Significance
The lactose permease of Escherichia coli (LacY), a model Major Facilitator Superfamily transporter, catalyzes galactoside/H+ symport by an alternating access mechanism that involves multiple conformational transitions. Nanobodies (Nbs) generated against a double mutant (LacYWW) that exists in an outward-open conformation stabilize the periplasmic-open conformer of wild-type LacY, block lactose transport, and lead to a 5–50-fold increase in the on-rate for galactoside binding to WT LacY. The galactoside-binding site in galactoside/LacYww/Nb9047 is superimposable with other sugar-bound LacYww structures and therefore is not perturbed due to crystal contacts or nanobody; thus, comparison with apo-LacYwwNb demonstrates that it most likely represents a transport intermediate primarily seen on the periplasmic side in response to the initial galactoside-binding.
Keywords: Major Facilitator Superfamily, X-ray structure, membrane protein, transport, conformational change
Abstract
The lactose permease of Escherichia coli (LacY), a dynamic polytopic membrane transport protein, catalyzes galactoside/H+ symport and operates by an alternating access mechanism that exhibits multiple conformations, the distribution of which is altered by sugar-binding. Camelid nanobodies were made against a double-mutant Gly46 → Trp/Gly262 → Trp (LacYWW) that produces an outward-open conformation, as opposed to the cytoplasmic open-state crystal structure of WT LacY. Nanobody 9047 (Nb9047) stabilizes WT LacY in a periplasmic-open conformation. Here, we describe the X-ray crystal structure of a complex between LacYWW, the high-affinity substrate analog 4-nitrophenyl-α-d-galactoside (NPG), and Nb9047 at 3-Å resolution. The present crystal structure demonstrates that Nb9047 binds to the periplasmic face of LacY, primarily to the C-terminal six-helical bundle, while a flexible loop of the Nb forms a bridge between the N- and C-terminal halves of LacY across the periplasmic vestibule. The bound Nb partially covers the vestibule, yet does not affect the on-rates or off-rates for the substrate binding to LacYWW, which implicates dynamic flexibility of the Nb–LacYWW complex. Nb9047-binding neither changes the overall structure of LacYWW with bound NPG, nor the positions of side chains comprising the galactoside-binding site. The current NPG-bound structure exhibits a more occluded periplasmic vestibule than seen in a previous structure of a (different Nb) apo-LacYWW/Nb9039 complex that we argue is caused by sugar-binding, with major differences located at the periplasmic ends of transmembrane helices in the N-terminal half of LacY.
The lactose permease from Escherichia coli (LacY) catalyzes stoichiometric symport of lactose and a proton across the membrane and is a member of the Major Facilitator Superfamily (MFS) of transporters. The protein is the most intensively studied secondary membrane transporter and is used as a template particularly in analysis of structural aspects of transport mechanisms. The transport of lactose is based on alternating access of galactoside- and H+-binding sites of LacY to either side of the membrane (1). The alternating access mechanism has been unequivocally documented for LacY by means of kinetics, accessibility, and structure determinations. The kinetic scheme of lactose transport includes multiple conformers, the distribution of which depends on sugar-binding. LacY is highly dynamic structurally, and multiple conformations have been characterized by biochemical and spectroscopic methods (2–9).
Initial X-ray structures were obtained for LacY in inward-open conformation (10–13). The molecule can be thought of as an N-terminal and a C-terminal six-helical bundle with a 35-amino-acid cytoplasmic loop between transmembrane helices (TMs) 6 and 7. The galactoside-binding site and the proton-binding site are located near the center of the protein and between the two six-helix bundles. The transporter cycles between states in which the binding site is accessible to solvent alternately from the cytoplasmic side alone, or to the periplasmic side. This was established by Cysteine-scanning mutagenesis, cross-linking, single-molecule fluorescence energy transfer, double-electron electron resonance (DEER) distance measurements, and stopped-flow galactoside-binding kinetics demonstrate that the periplasmic-open conformation is stabilized by binding of galactosides to purified LacY either in detergent solution, or after reconstitution into a phospholipid membrane (2–4, 6, 7, 9).
With the goal of generating an outward facing conformation, a double Gly-to-Trp mutant G46W/G262W (LacYWW) was constructed to introduce bulky side chains on the periplasmic side in the points of close contact between the N- and C-terminal six-helical halves of WT LacY (8). LacYWW is characterized by free access of the galactosides from the periplasmic side and high-affinity binding, while transport is abolished. The substrate-binding on-rate is as much as 50-fold faster to this mutant, and the Kd is fivefold lower than for WT LacY.
We obtained two X-ray structures of this mutant in the presence of higher-affinity analogs of lactose, β-d-galactopyranosyl-1-thio-β-d-galactopyranoside (TDG) (14), and with p-nitrophenyl-α-d-galactopyranoside (NPG) (15). The structure with bound TDG at 3.5-Å resolution (PDB ID code 4OAA) defined the periplasmic-open conformation with a tightly locked cytoplasmic side (14). The structure of LacYWW with bound NPG at 3.3-Å resolution (PDB ID code 4ZYR) showed the same periplasmic-open conformation with identical positions of helices (15). The interactions of amino acid residues with the key galactosyl moiety in both substrates share the same hydrogen bonds with some difference in orientation of hydrophobic groups Met23 and Phe27, consistent with the structural basis for primary specificity to the galactosyl moiety. These structures defined the roles of amino acid side chains in the sugar-binding site that had been identified biochemically, and they demonstrated the connectivity between the galactoside-binding site and the protonation site. They suggest that the substrate pathway and the protonation pathway proceed through physically separate regions that are connected by residues that must be capable of responding to the presence of water versus substrate in the binding site.
