Significance
When replication forks collapse at single-strand DNA breaks, the resulting single-ended DNA double-strand breaks (seDSBs) are repaired by sister chromatid recombination (SCR). The Mre11–Rad50–Nbs1 (MRN) protein complex and CtIP/Ctp1 nucleolytically resect the seDSB to initiate SCR, but MRN was also proposed to promote SCR by tethering the seDSB to the unbroken sister chromatid. Here, we show that MRN sequentially localizes with the seDSB and sister chromatid during SCR. Provision of an intrachromosomal repair template alleviates the requirement for the nonnucleolytic function of MRN in repair of a collapsed replication fork. Zinc-coordinating “hooks” in Rad50 that are linchpins for MRN intercomplexes are essential for SCR. These studies support the MRN tethering hypothesis.
Keywords: genome maintenance, double-strand break repair, recombination, Mre11, Rad50
Abstract
Collapsed replication forks, which are a major source of DNA double-strand breaks (DSBs), are repaired by sister chromatid recombination (SCR). The Mre11–Rad50–Nbs1 (MRN) protein complex, assisted by CtIP/Sae2/Ctp1, initiates SCR by nucleolytically resecting the single-ended DSB (seDSB) at the collapsed fork. The molecular architecture of the MRN intercomplex, in which zinc hooks at the apices of long Rad50 coiled-coils connect two Mre112–Rad502 complexes, suggests that MRN also structurally assists SCR. Here, Rad50 ChIP assays in Schizosaccharomyces pombe show that MRN sequentially localizes with the seDSB and sister chromatid at a collapsed replication fork. Ctp1, which has multivalent DNA-binding and DNA-bridging activities, has the same DNA interaction pattern. Provision of an intrachromosomal repair template alleviates the nonnucleolytic requirement for MRN to repair the broken fork. Mutations of zinc-coordinating cysteines in the Rad50 hook severely impair SCR. These data suggest that the MRN complex facilitates SCR by linking the seDSB and sister chromatid.
Efficient repair of collapsed DNA replication forks is essential for maintaining genome integrity. In eukaryotes, the most accurate repair of collapsed forks occurs by sister chromatid recombination (SCR) (1–3). In this replication-coupled mechanism of homologous recombination (HR) repair, the single-ended DNA double-strand break (seDSB) at the broken fork is first resected by the Mre11–Rad50–Nbs1 (MRN; also known as Mre11) protein complex, which has latent nuclease activities that are stimulated by CtIP/Ctp1/Sae2 (4–6). These activities displace the nonhomologous end-joining (NHEJ) factor Ku from the seDSB, which allows additional resection enzymes such as Exo1 to access the seDSB. Resection exposes a 3′ single-strand DNA (ssDNA) tail that is initially bound by Replication Protein A, which is then displaced by Rad51 with the aid of HR mediators such as Rad52 or BRCA2. These proteins also promote Rad51-nucleoprotein filament invasion and synapsis with homologous DNA sequences in the sister chromatid. The invading DNA strand primes DNA synthesis with the potential to reestablish the replication fork. The final step of replication-coupled SCR is resolution of the DNA joint molecule, a D-loop or Holliday junction, which is formed during strand invasion. Resolution completes sister chromatid exchange (SCE) and allows sister chromatid segregation during mitosis.
The resection activities of MRN-Ctp1 are well documented, as are its DNA damage signaling and telomere maintenance functions involving Tel1/ATM kinase (7, 8). MRN is also thought to promote DSB repair by tethering DNA molecules (9–13). Specifically, MRN was proposed to tether opposing DSBs to promote HR or NHEJ repair, or to tether DSBs to unbroken sister chromatids to facilitate HR. This latter activity might be especially important for repair of broken forks.
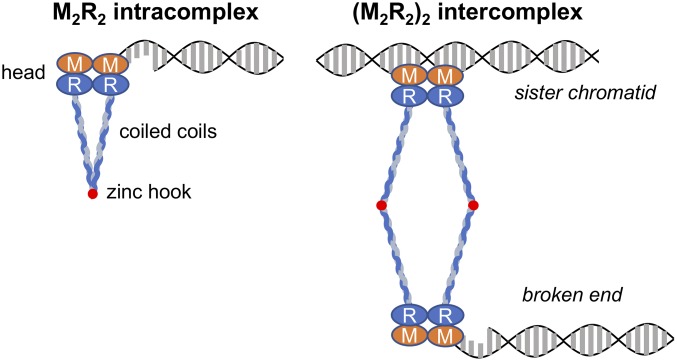
The molecular architecture of the Mre11–Rad50 subcomplex suggests how MRN might tether DNA molecules (9–14). Rad50, which resembles “structural maintenance of chromosomes” proteins, is an ATP-binding cassette (ABC)-type ATPase with Walker A and B ATP-binding motifs at its N and C termini. These domains bind together and are linked by a long antiparallel coiled-coil. Two Mre11 subunits, which contain nuclease active sites, associate with each other and two Rad50 ATPase domains to form a “head” domain, from which protrudes two Rad50 coiled-coils (Fig. 1). The apex of each coiled-coil has a CXXC motif that forms a zinc-mediated “hook” that dimerizes Rad50 molecules. Zinc hook-mediated dimerization within a dimeric assembly forms an M2R2 intracomplex, whereas dimerization between separate dimeric assemblies forms an (M2R2)2 intercomplex (Fig. 1). In principle, the (M2R2)2 intercomplex can coordinate long-range tethering of DNA molecules, for example, between a DSB and homologous sequences in the sister chromatid (Fig. 1). In Saccharomyces cerevisiae, efforts to test the tethering model revealed that mutational elimination of the Rad50 CXXC motif destabilizes the Mre11 complex and phenocopies rad50∆ (12, 15, 16). However, several missense mutations near the CXXC motif ablated Tel1 activity while only partially impairing HR repair in a plasmid-based assay, thus leaving uncertain whether MRN-mediated tethering promotes SCR (15).
