Significance
Extant protein sequences are the result of evolutionary pressure. Eliminating core packing interactions by mutation, regardless of position, generally results in similar perturbations to global stability. In contrast, we find that cavity creation has highly varied consequences for a protein folding landscape, depending upon the context in which the cavities are introduced. These observations have implications for interpreting evolutionary adaptation, as it is likely that proteins have evolved to exhibit optimal levels of conformational heterogeneity and dynamics. These results should also inform protein engineering efforts, as they provide insight into how sequence can modulate the population of functionally important excited states, as well as states that lead to secondary, undesirable reactions such as oligomerization, aggregation, surface activity, and phase separation.
Keywords: repeat protein folding, high pressure, NMR, SAXS, cooperativity
Abstract
The effect of introducing internal cavities on protein native structure and global stability has been well documented, but the consequences of these packing defects on folding free-energy landscapes have received less attention. We investigated the effects of cavity creation on the folding landscape of the leucine-rich repeat protein pp32 by high-pressure (HP) and urea-dependent NMR and high-pressure small-angle X-ray scattering (HPSAXS). Despite a modest global energetic perturbation, cavity creation in the N-terminal capping motif (N-cap) resulted in very strong deviation from two-state unfolding behavior. In contrast, introduction of a cavity in the most stable, C-terminal half of pp32 led to highly concerted unfolding, presumably because the decrease in stability by the mutations attenuated the N- to C-terminal stability gradient present in WT pp32. Interestingly, enlarging the central cavity of the protein led to the population under pressure of a distinct intermediate in which the N-cap and repeats 1–4 were nearly completely unfolded, while the fifth repeat and the C-terminal capping motif remained fully folded. Thus, despite modest effects on global stability, introducing internal cavities can have starkly distinct repercussions on the conformational landscape of a protein, depending on their structural and energetic context.
Folded proteins are extremely well packed, with densities similar to crystals of small organic molecules (1–3). This tight packing of folded proteins contributes significantly to their global stability (4–6). Nonetheless cavities exist in most proteins, occupying up to 2% of the total protein volume, and their size, shape, and distribution in protein structures are highly heterogeneous (7–9). It has been suggested that the existence of internal cavities imparts flexibility to folded proteins that may be important for function (10). While water molecules have been observed in some cavities of protein structures determined by X-ray crystallography, many rather large, hydrophobic cavities present no crystallographically determined solvent density (7, 11). Although under debate for some time, it appears now that, even in solution, the solvent occupancy of such hydrophobic cavities at ambient pressure is very low (12–14). Indeed, the strong correlation between internal cavity volume and the volume change for protein unfolding is consistent with solvent exclusion from deeply buried void volumes in proteins (15, 16).
Mutations that replace large hydrophobic amino acids, isoleucine, leucine, or valine, with alanine create internal cavities and have been shown to destabilize proteins by ∼1.1 kcal/mol/CH2 group, a bit less in smaller proteins (6). Indeed, the stability of cavity-containing mutants is linearly correlated with cavity size, with ∼24 cal/mol decrease in stability per cubic angstrom (5). Some deviation from this average perturbation can result from specific local features of protein structures. Cavities engineered into rigid regions of proteins tend to retain their expected shape based on the wild-type (WT) structure, whereas in more flexible regions of proteins, rearrangements can occur that decrease cavity volume (4, 17).
While the effect of cavity creation on protein global stability and native structure has been well documented, the consequences of introducing cavities on the population of excited states and folding intermediates have received less attention, despite the widespread application of cavity creation in protein engineering methods such as ϕ-value analysis (18, 19). If internal cavities contribute to protein conformational heterogeneity, it is conceivable that protein sequences have evolved to present internal packing characteristics required to define not only their folded structure and stability but also the population of less ordered states implicated in their function and proteostasis. To explore this notion, we set out to determine how the introduction of internal cavities in different regions of a protein affects its folding pathway and the population of intermediates on the folding landscape. We chose as a model system the leucine-rich repeat region of the Anp32 protein (pp32) since interpretation of contextual effects of the mutations is more straightforward in a protein exhibiting a repetitive linear architecture lacking sequence distant native contacts.
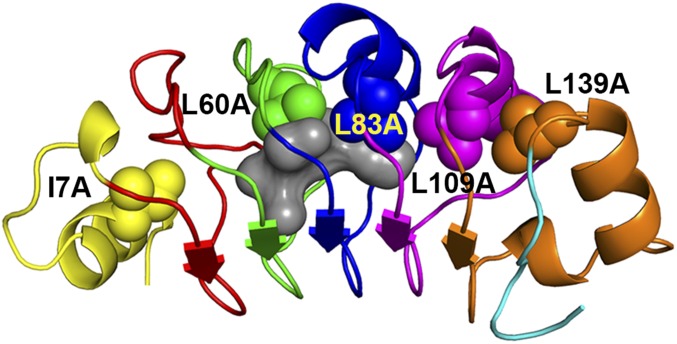
The pp32 protein is made up of five leucine-rich repeats flanked on the N and C termini by capping motifs (20) (Fig. 1). The global stability and folding mechanism of WT pp32 have been well characterized (21, 22). The ϕ-value analysis revealed an early transition state in which the C-terminal capping motif and the fifth repeat are ordered at the folding barrier. At least one partially structured intermediate was identified by double-jump denaturant kinetics and in high-pressure NMR experiments, in which the most stable C-terminal region of the protein is folded, while the N-terminal half remains disordered. Thus, there exists in WT pp32 an intrinsic energetic hierarchy with increasing stability from the N to the C terminus of the protein. We investigate here the effects of cavity creation on the folding landscape of pp32 using five of the cavity-containing variants engineered previously for ϕ-value analysis (21). These variants harbor cavities in the N-terminal capping motif (I7A), and the second (L60A), third (L83A), fourth (L109A), and fifth (L139A) repeats (Fig. 1). Previous equilibrium unfolding studies on these variants showed that they unfolded cooperatively in urea. The average decrease in global stability due to cavity creation for four of these variants was 1.2 ± 0.2 kcal/mol/CH2 group (21), in line with previous observations on other proteins (6). The N-terminal cavity-containing variant (I7A) was less perturbed by the mutation, with a decrease of 0.7 kcal/mol/CH2 group.
Fig. 1.
Schematic diagram of pp32. Leucine residues substituted by alanine in the five cavity-containing variants studied are rendered as spheres. The structure was rendered using Pymol (27). The central cavity (gray) was detected using Hollow and a 1.1-Å probe radius (28).
We used residue-specific NMR approaches to characterize the high-pressure and urea-induced unfolding transitions of these five cavity-containing variants of pp32. We found that despite a smaller global energetic perturbation, substitution of isoleucine by alanine at position 7 in the N-terminal capping motif resulted in very strong deviation from two-state unfolding behavior, amplifying the stability gradient already present in WT pp32. The intermediates of I7A had lost the native-state backbone amide resonance intensities in the perturbed N-terminal half of the protein, yet maintained some core packing. In contrast, cavity creation in the more stable C-terminal half of pp32 (L83A, L09A, and L139A) led to more cooperative unfolding than observed for the WT protein, presumably because the decrease in stability in the C-terminal repeats and capping motif attenuated the WT N- to C-terminal stability gradient. Interestingly, enlarging the central cavity of the protein (L60A) led to the population under pressure of a distinct intermediate, in which the N-cap and repeats 1–4 were nearly fully unfolded, while the fifth repeat and the C-cap retained their native structure. These results demonstrate that, despite similar effects on global stability, the effects of internal cavities on the conformational landscape of pp32 are strongly dependent upon their structural and energetic context.
