Significance
Many biological cofactors, including light-absorbing chromophores in photoreceptors, are modulated upon insertion into a protein. The steric contribution can cause structural distortions in the cofactor, and such effects are considered to be crucial for biological function. For example, the out-of-plane distortion of chromophores is a key factor in controlling their absorption spectra. In spite of the functional importance, such structural distortions are difficult to measure experimentally. In this study, we used a unique capability of Raman optical activity (ROA) to address the above-mentioned issue. This study applied ROA spectroscopy to a photoreceptor protein and indicates that a hydrogen out-of-plane ROA band provides a spectroscopic ruler for the out-of-plane distortion of the chromophore that is embedded in a protein environment.
Keywords: photoreceptor, chromophore, vibrational spectroscopy, density functional theory, molecular strain
Abstract
Photoactive yellow protein (PYP), from the phototrophic bacterium Halorhodospira halophila, is a small water-soluble photoreceptor protein and contains p-coumaric acid (pCA) as a chromophore. PYP has been an attractive model for studying the physical chemistry of protein active sites. Here, we explore how Raman optical activity (ROA) can be used to extract quantitative information on distortions of the pCA chromophore at the active site in PYP. We use 13C8-pCA to assign an intense signal at 826 cm−1 in the ROA spectrum of PYP to a hydrogen out-of-plane vibration of the ethylenic moiety of the chromophore. Quantum-chemical calculations based on density functional theory demonstrate that the sign of this ROA band reports the direction of the distortion in the dihedral angle about the ethylenic C=C bond, while its amplitude is proportional to the dihedral angle. These results document the ability of ROA to quantify structural deformations of a cofactor molecule embedded in a protein moiety.
Many biological cofactors, including light-absorbing chromophores in photoreceptors, are modulated upon insertion into a protein binding pocket by both electrostatic and steric interactions. The electrostatic component of these effects, including hydrogen bonding and charge–charge interactions, has been studied in some detail (1, 2). The steric contribution can cause structural distortions in the cofactor, and such effects have been considered to be crucial for biological function but are less well understood. Proposed functional roles for cofactor distortions include the out-of-plane distortion of chromophores as a key factor in controlling their absorption spectra (3, 4). Furthermore, photoexcitation of these proteins produces primary high-energy intermediates with structurally perturbed chromophores (5–8), which drive subsequent protein conformational changes (9, 10). Such structural distortions have proven difficult to measure experimentally.
Recent progress in Raman optical activity (ROA) spectroscopy has revealed this technique as a promising avenue to derive structural details on the distortion of a chromophore within a protein environment (11–15). ROA measures the difference in Raman scattering intensity between right (IR) and left (IL) circularly polarized incident light, which provides information on molecular chirality (16–18). The sum of IR and IL corresponds to the Raman spectrum. A protein environment can distort an achiral chromophore into a chiral conformation, and ROA spectroscopy provides an approach to derive detailed structural information of the chromophore in the protein under physiological solution conditions. This method can be extended to a structural studies of short-lived intermediate (15). These studies suggested that the hydrogen out-of-plane (HOOP) mode is especially sensitive to the distortion of the chromophore. We recently reported that preresonance conditions are ideal for measuring structurally informative ROA spectra, since chromophore signals are substantially enhanced without the disruption of the ROA effect that occurs under full-resonance conditions (14, 19). Here, we aim to further develop the use of preresonance ROA spectroscopy in determining chromophore distortions in photoactive yellow protein (PYP).