Thirty-two camelid nanobodies (Nbs) were generated against the periplasmic-open mutant LacYWW with the intention of obtaining Nbs that could favor, or trap, the so far unobserved outward facing structure of the highly dynamic WT LacY molecule (16). Biochemistry and kinetics of nine purified Nbs indicate that they bind stoichiometrically, exclusively to the periplasmic side of WT LacY, and abolish lactose transport in right-side-out membrane vesicles. We chose Nbs that increase the on-rate for galactoside-binding and binding affinity (lower Kd) with the goal of stabilizing an outward facing conformation of WT LacY. Thus, each Nb binds by conformational selection to the outward-open LacY molecule with exposed specific binding epitopes and traps a particular conformer. Remarkably, fluorescence-quenching studies with eight WT LacY–Nb complexes demonstrate that different Nbs appear to stabilize different outward-open states of WT LacY, which may represent transient conformers in the transport cycle (17).
The X-ray structure of the complex of apo-LacYWW with Nb9039 but without sugar-bound, obtained by crystallization in lipidic cubic phase (18), revealed an outward-open conformation with Nb9039 bound to the periplasmic surface of LacYWW. In the interface the majority of contacts are formed between the C-terminal half of LacY and complementarity-determining regions (CDRs) of the Nb. The long flexible CDR3 loop extends into the crevice between the N- and C-terminal halves, forming a bridge across the open periplasmic cavity. Relative to our galactoside-bound LacYWW (15), the periplasmic cavity is more open in sugar-free LacYWW (18), which is reflected in outward separation of the periplasmic ends of transmembrane helices, particularly in the N-terminal six-helical bundle of apo-LacYWW.
Here we report an X-ray structure of NPG-bound LacYWW in complex with a different Nb (Nb9047), determined to 3.0-Å resolution, which demonstrates specific multiple interactions of Nb9047 with an outward-open conformer and shows differences from the apo-LacYWW-Nb9039 due to, or commensurate with, galactoside-binding.
Results
Overall Structure of the LacYWW/NPG/Nb9047 Complex.
The mutant LacYWW was cocrystallized with Nb9047 in the presence of NPG, by vapor diffusion. Crystals were in space group P6522 with unit cell dimensions a = 153.6 Å, c = 193.7 Å and diffracted to 3.0-Å resolution (Fig. 1A and SI Appendix, Fig. S1). Determination was by molecular replacement using a modified version of our previous LacYWW structure (PDB ID code 4ZYR, with NPG removed). Data collection and refinement statistics are summarized in SI Appendix, Table S1.
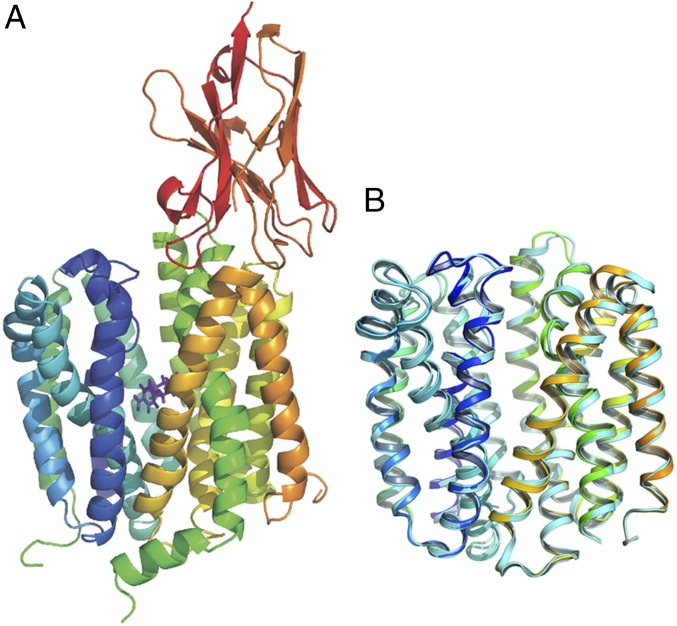
Fig. 1.
Overall structure of the LacYWW/NPG/Nb9047 complex. (A) Side view of the LacYWW/NPG/Nb9047 complex is presented as a cartoon, rainbow-colored from blue to orange for the backbone of LacYWW from N- to C-terminus with red-colored Nb9047 attached to the periplasmic side of LacYWW. NPG is shown as purple sticks. (B) Alignment of the LacYWW backbone shown in A (rainbow-colored, Nb and NPG removed) with our previous structure of the NPG-bound protein without Nb (cyan-colored, PDB ID code 4ZYR, NPG removed). Only the transmembrane helices and connecting loops are compared in both structures (aligned residues include 9–188 and 221–401 in N- and C-terminal halves). Residues in loop 6–7 and in N- and C-termini are omitted. Both molecules are oriented with their periplasmic side up.
