Importance of Membrane Fusion in Biology
Membrane fusion is ubiquitous in biology, both in natural cellular functions such as neurotransmission and in pathological processes such as viral infection. The fusion of lipid bilayer membranes involves membrane contact, merger, and formation of an aqueous fusion pore, allowing the merging of separate compartments (either extracellular or intracellular) and mixing of their contents. For example, in neurotransmission, the soluble N-ethylmaleimide–sensitive factor attachment receptor (SNARE) mediates fusion of synaptic vesicles with the plasma membrane to release neurotransmitters, driven by zipping of the four-helix bundle of the SNARE complex (1). Enveloped viruses such as the influenza virus and HIV infect target host cells through an analogous protein-mediated membrane fusion process (2), releasing the viral contents into the host cell. Hence, membrane fusion is of fundamental importance and also of practical interest for its potential role in efficient drug delivery, or the prevention of viral infection by blocking the pathway of entry into the cell. Membrane fusion has significant activation barriers associated with accessing high-energy, nonbilayer intermediate structures, arising from dehydration of membrane surfaces and bending the bilayer (1, 3, 4). The barriers for the two transitions from separate membranes to a hemifused intermediate and from there to pore formation have been estimated from experiments and simulations to range from 20 to 80 kBT, yielding a total energy barrier to achieve membrane fusion of greater than 40 kBT (4). Fusion proteins may lower these barriers and provide the necessary impetus to overcome them, for example by refolding with the release of substantial free energy.
Hemagglutinin as an Archetype
Influenza hemagglutinin (HA) has served as a paradigm for understanding the mechanism of protein-mediated membrane fusion (5–7), and it is also an important target for antiviral drug development (8–12). Various mechanisms have been proposed for HA-mediated membrane fusion, based on equilibrium prefusion and postfusion crystal structures and biochemical evidence, but the details of this highly dynamic process are largely unknown. In PNAS, Lin et al. (13) present important new insight into the energetic landscape and the dynamics of the pH-induced conformational changes that underlie HA-mediated membrane fusion based on all-atom molecular dynamics simulations of the full B-loop trimeric structure of HA2.
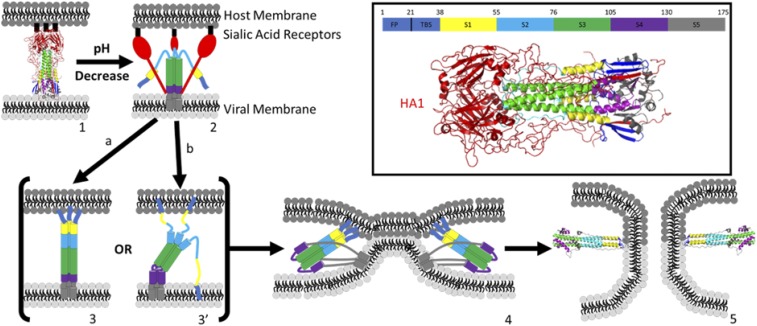
HA is postulated to undergo an astounding series of refolding reactions triggered by lowered pH (Fig. 1). The most remarkable aspect of the proposed mechanism is that it appears to be driven by HA refolding from a kinetically trapped, high-energy intermediate state (14). At neutral pH, the HA2 subdomain adopts a metastable structure having the fusion peptide (FP) buried in the interior of the trimer. The activated HA1 subdomain mediates attachment of the virion to the host cell by binding to sialic acid receptors on the host cell surface (Fig. 1, 1). This binding induces endocytosis and the virus is internalized in an endosome where the pH is lowered, causing the HA1 subdomain to dissociate from HA2 (Fig. 1, 2). Low pH in turn triggers refolding of the HA2 subdomain to extend the trimeric N-terminal coiled coil, exposing the FP, which then inserts into the host membrane (Fig. 1, 3). The protein is then anchored at either end into the viral and host membranes. An even more dramatic refolding of the C terminus of HA2 produces an antiparallel coiled coil, six-helix bundle scaffold (Fig. 1, 4). Finally, the long linker “leashes” to the viral transmembrane domain (TMD) refold in a zipper-like fashion against grooves in this scaffold, bringing the TMD and FPs to the same end of the rod-like structure (Fig. 1, 4). This action pulls the two membranes together and leads (somehow) to membrane fusion. The final step likely involves the cooperative merger of the TMD and FPs in the same membrane, to stabilize a fusion pore. A nonfusogenic state is produced if the second refolding step occurs before insertion of the FP into the host membrane, with the FP inserted instead into the viral membrane. Therefore, the timing of the two major refolding events is critically important. This model (and several close variants), termed the “spring-loaded” mechanism of membrane fusion, serves as a paradigm for understanding protein-mediated membrane fusion in general, but it remains untested at many levels. Despite the highly dynamic nature of this proposed mechanism, most of it has been inferred from equilibrium structures of HA fragments and from biochemical evidence (14–16). Thus, while the structures of putative intermediates suggest a mechanism for protein-mediated membrane fusion, the temporal and spatial evolution of the process has yet to be elucidated, and the mechanism remains hypothetical.