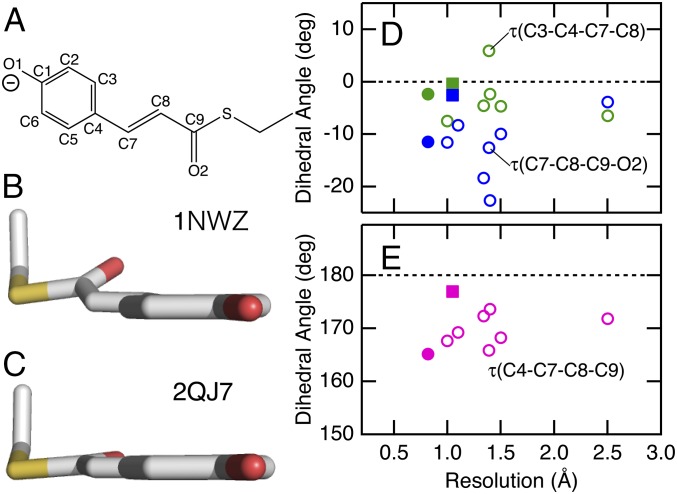
PYP is a small water-soluble blue light photoreceptor protein from the phototrophic bacterium Halorhodospira halophila, and it provides an attractive model system for studying the physical chemistry of protein active sites (20–22). It contains a p-coumaric acid (pCA) chromophore, which is covalently linked to Cys69 through a thiol ester bond (23, 24). As shown in Fig. 1A, the pCA is in the trans conformation and its phenolic oxygen is deprotonated in the initial dark state, pG. The chromophore is out-of-plane distorted in its active site, although the distortions of the pCA chromophore vary significantly among the available high-resolution crystal structures of the pG state (25–32). In Fig. 1 and SI Appendix, Table S1, we use three dihedral angles τ(C3–C4–C7–C8), τ(C4–C7–C8–C9), and τ(C7–C8–C9–O2) to characterize the chromophore distortions. The values of these angles exhibit a fair amount of scatter among these crystal structures, as displayed in D and E. The fact that the values do not appear to be converged even below 1-Å resolution documents that these dihedral angles are difficult to determine using state-of-the-art high-resolution X-ray and neutron diffraction approaches. PYP exhibits several intense ROA bands ascribed to the ethylenic HOOP modes (12, 14). Here, we report how these HOOP modes can be used to extract information on distortions of the pCA chromophore in its active site.
Fig. 1.
The structures of the pCA chromophore for PYP. (A) The structure and atom numbering of the chromophore. (B and C) The structures of the chromophore in two representative crystal structures (PDB ID codes 1NWZ and 2QJ7). (D and E) Dihedral angles τ(C3–C4–C7–C8), τ(C7–C8–C9–O2), and τ(C4–C7–C8–C9) as a function of crystallographic resolution. The data for 1NWZ and 2QJ7 are displayed as closed circles and squares, respectively.
Results and Discussion
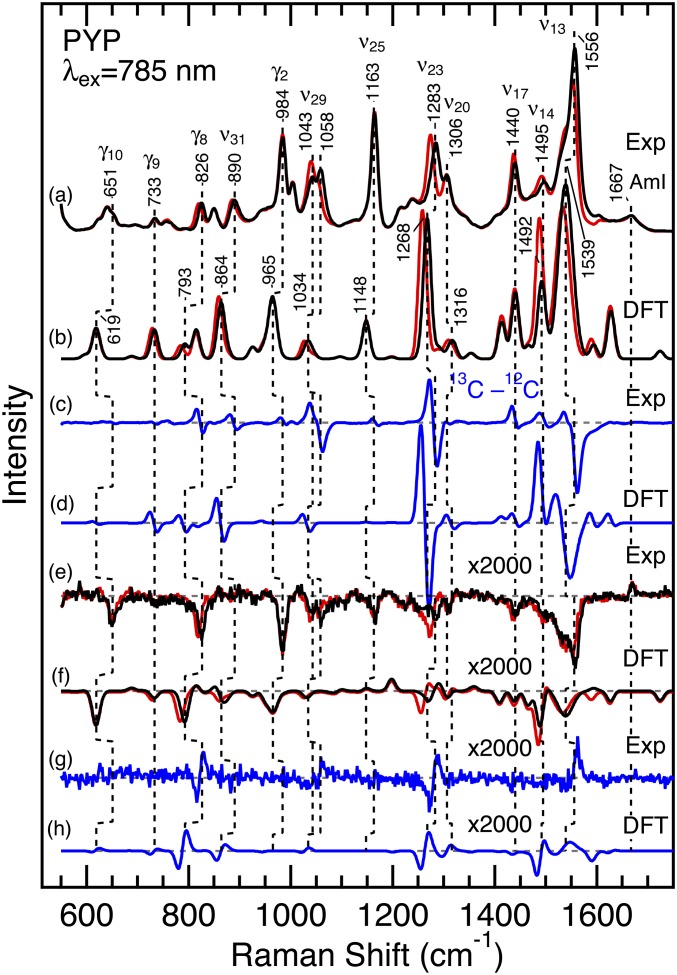
The published assignment of the proposed ethylenic HOOP modes of pCA was not confirmed, and the possibility that the observed ROA bands are due to the protein moiety remained open (12). Therefore, as a first step, we used isotope labeling to confirm the assignment of one of the important HOOP modes γ8. To conclusively test the proposed assignment to the γ8 mode, we prepared PYP whose chromophore is labeled with 13C at the C8 carbon atom. Fig. 2 shows the preresonance Raman and ROA spectra for the pG state of wild-type PYP with 785-nm excitation (black traces a and e, respectively). The resonance Raman spectra for a long-lived blue-shifted intermediate denoted pB (also called I2 or PYPM) are displayed in SI Appendix, Fig. S1. These spectra are consistent with those reported previously (12, 14, 33, 34). In Fig. 2, we also show the Raman and ROA spectra for 13C8-pCA PYP (red traces). In addition, the figure depicts the 12C/13C difference spectra (traces c and g). These data reveal that four different ROA signals, including the prominent band at 826 cm−1, are clearly affected by the isotope editing. This negative ROA band was tentatively assigned to γ8, which is a HOOP mode