Significance
Cardiac muscle contraction is powered by ATP hydrolysis during cycles of interaction between myosin-containing thick filaments and actin-containing thin filaments. This generates force in the cardiac muscle necessary for pumping blood through the body. Mutations in myosin alter this force generation leading to hypercontractility and hypertrophic cardiomyopathy (HCM). An energy-conserving, super relaxed state (SRX) of myosin, which has a very low ATPase activity, has previously been described in muscle fibers. Destabilization of the SRX has been proposed to be a chief cause of HCM. This work sheds light on the biochemical and molecular nature of SRX and demonstrates the mechanism of action of mavacamten, a cardiac inhibitor in phase 2 clinical trials. Mavacamten exerts its effects primarily by stabilizing the SRX of β-cardiac myosin.
Keywords: myosin, super relaxed state, interacting heads motif, cardiac inhibitor, mavacamten
Abstract
Mutations in β-cardiac myosin, the predominant motor protein for human heart contraction, can alter power output and cause cardiomyopathy. However, measurements of the intrinsic force, velocity, and ATPase activity of myosin have not provided a consistent mechanism to link mutations to muscle pathology. An alternative model posits that mutations in myosin affect the stability of a sequestered, super relaxed state (SRX) of the protein with very slow ATP hydrolysis and thereby change the number of myosin heads accessible to actin. Here we show that purified human β-cardiac myosin exists partly in an SRX and may in part correspond to a folded-back conformation of myosin heads observed in muscle fibers around the thick filament backbone. Mutations that cause hypertrophic cardiomyopathy destabilize this state, while the small molecule mavacamten promotes it. These findings provide a biochemical and structural link between the genetics and physiology of cardiomyopathy with implications for therapeutic strategies.
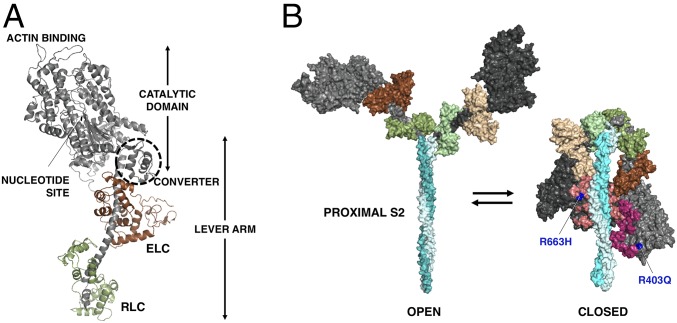
Muscle myosin is a hexamer consisting of two myosin heavy chains and two sets of light chains, the essential light chain (ELC) and the regulatory light chain (RLC). The myosin molecule can be divided into two parts, heavy meromyosin (HMM), which consists of two globular heads and the first ∼40% of the coiled-coil tail, and light meromyosin (LMM), which consists of the C-terminal ∼60% of the coiled-coil tail. LMM self-assembles creating the shaft of the myosin thick filament found in sarcomeres of the muscle. HMM can be further divided into subfragment 1 (S1), which is the globular head of the myosin that serves as the motor domain (1), and subfragment 2 (S2). S1 houses the ATP- and actin-binding sites followed by an ELC- and RLC-bound α-helix (lever arm) (Fig. 1A). S1 heads are arranged on the thick filament backbone in muscle in a quasihelical fashion. There is a 14.3-nm vertical spacing between two adjacent myosin molecules on the filament with a true repeat of 42.9 nm.
Fig. 1.
Human β-cardiac myosin structural models. (A) PyMol homology model of human β-cardiac myosin S1 in the prestroke state showing its various domains. The ELC is in brown, and the RLC is in green. Modeling was done as described previously (30, 45). (B) Structural models of the open on-state and the closed IHM off-state of human β-cardiac myosin. The motor domains are black (blocked head) and dark gray (free head), the ELCs are in shades of brown, and the RLCs are in shades of green. The positions of R403Q and R663H (blue) are shown in the IHM relative to the mesa residues (45) (blocked head: light pink; free head: dark pink). Modeling was done as described previously (30, 45).
Importantly, intramolecular interactions favoring a folded state of myosin (Fig. 1B) have been observed in both purified myosin preparations (2–6) and in myosin thick filaments isolated from striated muscle (7–10). Named the “interacting-heads motif” (IHM) (9), this structure has been proposed to be related to a super relaxed state (SRX) of muscle, defined as a state that has a much reduced basal ATPase rate of ∼0.003 s−1 (11). The SRX was initially discovered in rabbit skeletal muscle and thereafter was also seen in rabbit cardiac, tarantula exoskeletal, and mouse and human cardiac fibers (12–15). An appealing hypothesis is that the SRX state is related to the IHM myosin state in which the myosin S1 heads are interacting with one another and are folded back onto their own coiled-coil S2 tail (13, 16–19). Since the SRX state has been described only in fibers, it is possible that other sarcomeric proteins in the vicinity of the myosin are essential for establishing this state. There have been no studies that directly demonstrate a structural corollary of the SRX state with purified systems. Here we demonstrate biochemically that a decrease of basal ATPase to the levels seen in the SRX in fibers can be observed with purified human β-cardiac myosin alone.
Hypertrophic cardiomyopathy (HCM) is an autosomal dominant inherited disease of heart muscle (20–22) characterized by hypercontractility and subsequent hypertrophy of the ventricular walls. Systolic performance of the heart is preserved or even increased, but relaxation capacity is diminished. Patients with HCM are at increased risk of heart failure, atrial fibrillation, stroke, and sudden cardiac death. Current pharmacological management options are not disease specific and do not address the underlying HCM disease mechanism. It has been hypothesized that a small molecule that binds directly to myosin and normalizes the hypercontractility of the sarcomere may interrupt the development of downstream pathology (such as hypertrophy, fibrosis, and clinical complications) seen in this disease (23). Mavacamten (formerly MYK-461; MyoKardia Inc.) is an oral allosteric modulator of cardiac myosin and causes dose-dependent reductions in left ventricular contractility in healthy volunteers and HCM patients (24). This investigational drug is in phase 2 clinical trials and acts directly by interacting with the human β-cardiac myosin and normalizing its power output. Mavacamten was identified in a screen for actin-activated myosin ATPase inhibitors, and it lengthens the total cycle time (tc) of the ATPase cycle (25). This results in a reduction of the duty ratio (ts/tc) and therefore of the ensemble force, since Fens = fint⋅Na⋅duty ratio, where fint is the intrinsic force of a myosin molecule and Na is the total number of myosin heads in the sarcomere that are functionally accessible for interaction with actin. Here we show that mavacamten reduces Na by stabilizing purified human β-cardiac myosin in a state with a very slow release of bound nucleotide (SRX, an off-state of myosin). Using the same purified myosin, we then show by EM that mavacamtem also stabilizes a folded-back structural state of the protein that is reminiscent of the IHM.
The first HCM-causing mutation in β-cardiac myosin to be identified was R403Q (20). As with other HCM mutations, the R403Q mutation results in hypercontractility of the muscle (26, 27). We provide evidence here that the R403Q mutation results in a shift in the equilibrium toward an on-state of myosin and away from an off-state. These may correspond to the open and closed states shown in Fig. 1B. Such a shift in equilibrium would result in more heads being available for interaction with actin and the hypercontractility seen clinically. We see a similar phenotype in human cardiac samples with the HCM-causing R663H mutation. We further demonstrate that mavacamten reverses the destabilization of the SRX caused by these mutations.
A related and complementary body of work is described in Rohde et al. (28).
Results
Purified Human β-Cardiac Myosin Containing the Proximal S2 Portion of Its Coiled-Coil Tail Is in an Ionic Strength-Dependent SRX.
To test the hypothesis that the SRX observed in muscle fibers can be observed with purified β-cardiac myosin, we studied the single-turnover kinetics of myosin’s basal ATPase cycle for three purified human β-cardiac myosin constructs: 25-hep HMM (two-headed with the first 25 heptads of proximal S2), 2-hep HMM (two-headed with the first two heptads of proximal S2), and short S1 (sS1; single-headed with no S2 and truncated immediately after the ELC-binding domain, Fig. 1A). These constructs followed on the work of Trybus et al. (29) who showed that smooth muscle myosin with at least 15 heptads of the S2 region can achieve complete RLC phosphorylation regulation if it has a leucine zipper after the S2 region. A 25-hep HMM without a leucine zipper was not able to form a dimer completely and did not show complete regulation. However, a 25-hep HMM with a leucine zipper could form a stable dimer and showed complete regulation. Thus, the smooth muscle work showed that a stable dimer and at least 15 heptad repeats are essential for complete regulation of two-headed smooth muscle myosin (29). Trybus et al. (29) also showed that the 2-hep HMM of smooth muscle myosin can form a dimer only if a leucine zipper is present in the construct, and they confirmed this by native gels and EM images. Our 25-hep HMM construct has a GCN4 leucine zipper to ensure dimerization and shows ATPase regulation by RLC phosphorylation, as reported in earlier work (30). The ATPase activity of our 2-hep HMM, which again is stabilized as a dimer by insertion of a GCN4 domain, is not regulated by RLC phosphorylation (30). Both constructs form dimers as judged by a native acrylamide gel of 2-hep HMM (SI Appendix, Fig. S1) and EM images of 25-hep HMM (see Fig. 3G).