Fig. 1.
Proposed mechanism of HA-mediated membrane fusion: (1) prefusion HA structure at pH 7 (PDB ID code 1HGF); (1 → 2) low endosomal pH triggers refolding of HA, swinging the binding domain HA1 (red) away from HA2; (2 → 3) B loop refolds, extending coiled-coil structure (green), exposing the fusion peptides (FPs) (dark blue) to the host membrane; path a forms fully extended coiled coil, maintaining threefold symmetry; path b partially extends coiled coil, forming a symmetry broken intermediate [Lin et al. (13)]; (3 → 4) foldback of S4, S5 to form antiparallel coiled coil, six-helix bundle scaffold and zipping of linker leashes, bringing FP and TMD together and host and viral membranes into apposition to promote fusion; (4 → 5) pore formation, possibly facilitated by interaction between TMD and FP; structure (5) is the postfusion structure of HA (PDB ID code 1QU1). (Inset) Prefusion structure of HA (1HGF) with color legend identifying HA1 and each of the component structures of HA2, including the FP, two β-strands (TBS), helix A (S1), B loop (S2), coiled coil (S3), hinge region (S4), and ectodomain and TMD (S5).
Molecular Dynamics Simulations of HA Refolding
While the mechanism of HA refolding has been difficult to characterize experimentally, molecular dynamics simulations offer a tractable approach to the problem. Lin et al. (13) report all-atom, explicit-solvent molecular dynamics simulations to characterize the refolding of the trimeric B loop of the group 2 H3 HA. In the spring-loaded model, the B loop functions as a pH-dependent structural trigger that initiates and guides the initial HA refolding process. The key first step in this model is the refolding of the trimeric B loop from its prefusion loop structure to its postfusion coiled-coil structure. The folding energy landscape of the B loop derived by Lin et al. from thermodynamic sampling shows that at low pH it rapidly refolds to an intermediate state consisting of about one-half of the coiled-coil structure, but further folding to the fully extended coiled-coil structure is unfavorable. The simulations indicate that the N-terminal portion of the coiled coil is destabilized by the burial of a polar residue, Thr59. This residue breaks the canonical heptad repeat pattern of buried hydrophobic groups that normally stabilize a coiled-coil structure and causes the burial of water molecules in the hydrophobic core. Lin et al. (13) also performed microsecond timescale, constant-temperature kinetic simulations that show partial formation of the coiled-coil structure, but progress stops near Thr59, consistent with the thermodynamic simulations. The A helix is observed to stabilize the coiled-coil structure in experiments, but kinetic simulations of the B loop that include the A helix are similar to those without it and are also characterized by formation of significant nonnative β secondary structures near the C terminus.
The spring-loaded model postulates a downhill drive toward the formation of the extended coiled-coil structure, bringing the FP toward the host endosomal membrane to initiate fusion. The free-energy landscape derived from the simulations of Lin et al. indicates that this initial refolding event is only partially downhill to a half-formed coiled-coil structure and that the fully formed coiled-coil structure is unstable in this context. Previously, the Onuchic group (17, 18) proposed an alternative to the spring-loaded model from simulations of a minimally frustrated structure based on energy landscape theory. They discovered an order–disorder transition as the mechanism for the release of the FP from its burial site in the prefusion structure. This disorder quickly leads to a novel metastable intermediate with a broken threefold symmetry. Kinetic competition between the formation of the extended coiled coil and C-terminal melting results in two routes from this intermediate to the postfusion structure. On the energy landscape predicted by these simulations, the unfolding of S5 becomes decoupled from forming the HA2 coiled coil, allowing the S5 unfolding barrier to delay the foldback transition.
These results support formation of a long-lived intermediate state with broken threefold symmetry, having a frayed coiled coil with flexible C-terminal structures containing the FPs (Fig. 1, 3, path b). The frayed structure of this intermediate state might allow for FP insertion into both the viral and host membranes. A functional disadvantage of this intermediate is that it could lead to inactivation of some HA trimers, if all of the FPs are inserted into the viral membrane, consistent with experimental observation of a significant fraction of unproductive HAs (19). In contrast, a possible functional advantage of this fusion pathway is that it provides a greater free-energy release in the second refolding step (transition 3 → 4) to drive dehydration and bending of the two membranes to bring them into apposition. In path b of Fig. 1, the fully extended B-loop coiled coil is stabilized much later in the fusion process by interactions with the S1 coiled coil and contacts with S5. It is possible that this delayed formation of the extended coiled coil allows a greater release of free energy at the critical step in which the membranes are dehydrated and brought together.