Results
Cavity Creation Causes Context-Dependent Chemical Shift Perturbation Patterns.
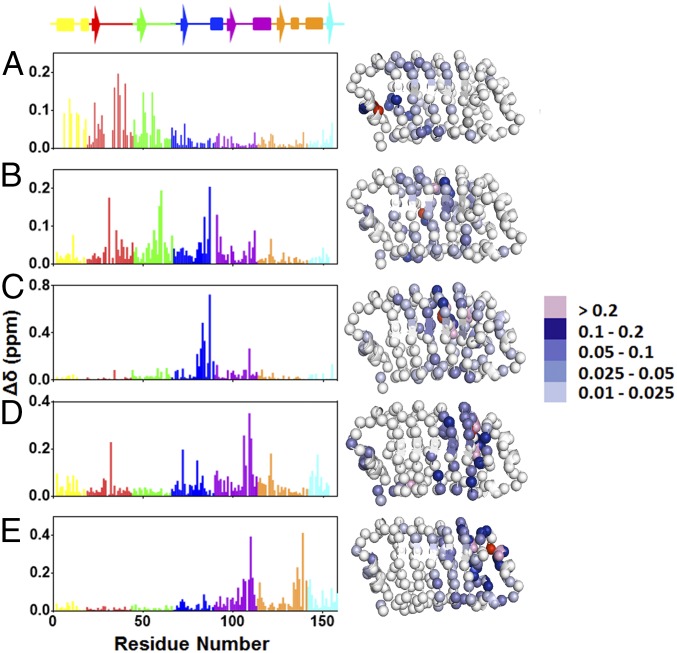
To examine the structural integrity of cavity variants of pp32, we measured and assigned backbone amide 1H and 15N chemical shifts. The chemical shift perturbations with respect to the WT, calculated as the geometric mean of the 1H and 15N chemical shift differences (17), reveal substantial chemical shifts in the vicinity of each of the five cavity sites. Overall, the extended cores of cavity variants show modest chemical shift changes, indicating sufficient rigidity to prevent large structural rearrangements. Comparing variants, we find that introduction of cavities in the C-terminal half of pp32 (repeats 4 and 5) results in larger amplitude of shift perturbations than the I7A and L60A substitutions, which result in smaller yet farther reaching chemical shift perturbations (Fig. 2). Perturbations appear to be larger in the loop regions connecting the β-strands and α-helices than in the secondary-structure elements themselves.
Fig. 2.
Chemical shift perturbations of the cavity-containing variants relative to WT pp32. (A) I7A, (B) L60A, (C) L83A, (D) L109A, and (E) L139A variants. The structures, based on the crystal structure of pp32 (19), depict the backbone nitrogen atoms as spheres. They were rendered using Pymol (23). The backbone nitrogen at the site of the mutation is colored in red. Residues are colored for the amplitude of the chemical shift as indicated in the scale to the Right of the structures.
Cavities in Different Structural Contexts Lead to Contrasting Effects on pp32 Pressure-Induced Unfolding Cooperativity.
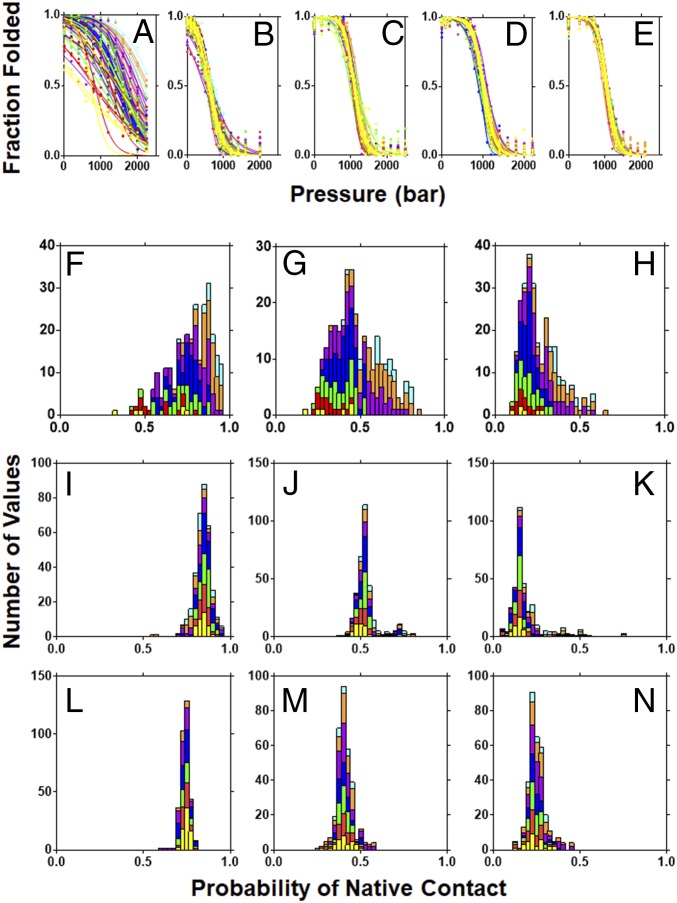
To examine the effects of cavities on the conformational landscape of pp32, we carried out high-pressure NMR experiments on the five cavity-containing variants. Pressure leads to protein unfolding due to a decrease in molar volume arising from the elimination upon unfolding of solvent excluded voids present in the folded states of proteins (16). Pressure-dependent 15N–1H heteronuclear single-quantum correlation NMR (HSQC NMR) spectra were acquired for the five cavity-containing variants of pp32 (SI Appendix, Fig. S1) and the intensity of the native-state amide peaks was plotted as a function of pressure (Fig. 3 A–E). The coincidence of all of the residue-specific pressure unfolding profiles is nearly perfect for the L139A. This coincidence diminishes slightly for variants with cavities in the fourth, third, and second repeats, while the residue-specific unfolding profiles observed for the I7A variant are highly dispersed. Fractional native contacts at several pressures were calculated, as previously described (22), as the geometric mean of the fractional HSQC amide peak intensities for each of the two residues in every pairwise native contact. Histograms of fractional native contacts for the L139A variant are narrow and symmetric, indicative of a nearly perfect two-state transition, whereas those for the I7A variant are quite broad and asymmetric, with the N-terminal repeats losing contacts at much lower pressure than the C-terminal regions (Fig. 3 F–N).
Fig. 3.
HSQC detected pressure-induced unfolding of the pp32 variants. (A–E) Pressure dependence of the native-state amide NH peaks for the pp32 variants: (A) I7A, (B) L60A, (C) L83A, (D) L109A, and (E) L139A variants. (F–N) fractional contact histograms for (F–H) I7A at 800, 1,600, and 2,000 bar, respectively; (I–K) L60A at 300, 600, and 900 bar, respectively; and (L–N) L139A at 700, 1,100, and 1,300 bar, respectively. Lines and bars are colored per repeat as in Fig. 1. Fractional contacts were calculated as described in Materials and Methods.