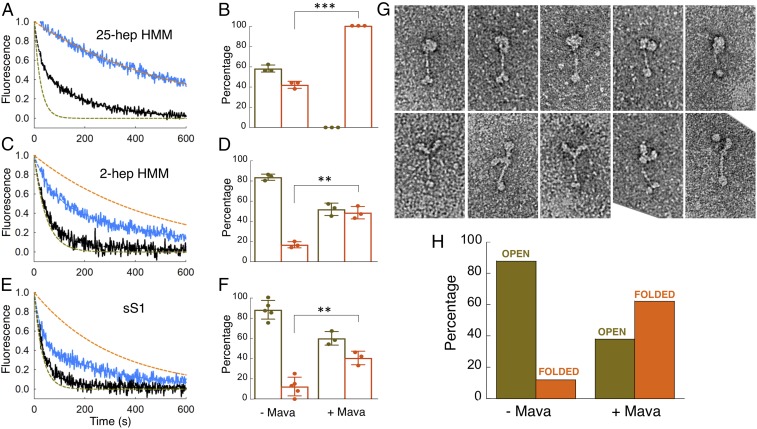
Fig. 3.
MANT-nucleotide release rates and percentage of folded states for purified human β-cardiac myosin fragments with and without mavacamten. (A, C, and E) Fluorescence decays of MANT-nucleotide release from 25-hep HMM, 2-hep HMM, and sS1 in 25 mM KAc with (blue curve) and without (black curve) mavacamten. Fitting the traces with mavacamten to exponential equations yielded the DRX and SRX rates of 0.014 and 0.0021 s−1 for 2-hep HMM and 0.035 and 0.0032 s−1 for sS1. The SRX rate for 25-hep HMM was 0.0017 s−1. The simulated orange and olive green dashed curves act as references for the single exponential fits of slow and fast phases, respectively, that derive from fitting the solid black data curves with the best two exponential fits. (B, D, and F) Percentage of myosin heads in the SRX (orange) versus DRX (olive green) states with and without mavacamten calculated from the double-exponential fits of the fluorescence decays corresponding to the MANT-nucleotide release from 25-hep HMM, 2-hep HMM, and sS1 at 25 mM KAc. The 25-hep and 2-hep data with and without mavacamten are from three measurements each. The measurements come from three individual protein preparations done on different days. sS1 data have three measurements with mavacamten and five measurements without mavacamten. The measurements come from three individual protein preparations done on different days. (G) Representative single-particle negatively stained EM images of the open and closed forms of the 25-hep HMM. Both the open and closed molecules were selected from a sample containing mavacamten cross-linked to 25-hep HMM at 25 mM KAc. (H) Percentage of 25-hep HMM molecules in the closed or open form in the presence and absence of mavacamten. A total of 622 and 183 molecules were counted for the with-mavacamtem and without-mavacamten samples, respectively. Ten micromolars mavacamten was used for single-turnover assays and 10 µM MYK-3046 (cross-linkable mavacamten) was used for EM imaging. Error bars denote SD. **P ≤ 0.01; ***P ≤ 0.001.
The basal ATPase rates of the three constructs were all within the range of 0.01–0.03 s−1 (SI Appendix, Fig. S2), consistent with high levels of actin activation of these human β-cardiac myosin constructs [about 100-fold activation by actin (30, 31)]. SRX is defined as a rate of nucleotide release from the myosin head that is even slower, about 0.003 s−1 (11).
To test the percentage of heads in the SRX for the three constructs, we adapted the single-nucleotide turnover assay, which is typically measured in a stopped-flow apparatus, to a plate-based measurement. The slow rates of normal myosin basal ATPase and the SRX-based nucleotide release make them amenable to a plate-based measurement. Moreover, owing to the difficulties in expressing high amounts of human cardiac proteins, the plate-based assay is an apt choice for this kind of experiment. This assay measured the fluorescent nucleotide release rates by loading the myosin heads with 2′/3′-O-(N-Methyl-anthraniloyl) (MANT)-ATP and then chasing with excess unlabeled ATP, as had been done in skinned fibers previously (11). As the MANT nucleotide is released from the myosin, its fluorescence decreases (11).
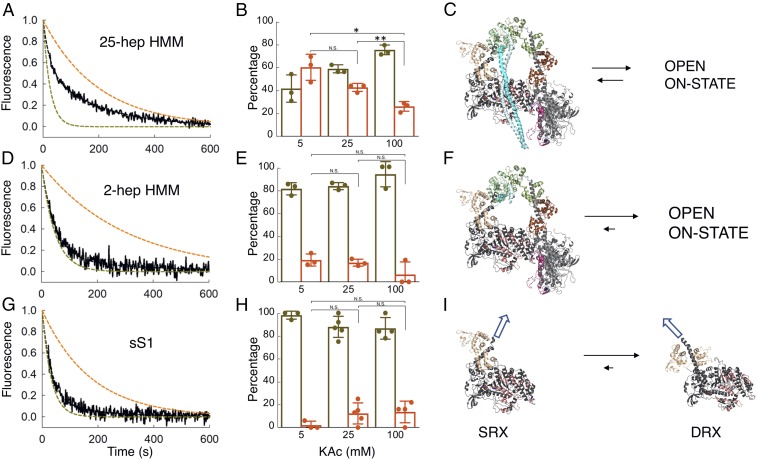
The decay rate of MANT-nucleotide fluorescence for 25-hep HMM was fit well by two exponential rate constants, one representing a basal ATPase rate of heads in a presumed open state (disordered relaxed state, DRX) of ∼0.03 s−1 and the other representing an SRX rate of ∼0.003 s−1 (Fig. 2A). In 100 mM potassium acetate (KAc), the amplitudes were 74 ± 2% of the basal DRX rate and 26 ± 2% of the SRX rate (Fig. 2B). This SRX level measured for the 25-hep HMM did not vary significantly from preparation to preparation, and we hypothesized that this fraction corresponded to myosin heads that are folded back onto their own proximal S2 tail, possibly in an IHM-like structural state (Fig. 2C). Consistent with this hypothesis, the fraction of myosin heads in the SRX increased from 26 ± 2% to 42 ± 2% (P ≤ 0.01) when the KAc was decreased from 100 mM to 25 mM and further increased from 26 ± 2% to 59 ± 7% (P ≤ 0.05) when the salt was decreased from 100 mM to 5 mM KAc (Fig. 2B). This would be expected since the IHM structure is thought to be held together largely by charge–charge interactions (9, 30, 32–34). The amplitudes of the fast (74 ± 2%) and slow (26 ± 2%) phases observed with the human β-cardiac 25-hep HMM at 100 mM KAc closely match the amplitudes observed in SRX experiments done with rabbit cardiac fibers (∼70% fast and ∼30% slow at 120 mM KAc) (13). There was no demonstrable photobleaching of the MANT nucleotide over the single-turnover data-collection time scales (SI Appendix, Fig. S3).
Fig. 2.
MANT-nucleotide release rates for purified human β-cardiac myosin fragments. (A, D, and G) Fluorescence decays of MANT-nucleotide release from 25-hep HMM, 2-hep HMM, and sS1, respectively, in 25 mM KAc (solid black curves). Fitting the traces to a double-exponential equation yielded the DRX and SRX rates of 0.040 and 0.0050 s−1 for 25-hep HMM, 0.021 and 0.0033 s−1 for 2-hep HMM, and 0.031 and 0.0057 s−1 for sS1. The simulated single-exponential orange dashed curves are the SRX rate (0.003–0.005 s−1), and the simulated single-exponential dashed olive green curves are the DRX rate (0.02–0.04 s−1). The simulated orange and olive green dashed curves act as references for the single exponential fits of slow and fast phases, respectively, that derive from fitting the solid black data curves with the best two exponential fits. Thus, the data (black lines) were fit with a combination of these two single exponentials. (B, E, and H) Percentage of myosin heads in the SRX (orange) versus DRX (olive green) states calculated from the amplitudes of the double-exponential fits of the fluorescence decays corresponding to the MANT-nucleotide release from the 25-hep HMM, 2-hep HMM, and sS1 constructs at various KAc concentrations. The 25-hep and 2-hep data are from three measurements each. The measurements come from three individual protein preparations done on different days. sS1 data have three, five, and four measurements at 5, 25, and 100 mM KAc, respectively. The measurements come from three individual protein preparations done on different days. (C) Homology model of folded-back human β-cardiac 25-hep HMM (MS03, https://spudlab.stanford.edu/homology-models/, showing only 13 heptad repeats of the S2) in equilibrium with an open on-state. (F) The 2-hep HMM (from MS03) in equilibrium with an open on-state. (I) The homology-modeled blocked head from the MS03 structure in equilibrium with the sS1 DRX structure from HBCprestrokeS1, downloadable at https://spudlab.stanford.edu/homology-models/. These two structures represent the two potential relaxed ATP- and/or ADP.Pi-bound states of the human β-cardiac myosin heads. The white arrows illustrate the different directions in which the light chain-binding regions point in the SRX vs. DRX states, where all alignments in C, F, and I are the same. Error bars denote SD. N.S., not significant, P > 0.05; *P ≤ 0.05; **P ≤ 0.01.