of the C8–H moiety (12, 35). It exhibits a 6 cm−1 downshift upon 13C8 substitution, supporting the assignment of this band to γ8. The effect of the isotopic substitution can also be seen at 1,556, 1,283, and ∼1,050 cm−1 in the difference ROA spectrum. These bands were ascribed to the C=C stretching mode ν13, HC7=C8H rocking ν23, and C8–C9 stretching ν29, respectively (12). All of these modes involve motions of the C8 atom, and the present observations are consistent with the previous assignment (12, 35).
Fig. 2.
Observed and calculated Raman and ROA spectra of PYP whose chromophore is unlabeled (black) and 13C8 labeled (red). (A) The observed Raman spectra. (B) The calculated Raman spectra. (C) The observed and (D) calculated 13C minus 12C difference Raman spectra. (E) The observed ROA spectra. (F) The calculated ROA spectra. (G) The observed and (H) calculated 13C minus 12C difference ROA spectra. PYP samples were dissolved in 10 mM Tris⋅HCl, pH 7.4, and the sample concentration was 4−5 mM. The spectra were obtained with 785-nm excitation (∼200 mW). The calculated spectra were based on model 1, and Gaussian band shapes with a 10 cm−1 width were used except a 20 cm−1 width for the highest band at 1,539 cm−1. Raman and ROA intensities for the highest intensity bands were reduced by a factor of 2 (1,319, 1,314, 1,310, 1,268, 1,266, and 1,259 cm−1) or 10 (1,633, 1,628, and 1,539 cm−1) to make the other bands visible in the figure. The ROA spectra are magnified by a factor of 2,000.
To provide a quantitative test of these band assignments, we performed density functional theory (DFT) calculations and examined the effects of the 13C8 substitution of the pCA chromophore. In these calculations, we used an active-site model (model 1) that consists of deprotonated pCA methyl thiol ester as a chromophore model (12). Model 1 also includes methanol, acetic acid, and methylamine to mimic surrounding Tyr42, Glu46, and Cys69 residues, respectively. These components were arranged on the basis of the crystal structure (SI Appendix, Fig. S3) (27, 32). In this model, six dihedral angles (listed in SI Appendix, Table S1) are constrained based on crystal structures of PYP (27, 32). These structural constraints have been shown to be important to reproduce the main features of the observed ROA spectrum (12). The hybrid functional B3LYP and the 6-31+G** basis set were used for these calculations, and a larger basis set with an additional diffused function (6-311++G**) only moderately affected the calculated spectra (SI Appendix, Fig. S4). In Fig. 2, we display the calculated Raman and ROA spectra (traces b and f). The 13C minus 12C difference Raman and ROA spectra are also calculated for model 1 (traces d and h), and most of the observed isotope effects are reproduced by the present calculation. Importantly, the assignment of the 826 cm−1 band to γ8 is confirmed by the observed shift of −6 cm−1 upon 13C8 substitution and the comparable shift of −8 cm−1 for model 1 (792–784 cm−1). In addition, the assignment of the other 13C8-senisitive bands such as ν13, ν23, and ν29 is also confirmed by the DFT calculations.