Each residue-specific pressure unfolding HSQC peak intensity profile for each variant was fit to a two-state transition, yielding residue-specific values for the apparent free-energy change at atmospheric pressure, ΔGf, and the volume change, ΔVf, of the transition (SI Appendix, Figs. S2 and S3). The HSQC native-state amide peak intensity for each residue reports on the transition from a conformational ensemble in which that residue resonates at its native-state frequency to another ensemble in which it does not, and is therefore, at least locally, two state. Analysis of the nearly coincident unfolding profiles from the amide HSQC peaks of the L139A, L109A, and L83A variants yielded symmetric and narrowly distributed ΔGf and ΔVf values (Table 1 and SI Appendix, Figs. S2 E–J, S3 E–J, and S4 E–J and Table S1), indicating that all residues reflect the complete transition from the folded to the unfolded state. The average ΔGf values are in good agreement with the values obtained for these variants previously from CD-detected urea unfolding (Table 1) (21). The volume changes for these variants are much larger (by 59–72 mL/mol) than that obtained previously for WT pp32 (Table 1 and SI Appendix, Fig. S4 F, H, and J and Table S1) (22), consistent with the creation of extra solvent-excluded void volume within the protein’s folded structure by the leucine-to-alanine mutations.
Table 1.
Thermodynamic parameters for WT pp32, I7A, and L139A unfolding by pressure and urea
| NMR observable | |||||||||
| Variant/perturbation | Parameter | U CH3 peak ∼0.92 ppm | U CH3 peak ∼0.85 ppm | Backbone –NH <all residues> | F and U Trp155 indole NH | Fluorescence | CD* | ||
| WT pressure† (1.4 M urea) | ΔG | −4.6 ± 0.4 (8.6 0 M U) | −5.4 ± 0.5 (9.4 0 M U) | −3.2 ± 0.4 (7.2 at 0 M U) | −3.4 ± 1.6 (7.4 at 0 M U) | −3.6 ± 0.1 (7.6 at 0 M U) | — | ||
| ΔV | 170 ± 15 | 166 ± 14 | 110 ± 12† | 109 ± 18 | 111 ± 7 | — | |||
| WT urea | ΔG | ND | ND | −6.2 ± 1.1 | −7.5 ± 0.5 | −7.9* | 7.9 ± 0.2* | ||
| m value | ND | ND | 2.3 ± 0.4 | 2.7 ± 0.2 | 2.86* | 2.86 ± 0.02* | |||
| I7A pressure | ΔG | −4.2 ± 0.1 | −3.2 ± 0.1 | −2.4 ± 0.3 | −3.7 ± 0.6 | −3.4 ± 0.1 4.25 at 0M U‡ | — | ||
| N-R4 | R5-C-cap | 2.0 ± 0.4 | 3.2 ± 0.2 | ||||||
| ΔV | 88 ± 8 | 67 ± 3 | 63 ± 8 | 64 ± 17 | 75 ± 3‡ | — | |||
| N-R4 | R5-C-cap | 59 ± 10 | 71 ± 4 | ||||||
| I7A urea | ΔG | −6.2 ± 0.3 | −6.6 ± 0.5 | −3.7 ± 2.3 | −6.0 ± 0.4 | — | −5.86 ± 0.06* | ||
| −2.8 ± 1.5 | −5.7 ± 2.7 | ||||||||
| m | 2.9 ± 0.2 | 3.0 ± 0.4 | 2.1 ± 1.0 | 2.9 ± 0.2 | — | 2.85 ± 0.04* | |||
| 1.7 ± 0.7 | 2.9 ± 1.3 | ||||||||
| L139A pressure (0 M urea) | ΔG | −4.6 ± 0.2 | −4.0 ± 0.2 | −4.5 ± 0.6 | −4.7 ± 0.3 | −4.0 ± 0.2 | |||
| ΔV | 180 ± 8 | 162 ± 8 | 178 ± 13 | 180 ± 11 | 153 ± 6 | — | |||
| L139A urea | ΔG | — | — | — | — | — | 4.4 ± 0.2* | ||
| m value | — | — | — | — | — | 3.4 ± 0.1* | |||
ΔG and ΔV values are in kilocalories⋅mole−1 and milliliter−1. m values are in kilocalorie−1⋅mole−1⋅molar−1. CD, circular dichroism. Individual values are the fitted value ± SE of the least-squares fit. Backbone NH are the mean ± SD over all residues. The Trp NH F and U peaks were fit globally assuming equal intensities for the unfolded and folded state peaks. Fluorescence is the average of least-square fits of three experiments ± SEM.
Dao et al. (21).
Fossat et al. (22).
Data were acquired in 0.3 M urea.
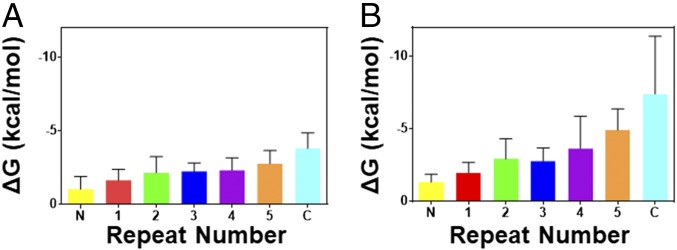
In contrast to the highly cooperative unfolding of the C-terminal cavity variants, the pressure-induced unfolding of I7A is clearly not globally two state. The residue-specific apparent ΔGf values increase in amplitude from the N to the C terminus (Fig. 4A, Table 1, and SI Appendix, Figs. S2A and S3A), and the histogram of these values is very broad and asymmetric (SI Appendix, Fig. S2B). The average apparent ΔGf values for all repeats of I7A are smaller than that observed in CD-detected urea denaturation (Table 1 and SI Appendix, Fig. S4). Likewise, the average apparent ΔVf values for each repeat are much smaller than those observed for WT pp32 (22), despite the introduction of a cavity in I7A. The small ΔGf and ΔVf values indicate that each residue is monitoring disruption of only a fraction of the protein. The ΔGf values can be reasonably grouped into three major bins (SI Appendix, Fig. S2K). Residues in the N-cap, parts of repeats 1 and 2, and a couple of residues in of the loop regions in repeats 3 and 4 (overlapping the pattern of chemical shift perturbations in Fig. 2A) exhibit ΔGf values below 1.8 kcal⋅mol−1, indicating local disruption to the N terminus, F → I1 (Table 1 and SI Appendix, Fig. S4 A and B). Average ΔGf and ΔVf values for most residues in repeats 2–4, as well as one to two residues in the loop regions of repeat 5 and the C-cap are larger than those in the N terminus and similar to each other (Fig. 4A and SI Appendix, Fig. S2 A, B, and K), suggesting that they reflect a transition, I1 → I2. The largest ΔGf and ΔVf values are found for the majority of residues in the extreme C terminus (repeat 5 and C-cap) and correspond to the final unfolding of the protein, I2 → U. Assuming for simplicity that this four-state model reasonably represents the unfolding of I7A under pressure, then the ΔGf and ΔVf values for each of these three transitions should add up to the total ΔGf and ΔVf for the complete F → U transition. Indeed, the total ΔVf is ∼180 mL/mol, similar to that found for the L139A cavity-containing variant. It is significant that each transition (F → I1, I1 → I2, and I2 → U), deduced from the residue-specific folded state HSQC peak profiles, exhibits a significant decrease in volume, 40–70 mL/mol (SI Appendix, Fig. S4B), indicating that solvent has replaced the cavity volume. Thus, the intermediate states are at least partially hydrated.
