOMED, Read codes, and UMLS in coding ambulatory family practice clinical records. Proc AMIA Annu Fall Symp. Philadelphia: Hanley and Belfus, 1996; 135-9. [PMC free article] [PubMed]
- 11.McCray AT, Razi AM, Bangalore AK, Browne AC, Stavri PZ. The UMLS Knowledge Source Server: A versatile Internet-based research tool. Cimino JJ (ed). Proceedings of the 1996 AMIA Annual Fall Symposium. Philadelphia: Hanley and Belfus, 1996; 164-8. [PMC free article] [PubMed]
- 12.McCray AT, Cheh ML, Bangalore AK, Rafei K, Razi AM, Divita G, Stavri PZ. Conducting the NLM/AHCPR Large Scale Vocabulary Test: A distributed Internet-based experiment. To appear in the proceedings of the 1997 AMIA Annual Fall Symposium. [PMC free article] [PubMed]
- 13.McCray AT, Srinivasan S, Browne AC. Lexical methods for managing variation in biomedical terminologies. Proc Annu Symp Comput Appl Med Care. 1994; 235-9. [PMC free article] [PubMed]
- 14.Aronson AR, Rindflesch TC, Browne AC. Exploiting a large thesaurus for information retrieval. Proceedings of RIAO. 1994; 197-216.
- 15.Fleiss JL. Measuring nominal scale agreement among many raters. Psychological Bulletin. 1971;76: 378-82. [Google Scholar]
- 16.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33: 159-74. [PubMed] [Google Scholar]
- 17.Chute CG, Elkin PL. A clinically derived terminology: qualification to reduction. To appear in Masys DM (ed). Proceedings of the 1997 AMIA Annual Fall Symposium. [PMC free article] [PubMed]