These Raman and ROA spectra of PYP containing 13C8-pCA allow the conclusive assignment of the ROA band at 826 cm−1 to the HOOP vibration of the ethylenic moiety. As displayed in Fig. 2, the γ8 band is one of the main ROA bands for PYP, and its intensity is more than 50% to that of the most intense ν13 band. This is in sharp contrast to the case of the Raman spectra, where the intensity of the γ8 band is only 15% of the ν13 intensity. The observation of the intense ROA band leads us to expect that the HOOP γ8 mode provides a good spectroscopic ruler for the chromophore distortions.
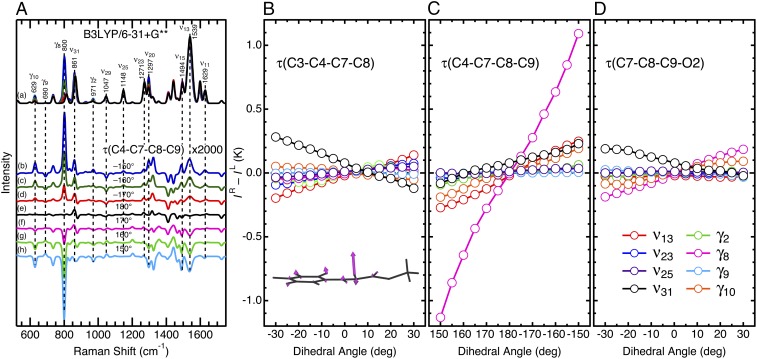
Thus, we aimed to extract information on structural deformations of the pCA from this band. We next performed systematic DFT calculations to explore what structural factors affect the ROA spectra, particularly the sign and intensity of HOOP bands. For this analysis, we focus on the three dihedral angles τ(C3–C4–C7–C8), τ(C4–C7–C8–C9), and τ(C7–C8–C9–O2) to characterize the out-of-plane distortions of the chromophore. We used the active-site models analogous to model 1 and varied these three dihedral angles up to ±30° from a planar structure, while the other structural parameters were not constrained. As an example, Fig. 3A displays the effects of varying τ(C4–C7–C8–C9) on the simulated Raman and ROA spectra. The dihedral twist about the C7=C8 bond does not affect the overall spectral features of the Raman spectra. An exception is the γ8 band near 800 cm−1, and its intensity increases when the chromophore is distorted, while its frequency exhibits a small 2 cm−1 upshift upon changing τ(C4–C7–C8–C9) by 30° from a planar structure. In contrast to the Raman spectra, many of the ROA bands change its signs and/or intensities as a function of τ(C4–C7–C8–C9). As illustrated in Fig. 3A, the γ8 band is especially sensitive to this dihedral twist. These results indicate that both the position and intensity of bands in Raman spectra generally are insensitive to changes in dihedral angle. On the other hand, the intensity of bands in ROA spectra are sensitive to dihedral angles, and thus provide an opportunity to study distortions of these angles in protein active sites.
Fig. 3.
Simulated Raman and ROA spectra of the active-site models for PYP. (A) Raman (a) and ROA (b–h) spectra are shown. The dihedral angle about the C7=C8 moiety was varied up to ±30° from a planar geometry of τ(C4–C7–C8–C9) = 180°. The ROA spectra are magnified by a factor of 2,000. (B–D) ROA intensities as a function of the dihedral twists for the chromophore model of PYP. K is a constant (SI Appendix). The Inset shows the normal mode γ8 for model 1.
Fig. 3 B–D illustrates the effects of chromophore distortions on the ROA band intensities, and the results can be summarized in the following two points. (i) The ROA intensities almost linearly change as a function of the dihedral angles. This implies that the ROA intensity reflects the extent of the out-of-plane distortions of the chromophore and its sign indicates the direction of the structural distortions. (ii) The γ8 mode is especially sensitive to τ(C4–C7–C8–C9). Because of this high sensitivity, even modest distortions of this angle can be experimentally detected. As summarized in SI Appendix, Table S1, the value of τ(C4–C7–C8–C9) for model 1 is 170.0°, which is consistent with both the direction and the degree of twist found in the crystal structures (27, 32). To confirm the dominant contribution of the twist about the C7=C8 bond, we have performed further DFT calculations for models with additional structural constraints. SI Appendix, Fig. S5 compares the γ8 intensities with and without a constraint of τ(C3–C4–C7–C8) = −10° or τ(C7–C8–C9–O2) = −10°. As seen in the figure, the additional structural constraints cause only minor effects on the γ8 intensities, implying that this ROA band can be used as a marker of the dihedral twist of the ethylenic C=C bond. We conclude that τ(C4–C7–C8–C9) is a major factor in determining the amplitude of the ROA band of the γ8 mode, and that the amplitude of this band at 826 cm−1 can therefore be used to gauge distortions over this angle at the PYP active site.