The overall structure of the NPG-bound LacYWW in the Nb9047 complex differs from NPG-bound LacYWW in absence of the Nb (PDB ID code 4ZYR) (15) only in the periplasmic interface with the Nb (Fig. 1B). The rmsd between Cα atoms is 0.9 Å. Therefore, Nb9047 (produced against an apo-form of LacYWW) binds to the NPG-bound conformation of LacY and does not cause any conformational change relative to NPG-LacYWW.
The structure of the LacYWW/NPG/Nb9047 complex has a stoichiometry of 1:1:1 and shows a more occluded periplasmic vestibule compared with the previously determined X-ray structure of an apo-LacYWW/Nb9039 complex (PDB ID code 5GXB) (18). In both cases the Nbs bind to the periplasmic surface of the LacYWW molecule with a partially occluded periplasmic cavity and are sealed against solvent on the cytoplasmic side (Fig. 2A). The rmsd between Cα atoms of the two LacYww structures (residues 9–188, 221–401; omitting the Nbs and the TM6-7 loop, N- and C-termini) is 1.5 Å, with larger differences clustered among helices at their periplasmic side where the vestibule is more occluded with NPG-bound (Fig. 2B). The positions of Cα atoms of these periplasmic ends of the helices and their connecting loops differ by 2–5.6 Å, while the cytoplasmic ends of the helices are virtually identical in both structures. The direction of helical shift is inward toward the vestibule in LacYWW/NPG/Nb9047 versus the apo-LacYWW/Nb9039 complex (Fig. 2A). The major shifts are observed from the kink of TM1 to the periplasmic end of TM2 (from Pro28 to Thr45) and in the periplasmic ends of TM3 and TM4 (from Pro89 to Gly111). For example, the distance between Cα atoms of Phe29 in the two aligned structures is about 5 Å, and the side chain is rotated significantly. No such differences exist between NPG-bound Nb9047/LacYWW and NPG-bound LacYWW without Nb (SI Appendix, Fig. S2), suggesting that we have visualized a partial structural closure of the periplasmic vestibule upon substrate-binding (Fig. 2).
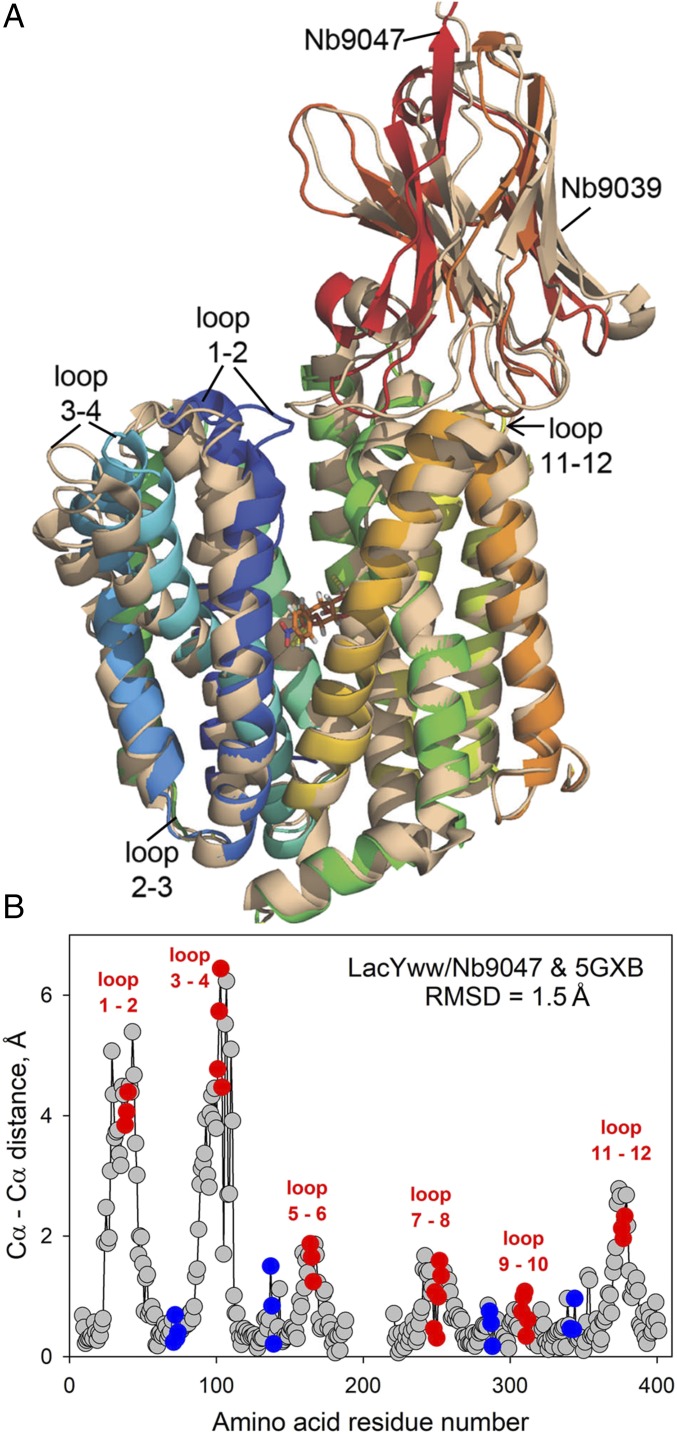
Fig. 2.
