Significance
The C–4 loop is a conserved structural motif present in all eukaryotic protein kinases. This study focuses on epidermal growth factor receptor (EGFR), a receptor tyrosine kinase that harbors many recurrent insertion mutations in the C–4 loop. We show that EGFR activity and drug sensitivity are strongly dependent on the nature, size, and location of mutations in the loop and that even subtle variations in the loop have a dramatic effect on kinase dynamics and free-energy landscape. These findings inform ongoing drug-discovery efforts on EGFR kinases and provide a structural framework for investigating the role of the understudied C–4 loop in kinase regulation, dynamics, and evolution.
Keywords: exon 20 insertion, conformational transition, drug resistance, personalized therapy, allostery
Abstract
Mutational activation of epidermal growth factor receptor (EGFR) in human cancers involves both point mutations and complex mutations (insertions and deletions). In particular, short in-frame insertion mutations within a conserved C–4 loop in the EGFR kinase domain are frequently observed in tumor samples and patients harboring these mutations are insensitive to first-generation EGFR inhibitors. Despite the prevalence and clinical relevance of insertion mutations, the mechanisms by which these mutations regulate EGFR activity and contribute to drug sensitivity are poorly understood. Using cell-based mutation screening, we find that the precise location, length, and sequence of the inserted segment are critical for ligand-independent EGFR activation and downstream signaling. We identify three insertion mutations (N771_P772insN, D770_N771insG, and D770GY) that activate EGFR in a unique way by relying more on the “acceptor” interface for kinase activation. Our drug inhibition studies indicate that these activating insertion mutations respond more favorably to osimertinib, a recently Food and Drug Administration-approved EGFR inhibitor for T790M-positive patients with lung cancer. Molecular dynamics simulations and umbrella sampling of WT and mutant EGFR suggest a model in which activating insertion mutations increase catalytic activity by relieving key autoinhibitory interactions associated with C-helix movement and by lowering the transition free energy () between active and inactive states. Our studies also identify a transition state sampled by activating insertion mutations that can be exploited in the design of mutant-selective EGFR inhibitors.
Many human cancers are caused by the accumulation of somatic mutations in oncogenes that confer selective growth advantage. The epidermal growth factor receptor (EGFR) kinase is one such oncogene that is mutationally activated in many human cancers, in particular lung cancer (1–4). Cancer genome sequencing studies have revealed numerous genomic alterations in EGFR, including point mutations, deletions, and insertions (5–7). A detailed understanding of these mutations is essential for developing effective therapeutic strategies (8). While some frequently occurring point mutations and deletion mutations, such as L858R, G719S, T790M, and E746_A750del, have been well studied (9–13), the structural and functional impact of many other recurrent mutations still remains largely unknown. In-frame insertions in exon 20 are a subcategory of understudied mutations that are frequently observed in patients with cancer (14–17). Most of the exon 20 insertions map to a short loop connecting the C-helix and 4 strand in the protein kinase domain, i.e., the C–4 loop (15). Exon 20 insertions confer resistance to first-generation EGFR inhibitors and are associated with poor clinical outcomes (6, 18). Recent studies suggest that the drug response profile of exon 20 insertions is heterogeneous and depends on the nature of the inserted segment (15, 16, 19). However, an incomplete understanding of how exon 20 insertion mutations regulate EGFR activity has hindered the development of therapies for patients harboring these mutations.
Structural and biochemical studies on human EGFR have provided considerable insight into their mechanisms of action in physiological and pathological states. Under physiological conditions, ligand binding to the extracellular domain induces dimerization and higher-order multimerization of the intracellular kinase domains (9, 20, 21). Structurally, the active state of the intracellular kinase domain is achieved through a “head-to-tail” configuration where one molecule of EGFR (the “donor” kinase) docks to the N lobe of another EGFR molecule (the “acceptor” kinase) in an asymmetric manner (9). Multimerization is established by forming a chain of asymmetric dimers, in which each EGFR protomer functions both as a donor and an acceptor kinase (21). The dimerized or multimerized EGFR complex is catalytically active due to the stabilization of key catalytic motifs that prime the kinase domain for autophosphorylation and downstream signaling (9, 21). Many activating oncogenic mutations, such as L858R, E746_A750del, G719S, G724S, R776H, and M766T, strictly rely on the asymmetric dimer for their activity (22–26). Intriguingly, some other complex mutations, including L747_A750P and D770_N771insNPG, are reported to be dimerization independent (23), although a low-resolution crystal structure of the D770_N771insNPG mutant [Protein Data Bank (PDB) ID: 4LRM] reveals an asymmetric dimer organization (15). The EGFR dimerization interface has been targeted by monoclonal antibody-based inhibitors such as cetuximab (27). Recently, several optimized stapled peptides that mimic the donor kinase interface were shown to be effective for EGFR inhibition (28). Thus, understanding the dimerization dependency and the mechanism of action of complex insertion mutations can inform ongoing drug-discovery efforts on EGFR kinases.