It is important to emphasize that SRX is defined as an off-state of myosin with a very low ATPase turnover rate and that our data do not establish that the off-state we are measuring is due to the IHM structural state. In this regard, the 2-hep HMM, which contains only two heptad repeats of the S2 tail, still showed 19 ± 3% SRX in 5 mM KAc (Fig. 2 D and E), but the difference in fractions of SRX at different KAc concentrations was not statistically significant (P > 0.05). It is possible that some IHM-like structure is still formed and that blocked head interactions with the free head help stabilize this level of SRX in the 2-hep HMM (Fig. 2F). However, our finding of a similar level of SRX (∼15%) in our sS1 preparations (Fig. 2 G and H) suggests an alternative explanation. It is important to note that myosin molecules in solution in any nucleotide state are likely to exist in a number of closely related conformational states in equilibrium. One possible explanation of this result is that ∼15% of the population of independent sS1 molecules in solution exist in a conformation that is similar to that stabilized in a folded off-state structure (Fig. 2I) in equilibrium with DRX heads (prestroke heads having ATP or ADP.Pi bound). We hypothesize that in the context of the 25-hep HMM this conformation favors the interactions with the proximal S2 and other head–head interactions that define an SRX-IHM state. Thus, our working hypothesis is that myosin heads can occupy an SRX without entering a folded state directly, but the SRX conformation of myosin heads is further stabilized by intramolecular interactions in an SRX-IHM. This idea gains strength by the mavacamten experiments described below.
To ensure that we were not missing any fast phase in our plate-based measurements, we performed the similar measurement with sS1 in a stopped-flow apparatus. The rates and amplitudes obtained were very similar to our plate-based measurements (SI Appendix, Fig. S4).
These results suggest that purified myosin can adopt a biochemical SRX that is stabilized by the S2 tail, especially under low ionic strength conditions that favor electrostatic interactions. These results directly link a folded structural state (possibly the IHM state seen by others) with the biochemical SRX using purified human β-cardiac myosin.
The Cardiac Myosin Inhibitor Mavacamten Stabilizes the SRX Folded-Back State of Purified Human β-Cardiac Myosin.
Using the MANT-nucleotide release rates as a measure of the level of SRX in the population, we examined whether the cardiac myosin inhibitor mavacamten has any effect on the distribution of ATPase states in the population of purified human β-cardiac myosin constructs. Mavacamten was selected on the basis that it inhibits the actin-activated activity of sS1 (25), but the mechanism of that inhibition is unknown. Here we show that it has a profound effect on stabilizing the SRX. Mavacamten induced a 20-fold reduction in the basal ATPase rate of the 25-hep HMM (SI Appendix, Fig. S2A). In the single-nucleotide turnover assay, in the presence of 10 µM mavacamten, the MANT-nucleotide release rates from 25-hep HMM at 25 mM KAc were fit well by a single exponential with a rate constant reflecting the SRX (SI Appendix, Table S1). At both 25 mM KAc (Fig. 3 A and B) and 100 mM KAc (SI Appendix, Fig. S5 and Table S2) essentially ∼100% of the myosin heads were in the SRX in the presence of mavacamten. In the absence of mavacamten, the fraction of 25-hep HMM in the SRX was ∼42% at 25 mM KAc (Figs. 2B and 3B) and ∼26% at 100 mM KAc (Fig. 2B). The effect of mavacamten on 2-hep HMM was less pronounced (Fig. 3 C and D), consistent with a role for the proximal S2 in stabilizing the SRX. However, 40–50% SRX was observed in the presence of mavacamten, suggesting either that the heads can interact with one another to some extent in the absence of proximal S2 and this is stabilized by mavacamten binding (shifting the equilibrium to the left in Fig. 2F) or that mavacamten stabilizes the SRX conformation of sS1 in equilibrium with DRX heads (e.g., shifting the equilibrium to the left in Fig. 2I).
Indeed, mavacamten increases the SRX levels of sS1 to nearly that of the 2-hep HMM (Fig. 3F vs. Fig. 3D). Mavacamten increased the SRX levels of 2-hep HMM to 48 ± 4% and sS1 to 40 ± 4%, which are not significantly different (P > 0.05). This is also reflected in a reduction of the basal myosin ATPase of the 2-hep HMM and sS1 constructs by the drug (SI Appendix, Fig. S2 B and C). Thus, we think it likely that the binding of mavacamten to sS1 alone stabilizes an SRX off-state, which is favored to fold into a more stabilized off-state structure, possibly the IHM state (SI Appendix, Fig. S6). Kawas et al. (35) reported no difference in the IC50 of mavacamten for S1, HMM, or myofibrils of bovine or human background and reported it to be in the range of 0.5–1 µM. The concentration of mavacamten used in the single-turnover assays was 10-fold higher than these reported IC50s. Thus, these assays were performed in the saturating regime of mavacamten, and we believe there is no artifact arising from the differential binding of mavacamten to different myosin constructs.
We used EM to provide evidence that the SRX observed with 25-hep HMM corresponds to folded-back (closed) heads. Fig. 3G shows representative images of folded-head structures (Upper) and open-head structures (Lower). After coupling the 25-hep HMM to a mavacamten analog containing a photoactivatable cross-linker, folded-head structures dominated the images, while the reverse was true in the absence of the mavacamten derivative (DMSO control) (Fig. 3H). Less than 20% of the myosin molecules were in a folded-head state in the absence of mavacamten, and ∼60% were in a folded-head state in the presence of mavacamten (Fig. 3H). The discrepancy between the fraction of closed-state myosin heads in the EM study (∼60%) and the fraction of SRX in the ATP turnover experiments (100%) is most likely due to well-known perturbation effects of the EM approach. Thus, interactions between the carbon substrate and the myosin tends to destabilize the folded-back structural state (6; see also refs. 36–40). Some groups have used a low concentration of glutaraldehyde to stabilize the folded state before grid preparation; we did not use glutaraldehyde or other cross-linking agents in our EM studies. These results more firmly link a folded-head state, possibly the IHM structural state, with a stabilized SRX.
The Cardiac Myosin Inhibitor Mavacamten Stabilizes an SRX Folded State of Porcine and Human Cardiac Muscle Fibers.
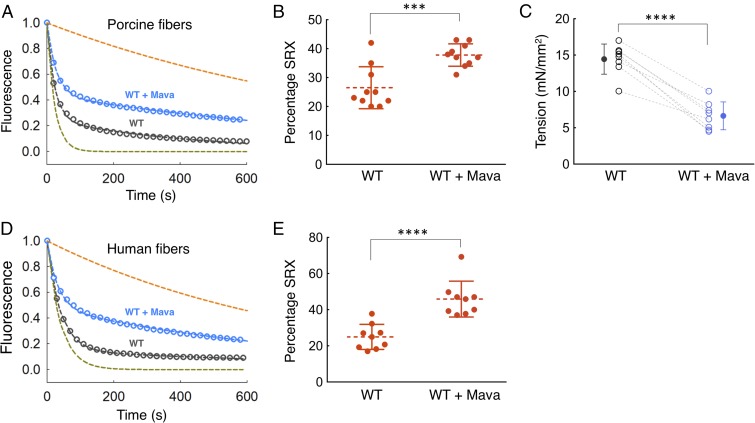
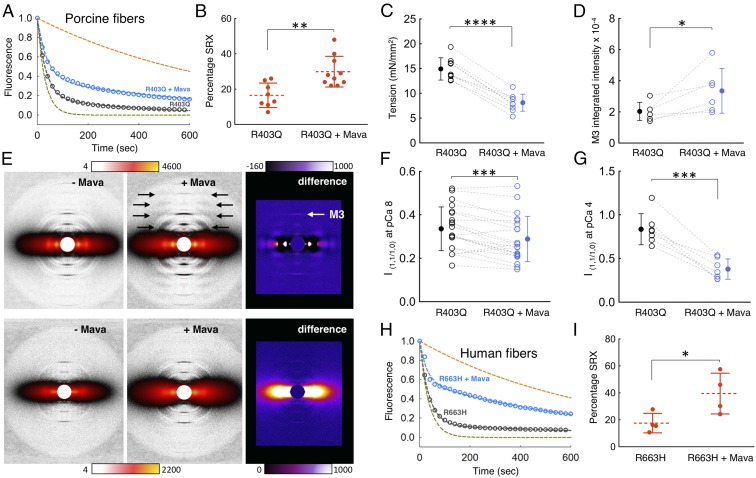
To examine whether the effects of mavacamten on purified myosin translate to an increase in the level of SRX in cardiac fibers, we carried out MANT-nucleotide release analyses on skinned fibers from relaxed minipig cardiac muscle. Mavacamten increased the percentage of the SRX in the porcine fibers from 26 ± 2% to 38 ± 1% (P ≤ 0.001) (Fig. 4 A and B and SI Appendix, Table S3 for rates). There was a corresponding decrease in tension measured in skinned porcine cardiac muscle fibers exposed to mavacamten from 14.4 ± 0.7 mN/mm2 to 6.6 ± 0.7 mN/mm2 [negative logarithm of calcium concentration (pCa) 5.8] (P ≤ 0.0001) (Fig. 4C). A similar mavacamten-mediated SRX stabilization effect was observed for WT human cardiac fibers. Mavacamten increased the SRX in the human fibers from 25 ± 2% to 46 ± 3% (P ≤ 0.0001) (Fig. 4 D and E and see SI Appendix, Table S4 for rates). Photobleaching of the MANT nucleotides was measured for the fiber-based SRX experiments and was found to be negligible (∼0.5%) (12).