Structural alignment of two Nb-bound LacYWW complexes reveals a partial closure on the periplasmic side associated with ligand-binding, suggesting two states on the transport pathway. (A) Cartoon demonstrating two superimposed structures (with and without bound NPG) aligned by Cα atoms: LacYWW/NPG/Nb9047 (rainbow-colored as in Fig. 1A) and apo-LacYWW/Nb9039 (wheat-colored, PDB ID code 5GXB). Nbs are attached to the periplasmic side of LacYWW. (B) Differences in positions of the main-chain Cα atoms of LacYWW between the two aligned structures based on residues 9–188 and 221–401 (Nbs and NPG removed). Amino acids in loop 6–7 and in N- and C-ends are omitted (rmsd = 1.5 Å). Red and blue dots represent residues in periplasmic and cytoplasmic loops, respectively.
These differences in positions of transmembrane helices in the current structure relative to the structure of the apo-LacYWW/Nb9039 complex are confirmed by measuring the angles of helical tilt axes. There are tilts of 5° in TM1, 8.5° in TM3, and 10° in TM4 and slight angular differences of 2° in TMs 5, 6, and 7 and 5° in TMs 8–12.
Structural variations within each six-helical bundle (TM1-6, TM7-12) in outward facing partially occluded LacYWW caused by substrate-binding are analyzed by independent alignment of N-terminal halves (residues from 9 to 188) and the C-terminal halves (residues from 221 to 401) in LacYWW/NPG/Nb9047 versus in apo-LacYWW/Nb9039 complexes. The differences are focused in the N-terminal six-helical bundles TM1–TM6 (rmsd 1.3 Å) versus 0.7 Å for TM7–TM12.
Area of Contact at the LacYWW-Nb9047 Interface.
All three CDR regions of Nb9047 are involved in the interactions with the periplasmic ends of helices and connecting loops of the C-terminal domain in LacYWW with a link to the N-terminal domain formed by CDR3 (Fig. 3). In total there are 12 hydrogen bonds formed between 10 residues of LacYWW and 11 residues of Nb9047 (SI Appendix, Figs. S3 and S4 and Table S2).
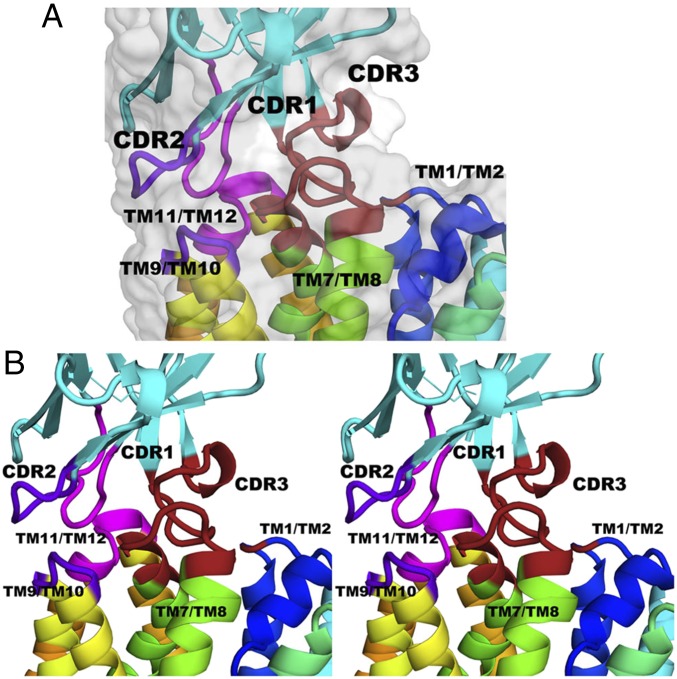
Fig. 3.
The area of contact between CDR loops of Nb9047 and the periplasmic surface of LacYWW with bound NPG. (A) A side view of the interface between LacYWW and Nb9047 shown as a cartoon structure with rainbow-colored LacYWW (from blue to orange for TM1 to TM12) and cyan-colored Nb9047. Variable loops of Nb9047 that contact LacYWW are in different colors: magenta for CDR1 and the loop between TM9/TM10; purple/blue for CDR2 and the loop between TM11/TM12; and brick red for CDR3 and the loops between TM7/TM8 and TM1/TM2. The surrounding surface is shown in gray. (B) A crossed-eye stereoview of the interface shown in A.
In the LacYWW/NPG/Nb9047 complex, part of the variable CDR3 sequence in the Nb (from Gln99 to Asn113) forms a loop linking Lys42 in the N-terminal half and Asn245 in the C-terminal half of LacY through a network of hydrogen bonds. The shape of the long CDR3 loop is formed by multiple contacts with residues from loop 7–8 and TM8 of LacY, so that the tip of the CDR3 loop protrudes into the partially open periplasmic cavity, while Asn102 and Gly103 in Nb form hydrogen bonds with Lys42 and Asn245 in LacY, respectively. The amino group of Nb Asn102 is hydrogen donor to the carbonyl oxygen of Lys42 (end of TM2 in LacYWW); carbonyl oxygen of neighboring Nb Gly103 is acceptor from the amino group of Asn245 (TM7 in LacYWW). These connections form a bridge across the periplasmic vestibule between the N- and C-terminal six-helical domains of LacYWW.