Fig. 4.
Average free-energy change per repeat for the I7A variant of pp32. Values were obtained from fits of the pressure-dependent (A) and urea-dependent (B) loss of native-state amide NH peak intensity of the I7A variant of pp32 as described in Materials and Methods. Bars are colored per repeat according to the schematic in Fig. 1.
The Pressure-Induced Unfolding Intermediates of the I7A Variant Retain Significant Core Packing.
To further characterize the intermediate and unfolded states populated in these high-pressure experiments, we monitored changes in intensity of two peaks in the 1D proton spectrum (∼0.85 and 0.92 ppm) corresponding to a composite of the unfolded state of all of the side-chain methyl groups. We also monitored the folded and unfolded state peaks of the tryptophan indole NH group in the 15N–1H HSQC spectrum for all five variants (SI Appendix, Fig. S5 A, C, E, G, and I). Analysis of the unfolded state methyl peak profiles for the L139A, L109A, and L83A variants (SI Appendix, Fig. S5 E, G, and I) yielded ΔGf values that were in good agreement with those obtained from the native-state amide peak intensities, as well as CD-detected urea-induced unfolding (Table 1 and SI Appendix, Table S1), consistent with highly cooperative unfolding. Likewise, the ΔVf values were similar to those obtained from the amide NH resonance profiles, and much larger than that observed for WT pp32, consistent with additional void volume resulting from the leucine-to-alanine mutations.
For the I7A variant, the composite profiles from the unfolded state methyl resonances did not reach a high-pressure plateau (SI Appendix, Fig. S5A), indicating that pressure does not completely unfold this variant in our pressure range. Moreover, ΔGf values were lower than those measured in CD-detected urea-induced unfolding experiments (Table 1), indicative of nonconcerted pressure-induced transitions. The onset of the appearance of the unfolded state methyl resonances begins above 1,000 bar, a pressure at which the native-state amide NH resonance intensities for residues in the N-terminal capping motif and repeats 1–3 have decreased by 30–60%. This observation suggests that the intermediates formed at lower pressures are partially folded, heterogeneous states in which the backbone amide resonances in the N-terminal regions of the protein are shifted with respect to the native state and exchange broadened so as to be undetectable, but in which significant side-chain core packing interactions are retained. Similar, although not identical, behavior has been observed for an N-terminal capping motif deletion variant of pp32 (23).
The folded and unfolded state peaks for the indole NH of Trp155 (in the extreme C terminus) of all variants were found to be in slow exchange (SI Appendix, Fig. S5 B, D, F, H, and J). The global stability extracted from analysis of the Trp155 indole NHs for the L83A, L109A, and L139A variants was in good agreement with that obtained in CD-detected urea-induced unfolding experiments (21) (Table 1 and SI Appendix, Table S1). In contrast, the profiles obtained for the I7A variant exhibited only ∼50% change by the highest pressure (2,500 bar) attainable by our HP NMR system (SI Appendix, Fig. S5B). Note that, although the tryptophan residue is not part of the natural sequence, but was added to all of the C terminus of all constructs to facilitate concentration determination, it has been shown to stabilize the folded LRR domain (21). The apparent ΔGf and ΔVf values from global analysis of the two profiles (F and U peaks of Trp155) were similar to those obtained from the average of the native-state amide NH resonances from the C-terminal capping motif (Table 1). These values predict that the transition monitored by the Trp155 F and U peaks to be complete by 4 kbar.
The unfolding profiles obtained from the increase in fluorescence of Trp155 as a function of pressure for L83A, L109A, and L139A variants yielded were similar to those obtained from Trp155 indole NH peaks (Table 1 and SI Appendix, Fig. S6 and Table S1). For complete fluorescence-detected unfolding of the I7A variant below 3,200 bar, 0.3 M urea was required (SI Appendix, Fig. S6). Analysis of the fluorescence unfolding profile yielded a small volume change of unfolding compared with WT (Table1), consistent with local unfolding of the C terminus. Extrapolation of the apparent free-energy change to 0 M urea using the m value obtained from the previously reported CD-detected urea experiments (21) yielded a ΔGf value that was larger than that obtained here from the Trp155 indole NH resonances of the I7A variant in absence of urea, indicating that pressure does not fully unfold the C terminus of the I7A variant in the absence of urea.
High-Pressure Small-Angle X-Ray Scattering Reveals I7A Is Not Completely Unfolded by Pressure.
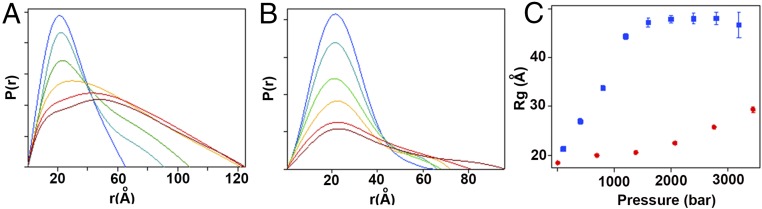
Previous pressure-dependent small-angle X-ray scattering (SAXS) measurements of WT pp32 in the presence of 1.4 M urea and a C-terminal destabilized variant in the presence of 0.5 M urea showed that these two proteins exhibited a radius of gyration (Rg) in the pressure-unfolded state equivalent to that obtained at atmospheric pressure in concentrated urea solution (23), ∼43 Å, which corresponds to a random coil polymer of the size of pp32 (23). High-pressure SAXS (HPSAXS) on the L139A and L7A pp32 variants (SI Appendix, Fig. S7 A and B) in the absence of urea yielded P(r) profiles and Rg values [from both P(r) and Guinier analysis] at atmospheric pressure (Fig. 5 A and C) similar to those obtained previously for WT pp32 in native conditions (23), and consistent with the folded state. As pressure increased, the P(r) plots for the L139A variant attained much larger Dmax values and the Rg increased to 47 Å, slightly larger than that obtained previously for WT pp32 under denaturing conditions, and the form of the Kratky plot at high pressure was consistent with a random coil configuration (SI Appendix, Fig. S7C). In previous work (23), a variant in which the N-terminal capping motif was deleted exhibited an Rg value at its high-pressure plateau (∼25 Å) that was much smaller than that obtained at high urea at atmospheric pressure (∼37 Å), this latter value corresponding to a random coil of the length of this N-cap deletion variant. As in the case of the capping variant, the Dmax (90 Å) and Rg (27 Å) values (Fig. 5 B and C) obtained for I7A at the highest pressures were much smaller than expected for a completely unfolded chain, and the Kratky plots (SI Appendix, Fig. S7D) indicate retention of some globular structure. Although the Rg vs. pressure plot for I7A did not reach a plateau by 3.5 kbar (Fig. 5C), unfolding is predicted from the C-terminal Trp155 indole NH NMR profiles to be 98% complete at this pressure. Hence, we conclude that the pressure-denatured state of this variant is not a random coil.
Fig. 5.