Fig. 4.
Effect of mavacamten on WT porcine and human cardiac fibers. Blue and dark gray curves denote experimental conditions with and without mavacamten, respectively. (A) SRX measurements with skinned cardiac fibers from WT pigs in the presence and absence of mavacamten. Fitting the traces to a double-exponential equation yielded the DRX and SRX rates of 0.043 and 0.0023 s−1 for porcine cardiac fibers in the absence of mavacamten and 0.036 and 0.0010 s−1 for porcine cardiac fibers in the presence of mavacamten. The simulated orange and green dashed curves act as references for the single exponential fits of slow and fast phases, respectively, that derive from fitting the solid blue and black data curves with the best two exponential fits. (B) Percentage of SRX detected in the porcine cardiac fibers with and without mavacamten. n = 10 unpaired measurements. (C) Maximum tension measurement in the skinned porcine cardiac fibers in the presence (1 µM) and absence of mavacamten. One pair of measurements was made per fiber; n = 8 paired measurements. Dashed lines represent a paired dataset with open circles. Solid circles denote the mean. (D) SRX measurements with skinned cardiac fibers from human hearts in the presence and absence of mavacamten. Fitting the traces to a double-exponential equation yielded the DRX and SRX rates of 0.046 and 0.0058 s−1 for human cardiac fibers in the absence of mavacamten and 0.057 and 0.0032 s−1 for human cardiac fibers in the presence of mavacamten. (E) The percentage of SRX in human cardiac fibers in the presence and absence of mavacamten. Fifty micromolars mavacamten was used for the fiber SRX assays. n = 9 unpaired measurements. Error bars for all measurements denote SD. ***P ≤ 0.001; ****P ≤ 0.0001.
To examine the spatial organization of the myosin heads, we used low-angle X-ray diffraction of porcine cardiac muscle fibers, which reveals information about the degree of order of the myosin heads along the myosin thick filaments as well as their proximity to the actin thin filaments. A representative low-angle X-ray diffraction image of a porcine cardiac muscle fiber shows the typical quasihelical diffraction pattern known for striated muscle (Fig. 5).
Fig. 5.
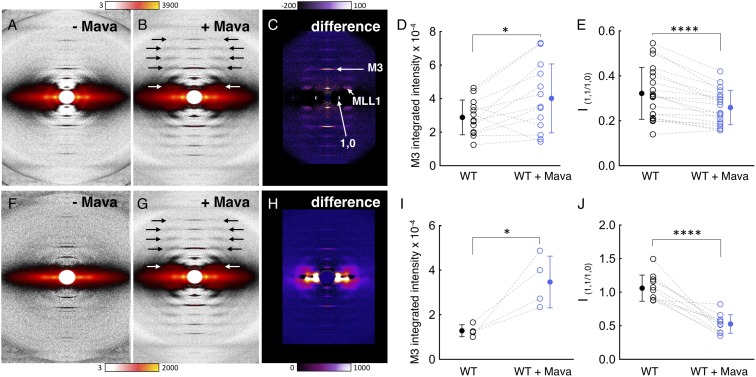
Effect of mavacamten on ordering of myosin heads onto the thick filaments in WT porcine fibers. One pair of measurements was made per fiber for all fiber diffraction experiments. Solid circles denote the mean. (A–C) Diffraction patterns of WT porcine fibers under relaxing conditions (pCa 8) without and with mavacamten and the difference in intensities. Color scales indicate increases in intensity. (D) Mavacamten increases the intensity of the meridional reflection M3 under relaxing conditions; n = 13 paired measurements. (E) Mavacamten decreases the equatorial intensity ratio, I1,1/1,0, under relaxing conditions. The intensity ratio (I1,1/I1,0) is the ratio of the integrated intensity of the 1,1 equatorial reflection arising from density in the plane containing both thick and thin filaments to that of the 1,0 equatorial reflection arising from density in the plane containing only thick filaments (72). Changes in the ratio I1,1/I1,0 are commonly used as a measure of shifts of mass, presumably cross-bridges, from the region of the thick filament to that of the thin filament; n = 20 paired measurements. (F–H) Diffraction patterns of WT porcine fibers under contracting conditions (pCa 4) without and with mavacamten and the difference in intensities. Color scales indicate increases in intensity. (I) Mavacamten increases the intensity of the meridional reflection M3 under contracting conditions; n = 4 paired measurements. (J) Mavacamten decreases the equatorial intensity ratio, I1,1/1,0, under contracting conditions. n = 11 paired measurements. In D, E, I, and J, blue and black colors denote experimental conditions with and without mavacamten, respectively. Dotted lines represent a paired dataset with open circles. Error bars for all measurements denote SD. *P ≤ 0.05; ****P ≤ 0.0001.
Mavacamten causes a dramatic ordering of myosin heads along the backbone of the thick filaments as observed by the substantial increase in the myosin-based helical layer line reflections for both relaxed (Fig. 5A vs. Fig. 5B, arrows) and contracting (Fig. 5F vs. Fig. 5G, arrows) porcine muscle fibers. The first myosin layer line (MLL1) intensity at 43 nm (Fig. 5 B and G, white arrows) increased about 40% with mavacamten treatment (MLL1+mava/MLL1−mava = 1.37 ± 0.13). While it is not known whether there is a correlation between the amount of ordered heads along the thick filament backbone and the ATPase kinetic state of the myosin heads, if we assume that only myosin heads in the SRX give rise to layer lines, a 40% increase in MLL1 intensity would imply 17 ± 6% more heads in the SRX. A prominent reflection is the M3 reflection on the meridian, which reports on the 14.3-nm repeat of the myosin heads along the longitudinal shaft of the thick filament in the relaxed state (Fig. 5C). The higher the intensity of this reflection, the more ordered are the heads along this 14.3-nm repeat. When the diffraction pattern of the relaxed muscle in the absence of mavacamten (Fig. 5A) is subtracted from the diffraction pattern in the presence of mavacamten (Fig. 5B), the difference (Fig. 5C) shows that the intensity of the M3 reflection is significantly increased by the addition of mavacamten (P ≤ 0.05). This is quantified in Fig. 5D where the results from 13 fibers are shown. An even more dramatic increase in the M3 reflection is seen when mavacamten is added to contracting fibers (P ≤ 0.05) (Fig. 5 F–I). Thus, for both relaxed and contracting fibers, mavacamten increases the quasihelical order of myosin heads on the thick filament backbone.
The 1,1 and 1,0 reflections on the equator provide information on the number of myosin heads that have moved away from the myosin thick filaments toward the actin thin filaments. The 1,1 lattice planes include the actin filament densities, while the 1,0 lattice planes include only the myosin thick filament densities. Thus, an increase in the intensity of the 1,0 reflection shows that more myosin heads have been sequestered near the shaft of the thick filaments, and the I1,1/I1,0 intensity ratio is reduced. As seen in Fig. 5 C and H, mavacamten causes an increase in the 1,0 reflection intensity. Furthermore, Fig. 5 E and J show that the I1,1/I1,0 intensity ratio is reduced (P ≤ 0.0001) by mavacamten in both relaxing and contracting conditions, suggesting that it causes more heads to become sequestered against the LMM backbone of the myosin thick filament.
In the discussion above, the SRX is defined by the fraction of MANT nucleotides that are released slowly from the fiber in a chase experiment. However, not all MANT nucleotides bind specifically to myosin in the fibers, and this complicates interpretation of the SRX signal. Hooijman et al. (13) showed that only 48 ± 3% of the MANT nucleotides are bound specifically to myosin in rabbit cardiac cells. Thus, when the nonspecific fraction is subtracted from the signal the calculated population of the myosin heads in the SRX is approximately twice the population calculated from the observed SRX signal. We will call this the “SRXcalc.”
Thus, the population of the SRXcalc in porcine cardiac cells goes from 54 ± 4% to 79 ± 2% upon the addition of mavacamten. This is an increase of 25%, compatible with the 17% increase observed in the small-angle X-ray fiber diffraction experiment. The DRXcalc, calculated as 1 − SRXcalc, goes from 46 ± 4% to 21 ± 8%, a decrease of 41%. This is almost exactly equal to the 46% decrease in tension, showing that, as expected, tension is proportional to the fraction of disordered myosin heads.
