Significance
The use of heterologous systems to express spider silk has become an attractive method. However, achieving cost-effective production and high yields is still challenging. Here, we describe the establishment of a targeted gene replacement system in Bombyx mori to express the major ampullate spidroin-1 gene (MaSp1) from the spider Nephila clavipes. With the aid of transcription activator-like effector nuclease-mediated homology-directed repair, we genetically replaced the silkworm fibroin heavy chain gene with MaSp1 with considerable transformation efficiency, and the chimeric MaSp1 yields reached up to 35.2% wt/wt of cocoon shells in transformed silkworms. The genetically modified silk fiber had significant changes in mechanical properties, with improved extensibility. This system will shed light on the future mass production of new biomaterials, including spider silk.
Keywords: genome editing, TALEN, spider silk, Bombyx mori
Abstract
Spider silk is one of the best natural fibers and has superior mechanical properties. However, the large-scale harvesting of spider silk by rearing spiders is not feasible, due to their territorial and cannibalistic behaviors. The silkworm, Bombyx mori, has been the most well known silk producer for thousands of years and has been considered an ideal bioreactor for producing exogenous proteins, including spider silk. Previous attempts using transposon-mediated transgenic silkworms to produce spider silk could not achieve efficient yields, due to variable promoter activities and endogenous silk fibroin protein expression. Here, we report a massive spider silk production system in B. mori by using transcription activator-like effector nuclease-mediated homology-directed repair to replace the silkworm fibroin heavy chain gene (FibH) with the major ampullate spidroin-1 gene (MaSp1) in the spider Nephila clavipes. We successfully replaced the ∼16-kb endogenous FibH gene with a 1.6-kb MaSp1 gene fused with a 1.1-kb partial FibH sequence and achieved up to 35.2% chimeric MaSp1 protein amounts in transformed cocoon shells. The presence of the MaSp1 peptide significantly changed the mechanical characteristics of the silk fiber, especially the extensibility. Our study provides a native promoter-driven, highly efficient system for expressing the heterologous spider silk gene instead of the transposon-based, random insertion of the spider gene into the silkworm genome. Targeted MaSp1 integration into silkworm silk glands provides a paradigm for the large-scale production of spider silk protein with genetically modified silkworms, and this approach will shed light on developing new biomaterials.
Natural silks, originating from silkworms or spiders, are ideal biomaterials for a wide range of applications not only in the silk industry but also in the military and medical industries (1, 2). Spider silk is considered one of the best silk fibers, with remarkable strength and extensibility (3). However, the large-scale production of silk by farming of spiders is not feasible, due to their territorial and cannibalistic behaviors (4, 5). In recent years, heterologous systems that produce spider silk proteins have been applied in different organisms, including bacteria, yeasts, mammalian cell lines, insect cells, and even transgenic animals and plants (6–11). Unfortunately, none of these expressed proteins can be naturally assembled into silk fibers, and extra manufacturing technologies are needed, which are extremely cost-inefficient. Furthermore, major spider silk proteins, such as the major ampullate silk protein (MaSp), have high molecular weights and highly repetitive sequences, leading to low protein yields when expressed in these heterologous systems (11).
As a lepidopteran model insect, the silkworm, Bombyx mori, has been the major contributor to the silk industry for thousands of years (12). The silkworm silk fiber is the only natural fiber that can be produced on a large scale for commercial use. The silkworm silk contains two types of major protein complexes: sericins and fibroins. Sericin proteins are hydrosoluble and enfold the fibroin when silkworms spin the silk (13). Fibroins are insoluble proteins that include the fibroin heavy chain (FibH), the fibroin light chain (FibL), and the fibrohexamerin protein (fhx/P25) in a molar ratio of 6:6:1, with molecular weights of 350 kDa (FibH), 25.8 kDa (FibL), and 25.7 kDa (P25) (14–16). The high molecular weight and repetitive sequences of the FibH gene make it the primary determiner of silk mechanical properties (17).
The similarities of high molecular weight and repetitive structure between FibH and MaSp provide the possibility of using the genetically modified silkworm as the host to produce spider silk. In recent years, advancements in genetic manipulation technologies, notably piggyBac-mediated germ line transformation, have been established in B. mori, and transposon-mediated transgenic silkworms have been used to express the synthetic MaSps of Nephila clavipes and Araneus ventricosus (11, 18). Although improved mechanical properties of these genetically modified silkworm silks have been reported, the spider silk protein amounts in transgenic silkworms were very low, varying from 0.37 to 0.61% to 2 to 5% of the total fibroin (11, 18). How to increase the protein amount of spider silk in transgenic silkworms is still a challenge. It is necessary to develop a highly efficient and more stable system to achieve the mass production of spider silk in genetically modified silkworms.
Recently, newly developed genome editing technologies, such as zinc finger nucleases, transcription activator-like effector nucleases (TALENs), and clustered regularly interspaced short-palindromic repeats (CRISPR) and CRISPR-associated protein (CRISPR/Cas) systems, have been successfully used for functional genomic analysis in many organisms, as well as in B. mori (19–22). These custom-designed nucleases induced sequence-specific double-stranded breaks (DSB) on the genome, subsequently repaired by either nonhomologous end joining or homology-directed repair (HDR) (23). In the case of HDR, exogenous DNA fragments flanking homologous sequences can be precisely integrated into targeted genomic loci (24). This phenomenon makes it possible to incorporate exogenous spider silk genes into a targeted locus of the silkworm genome and to control its expression with endogenous regulatory elements, such as the FibH promoter, to achieve mass protein production.