Fig. 1.
Mre11–Rad50 complexes. Composition of Mre11–Rad50 complexes and their proposed associations with DSBs and a SC repair template are shown.
Key points of the MRN tethering model remain untested, including the central idea that MRN interacts with the unbroken sister chromatid opposite a DSB. Sequence-directed meganucleases, which have been invaluable for many HR studies, suffer from the inherent limitation of cutting both sister chromatids. A system of efficiently collapsing replication forks at a specific site should overcome this investigative barrier. The fission yeast Schizosaccharomyces pombe uses such a system to switch between mating types (17). The mat1 locus, which contains either plus or minus cassettes, has a stable DNA lesion consisting of a DNA nick or a pair of labile ribonucleotides (18, 19). This single-strand break (SSB) is specific to both site and strand (SI Appendix, Fig. S1A). A replication termination site, located proximal to mat1, ensures that mat1 is always replicated by a replisome moving toward the centromere. The SSB forms on the neosynthesized lagging strand, and in the next cell cycle, leading-strand replication converts the SSB into a seDSB (18, 20). In the homothallic wild-type (WT) h90 strain, the silent donor alleles (mat2P and mat3M) provide templates for HR repair by gene conversion, which can proceed by intrachromosomal synthesis-dependent strand annealing (SDSA) (21) (SI Appendix, Fig. S1B). Selection of the silent donor alleles as repair templates occurs through the targeted recruitment of the Swi2/Swi5 recombination-promoting complex (RPC) to Swi2-dependent recombinational enhancer (SRE) DNA elements at mat2P and mat3M (22–24). The Swi2/Swi5 RPC directly interacts with Rad51 to promote intrachromosomal SDSA, effectively overriding interchromosomal SCR repair. However, in a mutant heterothallic “donorless” mat2,3∆ strain, which lacks the silent donor cassettes, repair occurs very efficiently by interchromosomal SCR (2, 25) (SI Appendix, Fig. S1B). Key recombinases such as Rad51 and Rad52, which are required for both SDSA and SCR, are essential for viability in both “donorplus” h90 (mat2P mat3M) and donorless mat2,3∆ cells. In contrast, Mus81-Eme1 Holliday junction resolvase, which is required to complete SCE, is fully dispensable in h90 but essential in mat2,3∆ cells (2, 26).
In this study, we use the fission yeast mating type switching system to test the MRN tethering hypothesis. Specifically, we determine whether MRN interacts with the unbroken sister chromatid during SCR repair of the broken fork, whether it has resection-independent functions that assist SCR, and whether the Rad50 zinc hook is important for this mechanism of DSB repair.
Results
Rad50 ChIP Assays with the seDSB and Sister Chromatid at a Collapsed Replication Fork.
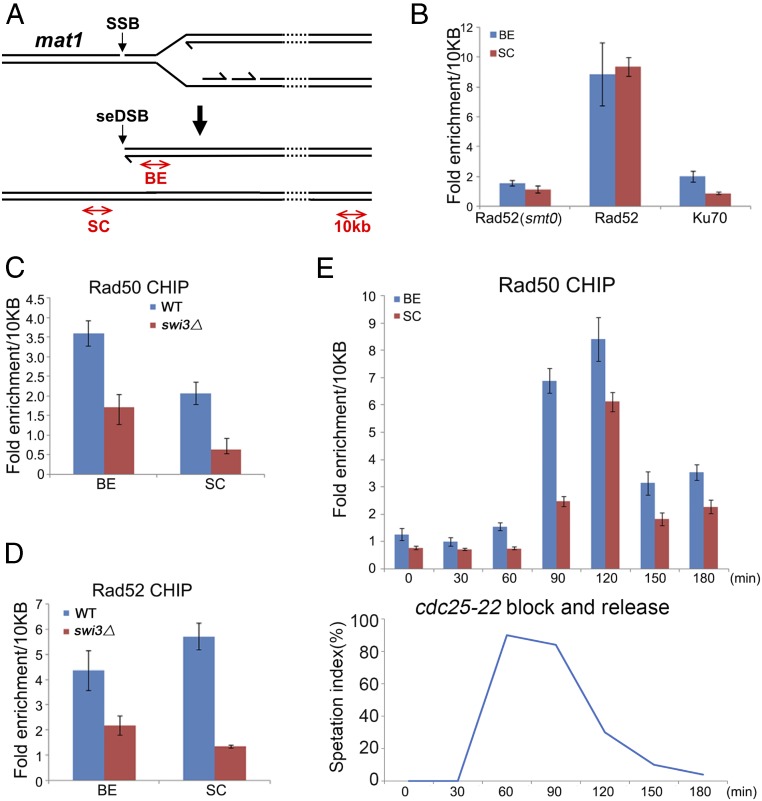
We performed ChIP with mat2,3∆ cells to assess localizations of DNA repair proteins at the mat1 broken fork (2). The “broken end” (BE) primers amplify DNA adjacent to the mat1 seDSB and in the sister chromatid, whereas the “sister chromatid” (SC) primers specifically amplify DNA in the unbroken sister chromatid (Fig. 2A). Signals were normalized to DNA located ∼10 kb from mat1. We reasoned that this region was unlikely to undergo substantial resection, as resection occurs at the rate of ∼4 kb⋅h−1 (6), whereas Mus81 binds recombination intermediates ∼30 min after the mat1 seDSB break is formed (2). Validation experiments with asynchronous cells revealed strong Rad52 ChIP enrichment of both BE and SC DNA (Fig. 2B), which was expected from Rad52’s role in strand invasion and synapsis (27, 28). To further validate these assays, we analyzed a mat1-M smt0 mat2,3∆ strain, which lacks the mat1 SSB that triggers fork collapse (2, 29). As predicted, BE and SC enrichment by Rad52 was abolished in this strain (Fig. 2B and SI Appendix, Fig. S2). The experimental system was further validated by analyzing interactions of the NHEJ factor Ku. As predicted, Ku70 ChIP specifically enriched BE DNA (Fig. 2B). This BE signal was weak relative to Rad52, which is consistent with the very transient association of Ku with DSBs formed in S-phase (6).