The level of SRXcalc in cells without mavacamten (54%) is similar to that of 25-hep HMM in low ionic strength (Fig. 2B), despite the higher ionic strength (120 mM KAc) used in the tissue experiments. This suggests that the SRX in cells is more stable than in purified protein preparations, consistent with the greater number of interactions that stabilize the IHM, including those with the thick filament backbone and additional proteins such as MyBP-C.
In both porcine and human cardiac cells SRXcalc goes to ∼80–95% upon the addition of mavacamten. This is similar to that with purified 25-hep HMM, which is ∼100% in both 25 mM KAc (Fig. 3 A and B) and 100 mM KAc (SI Appendix, Fig. S5).
These structural results, combined with the studies with purified human β-cardiac myosin described above, are consistent with mavacamten shifting the on–off state equilibrium toward the off-SRX folded-back state, which in muscle fibers is more ordered near and along the backbone of the myosin thick filament.
The Hypertrophic Cardiomyopathy Mutations R403Q and R663H Reduce the Level of SRX in Cardiac Muscle Fibers.
To better characterize the effects of mutations that cause HCM on cardiac structure and function, we generated a large-animal model of HCM in the Yucatan minipig (Methods and SI Appendix, Fig. S7). We generated a minipig (designated as “R403Q pigs”) that carries the heterozygous MYH7 R403Q mutation, which has been well characterized in mice and humans (20, 41).
Analysis of RNA from R403Q hearts at 2 mo of age confirmed the expression of mutant transcript at a ratio of ∼1:1 [47.9 ± 0.6 (mutant):52.1 ± 0.6 (WT)]. This ratio was similar throughout the left ventricle (samples from anteroseptum, basal septum, basal free wall, and apex). This is consistent with semiquantitative mass spectrometry (MS Bioworks, LLC; see Methods) performed on left ventricular biopsy samples of R403Q hearts at 3 mo of age, which identified R403Q and WT peptides at an ∼1:1 ratio [53% (mutant): 47% (WT)] (SI Appendix, Table S5). The R403Q piglets rapidly developed an in vivo phenotype with hypercontractility and hypertrophy consistent with HCM (42–44).
When SRX levels in cardiac fibers from the R403Q pigs were compared with those in fibers from the WT pigs, strikingly, the R403Q mutation caused a significant decrease in the percentage of SRX in the porcine fibers, from 26 ± 2% to 16 ± 2% (P ≤ 0.01) (compare Fig. 6 A and B with Fig. 4 A and B and see SI Appendix, Table S3 for rates). This result supports the hypothesis that myosin HCM mutations cause clinical hypercontractility by shifting the equilibrium from the closed off-state (SRX) to the open on-state in which the heads are free to interact with actin (30, 32, 45–47). The addition of mavacamten to the R403Q fibers returned the percentage of SRX back to the normal WT level (30 ± 3%, P ≤ 0.01) (Fig. 6B) and also reduced tension from 14.9 ± 0.7 mN/mm2 to 8.1 ± 0.6 mN/mm2 (pCa 5.8, P ≤ 0.0001) (Fig. 6C). The physiologically relevant pCa range for tension development in the heart is from ∼10−7 M Ca2+ to ∼10−6 M Ca2+ (red shaded area in SI Appendix, Fig. S8) (48), where the R403Q mutation induces hypercontractility and mavacamten reduces it toward normal (light blue to dark blue shaded area in SI Appendix, Fig. S8).
Fig. 6.
Mavacamten stabilizes the SRX in porcine and human cardiac fibers carrying HCM mutations. Blue and dark gray denote experimental conditions with and without mavacamten, respectively. (A) SRX measurement with skinned cardiac fibers from R403Q pigs in the presence and absence of mavacamten. Fitting the traces to a double-exponential equation yielded the DRX and SRX rates of 0.030 and 0.0024 s−1 for R403Q porcine cardiac fibers in the absence of mavacamten and 0.033 and 0.0025 s−1 for R403Q porcine cardiac fibers in the presence of mavacamten. The green and orange curves are simulated single-exponential fits of fast and slow phase, respectively. (B) Percentage of SRX detected in the R403Q porcine cardiac fibers with (n = 8 measurements) and without (n = 10 measurements) mavacamten. One measurement was made per fiber. (C) Maximum tension measurement in the skinned cardiac fibers from R403Q pigs in the presence (1 µM) and absence of mavacamten. One paired measurement was made per fiber; n = 9 paired measurements. Dotted lines represent a paired dataset with open circles. Solid circles denote the mean. (D) Mavacamten increases the intensity of the meridional reflection M3 under relaxing conditions. One paired measurement was made per fiber for all fiber diffraction experiments. Dotted lines represent a paired dataset with open circles. Solid circles denote the mean. n = 6 paired measurements. (E, Upper) Diffraction patterns of R403Q porcine fibers under relaxing conditions (pCa 8) without and with mavacamten and the difference in intensities. (Lower) Diffraction patterns of R403Q porcine fibers under contracting conditions (pCa 4) without and with mavacamten and the difference in intensities. Color scales indicate increases in intensity. (F) Mavacamten decreases the equatorial intensity ratio, I1,1/1,0, of R403Q porcine fibers under relaxing conditions; n = 21 paired measurements. (G) Mavacamten decreases the equatorial intensity ratio, I1,1/1,0, of R403Q porcine fibers under contracting conditions; n = 7 paired measurements. (H) SRX measurements with skinned cardiac fibers from R663H human hearts in the presence and absence of mavacamten. Fitting the traces to a double-exponential equation yielded the DRX and SRX rates of 0.050 and 0.0038 s−1 for R663H human cardiac fibers in the absence of mavacamten and 0.060 and 0.0037 s−1 for R663H human cardiac fibers in the presence of mavacamten. (I) The percentage of SRX with and without mavacamten in the R663H human cardiac fibers. Fifty micromolars mavacamten was used for the fiber SRX and diffraction experiments. One measurement was made per fiber; n = 4 unpaired measurements. Error bars for all measurements denote the SD. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001.
Treatment of R403Q skinned porcine fibers with mavacamten caused an increase in ordering of myosin heads along the backbone of the myosin thick filament, as judged by an increase in the myosin-based helical layer lines (Fig. 6E, black arrows) and the intensity of the M3 reflection (Fig. 6E, white arrow), consistent with a shift in equilibrium toward the myosin SRX folded-back off-state (Fig. 6 D and E). This mavacamten-dependent increase in the intensity of the M3 reflection was observed under relaxing conditions. Such an intensity change was difficult to quantify for the R403Q fibers under contracting conditions. Mavacamten also caused a significant decrease in the I1,1/I1,0 intensity ratio of the R403Q fibers (P ≤ 0.001) (Fig. 6 E–G), further supporting a shift in equilibrium toward the myosin SRX folded-back off-state more closely associated with the backbone of the myosin thick filament. This effect was quantified for the R403Q fibers under both relaxing and contracting conditions.
We also studied the impact of mavacamten on the SRX in a known human pathogenic HCM mutation, MYH7 R663H, with fibers isolated from human cardiac tissue. R663H had an SRX level of 18 ± 4%, and the addition of mavacamten to these fibers restored the SRX levels to 40 ± 8% (P ≤ 0.05), which is similar to the WT level (Fig. 6 H and I and see SI Appendix, Table S4 for rates). SRX in WT human cardiac fibers was 25 ± 2%, which was reduced to 18 ± 4% with the R663H mutation (P = 0.1). These changes are similar to those observed with the R403Q porcine samples and suggest that HCM mutations in myosin destabilize SRX in both pigs and humans.
Discussion
Over the past five decades our understanding of muscle physiology, biochemistry, and biophysics has greatly advanced. While not complete, we have a good understanding of the way many of the most important sarcomeric proteins function and their roles in muscle contractility. Now focus is shifting to understanding the molecular basis of disease states of these proteins and modern therapeutic approaches that can be applied to them. This study focuses on HCM mutations that cause hypercontractility of the heart and a small molecule, mavacamten, which resets this hypercontractility to normal.
Recent studies using human β-cardiac myosin support the view that many, if not most, myosin missense HCM mutations cause the hypercontractility observed clinically by shifting an equilibrium between a sequestered off-state of myosin heads to their on-state now able to interact with actin (30–32, 45, 46, 49–51). The majority of these mutations are highly localized to three surfaces of the myosin molecule: the converter domain (Fig. 1A) (32, 50), a relatively flat surface named the “myosin mesa” (Fig. 1B) (46), and the proximal part of S2 that interacts with the S1 heads in the IHM (Fig. 1B) (30–32, 45, 49–51).
While equating the folded-back structural SRX state studied here with the IHM state described by others will require further structural studies, the realization that the myosin mesa of the two S1 heads in the IHM are in fact cradling the proximal S2 in the IHM structural state (Fig. 1B) was striking (30, 31), as were the observations described above that the hot spots for HCM mutations all lie in locations that would potentially weaken an IHM structure. The mesa is highly enriched in HCM mutations in positively charged Arg residues (46), whereas the proximal S2 is highly enriched in HCM mutations in negatively charged Glu and Asp residues (50), and the converter HCM mutations are all in a position to potentially disrupt the IHM at a primary head–head interaction site (PHHIS) involving a particular surface of the blocked head of the IHM and the converter of the free head (30, 32, 51). We conclude that it is a reasonable hypothesis that myosin missense HCM mutations weaken an SRX folded-back state, resulting in more functionally accessible myosin heads for actin interaction, thus causing the hypercontractility observed for HCM clinically.