In the current study, we performed TALEN-mediated, targeted gene mutagenesis of the FibH gene in B. mori. At the same time, we incorporated a synthetic spider silk protein of 67 kDa, which combined the partial major ampullate spidroin-1 (MaSp1) gene of Nephila clavipes with the partial silkworm FibH sequence, with the aid of TALEN-mediated HDR. This system achieved complete gene replacement of the FibH gene with synthetic MaSp1 and reached a remarkable MaSp1 protein expression amount of up to 35.2% in the cocoon shells of transformed silkworms. The transformed silk fiber showed lower strength but higher extensibility than that of wild-type silkworm silk fibers. This study uses the endogenous promoter to drive exogenous gene expression in B. mori, and the mass production of exogenous spider silk protein will pave the way for developing new biomaterials using the silkworm as the bioreactor.
Results
TALEN-Mediated Gene Replacement Targeting FibH.
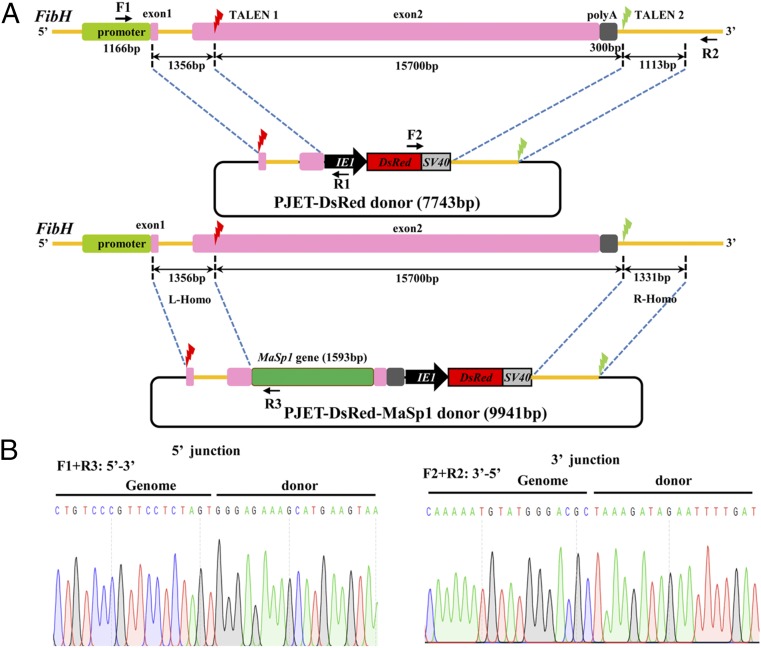
Since FibH is the major protein of the silk fiber and its protein amount reaches up to 70% wt/wt of the cocoon shell, modifying FibH will inevitably affect the mechanical characteristics of the silk fiber. The FibH gene cDNA is 15,381 bp and is highly repetitive, encoding a protein of 350 kDa in molecular weight. The genomic FibH sequence is 15,792 bp, including two exons and one 971-bp intron. N-terminal domains, including exon 1 (42 bp), intron (971 bp), and a part of the exon 2 (411 bp), are essential for FibH transcription, and the 180-bp C-terminal domain (CTD) is needed for silk spinning (25–28). To achieve FibH gene replacement, we designed two pairs of TALENs, one targeting exon 2 and the other targeting a sequence outside CTD (Fig. 1A). The TALEN activity in vitro was validated using the firefly luciferase single-strand annealing recombination assay in mammalian 293T cells (29). The luciferase activities of the two TALEN constructs were 13.9- and 43.3-fold higher than the control, indicating that they were efficient for in vivo experiments. For targeted integration of the FibH locus, two HDR donor plasmids were constructed. One plasmid (PJET-DsRed) contained a red fluorescent protein DsRed expression cassette and homologous left (1,356 bp) and right (1,331 bp) arms, while the other plasmid (PJET-DsRed-MaSp1) contained an additional chimeric MaSp1 expression cassette, which was designed to be controlled by the endogenous FibH promoter (Fig. 1A). DsRed expression was controlled by a baculovirus immediate early one (IE1) promoter to ubiquitously express red fluorescence, facilitating the screening of positive individuals from late embryonic to adult stages (30). Two TALEN target sites were introduced, flanking the homologous left and right arms to facilitate linearization in both plasmids. Expression of the endogenous FibH was completely disrupted after introducing both constructs, and MaSp1 expression was directed by the native FibH regulatory elements after introducing the PJET-DsRed-MaSp1 construct.
Fig. 1.