SAXS detected HP unfolding of the I7A and L139A variants of pp32. (A and B) Pressure-dependent pair distance distribution [P(r)] plots for pp32 L139A (A) and I7A (B), respectively, obtained from fits of the pressure-dependent scattering profiles Increasing pressures are from blue to red. For L139A, blue corresponds to atmospheric pressure, cyan to 0.4 kbar, green to 0.8 kbar, orange to 1.2 kbar, red to 1.6 kbar, and brown to 2.0 kbar. For I7A, blue corresponds to atmospheric pressure, cyan to 0.7 kbar, green to 1.4 kbar, orange to 2 kbar, pink to 2.8 kbar, and red to 3.5 kbar. (C) Radius of gyration as a function of pressure for L139A (blue) and I7A (red).
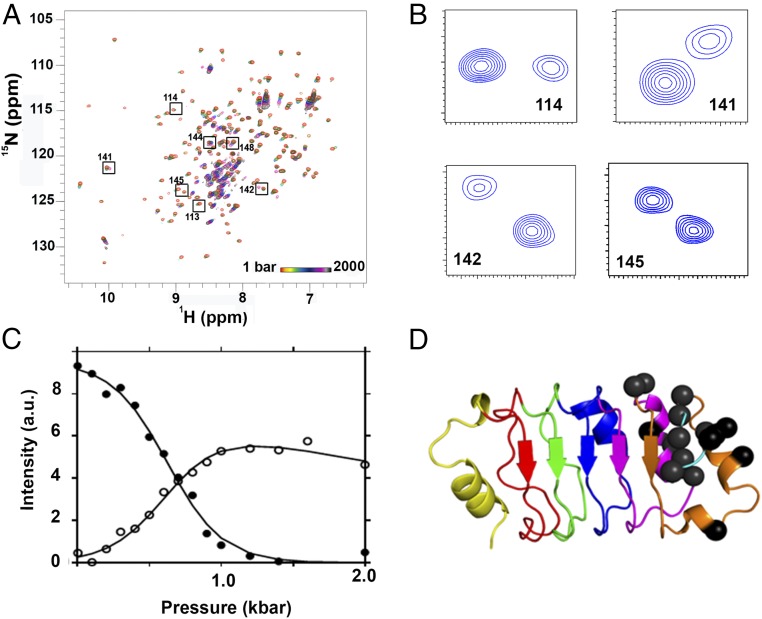
Under Pressure, the L60A Variant Populates a Unique Intermediate with a Folded C Terminus.
The L60A variant of pp32, with an enlarged cavity in the center of the protein, exhibited unique pressure-induced unfolding behavior, intermediate between the multistate transition observed for I7A, and the highly cooperative unfolding of the three C-terminal cavity-containing variants. While most of the native-state amide NH resonances showed sharp and reasonably concerted transitions (Fig. 3B), several residues in the C-terminal region exhibited clear three-state transitions, with the appearance at intermediate pressure of new peaks in slow exchange with and close in frequency to the folded state peaks (Fig. 6 A and B). The native-state peaks of L60A that did not exhibit three-state behavior yielded average free-energy values (Table 2 and SI Appendix, Fig. S2C) that were significantly smaller than that previously obtained in CD-detected urea unfolding experiments of this variant.
Fig. 6.
Pressure-dependent NMR of pp32 L60A. (A) HSQC spectra of pp32 L60A at multiple pressures from 1 bar (red) to 2 kbar (black) as indicated. Boxed and labeled residues exhibited pressure-dependent double peaks in slow exchange. (B) Double peaks in slow exchange at 600 bar for residues 114, 141, 142, and 145, as indicated. (C) Pressure dependence of the intensity of the two peaks, folded state (filled circles) and intermediate state (open circles) for residue 142. (D) Residues exhibiting double peaks in slow exchange are shown as black spheres on the pp32 structure.
Table 2.
Thermodynamic parameters for pp32 L60A unfolding by pressure and urea
| NMR observable | ||||||
| Perturbation/unfolding model | Unfolded state methyl peak ∼0.92 ppm | Unfolded state methyl peak ∼0.85 ppm | Folded state backbone –NH <all residues> | Trp155 indole NH unfolded state peak | CD* | |
| L60A pressure/two-state peaks | ΔG | −1.7 ± 0.2 | −1.7 ± 0.3 | −2.0 ± 0.4 | −1.8 ± 0.1 | — |
| ΔV | 88 ± 9 | 92 ± 12 | 138 ± 22 | 63 ± 3 | — | |
| L60A pressure/three-state NI† | ΔG | — | — | −2.0 ± 0.4 | — | |
| ΔV | — | — | 126 ± 18 | — | ||
| L60A pressure/three-state IU† | ΔG | — | — | −0.6 ± 0.4 | — | |
| ΔV | — | — | 24 ± 11 | — | ||
| L60A urea/two-state peaks | ΔG | −3.8 ± 0.4 | −4.0 ± 0.6 | −2.6 ± 0.5 | −3.6 ± 0.2 | −4.1 ± 0.1 |
| m value | 2.4 ± 0.3 | 2.5 ± 0.4 | 2.0 ± 0.3 | 2.3 ± 0.1 | 2.61 ± 0.01* | |
ΔG and ΔV values are in kilocalories⋅mole−1 and milliliter−1. m values are in kilocalorie−1⋅mole−1⋅molar−1. Individual values are the fitted value ± SE of the least-squares fit. Backbone NH are the mean ± SD over all residues. Fluorescence results are the average of least-square fits of three independent experiments ± SEM.
Dao et al. (21).
Only residues showing intermediate peaks were fit to three-state transition. Intensity profiles from all other residues were fit to a two-state model.
The residues of the L60A variant exhibiting three-state pressure-induced unfolding behavior are found in the fifth repeat and the C-terminal capping motif (Fig. 6C), corresponding to the most stable region of the WT protein, known to be ordered at the folding barrier (21). The first transition (native to intermediate, N → I) for these residues occurred fairly concomitantly with the concerted amide NH transitions, and the apparent average free-energy and volume changes for this N → I transition were equivalent to those for the concerted amide native-state peak loss profiles (Table 2 and SI Appendix, Table S2). Nonetheless, a few C-terminal native peaks and the intermediate state peaks retained some intensity at the highest pressures reached (Fig. 3 J and K). The apparent free-energy and volume changes for the second transition (intermediate to pressure unfolded state for these peaks, I → U) were much smaller than for the N → I transition (Table 2 and SI Appendix, Table S2), and the transition was not complete by 2.5 kbar. Since the chemical shifts of the intermediate peaks were very close to those of the folded state, we conclude that the fifth repeat and the C-cap remain folded in this intermediate, but that disruption of the rest of the protein alters their chemical environment. The total apparent free-energy change obtained from the fits of the two transitions (N → I → U) was significantly lower than that obtained previously in the urea-induced unfolding experiments, indicating that the pressure unfolded state of this variant was distinct from and not as destabilized as the urea-denatured state. However, the total volume change for the two transitions was similar to that observed for the C-terminal cavity-containing variants (Tables 1 and 2), and larger than that of WT pp32, consistent with an enlarged central cavity in the L60A variant.