In support of this model, several positively charged mesa HCM residues—R249Q, H251N, and R453C—significantly weaken the binding of proximal S2 to sS1 in biochemical experiments with purified myosin fragments, and D906G on the proximal S2 that is near the interface with the mesa in the human β-cardiac myosin folded-back homology models has the same effect (30, 31). Interestingly, R403Q has no effect on the affinity of S2 for sS1 (30), while here we show that it reduces the amount of SRX in muscle fibers.
So what is R403Q doing at the molecular level? R403Q is in a unique position on the myosin surface, residing at the tip of a pyramid of the IHM structure formed by three interacting faces: the actin-binding face, the mesa, and the PHHIS (30, 45). R403Q does not seem to have a significant effect on actin interaction (52), but it could be weakening the complex by weakening the interaction of the blocked head PHHIS with the free head converter (30, 45). Arg403 is also in a position to possibly bind to part of MyBP-C (30, 45), and the R403Q mutation may weaken a MyBP-C–SRX complex and in this way shift the equilibrium away from the SRX in fibers.
Similarly, it is not known which of these various interactions the mesa HCM mutation R663H is affecting, but we show here that it reduces the level of SRX in fibers. Biochemical experiments involving the effects of R403Q and R663H on the interactions between these human β-cardiac sarcomeric protein players will be necessary to elucidate the molecular mechanism by which these mutations reduce the SRX in the fiber.
The small molecule mavacamten, a cardiac inhibitor in phase 2 clinical trials, decreases contractility and suppresses the development of hypertrophy and fibrosis in HCM mouse models (25). How does this molecule affect the detailed structure of the myosin active site? The presence of ATP or ADP.Pi analogs in the active site of myosin causes the switch elements to adopt a closed state, which in turn has been inferred to stabilize the helical order of the thick filament (53–55). Blebbistatin, a small-molecule ATPase inhibitor, stabilizes a closed switch-2 state and also has been shown by EM (56) and fluorescence-based studies (57) to stabilize the IHM state of myosin. Conversely, omecamtiv mecarbil, a cardiac activator in phase 3 clinical trials, has been shown to stabilize the open state of myosin on the cardiac thick filament (57). Along similar lines, mavacamten has been previously shown to reduce the basal release rates of ADP and Pi (35), possibly by holding the switch elements in a closed state, which ultimately stabilizes the myosin heads in an SRX folded-back state. The high-resolution detailed structural basis of formation of this SRX folded-back state, presumed to be an SRX-IHM state, is an intriguing open question in the field. It is crucial now to obtain a high-resolution crystallographic or cryo-EM structure of the SRX folded-back state.
Here we provide evidence at the biochemical level that the cardiac SRX can occur with nothing more than an HMM-like molecule containing two S1 heads and the proximal part of the coiled-coil tail and that the level of SRX decreases with increasing ionic strength, consistent with many charge–charge interactions stabilizing the IHM. Our work demonstrates that the tail region of myosin is necessary to fully stabilize its SRX conformation, consistent with an SRX folded-back structure. Our 2-hep HMM, however, shows SRX behavior without the proximal tail, as does our sS1 construct, which shows a 15–20% population of SRX that increases to ∼50% SRX in the presence of mavacamten. We think it likely, therefore, that a small part of the population of independent sS1 molecules in solution exist in a conformation that is similar to that stabilized in the folded-back structure (SI Appendix, Fig. S6) and the binding of mavacamten shifts the equilibrium of sS1 heads in solution toward the structure of the heads in the SRX folded-back state. Structural studies will be necessary to assess this possibility further.
We hypothesize that the sS1 SRX conformation, stabilized by mavacamten, will have its light chain-binding helix tilted further in the prestroke direction (a pre-prestroke state) than normally achieved by the relaxed DRX prestroke state (SI Appendix, Fig. S6). Although a different conformation of the myosin head, this change in the direction of lever arm orientation is the same as expected for that of the actin-bound force-producing myosin head under load in contracting muscle, where the light chain-bound lever arm is pulled backward by the load and nucleotide release is slowed (58). The sS1 structure of Dominguez et al. (59), which is in an extreme pre-prestroke state, has indeed been suggested to fit into the blocked head of the IHM structure (3) and fits the MS03 homology-modeled structure from the J.A.S. laboratory (30, 45) and the 5TBY homology modeled structure from the Padron laboratory (32). Houdusse and coworkers (60), however, have proposed a newer model of the IHM structure, which has the blocked head lever arm much closer to the normal prestroke state than the Dominguez structure suggests. Since none of our work demonstrates that we are dealing with a bona fide IHM state, these structural concepts remain hypotheses that need to be tested by high-resolution EM or X-ray crystallographic structures of the human β-cardiac myosin constructs. Nonetheless, we speculate that a mavacamten-bound sS1 structure may represent a more realistic and higher-resolution version of the blocked head of the SRX-IHM, and possibly relate to the free head of the IHM as well.
The transition of the myosin heads in muscle from the SRX off-state to the DRX on-state is thought to be modulated by various factors. Activation of the sarcomere with Ca2+ clearly causes disordering of the myosin heads, as shown by the elegant low-angle X-ray diffraction studies of Hugh Huxley (61). Whether this reflects a transition between SRX and DRX heads with every beat of the heart is not clear. It is possible that SRX should be thought of as an economical way to park myosin heads when they are not needed. The equilibrium between parked and disordered heads may be determined by protein phosphorylations and may be a longer-term effect.
Stretch of the sarcomere leads to additional force which might come from recruitment of SRX-IHM heads leading to more heads in the DRX state (62–65). It has been suggested that this recruitment of SRX heads may explain the Frank–Starling effect (63, 66), long debated with regard to its mechanism of action (67).
Other factors modulating the transition of the SRX off-state to the DRX on-state could be phosphorylation of the RLC (30, 45, 68, 69) and/or phosphorylation of MyBP-C (30, 45, 70). There are reported phosphorylation sites on the cardiac ELC as well (71), whose possible role in the stabilization of the SRX is unknown. A great deal of work needs to be done to determine what factors modulate the transition from the SRX off-state to the DRX on-state.
Our work opens avenues for the development of novel therapeutics that specifically target the open–closed equilibrium of the myosin heads. It is conceivable that future cardiac activators can be designed that stabilize the open state, while inhibitors such as mavacamten stabilize the closed state of the myosin molecule in the thick filament. This forms an innovative basis for modulating cardiac contractility at the molecular level.
Materials and Methods
The human β-cardiac myosin sS1, 2-hep, and 25-hep HMM constructs were produced using a modified AdEasy Vector System (Qbiogene, Inc.). The MYH7 mutant pigs were generated by homologous recombination in fetal fibroblasts from Yucatan minipigs. The human protein constructs and the minipigs are described in further detail in SI Appendix. Human heart samples were obtained from a tissue bank maintained at the University of Michigan. Ventricular tissues taken from HCM patients at the time of myectomy (MYH7 R663H) or from nonfailing donor hearts (control) were snap-frozen in liquid nitrogen. The study had the approval of the University of Michigan Institutional Review Board, and subjects gave informed consent. Methods for basal ATPase measurements, single-turnover experiments, EM, fiber SRX measurements, tension measurements, and low-angle X-ray diffraction are described in further detail in SI Appendix.
Statistical significance was calculated using paired or unpaired t tests wherever applicable. Not significant, P > 0.05; *P ≤ 0.05; ** P ≤ 0.01; ***P ≤ 0.001; and ****P ≤ 0.0001. Error bars in figures denote SD; all errors reported in the text denote SEM.
Supplementary Material
Acknowledgments
We thank Dr. Sharlene Day of University of Michigan for providing human R663H and WT tissue samples; the MyoKardia medicinal chemistry group for preparation of mavacamten with a photoactivatable cross-linker; Dr. Roger Craig and Dr. Kyoung-Hwan Lee of the University of Massachusetts, Worcester for invaluable training and guidance during the initial phases of the EM work; Richard Jones of MS Bioworks for the mass spectroscopy analysis of the WT and R403 minipig cardiac tissues; John Perrino of the Stanford Microscopy Facility for technical help with the EM work; all members of the J.A.S. laboratory for discussions; Drs. Robert McDowell, Leslie Leinwand, Christine Seidman, Hector Rodriguez, Sadie Bartholomew Ingle, Carlos del Rio, and Kristina Green for useful discussions; and the reviewers, who made a number of useful suggestions that improved the paper. This work was funded by NIH Grants GM33289 and HL117138 (to J.A.S.), AR062279 (to R.C.), and HL084553 (to J.G.S.). D.V.T. is supported by Stanford Lucile Packard Child Health Research Institute Postdoctoral Award UL1 TR001085 and American Heart Association Postdoctoral Fellowship 17POST33411070. The EM project described was supported, in part, by American Recovery and Reinvestment Act Award 1S10RR026780-01 from the National Center for Research Resources (NCRR). This research used the Nikon Imaging Center, University of California, San Francisco and the resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357. This project was supported by Grant 9 P41 GM103622 from the National Institute of General Medical Sciences of the NIH (to T.C.I.). This work was previously deposited in BioRxiv (https://www.biorxiv.org/content/early/2018/03/10/266783). The content is solely the responsibility of the authors and does not necessarily reflect the official views of the NCRR, the National Institute of General Medical Sciences, or the NIH.