Schematic representation of the TALEN-mediated gene replacement system and the targeted integration of transgene constructs. (A) Schematic representation of the FibH gene and the TALEN targeting sites. The thin yellow line represents the FibH genomic locus, with the open boxes signifying the promoter, exons, and poly(A) signal. A 1,166-bp fragment (green box) located at the 5′ end represents the promoter region. The two 42-bp and 15,750-bp fragments (pink boxes) are exon 1 and exon 2, respectively. A 300-bp fragment (gray box) located at the 3′ end is the poly(A) signal. The red and green lightning icons indicate the two TALEN target sites. In the PJET-Red and PJET-MaSp1 donor constructs, a DsRed2 marker expression cassette driven by a baculovirus IE1 promoter was cloned into the PJET-1.2 vector. DNA fragments of 1,356 bp and 1,331 bp at the 5′ and 3′ ends flanking the TALEN sites were PCR-amplified, subcloned into vectors, and used as homologous arms (L-homo and R-homo, respectively). The 1,593-bp MaSp1 partial sequence is shown by the green box. Primer positions for amplification analyses of the integrated insertions in transformed silkworms are shown by arrows. Primer pairs of F1/R1 (or R3) and F2/R2 were used to amplify the 5′- and 3′-end insertion junctions, respectively. (B) Sequencing results of the integrated diagnostic DNA fragments to show 5′ and 3′ junction genome–donor integration.
Germ line transformation was performed by delivering mixed solutions of TALEN mRNAs and donor plasmids into preblastoderm embryos of B. mori. In total, 640 preblastoderm eggs in each group were microinjected, and the hatched embryos (G0) were reared to adult stage and sib-mated or crossed with wild-type (WT) moths. As a result, 15 DsRed-positive G1 broods were obtained from 116 broods of the PJET-DsRed group, and 10 DsRed-positive G1 broods were obtained from 129 broods of the PJET-DsRed-MaSp1 group (SI Appendix, Table S2). The transformation efficiency of 12.9% and 7.8% was defined for each group as the proportion of fertile injection survivors producing one or more transgenic offspring (30). Subsequently, PCR-based analysis was performed in DsRed-positive G1 animals to investigate the targeted integration of transgenes. As a result, both PJET-DsRed and PJET-DsRed-MaSp1 donors were precisely integrated into the FibH locus (Fig. 1B).
Physiological Consequences of FibH Disruption and Transgene Expression.
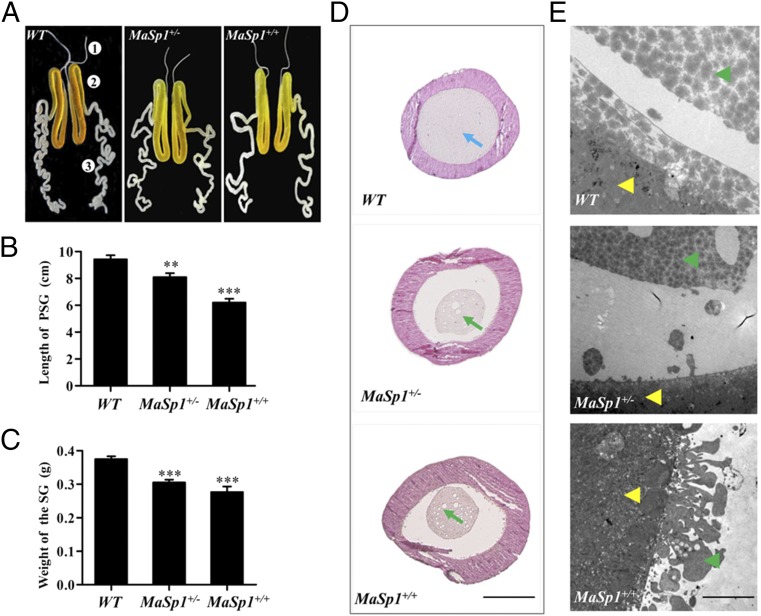
The animals carrying either PJET-DsRed or PJET-DsRed-MaSp1 transgenes grew normally, and no developmental defects appeared, suggesting that the transformation of the transgenes did not create deleterious consequences in silkworm physiology. Since FibH is predominantly expressed in the posterior silk gland (PSG) (31), we further investigated PSG development in the PJET-DsRed-MaSp1 animals. PSGs of both heterozygous (MaSp1+/−) and homozygous (MaSp1+/) animals at the fourth day of the fifth instar larval stage showed a significant reduction in weight and length compared with the WT animals (Fig. 2 A–C). Paraffin-embedded sections and subsequent hematoxylin–eosin staining revealed that PSG in the WT animals was dense and filled with proteins. However, MaSp1+/− and MaSp1+/+ PSGs displayed atrophic secretion, accompanied by small cavities (Fig. 2D). This result indicated that the total protein amounts significantly decreased in MaSp1+/− and MaSp1+/+ PSGs, probably due to the low molecular weight of MaSp1 (67 kDa) compared with the high molecular weight of FibH (350 kDa). The TEM results further showed that dense secretion in the WT animals formed bigger clumps than in the MaSp1+/− and MaSp1+/+ animals. In addition, the smooth boundary of the epidermal tissue layer in WT PSG became rough in MaSp1+/− and MaSp1+/+ PSGs (Fig. 2E).
Fig. 2.
Developmental phenotypes of silk glands in WT, MaSp1+/−, and MaSp1+/+ animals. (A) Gross morphology of the silk glands of WT, MaSp1+/−, and MaSp1+/+. The PSGs of MaSp1+/− and MaSp1+/+ were shorter in length than that in the WT animals. Numbers of 1, 2, and 3 indicate the anterior silk gland (ASG), middle silk gland (MSG), and PSG, respectively. (B) The statistical analysis of the length of the PSG (n = 10). Error bar: SD; ** and *** represent significant differences at the 0.01 and 0.001 level (t test) compared with the control. (C) The statistical analysis of the weight of the intact silk gland (n = 10). Error bar: SD; *** represents significant difference at the 0.001 level (t test) compared with the control. (D) Paraffin-embedded sections of the PSG from WT, MaSp1+/−, and MaSp1+/+ animals at the fourth day of the fifth instar stage. Tissues were stained with hematoxylin–eosin and were photographed under a bright field. Blue arrowheads indicate normal internal secretion structures, and green arrowheads indicate the atrophic secretion, which was smaller and vacuolated in MaSp1+/− and MaSp1+/+ animals. (Scale bar: 0.2 mm.) (E) The boundary of the epidermal tissue and secretion examined by the TEM. The yellow arrows show the epidermal tissue of the PSG, and the green arrows show the internal secretion. (Scale bar: 5 μm.)