Fig. 2.
Rad50 ChIP assays with the seDSB and unbroken sister chromatid at a collapsed replication fork. (A) Diagram of the mat1 locus showing the site of fork collapse and locations of PCR products used for ChIP assays. (B) Rad52 ChIP enriches the mat1 seDSB site (BE) and SC region, while Ku ChIP only enriches the BE region. Assays were performed with Rad52-5FLAG or pKu70-3HA expressed from their endogenous loci in mat2,3∆ backgrounds. The smt0− control strain has a 263-bp deletion at the mat1 locus that prevents SSB formation. (C) Rad50 ChIP enriches the BE and SC sites. ChIP assays were performed with TAP-Rad50 expressed from the endogenous locus. The swi3∆ strain is defective for formation of the SSB. (D) Rad52 ChIP enrichments at the BE and SC sites are decreased in swi3∆ cells. ChIP assays were performed with Rad52-5FLAG. (E) Rad50 ChIP enrichments at the BE and SC sites occur in S-phase. ChIP assays were performed with TAP-Rad50 in a mat2,3∆ cdc25-22 background. Cells were arrested in G2-phase by incubating at 35.5 °C for 4 h and were then shifted to 25 °C to resume cell cycle progression. The septation index correlates with entry into S-phase. Bars indicate mean ± SEM of triplicate samples. Similar results were observed in three (B and E) or two (C and D) independent experiments.
As predicted by tethering models, Rad50 ChIP enriched both BE and SC DNA in mat2,3∆ cells (Fig. 2C). To validate this result, we analyzed swi3∆ mat2,3∆ cells in which SSB formation is defective (30) and found there was only marginal BE enrichment and no SC enrichment by Rad50 ChIP (Fig. 2C). Rad52 ChIP enrichment of BE and SC DNA was similarly reduced in these cells (Fig. 2D). Next, we performed Rad50 ChIP as cell cycle-synchronized mat2,3∆ cultures passed through S-phase. Cells with the temperature-sensitive cdc25-22 mutation were arrested in late G2-phase and then allowed to resume cell cycle progression (2). In these cells, the replication fork collapses at the SSB starting ∼70 min after release from the G2 arrest (2). Consistent with these findings, Rad50 ChIP strongly enriched BE DNA at the 90-min and 120-min time points (Fig. 2E). Rad50 ChIP with the SC primers showed a similar pattern, with the peak signal appearing in the 120-min sample (Fig. 2E).
Rad50 BE and SC Interactions Occur Sequentially.
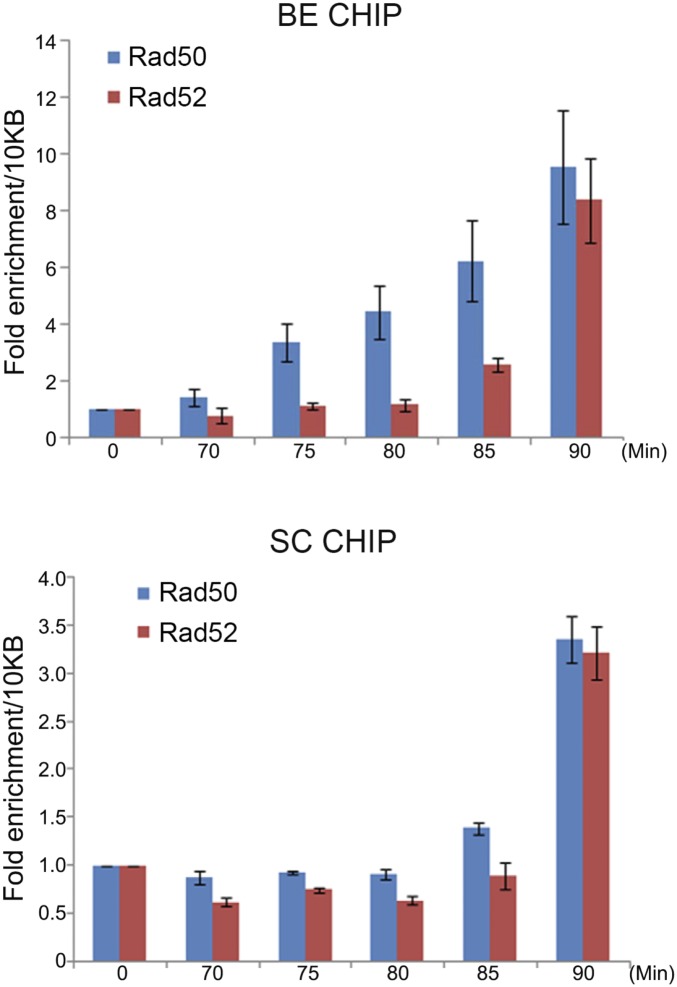
Tethering models predict sequential binding of MRN to the seDSB and sister chromatid. To test this prediction, we repeated the cell cycle ChIP experiment by taking samples at 5-min intervals. The Rad50 BE signal steadily increased in the 70- to 90-min period after release from the G2 arrest (Fig. 3). As predicted, the rise in the Rad50 SC signal was delayed about 10–15 min (Fig. 3). To provide an additional frame of reference, we performed Rad52 ChIP with the same samples. Here, the BE signal first increased at 85 min, followed about 5 min later by a substantial increase in the SC signal (Fig. 3). Thus, strand invasion into the sister chromatid appears to occur very quickly after Rad52 first binds the resected seDSB, and they occur about 15 min after MRN first binds the seDSB. Rad50 and Rad52 SC interactions occur at about the same time, although from these data alone, we cannot conclude whether they are concurrent. In any case, the sequential Rad50 interactions with BE and SC DNA are consistent with MRN coordinating SC interactions during SCR repair.