Computational structural modeling and molecular dynamics (MD) simulations have provided new atomic-level insights into EGFR regulation and activation. For example, long-timescale MD simulations of EGFR showed that the N lobe of the acceptor kinase is intrinsically disordered (11, 29) and oncogenic mutations, such as L858R, facilitate EGFR dimerization by quenching the intrinsic disorder (11). MD studies on other recurring mutations such as R776H in the C–4 loop have identified novel autoinhibitory interactions associated with kinase activation (25) and protonation-dependent changes in mutant EGFR functions (30). In addition, MD-based free-energy methods (26, 31), molecular mechanics calculations (32), and molecular modeling studies (33) have provided new insights into mutation-induced drug resistance mechanisms and the conformational transitions connecting active and inactive states (34–36).
In this study, we systematically analyzed exon 20 insertion mutations in EGFR, using a combination of experimental and computational approaches. We identified the C–4 loop, which is involved in kinase regulation (25, 37, 38), evolution (39), and chaperone-assisted folding (40, 41), as a hotspot for insertion mutations. Using a cell-based screen of 12 most commonly occurring insertions in the C–4 loop, we identified three activating mutations (N771_P772insN, D770_N771insG, and D770GY) that activate EGFR in a dimerization-dependent manner. These activating mutations rely more on the canonical acceptor interface than the donor interface for kinase activation. Drug response studies indicate that the three insertion mutations are insensitive toward reversible EGFR inhibitors (gefitinib, erlotinib, and lapatinib), but display a heterogeneous response toward irreversible inhibitors. In particular, all three insertion mutations respond to osimertinib more favorably. Likewise, D770GY is more sensitive to dacomitinib treatment compared with WT EGFR. To investigate the structural basis for mutational activation, we generated representative structural models for each of the insertion mutations and performed a total of 35 s MD simulations in various conformational states. MD simulations suggest that insertion mutations restrict the C-helix conformational freedom in the active state by hindering the formation of a critical autoinhibitory capping interaction associated with C-helix movement. Steered MD and umbrella sampling suggest that activating insertion mutations not only lower the free-energy difference between active and inactive states () but also stabilize key intermediate states between the two states. Taken together, these results suggest a unique mode of EGFR activation by insertion mutations that can be used in the development of EGFR-targeted cancer therapies.
Results
The C–4 Loop Is a Hotspot for Insertion Mutations in EGFR.
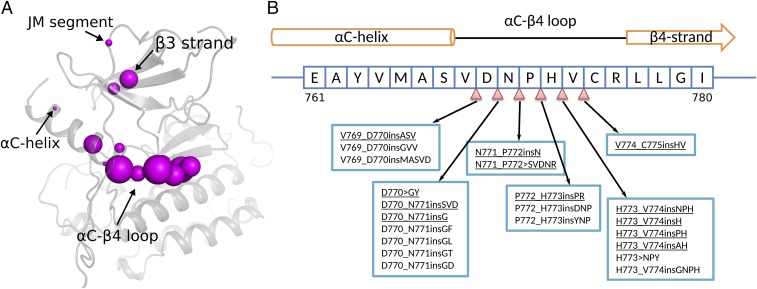
To comprehensively identify EGFR insertion mutations, we mined the Catalog of Somatic Mutations in Cancer (COSMIC v83) database and the Protein Kinase Ontology (ProKinO v2.0) (43). We identified a total of 97 distinct insertion mutations mapping to the EGFR kinase domain (Fig. 1A and SI Appendix, Dataset S1). The identified insertion mutations are heterogeneous in that they are variable in length and map to different regions of the protein. In particular, the C–4 loop, which connects the C-helix and 4 strand in the N-terminal ATP-binding lobe of the kinase domain, is an insertion hotspot accounting for 86.9% of unique insertion mutations in EGFR (Fig. 1A and SI Appendix, Dataset S1). Some of the frequently occurring insertion mutations in this loop include V769_D770insASV, D770_N771insSVD, H773_V774insNPH, H773_V774insH, D770_N771insG, D770GY, V774_C775insHV, H773_V774insPH, P772_H773insPR, N771_P772insN, H773_V774insAH, and N771_P772SVDNR (see Fig. 1 legend for insertion mutation nomenclature). These mutations are also referred to as exon 20 insertion mutations in the literature (6, 15). In addition to the C–4 loop, insertion mutations are also identified in the 3 strand (also referred to as exon 19 insertion mutations), the juxtamembrane segment, and the N terminus of the C-helix (Fig. 1A) (17, 44). These mutations, however, are not as frequent as the C–4 loop insertion mutations.