Targeted Mass Spider Silk Production.
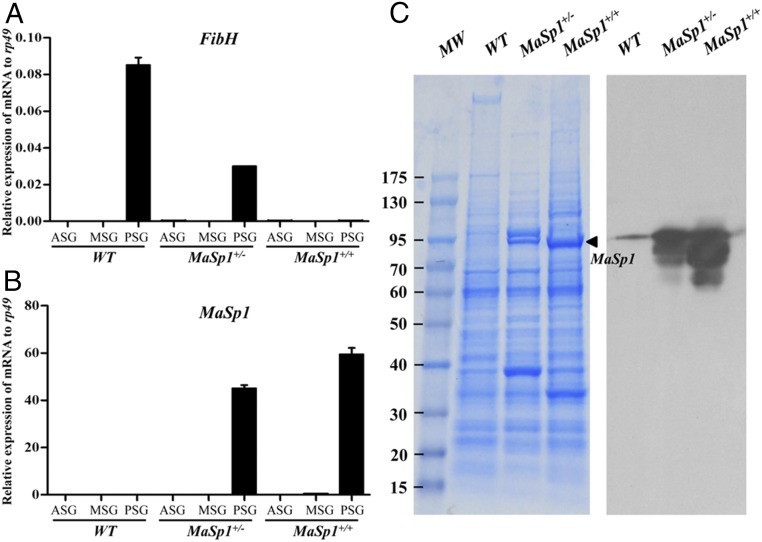
The relative mRNA levels of FibH and MaSp1 were investigated using qRT-PCR in the silk glands of WT, MaSp1+/−, and MaSp1+/+ animals. FibH was predominantly expressed in PSG of WT animals. Comparatively, its expression reduced to half in PSG of MaSp1+/− animals and was not detectable in MaSp1+/+ animals (Fig. 3A), indicating successful FibH disruption in MaSp1+/+ animals. In contrast, the relative mRNA levels of MaSp1 were undetectable in the PSGs of WT animals and were highly expressed in MaSp1+/− and MaSp1+/+ animals (Fig. 3B). These results suggested that the expression of MaSp1 in the transformed animals was controlled by the native FibH promoter. Abundant MaSp1 protein was detected by SDS/PAGE and the Western blotting analyses in the PSG of transformed silkworm larvae (Fig. 3C). These data suggested that MaSp1 was predominantly expressed in PSG and was mediated by the native FibH promoter.
Fig. 3.
MaSp1 expression in silk glands of transformed animals. (A) Relative mRNA expression of FibH determined using qRT-PCR in WT, MaSp1+/−, and MaSp1+/+ animals. ASG, MSG, and PSG were separated for investigation. The results are expressed as the means ± SD of three independent biological replicates. (B) Relative mRNA expression of MaSp1 determined using qRT-PCR in WT, MaSp1+/−, and MaSp1+/+ animals. (C) MaSp1 protein in PSG was subjected to SDS/PAGE analysis followed by Coomassie brilliant blue staining (Left) or Western blotting analysis (Right). Thin band in the WT lane was due to spillover from the large amount of MaSp1 present in the other lanes. The number in the left indicates the protein marker (kilodaltons). The black arrows show the MaSp1 protein.