Fig. 3.
Rad50 BE and SC ChIP interactions occur sequentially. ChIP assays were performed with TAP-Rad50 and Rad52-5FLAG in a mat2,3∆ cdc25-22 background. Samples were taken between 70 and 90 min after release from the cdc25-22 G2 arrest. Bars indicate mean ± SEM of triplicate samples. Similar results were observed in three independent experiments.
An Intrachromosomal Repair Template Partially Alleviates the Requirement for MRN.
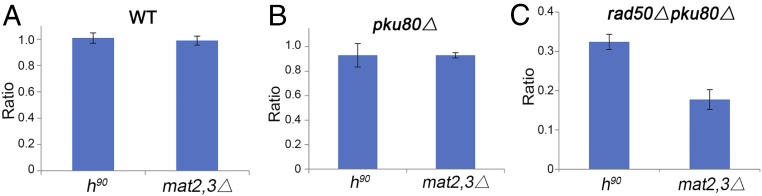
If MRN facilitates SC interactions required for SCR repair, then provision of an intrachromosomal repair template could be expected to alleviate this requirement. In h90 cells, the Swi2/Swi5 complex links Rad51 to the SRE elements at mat2P and mat3M, which bypasses interchromosomal SCR repair in favor of SDSA-mediated gene conversion (22–24). Both mechanisms normally require MRN to initiate resection, but this requirement can be partially suppressed by eliminating Ku, which allows Exo1 to initiate resection (2, 6, 11, 31). Importantly, h90 and mat2,3∆ strains experience equal rates of fork collapse (2, 25), and key recombinases such as Rad51 and Rad52 are essential for viability in both backgrounds (2). Accordingly, we compared the effects of eliminating Rad50 in the two backgrounds by measuring colony size, which, for HR-defective mutants, correlates well with viability (2). As expected, in the WT (rad50+ pku80+) background, colony sizes were equal in h90 and mat2,3∆ strains (Fig. 4A). The same relationship was observed in the pku80∆ background (Fig. 4B). However, in the rad50∆ pku80∆ background, colony size was substantially larger in the h90 strains (Fig. 4C). These data indicate there is an enhanced, resection-independent requirement for MRN in mat2,3∆ cells that repair the mat1 seDSB by interchromosomal SCR, compared with h90 cells that repair this seDSB by Swi2/Swi5-directed SDSA.
Fig. 4.
Provision of an intrachromosomal repair template partially suppresses the mat1 seDSB repair defect in rad50∆ pku80∆ cells. Colonies of the indicated genotypes were obtained by tetrad dissection of rad50∆ pku80∆ h− crossed to h90 or mat2,3∆ cells. Colony size (growth) was measured and normalized to WT h− as described in Materials and Methods. (A) Unimpaired growth of h90 and mat2,3∆ strains in the WT (rad50+ pku80+) background. (B) Equal growth of h90 and mat2,3∆ strains in the pku80∆ background indicates that intrachromosomal SDSA (h90) repair and interchromosomal SCR (mat2,3∆) repair of the mat1 collapsed fork are equally efficient in the absence of Ku. (C) Improved growth of h90 versus mat2,3∆ strains in the rad50∆ pku80∆ background indicates that the presence of the intrachromosomal repair template in h90 cells partially alleviates the requirement for Rad50 to repair the mat1 seDSB. Bars are mean ± SEM (n ≥ 3).