Fig. 1.
EGFR insertion mutations. (A) Structural location of insertion mutations in the EGFR kinase domain crystal structure (PDB ID: 2GS6). The size of the magenta sphere is proportional to the log of the number of patient samples containing the insertion mutation at the particular residue position. (B) Sequence location of insertion mutations within the EGFR C–4 loop. The nomenclature of insertion mutations follows the guidelines established previously (42). For example, V769_D770insASV indicates an Ala–Ser–Val insertion between residues V769 and D770, and D770GY indicates a complex insertion mutation in which D770 is replaced by a Gly–Tyr sequence. Insertion mutations characterized in this study are underlined. The complete list of patient-derived insertion mutations in EGFR can be found in SI Appendix, Dataset S1.
Identification of Activating Insertion Mutations.
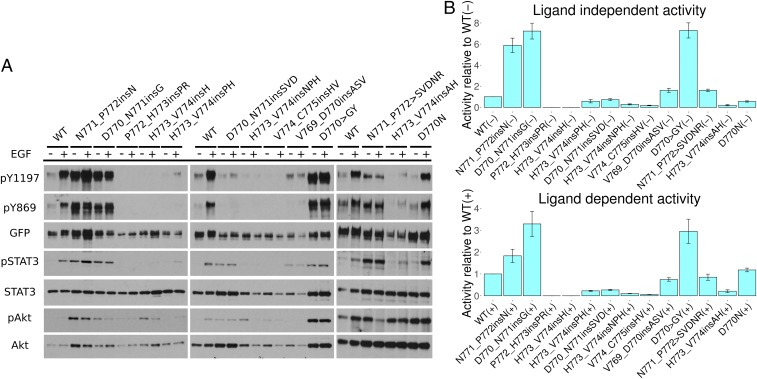
We next investigated the impact of C–4 loop insertion mutations on EGFR autophosphorylation activity and downstream signaling, using cell-based kinase assays (Materials and Methods). The insertion mutations exhibit varying levels of C-terminal tail tyrosine (Y1197) phosphorylation, activation loop (Y869) phosphorylation, and downstream protein phosphorylation in comparison with WT (Fig. 2 A and B). For most mutations, including the frequently occurring V769_D770insASV, D770_N771insSVD, and H773_V774insNPH mutations, the activity of the mutant form is comparable, or slightly reduced, relative to WT (Fig. 2 A and B). N771_P772insN, D770_N771insG, D770_N771insSVD, V769_D770insASV, D770GY, and N771_P772SVDNR exhibit similar Y1197 phosphorylation in the presence and absence of EGF ligand, suggesting that these insertion mutations are insensitive to EGF stimulation (Fig. 2A). This is different from the patient-derived point mutation (D770N) in the C–4 loop, which still shows ligand-dependent activation (Fig. 2A). Interestingly, three mutations (D770_N771insG, D770GY, and N771_P772insN) display increased autophosphorylation activity both in the presence and the absence of EGF (Fig. 2 A and B). Notably, downstream STAT3 phosphorylation is also enhanced for these mutants relative to WT (Fig. 2A), suggesting that N771_P772insN, D770_N771insG, and D770GY are activating gain-of-function mutations.
Fig. 2.
Cell-based screening of EGFR insertion mutations. (A) Western blot of 13 different EGFR mutations. The – and indicate the absence and presence of EGF ligand. Autophosphorylation activity is indicated by Y1197 and Y869 phosphorylation. GFP signal indicates the total expression level of EGFR. Downstream signaling of STAT3 and Akt is also shown. (B) Densitometry analysis of the relative phosphorylation activity of various EGFR insertion mutations compared with WT. The ratio between Y1197 and GFP signal is used as a measure of activity. The score for each insertion mutation is normalized to WT EGFR. Both ligand-independent activity (Top) and ligand-dependent activity (Bottom) are shown. SE bars represent three to five independent experiments.
Differential Dimerization Dependency of Activating C–4 Loop Insertion Mutations.