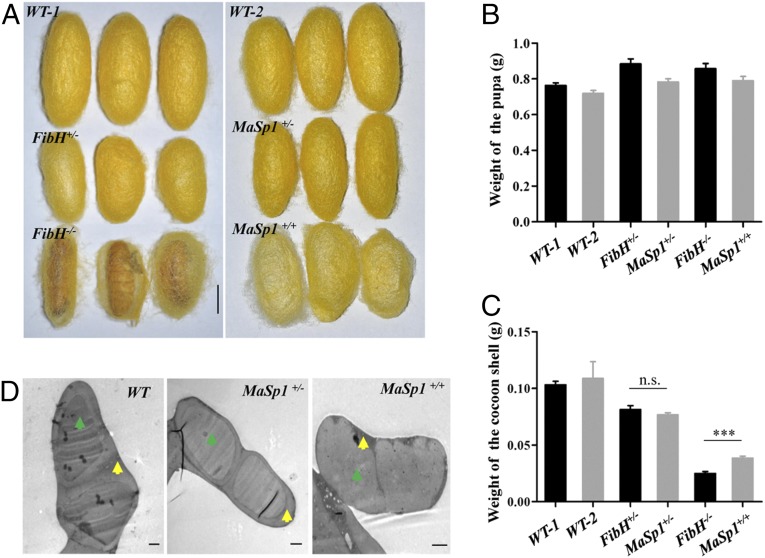
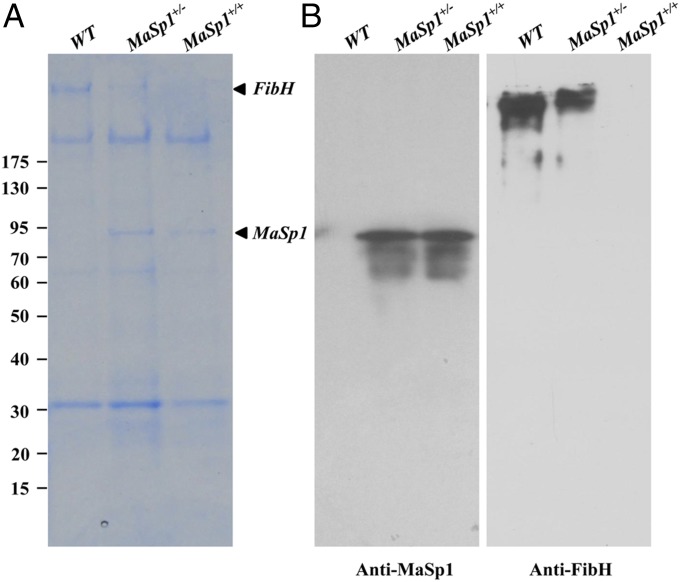
Disruption of FibH expression significantly reduced silk production, and the FibH−/− animals only formed very thin cocoon shells (Fig. 4A), similar to previous reports (32, 33). In comparison, both MaSp1+/− and MaSp1+/+ animals spun silk and formed elliptical cocoon shells (Fig. 4A). Both the PJET-DsRed and PJET-DsRed-MaSp1 groups showed increased pupa weight compared with the WT animals (Fig. 4B). However, the weight of the cocoon shells significantly decreased in both groups (Fig. 4C). TEM analysis revealed that the internal structure of the silk fiber and the sericin layer of MaSp1+/− and MaSp1+/+ silks became thinner. Particularly, the boundary of the sericin layer and fibroin was fuzzy in MaSp1+/+ silk fiber (Fig. 4D). The proteins extracted from cocoon shells were subjected to SDS/PAGE and Western blotting analyses. Abundant MaSp1 protein was detected in cocoon shells of both MaSp1+/− and MaSp1+/+ animals (Fig. 5), suggesting that chimeric MaSp1 was secreted into the cocoon shell. Therefore, the chimeric MaSp1 protein yields in MaSp1+/+ animals were calculated based on the average weight difference of a single cocoon shell between MaSp1+/+ (38.6 mg) and FibH−/− (25.0 mg) animals (Fig. 4C and Table 1). The result showed that the average MaSp1 protein amount reached to 13.6 mg, which was 35.2% wt/wt of a single cocoon shell in MaSp1+/+ animals (Table 1).
Fig. 4.
Analysis of transformed cocoons. (A) Morphology of the WT-1, FibH+/−, FibH−/−, WT-2, MaSp1+/−, and MaSp1+/+ cocoons. (Scale bar: 1 cm.) WT-1, FibH+/−, and FibH−/− are from the same brood. WT-2, MaSp1+/−, and MaSp1+/+ are from the same brood. (B) The statistical analysis of the weight of the pupa. (C) The statistical analysis of the weight of the cocoon shell. Error bar: SD; n.s. and *** represent significant differences at the nonsignificant and 0.001 level (t test) compared with the control. (D) The internal structure of the silk fiber examined by TEM. The yellow arrows show the sericin layer, and the green arrows represent fibroin. (Scale bars: 2 μm.)
Fig. 5.
MaSp1 expression in cocoon shells of transformed animals. (A) Results of Coomassie brilliant blue staining. (B) The results of Western blot using the antibodies for MaSp1 (Left) and FibH (Right). Numbers to the left indicate the protein molecular mass markers (kilodaltons).
Table 1.
Statistic analysis of the weight of cocoon and pupa
| Weight | WT-1 (n = 44) | FiBH+/− (n = 32) | FiBH−/− (n = 30) | WT-2 (n = 52) | MaSp1+/− (n = 52) | MaSp1+/+ (n = 38) |
| Cocoon weight, g | 0.103 ± 0.00323 | 0.0813 ± 0.00352 | 0.0250 ± 0.00174 | 0.109 ± 0.0148 | 0.0769 ± 0.00175 | 0.0386 ± 0.00168 |
| Pupa weight, g | 0.762 ± 0.0155 | 0.884 ± 0.0286 | 0.857 ± 0.0292 | 0.719 ± 0.0158 | 0.783 ± 0.0182 | 0.79 ± 0.0236 |
The table includes the number (n) of fibers tested in each group. WT-1, FibH+/−, and FibH−/− are from the same brood. WT-2, MaSp1+/−, and MaSp1+/+ are from the same brood.
Mechanical Properties of the MaSp1 Silk Fibers.