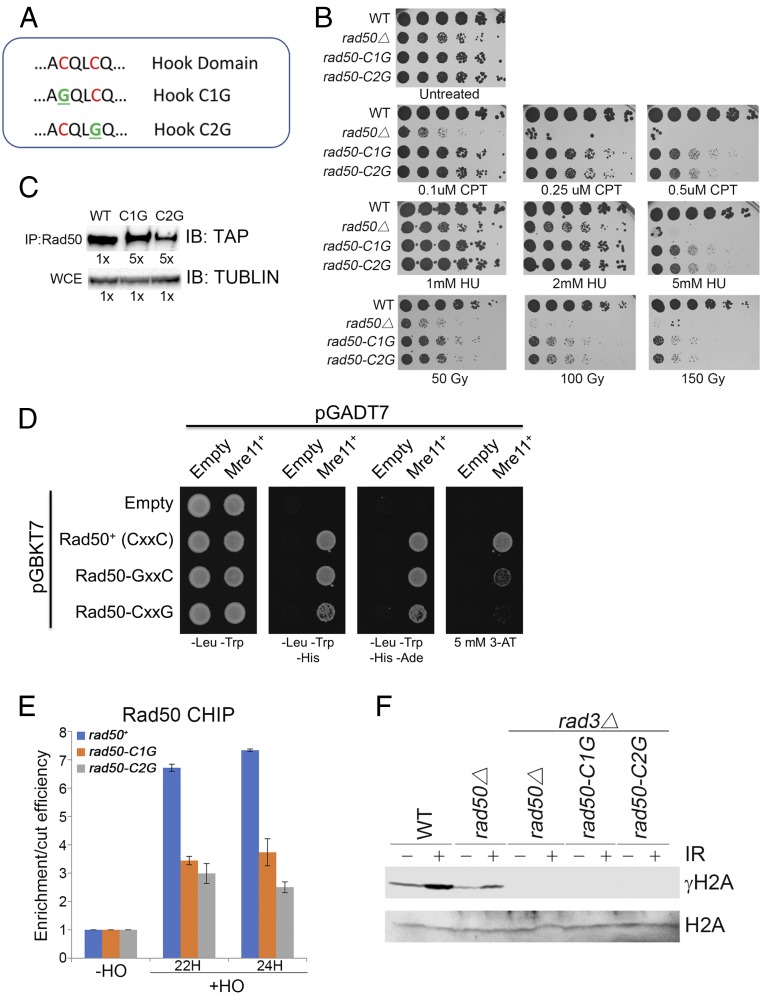
Rad50 Zinc Hook Is Critical for SCR.
The Rad50 zinc hook, which is the linchpin of the (M2R2)2 intercomplex, should be crucial for MRN-mediated tethering (9–14). S. cerevisiae studies established that the zinc-coordinating cysteines in the CXXC hook are essential for MRN complex stability and functions, whereas alterations of flanking residues can partially destabilize the hook interface while retaining substantial DSB repair activities (12, 15, 16). To provide another evolutionary perspective, we constructed S. pombe strains with glycine substitutions of either cysteine (C1G or C2G) in the CXXC motif (Fig. 5A). The rad50-C1G and rad50-C2G mutants retained partial resistance to several DNA damaging agents, including camptothecin, hydroxyurea, and ionizing radiation (IR) (Fig. 5B). These sensitivities might be partly explained by Rad50 instability, as immunoblotting revealed substantial reductions of Rad50 in both mutants (Fig. 5C). These effects made Mre11/Rad50 coimmunoprecipitations impractical; however, yeast two-hybrid assays indicated that the mutants retained weaken interactions with Mre11 (Fig. 5D). By ChIP, Rad50 in rad50-C1G and rad50-C2G cells was able to immunoprecipitate to a DSB created by HO endonuclease, although the signal was reduced compared with WT (Fig. 5E). However, the MRN-dependent phosphorylation of histone H2A by Tel1 was abolished in rad50-C1G and rad50-C2G backgrounds (Fig. 5F).
Fig. 5.
Analysis of Rad50 hook mutants. (A) Sequence alignment of the central portion of the Rad50 hook domain showing the conserved CXXC motif. The mutants are highlighted in green. (B) The rad50-C1G and rad50-C2G mutants retain partial resistance to DNA damaging agents. Fivefold serial dilutions of cells were plated on the indicated concentrations of camptothecin (CPT) or hydroxyurea (HU), or exposed to the indicated doses of IR before plating. (C) Reductions of Rad50 abundance in the rad50-C1G and rad50-C2G mutants. IP, immunoprecipitation; TUBLIN, tubulin. (Top) Immunoblot (IB) of immunoprecipitated TAP-Rad50. 5×, fivefold greater sample loading in mutants relative to WT. (Bottom) Antitubulin IB of whole-cell extracts (WCE). (D) Yeast two-hybrid interactions of WT and mutant Rad50 with Mre11. The leftmost panel (-Leu -Trp) selects for the reporter plasmids. The other panels indicate yeast two-hybrid interactions, with increasing stringency occurring right to left. (E) Rad50 hook mutants ChIP to a HO-induced DSB. Experiments were performed in ctp1∆ backgrounds to eliminate resection (6). Derepression of the thiamine (B1)-repressible nmt1 promoter that controls HO expression requires about 18–20 h of growth in −B1 media (media in which expression of HO endonuclease is induced). Samples were taken at 22 and 24 h. The ratio is shown for Rad50 enrichment normalized to cut efficiency as determined by PCR amplification across the HO cut site (6). The ratio in the presence of B1 was set to 1. Bars indicate mean ± SEM of triplicate samples. Similar results were observed in two independent experiments. (F) Zinc coordination by Rad50 is essential for Tel1 activity. Immunoblot of γ-H2A and total histone H2A with or without treatment with 90 Gy of IR. Assays were performed in rad3∆ backgrounds because both Rad3/ATR and Tel1/ATM phosphorylate histone H2A at DSBs (41).