CDR2 and CDR3 of Nb interact with residues in the periplasmic ends of TM7 and TM8 of LacYWW and the loop between them. Seven out of twelve hydrogen bonds between Nb9047 and LacYWW are concentrated in this region. Two of these are formed with Ser249 in loop 7–8 of LacYWW. The side-chain oxygen of Gln99 in CDR3 of Nb is acceptor from the hydroxyl group of Ser249; the guanidino group of Arg52 in CDR2 of Nb is a hydrogen-bond donor to the carbonyl oxygen of Ser249. Two residues in CDR3 are hydrogen-bonded to neighboring residues in loop 7–8 in LacYWW via backbone atoms; main-chain Nb N-Hs of Tyr106 and Ser107 are donors to the carbonyl oxygens of Phe251 and Ala252, respectively. The carbonyl oxygen of Gly104 and the carboxyl group of Glu108 in CDR3 of Nb are acceptors for the main-chain amine of Gly254 in TM8 of LacYWW.
Arg57 in CDR2 of Nb9047 is donor to two adjacent residues, Thr310 and hydroxyl group of Ser311 in the periplasmic loop 9–10 of LacYWW. Two hydrogen bonds link the periplasmic end of TM12 in LacYWW with CDR1 and CDR2 of Nb. The side-chain oxygen of Gln379 in LacYWW is a hydrogen bond acceptor from the indole N-H of Trp53 in CDR2 of Nb, and the main-chain amine of the neighboring Gly380 forms a hydrogen bond with the side-chain oxygen of Thr31 as acceptor in CDR1 of Nb. The interface also incorporates hydrophobic interactions between Ala252 (loop 7–8 in LacYWW) and Tyr106 in CDR3 of Nb, and possibly between Ile376 (loop 11–12 in LacYWW) and Phe32 in CDR1 of Nb.
Galactoside-Binding Site.
In the LacYWW/NPG/Nb9047 complex the electron densities of bound NPG and surrounding residues comprising the binding site are well-defined and clearly show the orientation of the ligand and its interactions (Fig. 4A and SI Appendix, Fig. S5). The NPG molecule in the middle of the binding pocket is in close proximity to the residues from TM1, TM4, TM5, TM8, and TM10; i.e., both six-helical halves of LacYWW are involved in ligand-binding (SI Appendix, Fig. S6 and Table S3). The network of NPG contacts includes multiple hydrogen bonds with amino acids Arg144 (TM5), Glu269 and Asn272 (TM8), and His322 (TM10), close connections with Trp151 (TM5) and Met23 (TM1), and also with Cys148 (TM5). Moreover, there are several tight intramolecular polar contacts between amino acid residues surrounding NPG that may be essential for coupling of substrate-binding with protonation/deprotonation and conformational transitions. Interaction between Glu269 and Asn272, which are separated by one helical turn in TM8, could be important for recognizing the relative configuration of O3 and O4 atoms in the galactoside, defining substrate specificity. Another contact is between Glu126 (TM4) and two residues: Arg144 (TM5), participating in galactoside-binding, and Tyr350 (TM11), which is part of the cluster of aromatic amino acids maintaining closure of the cytoplasmic vestibule (Phe140, Phe334, and Tyr350). The chain of contacts Arg144-Glu126-Tyr350 links the N- and C-terminal bundles in outward facing LacY and could be important for connecting galactoside-binding with conformational transitions. Additionally, His322 is in close proximity to Tyr236 and Glu325 in the protonation site.
Fig. 4.
The galactoside-binding site in the LacYWW/NPG/Nb9047 complex. (A) The molecule of NPG (yellow sticks) and surrounding side chains of LacYWW (yellow sticks), nitrogen atoms (blue), oxygen (red), sulfur (light yellow), and hydrogen (white). Dashed lines depict hydrogen bonds. (B) Crossed-eye stereoview of the structural alignment of substrate-binding sites of NPG-bound (LacYWW/NPG/Nb9047) and apo-LacYWW (LacYWW/Nb9039). Carbon atoms of the side chains in LacYWW/NPG/Nb9047 (yellow) and LacYWW/Nb9039 (PDB ID code 5GXB) (orange).
The current structure demonstrates almost no differences in configuration of the binding site versus our previous structure of NPG-bound LacYWW (PDB ID code 4ZYR) (15), yet the nitro-phenyl ring of NPG is tilted by about 30° (Fig. 4B). Clearly, Nb9047-binding does not change the architecture of the galactose-binding site in LacYWW or positions of the side chains in contact with NPG. This is consistent with practically identical kinetic parameters of NPG-binding measured with LacYWW in the absence or presence of bound Nb9047. Thus, the values of kon are 5.7 ± 0.2 or 5.8 ± 0.2 μM−1s−1, for LacYWW or its complex with Nb9047, respectively, and koff is 31 ± 1 s−1 in both cases; the calculated Kd is the same, 5.4 ± 0.3 μM versus 5.3 ± 0.3 μM (SI Appendix, Fig. S7) (16).