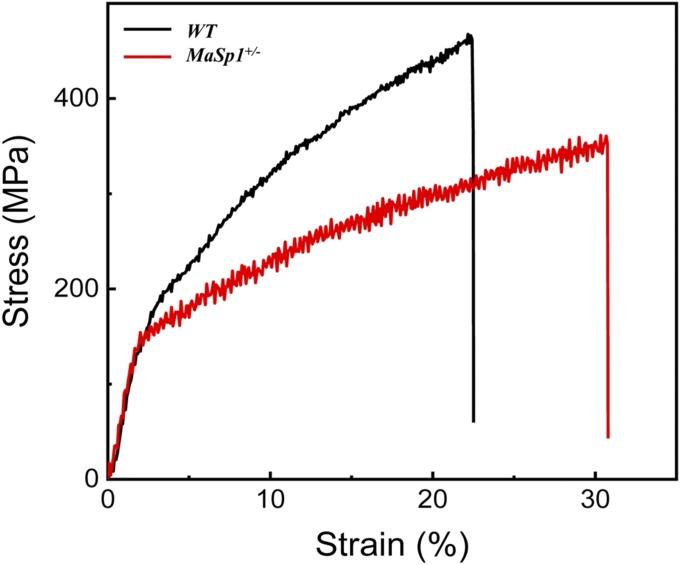
Due to difficulties in obtaining single fibers from the cocoon shells of the MaSp1+/+ animals, we investigated the mechanical properties of the single silk fibers from MaSp1+/− as well as WT animals. The average cross-section areas of the silk fibers were measured using Atlas software, and the results showed that the average cross-sectional areas of the MaSp1+/− fiber were 66.9 μm2, which was 15.8% smaller than that of the WT fibers of 79.5 μm2 (Table 2). The average breaking stress of the MaSp1+/− fiber was 371.5 MPa, which was 17.4% lower than that of the WT fibers of 449.5 MPa, indicating that the MaSp1 chimeric fiber was not as strong as the WT fiber. However, the MaSp1 chimeric fiber could be stretched to 32.2%, which was more elastic than the WT fiber, with 22.5% breaking strain. The average value of Young’s modulus showed no significant difference between the two kinds of fibers. Finally, the MaSp1+/− fiber showed a breaking energy of 84.8 MJ/m3, which was 22.5% higher than that of 69.2 MJ/m3 in the WT fiber (Fig. 6). These results indicated that the MaSp1 chimeric fiber had increased extensibility but decreased strength.
Table 2.
Mechanical properties of WT and MaSp1+/− silk fibers
| Group | Breaking stress, MPa | Breaking strain, % | Young’s modulus, GPa | Breaking energy, MJ/m3 | Average areas, μm2 |
| WT (n = 15) | 449.5 ± 27.6 | 22.5 ± 2.2 | 9.0 ± 1.7 | 69.2 ± 9.9 | 79.5 ± 9.5 |
| MaSp1+/− (n = 14) | 371.5 ± 27.5 | 32.2 ± 4.6 | 8.9 ± 1.3 | 84.8 ± 14.4 | 66.9 ± 8.4 |
The table includes the number (n) of fibers tested in each group.
Fig. 6.
Stress−strain curves of WT and MaSp1+/− silk fibers. The result of the WT fiber is shown in black, and the MaSp1+/− is shown in red respectively.
Discussion
In the current study, we report a genetic transformation strategy in B. mori using TALEN-mediated HDR, and we successfully achieved the targeted gene replacement of FibH with the synthesized spider silk gene MaSp1. This custom-designed TALEN system targeted exon 2 and the 3′ end of the FibH locus to induce DNA DSBs. Meanwhile, the presence of donor constructs resulted in the precise integration of the MaSp1 sequence to express the chimeric spider silk gene under the control of the endogenous FibH promoter as well as the IE1-derived expression of the DsRed selection marker. The transformation efficiency was 7.8 to 12.9%, which was similar to our previous work (24) and was comparable to transposon-based germ line transformation (34). Compared with transposon-based genetic transformation, which involves random insertion of multiple transgenes at different integration sites and leads to possible instability of integrated sequences (35), the current TALEN-mediated targeted gene integration provides a more stable expression system.
The FibH gene with high molecular weight (350 kDa) and a repetitive sequence is the main component of silkworm silk proteins and the primary determiner of silk mechanical properties. The use of the FibH promoter to drive spider silk gene expression in transgenic silkworms has been reported, and chimeric spider silk protein yields varied from 0.37 to 0.61% (18) to 2 to 5% (11) of the total fibroin, according to SDS/PAGE analysis. The low protein yield was likely due to endogenous FibH expression, which occupied most of the silk protein components. Another reason for this outcome was the exogenous FibH promoter used in these works, which did not function as efficiently as the endogenous FibH promoter. To achieve mass spider silk yields, we performed a FibH gene knockout and an MaSp1 gene knock-in at the same time. SDS/PAGE analysis clearly showed the presence of the MaSp1 protein in transformed silk glands and cocoon shells. We used a simpler method to measure the MaSp1 protein amount by comparing cocoon shell weights between MaSp1+/+ and FibH−/− animals, which both showed no endogenous FibH expression. As a result, the protein amounts of MaSp1 reached 35.2% wt/wt of a single cocoon shell. Considering the relatively small molecular weight of the chimeric MaSp1 (67 kDa) translated from the partial cDNA sequence, this remarkable yield will further increase when integrating proteins of higher molecular weights. This result will be of particular interest in the mass production of exogenous proteins, including intact spider silk protein. In the current study, smaller PSGs in the FibH−/− and MaSp1+/+ animals and the abnormal appearance of intracellular vesicles in PSG cells expressing the MaSp1 transgene were possible consequences of the MaSp1 protein having a smaller molecular weight than FibH. In addition, it was likely that there was an imbalance between the newly synthesized MaSp1 protein and FibL and P25, which are normally synthesized and then extruded in the spun thread in fixed molar quantities, resulting in failure to form an integrated complex. Such failure could disrupt the normal function of the secretory apparatus and possibly induce apoptotic events, giving rise to the observed abnormalities and reducing the ultimate amounts of all or most proteins. A CRISPR/Cas9-mediated FibH knockout analysis also reported similarly smaller silk glands, and that detailed analysis of changes in the transcriptional expression supports this idea (34).