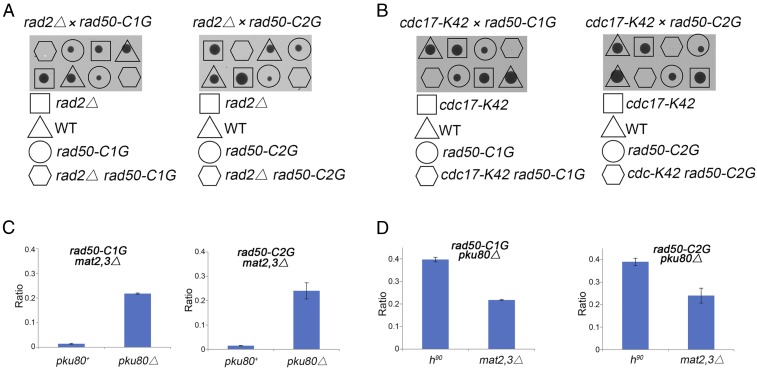
Mutants lacking the FEN-1 nuclease ortholog Rad2, or which are partially defective for the DNA ligase I ortholog Cdc17, accumulate SSBs that trigger replication fork collapse (32). Tetrad analysis revealed that the rad50-C1G and rad50-C2G mutations are lethal in these backgrounds, indicating that seDSB repair by SCR is critically impaired by these mutations (Fig. 6 A and B). We found that the rad50-C1G and rad50-C2G mutations were synthetic lethal with mat2,3∆, but this lethality was suppressed by pku80∆ (Fig. 6C). The growth of these mutants was significantly better in the h90 pku80∆ background compared with mat2,3∆ pku80∆ (Fig. 6D). Collectively, these data indicate that zinc coordination by Rad50 is critical for SCR repair.
Fig. 6.
Rad50 zinc hook is critical for SCR. (A) Tetrad analysis reveals synthetic lethality of rad50-C1G and rad50-C2G with mutants lacking Rad2, which is the FEN-1 nuclease ortholog. (B) Tetrad analysis reveals synthetic lethality of rad50-C1G and rad50-C2G with cdc17-K42. The DNA ligase I ortholog is cdc17. Spores were germinated at 25 °C, which is the permissive temperature for cdc17-K42. (C) The rad50-C1G and rad50-C2G mutations are synthetic lethal with mat2,3∆. This lethality is suppressed by pku80∆. (D) Improved growth of h90 versus mat2,3∆ strains in the rad50-C1G pku80∆ and rad50-C2G pku80∆ backgrounds indicates that the presence of the intrachromosomal repair template in h90 cells partially alleviates the requirement for the Rad50 zinc hook to repair the mat1 collapsed fork. Colony size (growth) was measured and normalized to WT h− as described in Materials and Methods. Bars are mean ± SEM (n ≥ 3).
Ctp1 Sequential ChIP Assays with the seDSB and Sister Chromatid.
Structural studies revealed that Ctp1 assembles into a tetramer with the potential for multivalent interactions with the MRN complex and DNA (33, 34). Biochemical assays confirmed a DNA-bridging activity for Ctp1 (7, 33). To provide additional insights, we analyzed Ctp1 localization at the mat1 broken fork using cell cycle-synchronized cells. We detected sequential BE and SC interactions in a pattern that closely correlated with the Rad50 ChIP studies (SI Appendix, Fig. S3). These data suggest that a Ctp1-mediated bridging function might be involved in SCR.
Discussion
The most important finding in this study is that MRN sequentially interacts with the seDSB and sister chromatid during SCR repair of a collapsed replication fork. These data support a key prediction of the MRN-mediated tethering hypothesis. However, in this model, the SC engagement of MRN is envisioned to precede Rad52’s association with the resected seDSB, whereas we observed by ChIP that Rad50 and Rad52 immunoprecipitated with the sister chromatid at approximately the same time. These unexpected results suggest that the SC associations of MRN and Rad52 might be mechanistically connected; for example, MRN might assist formation or stabilization of Rad51/Rad52-mediated synaptic junctions. These data cannot exclude the possibility that MRN is passively brought to the sister chromatid through a sustained affinity with the resected seDSB. However, this picture is inconsistent with microscopic studies of IR-induced DNA repair foci in S. cerevisiae, in which colocalization analyses indicated that Mre11 and Rad52 have mutually exclusive interactions with DSBs (35). Instances of colocalization were suggested to involve aggregation of DSBs in repair centers that are at different stages of processing. Repair centers are unlikely to explain our data because seDSB formation is genetically programmed and the viability of HR mutants suggests that spontaneous collapse of replication forks is relatively infrequent. If MRN and Rad52 have mutually exclusive interactions at seDSBs, then MRN’s association with the sister chromatid might involve a subset of slowly repaired breaks. This explanation is consistent with the persistence of a subset of Mre11 foci and unresected DSBs in S. cerevisiae studies (35–37). In either case, the physical proximity of MRN to the sister chromatid places it in a position to structurally assist SCR.
In a pku80∆ background, we found that provision of a Swi2/Swi5-directed SDSA repair template reduces the requirement for MRN to tolerate the mat1 seDSB, implying that a nonnucleolytic function of MRN is more critical for interchromosomal SCR. These data cannot exclude the hypothetical possibility that Swi2/Swi5-directed repair compensates for the partially impaired resection catalyzed by Exo1 in the rad50∆ pku80∆ background (6). However, arguing against this possibility, we observed that h90 and mat2,3∆ strains grow equally well in the absence of Exo1 (SI Appendix, Fig. S4A), which is required for efficient long-range resection (6). Moreover, the h90 genotype is not advantageous relative to mat2,3∆ in cells with the hypomorphic ctp1-6 and ctp1-25 mutations, which severely increase DNA damage sensitivity in the absence of Exo1 (32) (SI Appendix, Fig. S4B). These data are most straightforwardly explained by MRN promoting SCR by tethering the seDSB to the sister chromatid, but they do not exclude other nonnucleolytic functions of MRN as being important.