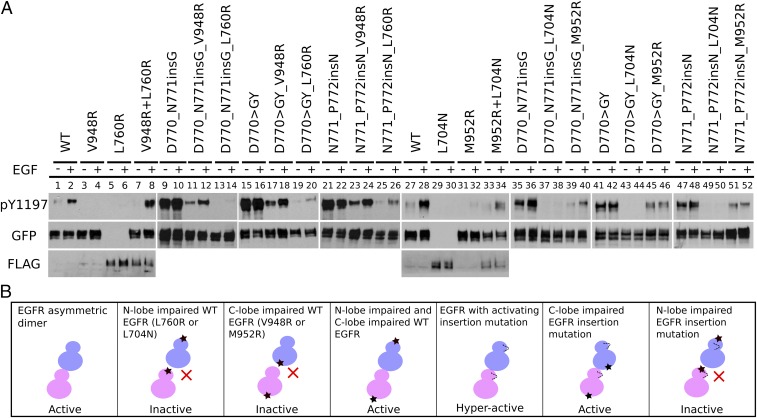
Previous studies have shown that activating point mutations in EGFR such as L858R/T790M and R776H enhance the acceptor activity in the asymmetric dimer (25, 45). To investigate the dimerization dependency of the activating C–4 loop insertion mutations (D770_N771insG, D770GY, and N771_P772insN), we introduced a C-lobe dimerization-deficient mutation (V948R or M952R) or an N-lobe dimerization-deficient mutation (L760R or L704N) in the insertion mutation plasmids. We selected two mutations in each interface to rule out the possibility that the mutations might alter the kinase domain itself rather than the dimer interaction. Western blot analysis indicates that insertion mutations are still active in the presence of C-lobe dimerization-deficient mutation (V948R or M952R) (Fig. 3A, lanes 11–12, 17–18, 23–24, 39–40, 45–46, and 51–52 and SI Appendix, Fig. S2). In contrast, the presence of the N-lobe dimerization-deficient mutation (L760R or L704N) results in significantly reduced Y1197 phosphorylation (Fig. 3A, lanes 13–14, 19–20, 25–26, 37–38, 43–44, and 49–50 and SI Appendix, Fig. S2). As a control, Y1197 phosphorylation activity is negligible for WT EGFR when dimerization-deficient mutations are present (Fig. 3A, lanes 3–6 and 29–33 and SI Appendix, Fig. S2); however, Y1197 phosphorylation can be rescued by cotransfecting the two mutants (V948RL760R or M952RL704N) (Fig. 3A, lanes 7–8 and 33–34 and SI Appendix, Fig. S1). These data indicate that activating C–4 loop insertion mutations rely more on the N-lobe interface than on the C-lobe interface for EGF-dependent and EGF-independent activation. In addition, we also tested the effect of introducing both V948R and M952R mutations in the insertion mutation background and observed comparable Y1197 autophosphorylation (SI Appendix, Figs. S2 and S3), suggesting that the C–4 loop insertion mutations could bypass the canonical C-lobe asymmetric dimer interface to achieve kinase activation.
Fig. 3.
Dimerization dependency of EGFR insertion mutations. (A) Western blot analysis of the N-lobe dimerization-deficient mutation (L760R or L704N) and the C-lobe dimerization-deficient mutation (V948R or M952R) on Y1197 autophosphorylation for WT and mutant (D770_N771insG, N771_P772insN, and D770GY) EGFR. All of the constructs used are tagged with GFP except for L760R and L704N, which are tagged with FLAG. Cotransfection of V948R and L760R is indicated by V948RL760R. Co-transfection of M952R and L704N is indicated by M952RL704N. Eight percent SDS/PAGE is used to resolve the cell lysate sample. Y1197 phosphorylation of V948RL760R and M952RL704N is also analyzed by 5% SDS/PAGE to separate GFP- and FLAG-tagged constructs (SI Appendix, Fig. S1 A and B). Densitometry analysis of three to five independent experiments is shown in SI Appendix, Fig. S2). (B) A schematic plot summarizing the observed differences in WT and mutant EGFR in the different dimerization-deficient mutant backgrounds.
Drug Sensitivity of Activating C–4 Loop Insertion Mutations to Reversible and Irreversible EGFR Inhibitors.