Fits of the appearance of the composite unfolded state side-chain methyl peaks in the 1D 1H spectrum (SI Appendix, Fig. S5C) of L60A yielded a ΔGf value within error of that obtained for the average of the concerted backbone NH residues (Table 2). These unfolded state methyl side-chain peaks begin to appear at ∼500 bar, just below the midpoint of the concerted loss of the native-state NH peaks and the N → I transition of the C-terminal residues, and continue until the limit of our pressure cell (2,500 bar), indicative of sequential contributions from side-chain methyl groups across the sequence. The unfolded state Trp155 indole NH peak begins to appear at ∼800 bar, at which pressure the concerted transition of most of the amide NH peaks is nearly complete. The free-energy change was about one-half that obtained in CD-detected urea denaturation experiments on this variant. In addition, the volume change was smaller than that obtained for the lower pressure N → I transition. These nonoverlapping transitions from multiple observables indicate the population of at least two intermediate species for L60A under pressure; the first is apparent around 500 bar in which the native-state amide intensity has been lost for residues in the N-cap and repeats 1–4, but the methyl side chains in this region are not yet in the unfolded state. This intermediate is similar to that observed for the I7A variant. A second intermediate is maximally populated around 1,200 bar. In this intermediate, the methyl side chains in the N-cap and repeats 1–4 are in the unfolded state, but repeat 5 and the C-cap remain folded.
Urea-Induced Unfolding of the I7A and L60A Variants Is Distinct from Pressure-Induced Unfolding.
The apparent ΔGf value obtained from the residue-specific high-pressure NMR profiles for the I7A and the L60A variants were much lower than those obtained previously from CD-detected urea-induced unfolding (21), indicating that the two perturbations lead to distinct conformational ensembles. We sought to ascertain how much of this difference was due to pressure vs. urea perturbation, and how much was a result of the differences in observables (global secondary structure by CD vs. local structural information by HSQC). We monitored urea-induced unfolding by HSQC for the WT, I7A, and L60A variants (SI Appendix, Figs. S8 and S9). Overall, the residue-specific urea-induced transitions observed for WT yielded apparent free-energy changes and m values (Table 1) that were close to those previously reported (21). Small deviations in the average values likely represent deviation from two-state behavior for the WT (Table 1 and SI Appendix, Figs. S8 H and I and S9 C and F).
Both the I7A and the L60A variants exhibited deviation from two-state unfolding in urea. Analysis of the profiles of the native-state amide NH peaks as a function of urea concentration for both variants yielded average apparent ΔGf and m values that were smaller than those obtained previously by CD, but larger than those obtained by pressure denaturation (Fig. 4, Tables 1 and 2, and SI Appendix, Fig. S8 F–I). The magnitude of these parameters values increased from the N to the C terminus for the I7A variant, and the ΔGf and m-value histograms were asymmetric for both variants, consistent with strong deviation from two-state behavior in urea-induced unfolding for the I7A variant and modest deviation for the L60A variant. The apparent ΔGf value extracted from the native-state amide NH profiles for residues in the fifth repeat and in the C-terminal capping motif of I7A were equivalent to that obtained by CD-detected urea denaturation, indicating complete unfolding of this variant at high urea. Comparison of repeat-based average ΔGf values (Fig. 4) and fractional contact histograms (SI Appendix, Figs. S10 and S11) obtained from the pressure-dependent HSQC spectra with those obtained from the urea-dependent HSQC showed smaller apparent free-energy changes and greater persistence of contacts in the C terminus for both the I7A and the L60A variants at high pressure compared with high urea, although the pattern of this persistence was distinct for the two variants. The difference between pressure and urea denaturation was more pronounced for the I7A variant. It is noteworthy that, unlike the situation under pressure, no intermediate peaks were apparent in the HSQC spectra of L60A in urea-induced unfolding. In contrast to the results from the HSQC spectra, for all three variants (WT, I7A, and L60A), the apparent free energies and m values extracted from the unfolded-state side-chain methyl peaks and tryptophan indole NH peaks were equivalent to or even slightly larger than those extracted from the CD-detected unfolding (Table 1). These observations indicate, first, that heterogeneous intermediate states are populated in the urea denaturation of the I7A and L60A variants. However, given the differences in ΔGf values, the intermediates populated using these two different perturbation approaches are distinct.
Discussion
We investigated the relationship between the changes in global stability and folding cooperativity that result from disruption of core packing by the introduction of internal cavities in a protein. The linear architecture of the leucine-rich repeat protein pp32 facilitates interpretation of how structural context modulates this relationship. We found that a substitution known to cause the smallest perturbation to the global stability of pp32, the I7A mutation, led to the largest perturbation of the folding landscape. In this variant, the N- to C-terminal stability gradient of the WT protein is accentuated, leading to a high-equilibrium population of multiple intermediates in both pressure and urea denaturation experiments. Introduction of a cavity in the N-terminal capping motif leads to destabilization of that motif, but also energetic repercussions through repeat 4, highlighting the energetic coupling between repeats. These intermediates retain some core packing in heterogeneous conformational ensembles and are increasingly disrupted from the N to C terminus as the denaturing perturbation increases. It is interesting to note that, in a previous investigation of cavity effects on folding landscapes in a globular protein (a hyperstable variant of staphylococcal nuclease), the mutation that caused the largest decrease in folding cooperativity and increased the population of excited states was in the most stable hydrophobic core of the protein (16, 24, 25). Hence, the impact of packing defects depends not only on the local stability of the region in which they are introduced, but on the structural/energetic hierarchy of the entire protein. The intermediate and final states attained by the I7A variant of pp32 in pressure experiments are less disrupted than those obtained using urea. The distinct unfolded states observed for this variant vis à vis these two denaturants arise because pressure has no further effects once the structure has been disrupted sufficiently to allow solvation of the internal solvent excluded void volume, while urea favors extended states that expose a maximal amount of surface area.
In contrast to the complexity resulting from cavity creation in the N-terminal capping repeat, cavities introduced at three positions in the C-terminal half of the protein (L83A, L109A, and L139A) all have similar consequences on global stability and led to highly cooperative (two-state) unfolding transitions, increasingly so as the mutation is moved toward the extreme C terminus. These mutations destabilize the C-terminal half of the protein relative to the N-terminal half, essentially abolishing the stability gradient present in WT pp32. In this case, since all regions of the protein exhibit very similar local stability, unfolding occurs in a concerted fashion. Finally, the L60A mutation enlarges the main cavity and destabilizes the central core of the protein. Repercussions of this central destabilization propagate throughout the protein. In this variant, in addition to molten-globule–like intermediates, a stable intermediate species is clearly identified under pressure, which is nearly completely unfolded in the N-terminal capping motif and repeats 1–4, while retaining nearly native structure in repeat 5 and the C-terminal capping motif.
The present results demonstrate that the context of packing defects inside a proteins’ core, while leading to very similar effects on the global stability and relatively minor adjustments to the global structure, can have profoundly distinct effects on the folding landscape of the protein. This observation has implications for understanding the evolutionary pressures on protein sequence, to the extent that dynamics, excited states, and folding intermediates are implicated in protein function and homeostasis. These observations are of considerable importance as well to efforts in protein design for biotechnological and pharmaceutical applications. They suggest that simple screens for stability and function may not be sufficient to select for optimal properties of the folding landscape that maximize function and minimize side reactions such as oligomerization, surface activity, phase separation, and aggregation.
Materials and Methods
Protein Purification.
Cavity variants of pp32 were produced as C-terminal His-tag fusions and purified as previously described (21). For NMR measurements, proteins were expressed in minimal media augmented with 15N-labeled (NH4)2SO4 as previously described (22).
NMR.