The Rad50 zinc hook is the linchpin of the (M2R2)2 intercomplex (12). We found that mutating critical zinc-coordinating cysteines does not fully inactivate the MRN DNA repair functions or block the ability of MRN DNA to bind a DSB, and yet these mutants are profoundly defective for repair of the mat1 broken fork. A more extensive analysis will be needed to fully understand the consequences of these mutations; however, at this stage, the data support the hypothesis that zinc hook-mediated coordination of the (M2R2)2 intercomplex is critical for SCR repair. Finally, our studies indicate that Ctp1 also associates with the unbroken sister chromatid at the mat1 collapsed replication fork, suggesting that the multivalent DNA-bridging activity of tetrameric Ctp1 detected in vitro (33) may promote SC interactions during SCR repair.
Materials and Methods
General Methods.
Standard methods were used for constructing and culturing strains and synchronizing cultures (38). Strains are listed in SI Appendix, Table S1. Yeast extract, glucose, and supplements (YES) or versions of Edinburgh minimal media were used for all experiments. DNA damage sensitivity assays were performed by using fivefold serial dilutions of log-phase cells plated on YES agar. Colony size comparisons were performed from tetrad dissections Colonies were photographed after 3–6 d of growth at 30 °C on YES media. ImageJ (NIH) was used to determine colony size. All measurements were normalized to WT h− colonies derived from the same cross. Genetic crosses involving self-fertile h90 were performed at a 100:1 mating ratio (h−/h90). Plasmids and methods for Mre11/Rad50 yeast two-hybrid assays have been described elsewhere (11).
ChIP Assays.
ChIP experiments were performed as described elsewhere (2, 6, 11). Rabbit IgG conjugated to tosylactivated magnetic beads (Dynal; Invitrogen) was used to precipitate tandem affinity purification (TAP)-Rad50 and Ctp1-TAP, while mouse anti-FLAG (Sigma) antibody and mouse anti-HA (Roche Applied Science) conjugated to anti-mouse magnetic beads (Dynal; Invitrogen) were used to precipitate Rad52-FLAG and Ku70-HA, respectively. The BE primers amplify a 188-bp DNA fragment centered 503 bp from the break site. The SC primers amplify a 142-bp DNA fragment centered 274 bp from the break site. The “10-kb” primers amplify a 90-bp DNA fragment located about 9,600 bp from the break site. The ChIP experiments using HO endonuclease expressed from the thiamine-repressible nmt1 promoter were performed as described elsewhere (11). Primers are listed in SI Appendix, Table S2.
Western Blots.
Whole-cell extracts were prepared from exponentially growing cells, followed by standard lysis. For Rad50 blots, proteins were resolved in 8% Tris-Glycine Gels (Life Technologies). Membranes were probed with peroxidase antiperoxidase (PAP) (P1291; Sigma) antibodies. The phosphorylated histone H2A (γ-H2A) blots were performed as described elsewhere (39). Membranes were probed with phosphospecific γ-H2A antibody (40), courtesy of Christophe Redon, National Cancer Institute, Bethesda, MD, and total histone H2A antibody (39235; Active Motif).
Supplementary Material
Acknowledgments
We thank Nick Boddy for helpful discussions and Tamara Johnson for preliminary experiments. This work was supported by NIH Grants GM059447, CA077325, and CA117638.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1808189115/-/DCSupplemental.
References
- 1.Aguilera A, García-Muse T. Causes of genome instability. Annu Rev Genet. 2013;47:1–32. doi: 10.1146/annurev-genet-111212-133232. [DOI] [PubMed] [Google Scholar]
- 2.Roseaulin L, et al. Mus81 is essential for sister chromatid recombination at broken replication forks. EMBO J. 2008;27:1378–1387. doi: 10.1038/emboj.2008.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mayle R, et al. DNA REPAIR. Mus81 and converging forks limit the mutagenicity of replication fork breakage. Science. 2015;349:742–747. doi: 10.1126/science.aaa8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kowalczykowski SC. An overview of the molecular mechanisms of recombinational DNA repair. Cold Spring Harb Perspect Biol. 2015;7:a016410. doi: 10.1101/cshperspect.a016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Symington LS, Rothstein R, Lisby M. Mechanisms and regulation of mitotic recombination in Saccharomyces cerevisiae. Genetics. 2014;198:795–835. doi: 10.1534/genetics.114.166140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Langerak P, Mejia-Ramirez E, Limbo O, Russell P. Release of Ku and MRN from DNA ends by Mre11 nuclease activity and Ctp1 is required for homologous recombination repair of double-strand breaks. PLoS Genet. 2011;7:e1002271. doi: 10.1371/journal.pgen.1002271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andres SN, Williams RS. CtIP/Ctp1/Sae2, molecular form fit for function. DNA Repair (Amst) 2017;56:109–117. doi: 10.1016/j.dnarep.2017.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gobbini E, Cassani C, Villa M, Bonetti D, Longhese MP. Functions and regulation of the MRX complex at DNA double-strand breaks. Microb Cell. 2016;3:329–337. doi: 10.15698/mic2016.08.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stracker TH, Petrini JH. The MRE11 complex: Starting from the ends. Nat Rev Mol Cell Biol. 2011;12:90–103. doi: 10.1038/nrm3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Jager M, et al. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol Cell. 2001;8:1129–1135. doi: 10.1016/s1097-2765(01)00381-1. [DOI] [PubMed] [Google Scholar]
- 11.Williams RS, et al. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hopfner KP, et al. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418:562–566. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- 13.Moreno-Herrero F, et al. Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature. 2005;437:440–443. doi: 10.1038/nature03927. [DOI] [PubMed] [Google Scholar]