The simulations of Lin et al. provide a glimpse of the initial events in the refolding of HA. Additional work will be required to fully elucidate this complex process. Experimental approaches have been developed to initiate membrane protein dynamics of a related influenza protein, M2, using a laser-induced pH jump coupled with time-resolved fluorescence and infrared spectroscopy (20). These methods can be applied to resolve the dynamics of HA refolding initiated by rapidly lowering the pH. More extensive simulations involving the complete HA2 structure are likely to become feasible as methodology and computing power continue to improve, allowing a better description of the entire refolding process.
Acknowledgments
Our research is supported by NIH/National Institute of General Medical Science Grants R01 GM053640 and P01 GM069036.
Footnotes
The authors declare no conflict of interest.
See companion article on page E7905 in issue 34 of volume 115.
References
- 1.Blumenthal R, Clague MJ, Durell SR, Epand RM. Membrane fusion. Chem Rev. 2003;103:53–69. doi: 10.1021/cr000036+. [DOI] [PubMed] [Google Scholar]
- 2.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: The influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 3.Aeffner S, Reusch T, Weinhausen B, Salditt T. Energetics of stalk intermediates in membrane fusion are controlled by lipid composition. Proc Natl Acad Sci USA. 2012;109:E1609–E1618. doi: 10.1073/pnas.1119442109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boonstra S, et al. Hemagglutinin-mediated membrane fusion: A biophysical perspective. Annu Rev Biophys. 2018;47:153–173. doi: 10.1146/annurev-biophys-070317-033018. [DOI] [PubMed] [Google Scholar]
- 5.Jahn R, Lang T, Südhof TC. Membrane fusion. Cell. 2003;112:519–533. doi: 10.1016/s0092-8674(03)00112-0. [DOI] [PubMed] [Google Scholar]
- 6.Söllner TH. Intracellular and viral membrane fusion: A uniting mechanism. Curr Opin Cell Biol. 2004;16:429–435. doi: 10.1016/j.ceb.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 7.Tamm LK, Crane J, Kiessling V. Membrane fusion: A structural perspective on the interplay of lipids and proteins. Curr Opin Struct Biol. 2003;13:453–466. doi: 10.1016/s0959-440x(03)00107-6. [DOI] [PubMed] [Google Scholar]
- 8.Ekiert DC, et al. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science. 2011;333:843–850. doi: 10.1126/science.1204839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corti D, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 10.Fleishman SJ, et al. Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science. 2011;332:816–821. doi: 10.1126/science.1202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chevalier A, et al. Massively parallel de novo protein design for targeted therapeutics. Nature. 2017;550:74–79. doi: 10.1038/nature23912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kadam RU, et al. Potent peptidic fusion inhibitors of influenza virus. Science. 2017;358:496–502. doi: 10.1126/science.aan0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin X, Noel JK, Wang Q, Ma J, Onuchic JN. Atomistic simulations indicate the functional loop-to-coiled-coil transition in influenza hemagglutinin is not downhill. Proc Natl Acad Sci USA. 2018;115:E7905–E7913. doi: 10.1073/pnas.1805442115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carr CM, Chaudhry C, Kim PS. Influenza hemagglutinin is spring-loaded by a metastable native conformation. Proc Natl Acad Sci USA. 1997;94:14306–14313. doi: 10.1073/pnas.94.26.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 16.Carr CM, Kim PS. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 17.Lin X, et al. Order and disorder control the functional rearrangement of influenza hemagglutinin. Proc Natl Acad Sci USA. 2014;111:12049–12054. doi: 10.1073/pnas.1412849111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin X, Noel JK, Wang Q, Ma J, Onuchic JN. Lowered pH leads to fusion peptide release and a highly dynamic intermediate of influenza hemagglutinin. J Phys Chem B. 2016;120:9654–9660. doi: 10.1021/acs.jpcb.6b06775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivanovic T, Harrison SC. Distinct functional determinants of influenza hemagglutinin-mediated membrane fusion. eLife. 2015;4:e11009. doi: 10.7554/eLife.11009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeong BS, Dyer RB. Proton transport mechanism of M2 proton channel studied by laser-induced pH jump. J Am Chem Soc. 2017;139:6621–6628. doi: 10.1021/jacs.7b00617. [DOI] [PMC free article] [PubMed] [Google Scholar]