The targeted integration of MaSp1 as well as the disruption of FibH significantly changed the mechanical properties of silkworm silk fiber. The breaking stress of the chimeric MaSp1+/− fiber was inferior to that of the WT silk fiber. We presume that this result was due to (i) the chimeric MaSp1 protein having a less repetitive structure and a smaller molecular weight (67 kDa) than that of FibH (350 kDa) and (ii) unlike transposon-derived transgenic silkworms, the endogenous FibH amount being reduced to half in the MaSp1+/− animals. However, the breaking strain and breaking energy significantly increased in MaSp1+/− fiber, indicating that the incorporation of MaSp1 into the silkworm silk fiber increased the extensibility. Improving toughness and extensibility of the fused spider/silkworm fiber will be largely dependent on integrating spider genes with different secondary structures and higher molecular weights containing repetitive structures. Nonetheless, our data provide an example of the considerable mass production of spider silk in a heterologous expression system, and they shed light on the future application of natural biomaterials, including spider silk fiber.
Materials and Methods
Details about silkworm materials are described in SI Appendix, Materials and Methods. TALENs vector and donor plasmid construction, silkworm germ line transformation, targeted integration analysis, gene expression, morphology analysis, Western blotting analysis, and mechanical testing of silk fibers were carried out according to protocols described in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Hui Xiang for her generous help in TALENs vector construction. This work was supported by Chinese Academy of Sciences Grant KJZD-EW-L12-02, National Postdoctoral Program for Innovative Talents Grant BX201700268, China Postdoctoral Science Foundation Grant 2017M621548, Zhejiang Public Welfare Technologies Research Projects Grant 2017C32058, and Projects of Zhejiang Provincial Science and Technology Plans Grant 2016C02054-19.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1806805115/-/DCSupplemental.
References
- 1.Tao H, Kaplan DL, Omenetto FG. Silk materials—A road to sustainable high technology. Adv Mater. 2012;24:2824–2837. doi: 10.1002/adma.201104477. [DOI] [PubMed] [Google Scholar]
- 2.Tokareva O, Jacobsen M, Buehler M, Wong J, Kaplan DL. Structure-function-property-design interplay in biopolymers: Spider silk. Acta Biomater. 2014;10:1612–1626. doi: 10.1016/j.actbio.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blamires SJ, Blackledge TA, Tso IM. Physicochemical property variation in spider silk: Ecology, evolution, and synthetic production. Annu Rev Entomol. 2017;62:443–460. doi: 10.1146/annurev-ento-031616-035615. [DOI] [PubMed] [Google Scholar]
- 4.Scheibel T. Spider silks: Recombinant synthesis, assembly, spinning, and engineering of synthetic proteins. Microb Cell Fact. 2004;3:14. doi: 10.1186/1475-2859-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yip EC, Rayor LS. Maternal care and subsocial behaviour in spiders. Biol Rev Camb Philos Soc. 2014;89:427–449. doi: 10.1111/brv.12060. [DOI] [PubMed] [Google Scholar]
- 6.Andersson M, et al. Biomimetic spinning of artificial spider silk from a chimeric minispidroin. Nat Chem Biol. 2017;13:262–264. doi: 10.1038/nchembio.2269. [DOI] [PubMed] [Google Scholar]
- 7.Jansson R, et al. Functionalized silk assembled from a recombinant spider silk fusion protein (Z-4RepCT) produced in the methylotrophic yeast Pichia pastoris. Biotechnol J. 2016;11:687–699. doi: 10.1002/biot.201500412. [DOI] [PubMed] [Google Scholar]
- 8.Lazaris A, et al. Spider silk fibers spun from soluble recombinant silk produced in mammalian cells. Science. 2002;295:472–476. doi: 10.1126/science.1065780. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, et al. Expression of EGFP-spider dragline silk fusion protein in BmN cells and larvae of silkworm showed the solubility is primary limit for dragline proteins yield. Mol Biol Rep. 2008;35:329–335. doi: 10.1007/s11033-007-9090-6. [DOI] [PubMed] [Google Scholar]
- 10.Hauptmann V, et al. Native-sized spider silk proteins synthesized in planta via intein-based multimerization. Transgenic Res. 2013;22:369–377. doi: 10.1007/s11248-012-9655-6. [DOI] [PubMed] [Google Scholar]
- 11.Teulé F, et al. Silkworms transformed with chimeric silkworm/spider silk genes spin composite silk fibers with improved mechanical properties. Proc Natl Acad Sci USA. 2012;109:923–928. doi: 10.1073/pnas.1109420109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldsmith MR, Shimada T, Abe H. The genetics and genomics of the silkworm, Bombyx mori. Annu Rev Entomol. 2005;50:71–100. doi: 10.1146/annurev.ento.50.071803.130456. [DOI] [PubMed] [Google Scholar]
- 13.Mondal M, Trivedy K, Kumar SN. The silk proteins, sericin and fibroin in silkworm, Bombyx mori Linn.—A review. Caspian J Environ Sci. 2007;5:63–76. [Google Scholar]