The HOOP modes in Raman spectra for a photoreceptor protein have been used to probe the chromophore distortions (36, 37). Thus, we next examine the effects of the structural distortion on the Raman intensities of γ8 in SI Appendix, Fig. S6. In this work, we aimed to identify the most valuable spectroscopic readout for obtaining information on dihedral angles. In contrast to the ROA intensities, the Raman intensities exhibit a nonlinear dependence on the dihedral twist about the C7=C8 bond. In fact, the data for the γ8 Raman band can be fitted with a parabolic curve as illustrated as a dashed line in the figure. On the basis of these data in A, we also calculated circular intensity difference (CID) ∆, which is defined by Eq. 1:
| [1] |
SI Appendix, Fig. S6B demonstrates that the CID value exhibits somewhat complicated dependence on τ(C4–C7–C8–C9). This behavior of the ∆ value is due to the fact that the ROA signal is roughly proportional to a dihedral twist, while the Raman intensity exhibits a parabolic dependency. The theoretical basis for these differences is discussed in SI Appendix. These theoretical considerations also show that the ROA invariants for the distorted structure are proportional to the Raman polarizability. Thus, the large Raman intensity for γ8 explains the high sensitivity of the corresponding ROA band. These considerations indicate that the amplitude of the ROA band of γ8 is the most valuable experimental parameter for examining distortion of τ(C4–C7–C8–C9). The ROA intensity of this mode is directly proportional to this dihedral angle, providing a spectroscopic ruler for distortions of this bond.
As discussed above, the ROA intensities are roughly proportional to the three dihedral twists (Fig. 3). It is, therefore, expected that we can estimate a relative value of a dihedral angle from the observed spectra. For instance, if the γ8 ROA intensity of a mutant PYP is one-half of that of wild type, we expect that τ(C4–C7–C8–C9) of the mutant is also one-half compared with that of wild type. Ideally, the amplitude of the ROA band of γ8 would allow the determination of the absolute value of this angle. Two approaches can be considered for obtaining the absolute value of the dihedral angle of τ(C4–C7–C8–C9). First, the experimental determination of the absolute Raman and ROA cross-sections of this mode would allow the absolute value of this dihedral angle to be derived. This approach therefore would require high-accuracy measurements as well as theoretical predictions of the absolute cross-section. Second, the CID for the HOOP mode could be used. Since this is a dimensionless value, measurement of the absolute Raman and ROA cross-sections is not needed for an experimental determination. Care should be taken in this approach, since the dependence of the CID on the dihedral twist is not linear (SI Appendix, Fig. S6B). This approach would rely on the accurate computation of the intensities of the Raman and ROA bands of a vibrational mode. The calculated CID value for γ8 at a dihedral angle of 170° is −1.3 × 10−3, while the experimentally determined value reported here is −0.82 × 10−3. While an accuracy of within a factor ∼1.5 is reasonable for current quantum-chemical calculations, the estimated value of τ(C4–C7–C8–C9) would have a relatively large error. Therefore, this approach will require improvements in computational methods for deriving absolute intensities of Raman and ROA signals.
Finally, we briefly discuss energies associated with the chromophore distortions. SI Appendix, Fig. S7 displays the relative energies ΔE compared with a planar geometry for the active-site models used in Fig. 3 and SI Appendix, Fig. S5. It is seen from the figure that the distortion energy for the C7=C8 double bond is higher than that for the C4–C7 or C8–C9 single bond. For the higher-energy C=C bond, a twist by 30° causes a distortion energy of ∼16 kJ⋅mol−1, which is somewhat smaller than a typical energy for a hydrogen bond (20–60 kJ⋅mol−1) (38, 39). This analysis indicates that ROA has an ability to detect subtle structural distortions of the chromophore that can be induced by hydrogen bonding interactions with the surrounding protein moiety.