Chemical shift differences were calculated with respected to WT pp32 as the geometric mean of the chemical shift difference in the 1H and 15N dimensions as previously described (17). Folded state peaks in the 1H−15N HSQC spectra of the pp32 variants were assigned by standard 15N-NOESY-HSQC and 15N-TOCSY-HSQC experiments and by comparison with WT pp32 assignments (4, 18). High-pressure NMR was performed using a commercial zirconia high-pressure NMR cell and an Xtreme-60 Syringe Pump (Daedalus Innovations). 1H−15N HSQC and 1D proton spectra were measured as a function of pressure on 15N-labeled pp32 cavity-containing variants in 20 mM Bis-Tris, 10 mM NaCl, 5 mM DTT, 10% D2O, and 90% H2O at pH 6.8 and 20 °C. Bis-Tris buffer was used because it has negligible pressure dependence of its pKa (26). HP-NMR experiments were performed on a 600-MHz Avance III Wide-Bore NMR spectrometer (Bruker) at the NMR Core Facility, Rensselaer Polytechnic Institute, as previously described (18). Urea-dependent experiments were carried out on a 600-MHz Avance III Standard-Bore NMR spectrometer (Bruker), equipped with a 5-mm 1H/2H/13C/15N cryoprobe with z-axis gradients. D1 was set to >5 × T1 for the longest T1 value measured. Samples for urea-dependent NMR experiments were made by mixing varying volumes of a stock of 200 μM pp32 variant, 20 mM Bis-Tris, 10 mM NaCl, 5 mM DTT, and 10% D2O, pH 6.8, in absence of urea with a stock of 200 μM pp32 variant at 3 M urea in the same buffers. In addition to the 2D NMR, equilibrium 1D proton spectra were collected at each pressure and urea concentration. Spectra were processed with Topspin and analyzed with CcpNmr (24). We note that peak volumes were used for the intensity values, although no pressure-dependent broadening of the native-state peaks was observed for any of the variants.
NMR Data Analysis.
From the 1H−15N HSQC spectra, folded state peak intensities (I) for each backbone amide group were individually fit into a two-state model as a function of pressure (p):
| [1] |
The value, IF, represents the maximal native-state peak intensity, while the value of the native-state peak intensity at the end of the transition, Iu, was constrained to be zero. From these fits, we obtained residue-specific apparent folding free energies at ambient pressure (ΔGf = −ΔGu) and apparent volume change of folding (∆Vf = −ΔVu). The pressure-dependent intensities of the two denatured state methyl proton resonances were also fit to the two-state model (Eq. 1). In this case, the value IF was the native-state plateau value, since some contribution from nearby peaks can render this value nonzero, while the value of Iu at the high-pressure or high-denaturant plateau represents the maximal intensity of these peaks in the unfolded state. Additionally, the pressure dependence of the intensities of the folded and unfolded peaks of the Trp155 side-chain indole NH were fit globally using the same two-state model (Eq. 1) with shared thermodynamic parameters (ΔGf and ΔVf and interconverting high- and low-pressure plateau values between the two datasets). Fractional contact maps and histograms were generated as a function of pressure as previously described (22) using the native contact map calculated from WT pp32 crystal structure (20) [Protein Data Bank (PDB) ID code 2JE0], using the SMOG Shadow Contact Map web server with a threshold of 6 Å around the Cα of each residue PDB:2JE0 (16, 25). Schematics of protein structures were rendered using Pymol (27), and cavities were detected using Hollow and a 1.1-Å probe radius (28).
HPSAXS.
The HPSAXS data were collected at the G-1 station of the Cornell High Energy Synchrotron Source (CHESS) (29). The HPSAXS cell body was machined out of Inconel 725 alloy and fitted with two 0.6-mm-thick diamond X-ray windows (30). The cell was encased in a circulating water-cooled brass jacket to maintain cell temperature at 20 °C. Pressure in the range of 1–3.5 kbar was maintained by a Barocycler HUB440 high-pressure pump (Pressure BioSciences). Samples were irradiated with a 250 × 250-µm X-ray beam of wavelength λ = 1.25 Å, with a flux of ∼5 × 1011 photons/s. Scattering was measured with a Pilatus 100K-S detector (Dectris) with a pixel size of 172 × 172 µm and an active area of 83.8 × 33.5 mm (487 × 195 pixels), over the q range of ∼0.01–0.26 Å−1 [where q is the wave vector defined as q = (4π sinθ/λ) and 2θ is the scattering angle].
The samples were stored at −80 °C. Before use, the samples were equilibrated at room temperature for about 30 min and then centrifuged at 5,000 rpm (VWR 1816 Micro; VWR International) for 10 min at 4 °C. Samples were then loaded into a disposable plastic sample holder that fits into the HPSAXS body, as described in ref. 30. The sample holder is equipped with two 7.5-µm-thick kapton X-ray windows. The sample was isolated in this cell from the surrounding water pressurization medium and pressure is transmitted by a plug of vacuum grease (High Vacuum Grease; Dow Corning) that acts as a liquid-like pressure equilibrating piston. HPSAXS measurements on pp32 samples and buffer were performed at 20 °C with 20 exposures of 0.5 s each (for a total of 10-s exposure at each dataset). The 2D SAXS images were azimuthally integrated about the beam center and normalized by the transmitted intensities via standard image correction procedures using the BioXTAS RAW 1.3.0 software package (31). The specimen-to-detector distance, determined by acquiring diffraction rings from a silver behenate powder standard, was 1,487.2 mm. The pair distribution plots, P(r), were calculated using the GNOM software package (32). Kratky plots were derived from the GNOM results using Igor Pro-6.37 (WaveMetrics). Radius of gyration values were calculated from the Guinier plots using the BioXTAS RAW 1.4.0 software (31) and from the P(r) plots using the GNOM software package (32). A sensitivity analysis of the effect of Dmax on the Rg value calculated from the P(r) plots was carried out, and the value that was in closest agreement with that obtained from the Guinier plots was chosen. Guinier plots were linear, even at very low q values. All Guinier fits were done with qmaxRg of 1.3.
High-Pressure Fluorescence.
HP-fluorescence experiments were carried out on the HP-fluorescence apparatus (ISS). Samples contained 200–400 μM pp32 and buffer given above. For the pp32 I7A sample, 0.3 M urea was used to promote unfolding in the accessible pressure range. Protein sample was added to a 500-μL glass cuvette sealed with a DuraSeal cap and O-ring, which was then placed into a stainless-steel high-pressure vessel with four sapphire windows [described in Dellarole and Royer (33)]. The vessel was connected to automatic pressure pump (Pressure Bioscience) through high-pressure resistant stainless-steel pipes filled with Millipore water. Hydrostatic pressure was applied incrementally, in the range of 1–3,500 bar. Temperature was maintained 20 °C by circulating water bath surrounding the vessel. The excitation light was provided from a xenon lamp at 280 nm using a monochromator. Emission intensity at 340 nm was collected on a photomultiplier tube oriented at 90° to the excitation path in photon counting mode. After each pressure increase, fluorescence was monitored continuously to ensure equilibration before measurement. Values over several seconds at the plateau were averaged at each pressure and fit to version of the two-state model (Eq. 1) that was modified to include sloping high-pressure plateau baselines, where necessary.