Footnotes
Conflict of interest statement: J.A.S. is a cofounder of MyoKardia, a biotechnology company developing small molecules that target the sarcomere for the treatment of inherited cardiomyopathies, and of Cytokinetics and is a member of their scientific advisory boards. J.G.S. is a cofounder of MyoKardia and a member of its scientific advisory board. K.M.R. and R.C. are members of the MyoKardia scientific advisory board. R.L.A., M.H., and F.L.W. are employees of and own shares in MyoKardia. E.M.G. owns shares in MyoKardia.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1809540115/-/DCSupplemental.
References
- 1.Toyoshima YY, et al. Myosin subfragment-1 is sufficient to move actin filaments in vitro. Nature. 1987;328:536–539. doi: 10.1038/328536a0. [DOI] [PubMed] [Google Scholar]
- 2.Wendt T, Taylor D, Messier T, Trybus KM, Taylor KA. Visualization of head-head interactions in the inhibited state of smooth muscle myosin. J Cell Biol. 1999;147:1385–1390. doi: 10.1083/jcb.147.7.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wendt T, Taylor D, Trybus KM, Taylor K. Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2. Proc Natl Acad Sci USA. 2001;98:4361–4366. doi: 10.1073/pnas.071051098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burgess SA, et al. Structures of smooth muscle myosin and heavy meromyosin in the folded, shutdown state. J Mol Biol. 2007;372:1165–1178. doi: 10.1016/j.jmb.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 5.Jung HS, et al. Role of the tail in the regulated state of myosin 2. J Mol Biol. 2011;408:863–878. doi: 10.1016/j.jmb.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jung HS, Komatsu S, Ikebe M, Craig R. Head-head and head-tail interaction: A general mechanism for switching off myosin II activity in cells. Mol Biol Cell. 2008;19:3234–3242. doi: 10.1091/mbc.E08-02-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woodhead JL, et al. Atomic model of a myosin filament in the relaxed state. Nature. 2005;436:1195–1199. doi: 10.1038/nature03920. [DOI] [PubMed] [Google Scholar]
- 8.Zoghbi ME, Woodhead JL, Moss RL, Craig R. Three-dimensional structure of vertebrate cardiac muscle myosin filaments. Proc Natl Acad Sci USA. 2008;105:2386–2390. doi: 10.1073/pnas.0708912105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alamo L, et al. Three-dimensional reconstruction of tarantula myosin filaments suggests how phosphorylation may regulate myosin activity. J Mol Biol. 2008;384:780–797. doi: 10.1016/j.jmb.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al-Khayat HA, Kensler RW, Squire JM, Marston SB, Morris EP. Atomic model of the human cardiac muscle myosin filament. Proc Natl Acad Sci USA. 2013;110:318–323. doi: 10.1073/pnas.1212708110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McNamara JW, Li A, Dos Remedios CG, Cooke R. The role of super-relaxed myosin in skeletal and cardiac muscle. Biophys Rev. 2015;7:5–14. doi: 10.1007/s12551-014-0151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stewart MA, Franks-Skiba K, Chen S, Cooke R. Myosin ATP turnover rate is a mechanism involved in thermogenesis in resting skeletal muscle fibers. Proc Natl Acad Sci USA. 2010;107:430–435. doi: 10.1073/pnas.0909468107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hooijman P, Stewart MA, Cooke R. A new state of cardiac myosin with very slow ATP turnover: A potential cardioprotective mechanism in the heart. Biophys J. 2011;100:1969–1976. doi: 10.1016/j.bpj.2011.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naber N, Cooke R, Pate E. Slow myosin ATP turnover in the super-relaxed state in tarantula muscle. J Mol Biol. 2011;411:943–950. doi: 10.1016/j.jmb.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McNamara JW, et al. Ablation of cardiac myosin binding protein-C disrupts the super-relaxed state of myosin in murine cardiomyocytes. J Mol Cell Cardiol. 2016;94:65–71. doi: 10.1016/j.yjmcc.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooke R. The role of the myosin ATPase activity in adaptive thermogenesis by skeletal muscle. Biophys Rev. 2011;3:33–45. doi: 10.1007/s12551-011-0044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson C, Naber N, Pate E, Cooke R. The myosin inhibitor blebbistatin stabilizes the super-relaxed state in skeletal muscle. Biophys J. 2014;107:1637–1646. doi: 10.1016/j.bpj.2014.07.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alamo L, et al. Conserved intramolecular interactions maintain myosin interacting-heads motifs explaining tarantula muscle super-relaxed state structural basis. J Mol Biol. 2016;428:1142–1164. doi: 10.1016/j.jmb.2016.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nogara L, et al. Spectroscopic studies of the super relaxed state of skeletal muscle. PLoS One. 2016;11:e0160100. doi: 10.1371/journal.pone.0160100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geisterfer-Lowrance AA, et al. A molecular basis for familial hypertrophic cardiomyopathy: A beta cardiac myosin heavy chain gene missense mutation. Cell. 1990;62:999–1006. doi: 10.1016/0092-8674(90)90274-i. [DOI] [PubMed] [Google Scholar]
- 21.Seidman CE, Seidman JG. Hypertrophic cardiomyopathy. In: Scriver CR, et al., editors. The Metabolic and Molecular Bases of Inherited Disease. McGraw-Hill; 2000. pp. 5532–5452. [Google Scholar]
- 22.Seidman JG, Seidman C. The genetic basis for cardiomyopathy: From mutation identification to mechanistic paradigms. Cell. 2001;104:557–567. doi: 10.1016/s0092-8674(01)00242-2. [DOI] [PubMed] [Google Scholar]
- 23.Spudich JA. Hypertrophic and dilated cardiomyopathy: Four decades of basic research on muscle lead to potential therapeutic approaches to these devastating genetic diseases. Biophys J. 2014;106:1236–1249. doi: 10.1016/j.bpj.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maron M, et al. Abstract 16842: Obstructive hypertrophic cardiomyopathy: Initial single ascending dose data in healthy volunteers and patients. Circulation. 2016;134:A16842. [Google Scholar]
- 25.Green EM, et al. A small-molecule inhibitor of sarcomere contractility suppresses hypertrophic cardiomyopathy in mice. Science. 2016;351:617–621. doi: 10.1126/science.aad3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson WS, Criley JM, Ross RS. Dynamics of left ventricular emptying in hypertrophic subaortic stenosis. A cineangiographic and hemodynamic study. Am Heart J. 1967;73:4–16. doi: 10.1016/0002-8703(67)90303-1. [DOI] [PubMed] [Google Scholar]
- 27.Tyska MJ, et al. Single-molecule mechanics of R403Q cardiac myosin isolated from the mouse model of familial hypertrophic cardiomyopathy. Circ Res. 2000;86:737–744. doi: 10.1161/01.res.86.7.737. [DOI] [PubMed] [Google Scholar]
- 28.Rohde JA, Roopnarine O, Thomas DD, Muretta JM. Mavacamten stabilizes an autoinhibited state of two-headed cardiac myosin. Proc Natl Acad Sci USA. July 17, 2018;115:E7486–E7494. doi: 10.1073/pnas.1720342115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trybus KM, Freyzon Y, Faust LZ, Sweeney HL. Spare the rod, spoil the regulation: Necessity for a myosin rod. Proc Natl Acad Sci USA. 1997;94:48–52. doi: 10.1073/pnas.94.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nag S, et al. The myosin mesa and the basis of hypercontractility caused by hypertrophic cardiomyopathy mutations. Nat Struct Mol Biol. 2017;24:525–533. doi: 10.1038/nsmb.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adhikari AS, et al. Early-onset hypertrophic cardiomyopathy mutations significantly increase the velocity, force, and actin-activated ATPase activity of human β-cardiac myosin. Cell Rep. 2016;17:2857–2864. doi: 10.1016/j.celrep.2016.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alamo L, et al. Effects of myosin variants on interacting-heads motif explain distinct hypertrophic and dilated cardiomyopathy phenotypes. eLife. 2017;6:e24634. doi: 10.7554/eLife.24634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore JR, Leinwand L, Warshaw DM. Understanding cardiomyopathy phenotypes based on the functional impact of mutations in the myosin motor. Circ Res. 2012;111:375–385. doi: 10.1161/CIRCRESAHA.110.223842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blankenfeldt W, Thomä NH, Wray JS, Gautel M, Schlichting I. Crystal structures of human cardiac beta-myosin II S2-Delta provide insight into the functional role of the S2 subfragment. Proc Natl Acad Sci USA. 2006;103:17713–17717. doi: 10.1073/pnas.0606741103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kawas RF, et al. A small-molecule modulator of cardiac myosin acts on multiple stages of the myosin chemomechanical cycle. J Biol Chem. 2017;292:16571–16577. doi: 10.1074/jbc.M117.776815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knight P, Trinick J. Structure of the myosin projections on native thick filaments from vertebrate skeletal muscle. J Mol Biol. 1984;177:461–482. doi: 10.1016/0022-2836(84)90295-x. [DOI] [PubMed] [Google Scholar]