In summary, we have measured the Raman and ROA spectra of 13C8-pCA PYP. The reported results provide clear evidence that the ethylenic HOOP mode γ8 exhibits an intense ROA band for PYP. Further DFT calculations using the active-site models demonstrated that the γ8 ROA band is specifically sensitive to an out-of-plane distortion of the ethylenic C7=C8 bond. The sign of the ROA band reflects the direction of the distortion, and its intensity is proportional to a dihedral twist of τ(C4–C7–C8–C9). These observations indicate that a HOOP ROA band provides a spectroscopic ruler for the out-of-plane distortion of the chromophore that is embedded in a protein environment. Since structural distortions of cofactors are considered to be important for many biological functions (3, 4, 8–10), ROA spectroscopy will be useful to detect this functionally important structural information.
Materials and Methods
We prepared p-coumaric-8-13C-acid (13C8-pCA) from p-hydroxybenzaldehyde and triethylphosphonoacetate-1-13C followed by alkaline hydrolysis of the ester (SI Appendix). Production of wild-type PYP apoprotein from Escherichia coli, reconstitution of the holoprotein with the chromophore, and the subsequent protein purification were performed as reported in a previous study (40). The near-infrared ROA instrument used in this study is based on an incident circular polarization scheme described previously (11, 12). Preresonance Raman and ROA spectra with 785-nm excitation were calculated using the DFT method via the Gaussian 09 program (41). The hybrid functional B3LYP and the 6-31+G** basis set were used for these calculations. In some cases, a larger basis set with an additional diffused function (6-311++G**) was used. The calculated frequencies were scaled using a factor of 0.9648 (42).
Supplementary Material
Acknowledgments
We thank T. Horiuchi for his assistance in preparing 13C-labeled compound. This work was supported by Japan Society for the Promotion of Science KAKENHI Grants 17K05756 (to M.U.) and 16K17859 (to T.F.), and National Science Foundation Grant CHE-1413739 (to W.D.H.). A portion of the computations was performed at the Research Center for Computational Science in Okazaki, Japan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. Q.Y. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1806491115/-/DCSupplemental.
References
- 1.Spudich JL, Yang C-S, Jung K-H, Spudich EN. Retinylidene proteins: Structures and functions from archaea to humans. Annu Rev Cell Dev Biol. 2000;16:365–392. doi: 10.1146/annurev.cellbio.16.1.365. [DOI] [PubMed] [Google Scholar]
- 2.Boulanger E, Harvey JN. QM/MM methods for free energies and photochemistry. Curr Opin Struct Biol. 2018;49:72–76. doi: 10.1016/j.sbi.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 3.Rocha-Rinza T, Sneskov K, Christiansen O, Ryde U, Kongsted J. Unraveling the similarity of the photoabsorption of deprotonated p-coumaric acid in the gas phase and within the photoactive yellow protein. Phys Chem Chem Phys. 2011;13:1585–1589. doi: 10.1039/c0cp01075h. [DOI] [PubMed] [Google Scholar]
- 4.Sekharan S, Morokuma K. QM/MM study of the structure, energy storage, and origin of the bathochromic shift in vertebrate and invertebrate bathorhodopsins. J Am Chem Soc. 2011;133:4734–4737. doi: 10.1021/ja200322w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schotte F, et al. Watching a signaling protein function in real time via 100-ps time-resolved Laue crystallography. Proc Natl Acad Sci USA. 2012;109:19256–19261. doi: 10.1073/pnas.1210938109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jung YO, et al. Volume-conserving trans-cis isomerization pathways in photoactive yellow protein visualized by picosecond X-ray crystallography. Nat Chem. 2013;5:212–220. doi: 10.1038/nchem.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuramochi H, et al. Probing the early stages of photoreception in photoactive yellow protein with ultrafast time-domain Raman spectroscopy. Nat Chem. 2017;9:660–666. doi: 10.1038/nchem.2717. [DOI] [PubMed] [Google Scholar]