- 14.Chevillard M, Couble P, Prudhomme JC. Complete nucleotide sequence of the gene encoding the Bombyx mori silk protein P25 and predicted amino acid sequence of the protein. Nucleic Acids Res. 1986;14:6341–6342. doi: 10.1093/nar/14.15.6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamaguchi K, et al. Primary structure of the silk fibroin light chain determined by cDNA sequencing and peptide analysis. J Mol Biol. 1989;210:127–139. doi: 10.1016/0022-2836(89)90295-7. [DOI] [PubMed] [Google Scholar]
- 16.Zhou CZ, et al. Fine organization of Bombyx mori fibroin heavy chain gene. Nucleic Acids Res. 2000;28:2413–2419. doi: 10.1093/nar/28.12.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanaka K, et al. Determination of the site of disulfide linkage between heavy and light chains of silk fibroin produced by Bombyx mori. Biochim Biophys Acta. 1999;1432:92–103. doi: 10.1016/s0167-4838(99)00088-6. [DOI] [PubMed] [Google Scholar]
- 18.Kuwana Y, Sezutsu H, Nakajima K, Tamada Y, Kojima K. High-toughness silk produced by a transgenic silkworm expressing spider (Araneus ventricosus) dragline silk protein. PLoS One. 2014;9:e105325. doi: 10.1371/journal.pone.0105325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takasu Y, et al. Targeted mutagenesis in the silkworm Bombyx mori using zinc finger nuclease mRNA injection. Insect Biochem Mol Biol. 2010;40:759–765. doi: 10.1016/j.ibmb.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 20.Ma S, et al. Highly efficient and specific genome editing in silkworm using custom TALENs. PLoS One. 2012;7:e45035. doi: 10.1371/journal.pone.0045035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sajwan S, et al. Efficient disruption of endogenous Bombyx gene by TAL effector nucleases. Insect Biochem Mol Biol. 2013;43:17–23. doi: 10.1016/j.ibmb.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, et al. The CRISPR/Cas system mediates efficient genome engineering in Bombyx mori. Cell Res. 2013;23:1414–1416. doi: 10.1038/cr.2013.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, et al. Site-specific, TALENs-mediated transformation of Bombyx mori. Insect Biochem Mol Biol. 2014;55:26–30. doi: 10.1016/j.ibmb.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He YX, et al. N-Terminal domain of Bombyx mori fibroin mediates the assembly of silk in response to pH decrease. J Mol Biol. 2012;418:197–207. doi: 10.1016/j.jmb.2012.02.040. [DOI] [PubMed] [Google Scholar]
- 26.Shimizu K, et al. Structure and function of 5′-flanking regions of Bombyx mori fibroin heavy chain gene: Identification of a novel transcription enhancing element with a homeodomain protein-binding motif. Insect Biochem Mol Biol. 2007;37:713–725. doi: 10.1016/j.ibmb.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 27.Wang SP, Guo TQ, Guo XY, Huang JT, Lu CD. Structural analysis of fibroin heavy chain signal peptide of silkworm Bombyx mori. Acta Biochim Biophys Sin (Shanghai) 2006;38:507–513. doi: 10.1111/j.1745-7270.2006.00189.x. [DOI] [PubMed] [Google Scholar]
- 28.Wang SP, Guo TQ, Guo XY, Huang JT, Lu CD. In vivo analysis of fibroin heavy chain signal peptide of silkworm Bombyx mori using recombinant baculovirus as vector. Biochem Biophys Res Commun. 2006;341:1203–1210. doi: 10.1016/j.bbrc.2006.01.081. [DOI] [PubMed] [Google Scholar]
- 29.Huang P, et al. Heritable gene targeting in zebrafish using customized TALENs. Nat Biotechnol. 2011;29:699–700. doi: 10.1038/nbt.1939. [DOI] [PubMed] [Google Scholar]
- 30.Tan A, et al. Transgene-based, female-specific lethality system for genetic sexing of the silkworm, Bombyx mori. Proc Natl Acad Sci USA. 2013;110:6766–6770. doi: 10.1073/pnas.1221700110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oyama F, Mizuno S, Shimura K. Predominant synthesis of fibroin heavy and light chains on the membrane-bound polysomes prepared from the posterior silk gland of the silkworm, Bombyx mori. J Biochem. 1984;96:1143–1153. doi: 10.1093/oxfordjournals.jbchem.a134932. [DOI] [PubMed] [Google Scholar]
- 32.Ma S, et al. Genome editing of BmFib-H gene provides an empty Bombyx mori silk gland for a highly efficient bioreactor. Sci Rep. 2014;4:6867. doi: 10.1038/srep06867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui Y, et al. New insight into the mechanism underlying the silk gland biological process by knocking out fibroin heavy chain in the silkworm. BMC Genomics. 2018;19:215. doi: 10.1186/s12864-018-4602-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tamura T, et al. Germline transformation of the silkworm Bombyx mori L. using a piggyBac transposon-derived vector. Nat Biotechnol. 2000;18:81–84, and erratum (2000) 18:559. doi: 10.1038/71978. [DOI] [PubMed] [Google Scholar]
- 35.Fraser MJ., Jr Insect transgenesis: Current applications and future prospects. Annu Rev Entomol. 2012;57:267–289. doi: 10.1146/annurev.ento.54.110807.090545. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.