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation (NSF) Grants MCB 105966 and MCB 1514575 (to C.A.R.) and NIH Grant GM068462 (to D.B.). CHESS is supported by the NSF and NIH/National Institute of General Medical Sciences (NIGMS) via NSF Award DMR-1332208, and the MacCHESS resource is supported by NIH/NIGMS Award GM-103485.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. G.H. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1807379115/-/DCSupplemental.
References
- 1.Richards FM. The interpretation of protein structures: Total volume, group volume distributions and packing density. J Mol Biol. 1974;82:1–14. doi: 10.1016/0022-2836(74)90570-1. [DOI] [PubMed] [Google Scholar]
- 2.Chothia C. Principles that determine the structure of proteins. Annu Rev Biochem. 1984;53:537–572. doi: 10.1146/annurev.bi.53.070184.002541. [DOI] [PubMed] [Google Scholar]
- 3.Liang J, Dill KA. Are proteins well-packed? Biophys J. 2001;81:751–766. doi: 10.1016/S0006-3495(01)75739-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eriksson A, et al. Response of a protein structure to cavity-creating mutations and its relation to the hydrophobic effect. Science. 1992;255:178–183. doi: 10.1126/science.1553543. [DOI] [PubMed] [Google Scholar]
- 5.Baase WA, Liu L, Tronrud DE, Matthews BW. Lessons from the lysozyme of phage T4. Protein Sci. 2010;19:631–641. doi: 10.1002/pro.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pace CN, et al. Contribution of hydrophobic interactions to protein stability. J Mol Biol. 2011;408:514–528. doi: 10.1016/j.jmb.2011.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rashin AA, Iofin M, Honig B. Internal cavities and buried waters in globular proteins. Biochemistry. 1986;25:3619–3625. doi: 10.1021/bi00360a021. [DOI] [PubMed] [Google Scholar]
- 8.Pereira B, Jain S, Garde S. Quantifying the protein core flexibility through analysis of cavity formation. J Chem Phys. 2006;124:74704. doi: 10.1063/1.2149848. [DOI] [PubMed] [Google Scholar]
- 9.Graziano G. Cavity size distribution in the interior of globular proteins. Chem Phys Lett. 2007;434:316–319. [Google Scholar]
- 10.Maeno A, et al. Cavity as a source of conformational fluctuation and high-energy state: High-pressure NMR study of a cavity-enlarged mutant of T4 lysozyme. Biophys J. 2015;108:133–145. doi: 10.1016/j.bpj.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matthews BW, Liu L. A review about nothing: Are apolar cavities in proteins really empty? Protein Sci. 2009;18:494–502. doi: 10.1002/pro.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin H, Feng G, Clore GM, Hummer G, Rasaiah JC. Water in the polar and nonpolar cavities of the protein interleukin-1β. J Phys Chem B. 2010;114:16290–16297. doi: 10.1021/jp108731r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qvist J, Davidovic M, Hamelberg D, Halle B. A dry ligand-binding cavity in a solvated protein. Proc Natl Acad Sci USA. 2008;105:6296–6301. doi: 10.1073/pnas.0709844105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nucci NV, Fuglestad B, Athanasoula EA, Wand AJ. Role of cavities and hydration in the pressure unfolding of T4 lysozyme. Proc Natl Acad Sci USA. 2014;111:13846–13851. doi: 10.1073/pnas.1410655111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ando N, et al. Structural and thermodynamic characterization of T4 lysozyme mutants and the contribution of internal cavities to pressure denaturation. Biochemistry. 2008;47:11097–11109. doi: 10.1021/bi801287m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roche J, et al. Cavities determine the pressure unfolding of proteins. Proc Natl Acad Sci USA. 2012;109:6945–6950. doi: 10.1073/pnas.1200915109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roche J, et al. Structural, energetic, and dynamic responses of the native state ensemble of staphylococcal nuclease to cavity-creating mutations. Proteins. 2013;81:1069–1080. doi: 10.1002/prot.24231. [DOI] [PubMed] [Google Scholar]
- 18.Sosnick TR, Barrick D. The folding of single domain proteins—have we reached a consensus? Curr Opin Struct Biol. 2011;21:12–24. doi: 10.1016/j.sbi.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sosnick TR, Krantz BA, Dothager RS, Baxa M. Characterizing the protein folding transition state using psi analysis. Chem Rev. 2006;106:1862–1876. doi: 10.1021/cr040431q. [DOI] [PubMed] [Google Scholar]
- 20.Huyton T, Wolberger C. The crystal structure of the tumor suppressor protein pp32 (Anp32a): Structural insights into Anp32 family of proteins. Protein Sci. 2007;16:1308–1315. doi: 10.1110/ps.072803507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dao TP, Majumdar A, Barrick D. Highly polarized C-terminal transition state of the leucine-rich repeat domain of PP32 is governed by local stability. Proc Natl Acad Sci USA. 2015;112:E2298–E2306. doi: 10.1073/pnas.1412165112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fossat MJ, et al. High-resolution mapping of a repeat protein folding free energy landscape. Biophys J. 2016;111:2368–2376. doi: 10.1016/j.bpj.2016.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, et al. High-pressure NMR and SAXS reveals how capping modulates folding cooperativity of the pp32 leucine-rich repeat protein. J Mol Biol. 2018;430:1336–1349. doi: 10.1016/j.jmb.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 24.Roche J, et al. Remodeling of the folding free energy landscape of staphylococcal nuclease by cavity-creating mutations. Biochemistry. 2012;51:9535–9546. doi: 10.1021/bi301071z. [DOI] [PubMed] [Google Scholar]
- 25.Roche J, et al. Effect of internal cavities on folding rates and routes revealed by real-time pressure-jump NMR spectroscopy. J Am Chem Soc. 2013;135:14610–14618. doi: 10.1021/ja406682e. [DOI] [PubMed] [Google Scholar]
- 26.Quinlan RJ, Reinhart GD. Baroresistant buffer mixtures for biochemical analyses. Anal Biochem. 2005;341:69–76. doi: 10.1016/j.ab.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 27.Schrödinger LLC 2015. The PyMOL Molecular Graphics System, Version 1.7.4 (Schrödinger, LLC, New York)
- 28.Ho BK, Gruswitz F. HOLLOW: Generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct Biol. 2008;8:49. doi: 10.1186/1472-6807-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Acerbo AS, Cook MJ, Gillilan RE. Upgrade of MacCHESS facility for X-ray scattering of biological macromolecules in solution. J Synchrotron Radiat. 2015;22:180–186. doi: 10.1107/S1600577514020360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ando N, Chenevier P, Novak M, Tate MW, Gruner SM. High hydrostatic pressure small-angle X-ray scattering cell for protein solution studies featuring diamond windows and disposable sample cells. J Appl Cryst. 2008;41:167–175. [Google Scholar]
- 31.Hopkins JB, Gillilan RE, Skou S. BioXTAS RAW: Improvements to a free open-source program for small-angle X-ray scattering data reduction and analysis. J Appl Cryst. 2017;50:1545–1553. doi: 10.1107/S1600576717011438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Semenyuk AV, Svergun DI. GNOM. A program package for small-angle scattering data processing. J Appl Cryst. 1991;24:537–540. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dellarole M, Royer CA. High-pressure fluorescence applications. Methods Mol Biol. 2014;1076:53–74. doi: 10.1007/978-1-62703-649-8_4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.