- 8.Gamiz-Hernandez AP, Kaila VR. Conversion of light-energy into molecular strain in the photocycle of the photoactive yellow protein. Phys Chem Chem Phys. 2016;18:2802–2809. doi: 10.1039/c5cp05244k. [DOI] [PubMed] [Google Scholar]
- 9.Warshel A, Barboy N. Energy storage and reaction pathways in the first step of the vision process. J Am Chem Soc. 1982;104:1469–1476. [Google Scholar]
- 10.Birge RR, Cooper TM. Energy storage in the primary step of the photocycle of bacteriorhodopsin. Biophys J. 1983;42:61–69. doi: 10.1016/S0006-3495(83)84369-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Unno M, Kikukawa T, Kumauchi M, Kamo N. Exploring the active site structure of a photoreceptor protein by Raman optical activity. J Phys Chem B. 2013;117:1321–1325. doi: 10.1021/jp4001187. [DOI] [PubMed] [Google Scholar]
- 12.Shingae T, Kubota K, Kumauchi M, Tokunaga F, Unno M. Raman optical activity probing structural deformations of the 4-hydroxycinnamyl chromophore in photoactive yellow protein. J Phys Chem Lett. 2013;4:1322–1327. doi: 10.1021/jz400454j. [DOI] [PubMed] [Google Scholar]
- 13.Kubota K, et al. Active site structure of photoactive yellow protein with a locked chromophore analogue revealed by near-infrared Raman optical activity. J Phys Chem Lett. 2013;4:3031–3038. doi: 10.1021/jz400454j. [DOI] [PubMed] [Google Scholar]
- 14.Haraguchi S, et al. Experimental detection of the intrinsic difference in Raman optical activity of a photoreceptor protein under preresonance and resonance conditions. Angew Chem Int Ed Engl. 2015;54:11555–11558. doi: 10.1002/anie.201505466. [DOI] [PubMed] [Google Scholar]
- 15.Fujisawa T, Leverenz RL, Nagamine M, Kerfeld CA, Unno M. Raman optical activity reveals carotenoid photoactivation events in the orange carotenoid protein in solution. J Am Chem Soc. 2017;139:10456–10460. doi: 10.1021/jacs.7b05193. [DOI] [PubMed] [Google Scholar]
- 16.Blanch EW, Hecht L, Barron LD. Vibrational Raman optical activity of proteins, nucleic acids, and viruses. Methods. 2003;29:196–209. doi: 10.1016/s1046-2023(02)00310-9. [DOI] [PubMed] [Google Scholar]
- 17.He Y, Wang B, Dukor RK, Nafie LA. Determination of absolute configuration of chiral molecules using vibrational optical activity: A review. Appl Spectrosc. 2011;65:699–723. doi: 10.1366/11-06321. [DOI] [PubMed] [Google Scholar]
- 18.Parchaňský V, Kapitán J, Bouř P. Inspecting chiral molecules by Raman optical activity spectroscopy. RSC Adv. 2014;4:57125–57136. [Google Scholar]
- 19.Nafie LA. Theory of resonance Raman optical activity: The single electronic state limit. Chem Phys. 1996;205:309–322. [Google Scholar]
- 20.Hellingwerf KJ, Hendriks J, Gensch T. Photoactive yellow protein, a new type of photoreceptor protein: Will this “yellow lab” bring us where we want to go? J Phys Chem A. 2003;107:1082–1094. [Google Scholar]
- 21.Imamoto Y, Kataoka M. Structure and photoreaction of photoactive yellow protein, a structural prototype of the PAS domain superfamily. Photochem Photobiol. 2007;83:40–49. doi: 10.1562/2006-02-28-IR-827. [DOI] [PubMed] [Google Scholar]
- 22.Kottke T, Hoff WD, Xie A, Larsen DS. Photoreceptors take charge: Emerging principles for light sensing. Annu Rev Biophys. 2018;47:291–313. doi: 10.1146/annurev-biophys-070317-033047. [DOI] [PubMed] [Google Scholar]
- 23.Hoff WD, et al. Thiol ester-linked p-coumaric acid as a new photoactive prosthetic group in a protein with rhodopsin-like photochemistry. Biochemistry. 1994;33:13959–13962. doi: 10.1021/bi00251a001. [DOI] [PubMed] [Google Scholar]
- 24.Baca M, et al. Complete chemical structure of photoactive yellow protein: Novel thioester-linked 4-hydroxycinnamyl chromophore and photocycle chemistry. Biochemistry. 1994;33:14369–14377. doi: 10.1021/bi00252a001. [DOI] [PubMed] [Google Scholar]
- 25.Borgstahl GEO, Williams DR, Getzoff ED. 1.4 Å structure of photoactive yellow protein, a cytosolic photoreceptor: Unusual fold, active site, and chromophore. Biochemistry. 1995;34:6278–6287. doi: 10.1021/bi00019a004. [DOI] [PubMed] [Google Scholar]