Comparison between the sugar-binding site in NPG-bound LacYWW (in LacYWW/NPG/Nb9047, or without Nb in LacYWW/NPG, PDB ID code 4ZYR), and the apo-LacYWW/Nb9039 complex (PDB ID code 5GXB) shows that galactoside-binding causes structural movements of the periplasmic helical ends that move toward the central axis and side chains that in all but one case move toward the sugar. Residues move closer to the center of the binding site (by the distances given in parentheses) as follows: Met23 (2.9 Å), Asn119 (3.6 Å), Trp151 (2.0 Å), Arg144 (1.3 Å), Glu126 (1.9 Å), Asn272 (1.0 Å), and Glu269 (0.9 Å) to coordinate NPG in ligand-bound structures, while His322, hydrogen-bonded to the galactose moiety, moves away from the binding site and 1.6 Å closer to the critical protonation-sensitive Glu325 (Fig. 4B; compare yellow- vs. orange-colored side chains). These structure changes suggest that the first response of the protein to binding of specific substrates results in inward motion of the transmembrane helices on the periplasmic side.
Discussion
Different Nbs developed against LacYWW as an antigen exhibit different extents of inhibition of lactose transport catalyzed by WT LacY, diverse effects on sugar-binding kinetics, variable binding affinity of Nbs, and trapping of different outward-open conformations of the transporter (16, 17). These differences indicate that Nbs may recognize several transient conformers that could represent natural intermediates in the transport cycle.
There are two Nb-bound structures available now for periplasmic-open LacYWW, of which in one (the current structure) at 3.0-Å resolution, Nb9047 is bound to LacYWW/NPG, while in the other, Nb9039 is targeted to the apo-LacYWW at 3.3 Å (PDB ID code 5GXB) (18). We tried to crystallize 12 Nbs (out of 32) in complex with LacY, and with LacYWW and Nb9039 did cocrystallize with LacYWW/NPG but gave only 6–7-Å resolution diffraction, while LacYWW/Nb9047 did not form crystals in the absence of galactosides. These Nbs recognize different epitopes on the periplasmic surface of LacY and in each case (Nb9047 and Nb9039) interact mostly with loops in the C-terminal half of LacYWW. There are 12 hydrogen bonds observed in the interface between Nb9047 and LacYWW, and 11 in LacYWW/Nb9039. All three CDR loops of Nb9047 participate in H-bond formation, whereas the Nb9039 interface to LacYWW is primarily via its CDR3 loop. Remarkably, in both structures, CDR3 loops of Nbs create a bridge across the open periplasmic cavity between C- and N-terminal halves of LacY, and this contact may explain preferences for different conformers. The variable CDR3 loops in Nb9047 and Nb9039 are different in length and sequence. Nevertheless, both CDR3 loops form contacts with the same residues, Lys42 in the N-terminal half and Asn245 in the C-terminal half of LacYWW, although the nature and the length of these links differ. Thus, the amino group of Asn102 in Nb9047 contacts with the main-chain backbone carbonyl oxygen of Lys42, while the side chain of Tyr104 in Nb9039 forms a hydrogen bond with the amino group of the side chain of Lys42. Connections with Asn245 in the C-terminal half of LacYWW are formed by Gly103 in Nb9047 (adjacent to Asn102), and by Ser103 in Nb9039 (next residue to Tyr104), so that only two residues in CDR3 loops of both Nbs are positioned between N- and C-terminal bundles. However, the configuration of connections is different, and the entire distance between N- and C-terminal bundles turns out to be much shorter in the current structure with bound NPG than in apo-LacYWW/Nb9039 complex (the Cα atoms of Lys42 and Asn245 are separated by 11 Å and 16 Å, respectively), indicating that these two Nbs were developed against different conformers of LacY.
Although made against the outward facing conformation of LacYWW, Nb9047, like Nb9039, binds WT LacY with about the same Kd values as for LacYWW, in the nM range. The kon values for NPG binding to WT LacY increased dramatically after formation of complexes with Nb9047 or Nb9039 (34 or 22 times, respectively) to the same as for LacYWW, indicating that the Nb indeed stabilizes the outward facing conformation of WT LacY in solution (SI Appendix, Fig. S7) (16, 17). This validates the notion that nanobodies made against mutationally stabilized states do bind to and stabilize the same state in the wild-type LacY. Although we did not yet succeed in crystallizing these wild-type complexes, perhaps because they still retain some flexibility, the approach is valid to understanding the structures of states through the transport cycle.
Several of the Nbs completely block lactose transport activity of WT LacY, bind tightly to the periplasmic side of purified transporter, and stabilize outward-open conformations (16, 17). The CDR loops of these Nbs may interact with WT LacY in a manner similar to the two Nb–LacYWW X-ray structures by inserting a bridge that separates N- and C-terminal halves of LacY. Hence, different Nbs may stabilize different conformers by formation a bridge of a distinctive length and, therefore, could help to obtain structures of transient intermediates with various extents of periplasmic side-opening. This emphasizes that there are shallow energy differences between states necessary for the transport cycle that can be stabilized.
The X-ray structure reported here shows that Nb9047 binds to the preexisting conformation of LacYWW and does not change its structure. The rmsd between Nb9047/LacYWW/NPG and LacYWW/NPG (PDB ID code 4ZYR) is 0.9 Å. The alignment was based on all Cα atoms in the transmembrane helices (Fig. 1B and SI Appendix, Fig. S2). The configuration of amino acid residues participating in NPG-binding is identical in both structures (with or without bound Nb9047; Fig. 4B). Importantly, high accessibility of the sugar-binding site in LacYWW is not affected by partial closure of the open periplasmic vestibule by the Nb CDR3; binding of NPG has the same high kon value (5.7 vs. 5.8 μM−1s−1). The kon value for NPG-binding increases dramatically in the WT LacY–Nb9047 complex (from 0.2 to 6.8 μM−1s−1) due to Nb9047 stabilizing a conformer with greater substrate accessibility (SI Appendix, Fig. S7).