- 14.Schiller CB, Seifert FU, Linke-Winnebeck C, Hopfner KP. Structural studies of DNA end detection and resection in homologous recombination. Cold Spring Harb Perspect Biol. 2014;6:a017962. doi: 10.1101/cshperspect.a017962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hohl M, et al. Interdependence of the rad50 hook and globular domain functions. Mol Cell. 2015;57:479–491. doi: 10.1016/j.molcel.2014.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiltzius JJ, Hohl M, Fleming JC, Petrini JH. The Rad50 hook domain is a critical determinant of Mre11 complex functions. Nat Struct Mol Biol. 2005;12:403–407. doi: 10.1038/nsmb928. [DOI] [PubMed] [Google Scholar]
- 17.Arcangioli B. A site- and strand-specific DNA break confers asymmetric switching potential in fission yeast. EMBO J. 1998;17:4503–4510. doi: 10.1093/emboj/17.15.4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaykov A, Arcangioli B. A programmed strand-specific and modified nick in S. pombe constitutes a novel type of chromosomal imprint. Curr Biol. 2004;14:1924–1928. doi: 10.1016/j.cub.2004.10.026. [DOI] [PubMed] [Google Scholar]
- 19.Vengrova S, Dalgaard JZ. The wild-type Schizosaccharomyces pombe mat1 imprint consists of two ribonucleotides. EMBO Rep. 2006;7:59–65. doi: 10.1038/sj.embor.7400576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holmes AM, Kaykov A, Arcangioli B. Molecular and cellular dissection of mating-type switching steps in Schizosaccharomyces pombe. Mol Cell Biol. 2005;25:303–311. doi: 10.1128/MCB.25.1.303-311.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamada-Inagawa T, Klar AJ, Dalgaard JZ. Schizosaccharomyces pombe switches mating type by the synthesis-dependent strand-annealing mechanism. Genetics. 2007;177:255–265. doi: 10.1534/genetics.107.076315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akamatsu Y, Dziadkowiec D, Ikeguchi M, Shinagawa H, Iwasaki H. Two different Swi5-containing protein complexes are involved in mating-type switching and recombination repair in fission yeast. Proc Natl Acad Sci USA. 2003;100:15770–15775. doi: 10.1073/pnas.2632890100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jia S, Yamada T, Grewal SI. Heterochromatin regulates cell type-specific long-range chromatin interactions essential for directed recombination. Cell. 2004;119:469–480. doi: 10.1016/j.cell.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 24.Jakočiūnas T, Holm LR, Verhein-Hansen J, Trusina A, Thon G. Two portable recombination enhancers direct donor choice in fission yeast heterochromatin. PLoS Genet. 2013;9:e1003762. doi: 10.1371/journal.pgen.1003762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klar AJ, Miglio LM. Initiation of meiotic recombination by double-strand DNA breaks in S. pombe. Cell. 1986;46:725–731. doi: 10.1016/0092-8674(86)90348-x. [DOI] [PubMed] [Google Scholar]
- 26.Boddy MN, et al. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107:537–548. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- 27.Sugawara N, Wang X, Haber JE. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol Cell. 2003;12:209–219. doi: 10.1016/s1097-2765(03)00269-7. [DOI] [PubMed] [Google Scholar]
- 28.Wolner B, van Komen S, Sung P, Peterson CL. Recruitment of the recombinational repair machinery to a DNA double-strand break in yeast. Mol Cell. 2003;12:221–232. doi: 10.1016/s1097-2765(03)00242-9. [DOI] [PubMed] [Google Scholar]
- 29.Styrkársdóttir U, Egel R, Nielsen O. The smt-0 mutation which abolishes mating-type switching in fission yeast is a deletion. Curr Genet. 1993;23:184–186. doi: 10.1007/BF00352020. [DOI] [PubMed] [Google Scholar]
- 30.Dalgaard JZ, Klar AJ. swi1 and swi3 perform imprinting, pausing, and termination of DNA replication in S. pombe. Cell. 2000;102:745–751. doi: 10.1016/s0092-8674(00)00063-5. [DOI] [PubMed] [Google Scholar]
- 31.Tomita K, et al. Competition between the Rad50 complex and the Ku heterodimer reveals a role for Exo1 in processing double-strand breaks but not telomeres. Mol Cell Biol. 2003;23:5186–5197. doi: 10.1128/MCB.23.15.5186-5197.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jensen KL, Russell P. Ctp1-dependent clipping and resection of DNA double-strand breaks by Mre11 endonuclease complex are not genetically separable. Nucleic Acids Res. 2016;44:8241–8249. doi: 10.1093/nar/gkw557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andres SN, et al. Tetrameric Ctp1 coordinates DNA binding and DNA bridging in DNA double-strand-break repair. Nat Struct Mol Biol. 2015;22:158–166. doi: 10.1038/nsmb.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davies OR, et al. CtIP tetramer assembly is required for DNA-end resection and repair. Nat Struct Mol Biol. 2015;22:150–157. doi: 10.1038/nsmb.2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: Spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 36.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Forsburg SL, Rhind N. Basic methods for fission yeast. Yeast. 2006;23:173–183. doi: 10.1002/yea.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Limbo O, Porter-Goff ME, Rhind N, Russell P. Mre11 nuclease activity and Ctp1 regulate Chk1 activation by Rad3ATR and Tel1ATM checkpoint kinases at double-strand breaks. Mol Cell Biol. 2011;31:573–583. doi: 10.1128/MCB.00994-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakamura TM, Du LL, Redon C, Russell P. Histone H2A phosphorylation controls Crb2 recruitment at DNA breaks, maintains checkpoint arrest, and influences DNA repair in fission yeast. Mol Cell Biol. 2004;24:6215–6230. doi: 10.1128/MCB.24.14.6215-6230.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.