- 26.van Aalten DMF, Crielaard W, Hellingwerf KJ, Joshua-Tor L. Conformational substates in different crystal forms of the photoactive yellow protein–Correlation with theoretical and experimental flexibility. Protein Sci. 2000;9:64–72. doi: 10.1110/ps.9.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Getzoff ED, Gutwin KN, Genick UK. Anticipatory active-site motions and chromophore distortion prime photoreceptor PYP for light activation. Nat Struct Biol. 2003;10:663–668. doi: 10.1038/nsb958. [DOI] [PubMed] [Google Scholar]
- 28.Anderson S, Crosson S, Moffat K. Short hydrogen bonds in photoactive yellow protein. Acta Crystallogr D Biol Crystallogr. 2004;60:1008–1016. doi: 10.1107/S090744490400616X. [DOI] [PubMed] [Google Scholar]
- 29.Shimizu N, Kamikubo H, Yamazaki Y, Imamoto Y, Kataoka M. The crystal structure of the R52Q mutant demonstrates a role for R52 in chromophore pKa regulation in photoactive yellow protein. Biochemistry. 2006;45:3542–3547. doi: 10.1021/bi051430a. [DOI] [PubMed] [Google Scholar]
- 30.Fisher SZ, et al. Neutron and X-ray structural studies of short hydrogen bonds in photoactive yellow protein (PYP) Acta Crystallogr D Biol Crystallogr. 2007;63:1178–1184. doi: 10.1107/S0907444907047646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coureux P-D, Fan ZP, Stojanoff V, Genick UK. Picometer-scale conformational heterogeneity separates functional from nonfunctional states of a photoreceptor protein. Structure. 2008;16:863–872. doi: 10.1016/j.str.2008.02.022. [DOI] [PubMed] [Google Scholar]
- 32.Yamaguchi S, et al. Low-barrier hydrogen bond in photoactive yellow protein. Proc Natl Acad Sci USA. 2009;106:440–444. doi: 10.1073/pnas.0811882106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Unno M, Kumauchi M, Sasaki J, Tokunaga F, Yamauchi S. Evidence for a protonated and cis configuration chromophore in the photobleached intermediate of photoactive yellow protein. J Am Chem Soc. 2000;122:4233–4234. [Google Scholar]
- 34.Unno M, Kumauchi M, Sasaki J, Tokunaga F, Yamauchi S. Assignment of resonance Raman spectrum of photoactive yellow protein in its long-lived blue-shifted intermediate. J Phys Chem B. 2003;107:2837–2845. [Google Scholar]
- 35.Unno M, Kumauchi M, Tokunaga F, Yamauchi S. Vibrational assignment of the 4-hydroxycinnamyl chromophore in photoactive yellow protein. J Phys Chem B. 2007;111:2719–2726. doi: 10.1021/jp066434j. [DOI] [PubMed] [Google Scholar]
- 36.Eyring G, Curry B, Broek A, Lugtenburg J, Mathies R. Assignment and interpretation of hydrogen out-of-plane vibrations in the resonance Raman spectra of rhodopsin and bathorhodopsin. Biochemistry. 1982;21:384–393. doi: 10.1021/bi00531a028. [DOI] [PubMed] [Google Scholar]
- 37.Kukura P, McCamant DW, Yoon S, Wandschneider DB, Mathies RA. Structural observation of the primary isomerization in vision with femtosecond-stimulated Raman. Science. 2005;310:1006–1009. doi: 10.1126/science.1118379. [DOI] [PubMed] [Google Scholar]
- 38.Kaledhonkar S, Hara M, Stalcup TP, Xie A, Hoff WD. Strong ionic hydrogen bonding causes a spectral isotope effect in photoactive yellow protein. Biophys J. 2013;105:2577–2585. doi: 10.1016/j.bpj.2013.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steiner T. The hydrogen bond in the solid state. Angew Chem Int Ed Engl. 2002;41:49–76. doi: 10.1002/1521-3773(20020104)41:1<48::aid-anie48>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 40.Kort R, et al. The xanthopsins: A new family of eubacterial blue-light photoreceptors. EMBO J. 1996;15:3209–3218. [PMC free article] [PubMed] [Google Scholar]
- 41.Frisch MJ, et al. Gaussian 09. Gaussian, Inc.; Wallingford, CT: 2009. [Google Scholar]
- 42.Merrick JP, Moran D, Radom L. An evaluation of harmonic vibrational frequency scale factors. J Phys Chem A. 2007;111:11683–11700. doi: 10.1021/jp073974n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.