- 37.Trinick J, Elliott A. Effect of substrate on freeze-dried and shadowed protein structures. J Microsc. 1982;126:151–156. doi: 10.1111/j.1365-2818.1982.tb00364.x. [DOI] [PubMed] [Google Scholar]
- 38.Craig R, Smith R, Kendrick-Jones J. Light-chain phosphorylation controls the conformation of vertebrate non-muscle and smooth muscle myosin molecules. Nature. 1983;302:436–439. doi: 10.1038/302436a0. [DOI] [PubMed] [Google Scholar]
- 39.Trybus KM, Lowey S. Conformational states of smooth muscle myosin. Effects of light chain phosphorylation and ionic strength. J Biol Chem. 1984;259:8564–8571. [PubMed] [Google Scholar]
- 40.Stafford WF, et al. Calcium-dependent structural changes in scallop heavy meromyosin. J Mol Biol. 2001;307:137–147. doi: 10.1006/jmbi.2000.4490. [DOI] [PubMed] [Google Scholar]
- 41.Geisterfer-Lowrance AA, et al. A mouse model of familial hypertrophic cardiomyopathy. Science. 1996;272:731–734. doi: 10.1126/science.272.5262.731. [DOI] [PubMed] [Google Scholar]
- 42.Green EM, et al. Abstract 14467: A minipig genetic model of hypertrophic cardiomyopathy. Circulation. 2016;134:A14467. [Google Scholar]
- 43.del Rio CL, et al. Abstract 20770: A novel mini-pig genetic model of hypertrophic cardiomyopathy: Altered myofilament dynamics, hyper-contractility, and impaired systolic/diastolic functional reserve in vivo. Circulation. 2017;136:A20770. [Google Scholar]
- 44.Henze M, et al. Cardiac muscle function across the natural history of a genetic minipig model of hypertrophic cardiomyopathy. J Mol Cell Cardiol. 2017;112:166. [Google Scholar]
- 45.Trivedi DV, Adhikari AS, Sarkar SS, Ruppel KM, Spudich JA. Hypertrophic cardiomyopathy and the myosin mesa: Viewing an old disease in a new light. Biophys Rev. 2018;10:27–48. doi: 10.1007/s12551-017-0274-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spudich JA. The myosin mesa and a possible unifying hypothesis for the molecular basis of human hypertrophic cardiomyopathy. Biochem Soc Trans. 2015;43:64–72. doi: 10.1042/BST20140324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nag S, et al. 2016 Beyond the myosin mesa: A potential unifying hypothesis on the underlying molecular basis of hyper-contractility caused by a majority of hypertrophic cardiomyopathy mutations. bioRxiv:10.1101/065508. Preprint, posted July 24, 2016.
- 48.Previs MJ, et al. Myosin-binding protein C corrects an intrinsic inhomogeneity in cardiac excitation-contraction coupling. Sci Adv. 2015;1:e1400205. doi: 10.1126/sciadv.1400205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spudich JA, et al. Effects of hypertrophic and dilated cardiomyopathy mutations on power output by human β-cardiac myosin. J Exp Biol. 2016;219:161–167. doi: 10.1242/jeb.125930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Homburger JR, et al. Multidimensional structure-function relationships in human β-cardiac myosin from population-scale genetic variation. Proc Natl Acad Sci USA. 2016;113:6701–6706. doi: 10.1073/pnas.1606950113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kawana M, Sarkar SS, Sutton S, Ruppel KM, Spudich JA. Biophysical properties of human β-cardiac myosin with converter mutations that cause hypertrophic cardiomyopathy. Sci Adv. 2017;3:e1601959. doi: 10.1126/sciadv.1601959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nag S, et al. Contractility parameters of human β-cardiac myosin with the hypertrophic cardiomyopathy mutation R403Q show loss of motor function. Sci Adv. 2015;1:e1500511. doi: 10.1126/sciadv.1500511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu S, et al. The M.ADP.Pi state is required for helical order in the thick filaments of skeletal muscle. Biophys J. 1999;77:2665–2676. doi: 10.1016/s0006-3495(99)77101-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu S, Offer G, Gu J, White HD, Yu LC. Temperature and ligand dependence of conformation and helical order in myosin filaments. Biochemistry. 2003;42:390–401. doi: 10.1021/bi026085t. [DOI] [PubMed] [Google Scholar]
- 55.Zoghbi ME, Woodhead JL, Craig R, Padrón R. Helical order in tarantula thick filaments requires the “closed” conformation of the myosin head. J Mol Biol. 2004;342:1223–1236. doi: 10.1016/j.jmb.2004.07.037. [DOI] [PubMed] [Google Scholar]
- 56.Zhao FQ, Padrón R, Craig R. Blebbistatin stabilizes the helical order of myosin filaments by promoting the switch 2 closed state. Biophys J. 2008;95:3322–3329. doi: 10.1529/biophysj.108.137067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kampourakis T, Zhang X, Sun YB, Irving M. Omecamtiv mercabil and blebbistatin modulate cardiac contractility by perturbing the regulatory state of the myosin filament. J Physiol. 2018;596:31–46. doi: 10.1113/JP275050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu C, Kawana M, Song D, Ruppel KM, Spudich J. 2018. Controlling load-dependent contractility of the heart at the single molecule level. bioRxiv:10.1101/258020. Preprint, posted January 31, 2018.
- 59.Dominguez R, Freyzon Y, Trybus KM, Cohen C. Crystal structure of a vertebrate smooth muscle myosin motor domain and its complex with the essential light chain: Visualization of the pre-power stroke state. Cell. 1998;94:559–571. doi: 10.1016/s0092-8674(00)81598-6. [DOI] [PubMed] [Google Scholar]
- 60.Robert-Paganin J, Auguin D, Houdusse A. 2018. Hypertrophic cardiomyopathy disease results from disparate impairments of cardiac myosin function and auto-inhibition. bioRxiv:10.1101/324830. Preprint, posted May 17, 2018. [DOI] [PMC free article] [PubMed]
- 61.Huxley HE, Faruqi AR, Kress M, Bordas J, Koch MH. Time-resolved X-ray diffraction studies of the myosin layer-line reflections during muscle contraction. J Mol Biol. 1982;158:637–684. doi: 10.1016/0022-2836(82)90253-4. [DOI] [PubMed] [Google Scholar]
- 62.Linari M, et al. Force generation by skeletal muscle is controlled by mechanosensing in myosin filaments. Nature. 2015;528:276–279. doi: 10.1038/nature15727. [DOI] [PubMed] [Google Scholar]
- 63.Reconditi M, et al. Myosin filament activation in the heart is tuned to the mechanical task. Proc Natl Acad Sci USA. 2017;114:3240–3245. doi: 10.1073/pnas.1619484114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reconditi M, et al. Motion of myosin head domains during activation and force development in skeletal muscle. Proc Natl Acad Sci USA. 2011;108:7236–7240. doi: 10.1073/pnas.1018330108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Irving M. Regulation of contraction by the thick filaments in skeletal muscle. Biophys J. 2017;113:2579–2594. doi: 10.1016/j.bpj.2017.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ait-Mou Y, et al. Titin strain contributes to the Frank-Starling law of the heart by structural rearrangements of both thin- and thick-filament proteins. Proc Natl Acad Sci USA. 2016;113:2306–2311. doi: 10.1073/pnas.1516732113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Solaro RJ. Mechanisms of the Frank-Starling law of the heart: The beat goes on. Biophys J. 2007;93:4095–4096. doi: 10.1529/biophysj.107.117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kampourakis T, Irving M. Phosphorylation of myosin regulatory light chain controls myosin head conformation in cardiac muscle. J Mol Cell Cardiol. 2015;85:199–206. doi: 10.1016/j.yjmcc.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kampourakis T, Sun YB, Irving M. Myosin light chain phosphorylation enhances contraction of heart muscle via structural changes in both thick and thin filaments. Proc Natl Acad Sci USA. 2016;113:E3039–E3047. doi: 10.1073/pnas.1602776113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kampourakis T, Yan Z, Gautel M, Sun YB, Irving M. Myosin binding protein-C activates thin filaments and inhibits thick filaments in heart muscle cells. Proc Natl Acad Sci USA. 2014;111:18763–18768. doi: 10.1073/pnas.1413922112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang W, Szczesna-Cordary D. Molecular mechanisms of cardiomyopathy phenotypes associated with myosin light chain mutations. J Muscle Res Cell Motil. 2015;36:433–445. doi: 10.1007/s10974-015-9423-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Millman BM. The filament lattice of striated muscle. Physiol Rev. 1998;78:359–391. doi: 10.1152/physrev.1998.78.2.359. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.