The comparison of NPG-bound LacYWW with apo-LacYWW reveals that galactoside-binding causes distinct structural changes on the periplasmic side of the molecule. Overall, the rmsd between all Cα atoms in transmembrane helices alone of NPG-bound LacYWW/Nb9047 (the current structure) and apo-LacYWW/Nb9039 (PDB ID code 5GXB) is 1.5 Å. The principle contributions to this overall difference are focused on the periplasmic portions of the N-terminal six-helical bundle that combine to give a vestibule more closed around the substrate (Fig. 2). The comparison of six-helical halves separately in the current structure versus in the apo-LacYWW/Nb9039 complex shows that the changes within each bundle are greater in the N-terminal six-helical bundles (TM1–TM6) with rmsd = 1.3 Å than in the C-terminal six-helical bundles (TM7–TM12) with rmsd = 0.7 Å. This is consistent with the abundance of kinks and bends in helices of the N-terminal half, particularly in the first four helices (TM1–TM4), which may encode additional flexibility in this region. Therefore, regarding the ordered occurrence of the intermediates in transport cycle, the structures show that movement at the periplasmic side in the N-terminal domain (especially in TM1–TM2 and TM3–TM4) could play an important role in the sequence of events triggered by sugar-binding from that side. While the cytoplasmic side remains tightly closed, binding of galactoside to the outward facing LacYWW causes closure initially at that side to move in the direction of sequestering the substrate on the way to an occluded transition state.
Materials and Methods
Materials.
NPG and buffers were from Sigma-Aldrich. Talon superflow resin was from BD Clontech. Dodecyl-β-d-maltopyranoside (DDM) and n-nonyl-β-d-glucoside (NG) were from Affymetrix. All other materials were of reagent grade and obtained from commercial sources.
Preparation of Nb9047 was as previously described (16). Inducible periplasmic expression of His-tagged Nb in E. coli yields >95% pure protein using immobilized metal ion affinity chromatography (Talon resin). Purified Nb9047 (20 mg/mL) in 100 mM potassium phosphate (KPi, pH 7.5) were frozen in liquid nitrogen and stored at −80 °C before use.
Growth, Expression, and Purification.
Plasmid pT7-5 encoding LacYWW with a six-His-tag was expressed in E. coli C41. The protein purification was as described earlier (15). Briefly, membranes were prepared and solubilized in 2% DDM, and LacY was purified on a Talon column. Protein was eluted in 20 mM Hepes/0.2% NG/200 mM imidazole buffer (pH 6.5). Purified Nb9047 was added to LacY in a 3:1 molar ratio and subjected to size-exclusion chromatography. A homogeneous peak containing LacYWW/Nb9047 complex was eluted in 20 mM Hepes/0.2% NG (pH 6.5), 50 mm NaCl and used for crystallization.
Crystallization, Data Collection, and Structure Determination.
NPG was added (1 mM, final concentration) to a protein solution (10 mg/mL in 20 mM Hepes/0.2% NG, pH 6.5, 50 mm NaCl) before crystallization trials. Crystallization screens were performed using the hanging-drop vapor diffusion method on a Mosquito Crystal Robot (TTP Labtech) in a 96-well plate with a drop ratio of 100-nL–well solution to 300 nL protein. Crystals appeared in 24 h and grew to the size of 60 µm in a week. Crystals were harvested and screened for diffraction. Crystals were reproducible and appeared in a wide range of PEG concentrations, at different pHs and with different buffers. The best diffracting crystals grew in 0.1 M MgCl2, 28% PEG1000, 0.1 M Tris⋅HCl, pH 8.5. No cryoprotectant was added before data collection.
Diffraction data were collected at the Lawrence Berkeley National Laboratory Advanced Light Source Beamline 8.3.1, at −170 °C at a wavelength of 1.115 Å. The highest-resolution crystals diffracted to 2.9 Å. Data were indexed with HKL2000 (19) and processed with XDS (20).
The structure was determined by molecular replacement using a ligand-stripped version of LacYWW-NPG (PDB ID code 4ZYR) as the search model (75% of the mass of the complex). To interpret bound Nb9047, the available structure of another camelid Nb (PDB ID code 5BOP) was placed into the 2Fo-Fc density and the sequence substituted by that of Nb9047, and the binary complex was refined for several cycles. The NPG ligand was added and followed by further refinement. We used a resolution cutoff of 3 Å for reporting data and refinement statistics in SI Appendix, Table S1, based on the statistically significant correlation coefficient CC1/2 (21) for data in the 3.1- to 3.0-Å range. We refined the structure using the data to 2.9 Å, since the 3.0- to 2.9-Å–resolution shell included observed reflections. The program COOT was used for density-fitting (22), and refinement was carried out using PHENIX (23) and Refmac from the CCP4 suite (24). Hydrogen atoms were included in expected positions. The data and refinement statistics are shown in SI Appendix, Table S1. Refinement proceeded with manual rebuilding in COOT with the assistance of sigmaA-weighted 2Fo-Fc and Fo-Fc and complete Omit maps by maximum-likelihood–based energy minimization and isotropic B-factor refinement in PHENIX and Refmac. The resulting model had improved R-work and R-free to 0.276 and 0.295, respectively. The MolProbity server (25) was employed for structural assessment and validation. The final round improved stereochemistry and geometry of the structure with 92% residues in favored regions, 6.8% in allowed regions. The clashes between atoms came down to 0.5%. Structural figures were drawn using PyMOL.
Supplementary Material
Acknowledgments
We thank Dr. James Holton and George Meigs for their counsel and development of beamline 8.3.1 of the Advanced Light Source, Berkeley. We are indebted to Jun-Yong Choe from Rosalind Franklin University of Medicine and Science for the suggestion to utilize graphical image of distance differences between Cα atoms in aligned structures. We are grateful to University of California Office of the President, Multicampus Research Programs and Initiatives Grant MR‐15‐328599, and the “Program for Breakthrough Biomedical Research,” which is partially funded by the Sandler Foundation, for support of beamline 8.3.1. This work was supported by National Institute of Health Grant R01 GM024485 (to R.M.S.). H.R.K. was supported by NIH Grant GM120043, National Science Foundation Eager Grant MCB1747705, and a gift from Ruth and Bucky Stein (to H.R.K.). We thank INSTRUCT-ERIC, part of the European Strategy Forum on Research Infrastructures and the Research Foundation-Flanders (FWO) for funding nanobody discovery (J.S.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 6C9W).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1801774115/-/DCSupplemental.
References
- 1.Kaback HR. A chemiosmotic mechanism of symport. Proc Natl Acad Sci USA. 2015;112:1259–1264. doi: 10.1073/pnas.1419325112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaback HR, et al. Site-directed alkylation and the alternating access model for LacY. Proc Natl Acad Sci USA. 2007;104:491–494. doi: 10.1073/pnas.0609968104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Majumdar DS, et al. Single-molecule FRET reveals sugar-induced conformational dynamics in LacY. Proc Natl Acad Sci USA. 2007;104:12640–12645. doi: 10.1073/pnas.0700969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smirnova I, et al. Sugar binding induces an outward facing conformation of LacY. Proc Natl Acad Sci USA. 2007;104:16504–16509. doi: 10.1073/pnas.0708258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou Y, Guan L, Freites JA, Kaback HR. Opening and closing of the periplasmic gate in lactose permease. Proc Natl Acad Sci USA. 2008;105:3774–3778. doi: 10.1073/pnas.0800825105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smirnova I, Kasho V, Sugihara J, Kaback HR. Probing of the rates of alternating access in LacY with Trp fluorescence. Proc Natl Acad Sci USA. 2009;106:21561–21566. doi: 10.1073/pnas.0911434106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smirnova I, Kasho V, Sugihara J, Kaback HR. Opening the periplasmic cavity in lactose permease is the limiting step for sugar binding. Proc Natl Acad Sci USA. 2011;108:15147–15151. doi: 10.1073/pnas.1112157108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smirnova I, Kasho V, Sugihara J, Kaback HR. Trp replacements for tightly interacting Gly-Gly pairs in LacY stabilize an outward-facing conformation. Proc Natl Acad Sci USA. 2013;110:8876–8881. doi: 10.1073/pnas.1306849110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smirnova I, Kasho V, Kaback HR. Real-time conformational changes in LacY. Proc Natl Acad Sci USA. 2014;111:8440–8445. doi: 10.1073/pnas.1408374111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abramson J, et al. The lactose permease of Escherichia coli: Overall structure, the sugar-binding site and the alternating access model for transport. FEBS Lett. 2003;555:96–101. doi: 10.1016/s0014-5793(03)01087-1. [DOI] [PubMed] [Google Scholar]
- 11.Mirza O, Guan L, Verner G, Iwata S, Kaback HR. Structural evidence for induced fit and a mechanism for sugar/H+ symport in LacY. EMBO J. 2006;25:1177–1183. doi: 10.1038/sj.emboj.7601028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guan L, Mirza O, Verner G, Iwata S, Kaback HR. Structural determination of wild-type lactose permease. Proc Natl Acad Sci USA. 2007;104:15294–15298. doi: 10.1073/pnas.0707688104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaptal V, et al. Crystal structure of lactose permease in complex with an affinity inactivator yields unique insight into sugar recognition. Proc Natl Acad Sci USA. 2011;108:9361–9366. doi: 10.1073/pnas.1105687108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar H, et al. Structure of sugar-bound LacY. Proc Natl Acad Sci USA. 2014;111:1784–1788. doi: 10.1073/pnas.1324141111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar H, Finer-Moore JS, Kaback HR, Stroud RM. Structure of LacY with an α-substituted galactoside: Connecting the binding site to the protonation site. Proc Natl Acad Sci USA. 2015;112:9004–9009. doi: 10.1073/pnas.1509854112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smirnova I, et al. Outward-facing conformers of LacY stabilized by nanobodies. Proc Natl Acad Sci USA. 2014;111:18548–18553. doi: 10.1073/pnas.1422265112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smirnova I, et al. Transient conformers of LacY are trapped by nanobodies. Proc Natl Acad Sci USA. 2015;112:13839–13844. doi: 10.1073/pnas.1519485112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang X, et al. Crystal structure of a LacY-nanobody complex in a periplasmic-open conformation. Proc Natl Acad Sci USA. 2016;113:12420–12425. doi: 10.1073/pnas.1615414113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 20.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.