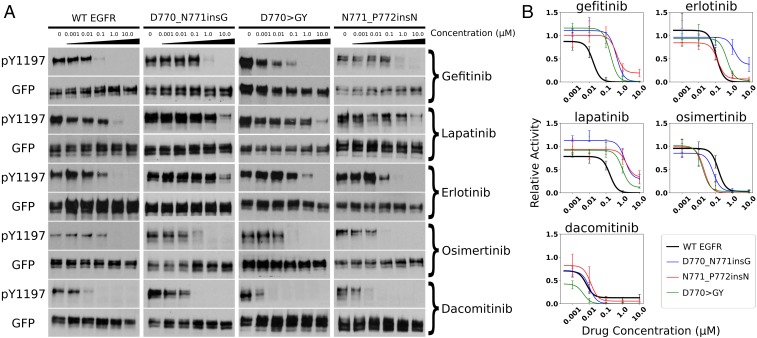
Exon 20 insertion mutations are generally associated with poor clinical response to first-generation EGFR inhibitors, such as gefitinib, erlotinib, and lapatinib (6, 14). We tested the dose-dependent response of the three activating C–4 loop insertion mutations against these inhibitors. Our data indicate that the activating insertion mutants (D770_N771insG, D770GY, and N771_P772insN) are insensitive to these inhibitors as the inhibition profile is either unaltered or resistant with increasing drug concentration (Fig. 4A), suggesting that patients harboring these mutations might not benefit from first-generation EGFR inhibitors. Recently, second-generation nonreversible covalent bond inhibitors, such as osimertinib, dacomitinib, and afatinib, have been developed to overcome drug resistance conferred by the gatekeeper (T790M) mutation (46–48). We tested the responsiveness of activating C–4 loop insertion mutations to these nonreversible EGFR inhibitors (osimertinib and dacomitinib). Interestingly, we find that the three insertion mutations respond to osimertinib more favorably, as the Y1197 autophosphorylation is inhibited to various degrees at 0.1 M drug concentration for insertion mutations but not for WT EGFR (Fig. 4 A and B). In addition, we find that the D770GY mutant displays a sensitive profile toward dacomitinib (Fig. 4 A and B). In conclusion, our drug sensitivity assays demonstrate a heterogeneous response to irreversible EGFR inhibitors by activating C–4 loop insertion mutations.
Fig. 4.
Drug inhibition assay of five EGFR inhibitors (gefitinib, lapatinib, erlotinib, osimertinib, and dacomitinib). (A) The amount of Y1197 phosphorylation at drug concentration (0, 0.001 M, 0.01 M, 0.1 M, 1.0 M, and 10.0 M) is tested for WT and three activating insertion mutations. (B) Densitometry quantification of the drug treatment experiments with three to five replicates. SE bars represent three to five independent experiments.
Activating Insertion Mutations Restrict Kinase Conformational Freedom.
To investigate the structural basis for kinase domain activation by C–4 loop insertion mutations, we used homology modeling and loop refinement strategies to generate an ensemble of structural models for each activating insertion mutation (Materials and Methods). The generated models were clustered based on the root-mean-square deviation (rmsd) values. Representative models from each cluster were used for MD simulations. The number of clusters identified for each insertion mutation and the Rosetta energy distribution for each cluster are shown in SI Appendix, Figs. S4 and S5. Comparisons of the MD trajectories indicate that the overall thermal fluctuations of WT and mutant EGFR are comparable during the course of the simulations (SI Appendix, Figs. S6–S8). However, the intrinsic dynamics of the kinase domain, particularly for regions involved in kinase regulation, are significantly different, as described below.
Restricted conformational sampling by activating C–4 loop insertions in the active state.
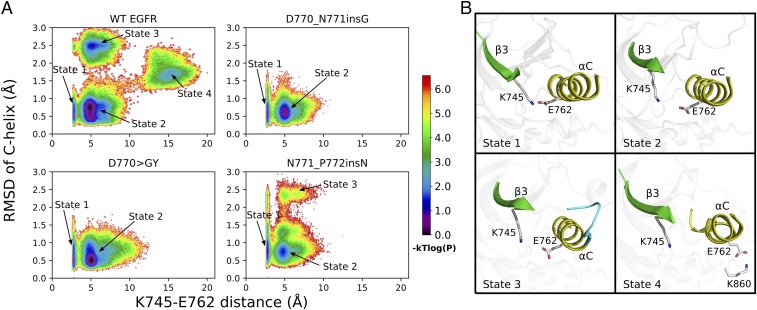
We used the rmsd of the regulatory C-helix and the K745–E762 salt bridge distance as proxies (variables) to compare the conformational freedom of WT and mutant EGFR in the active state. A joint probability distribution plot of these two variables in energy units is shown in Fig. 5A, where each point represents the log-transformed frequency of MD snapshots adopting the corresponding C-helix rmsd and K745–E762 distance. Based on the plot, distinct clusters/states can be identified for WT and mutant EGFR. In general, WT is more flexible and samples four distinct states/clusters (indicated by the arrows in Fig. 5A), while the three activating insertion mutations explore only a subset of these states. State 1 corresponds to a native-like “active” conformation shared by WT and mutant EGFR with an intact C-helix and a well-established K745–E762 salt bridge (Fig. 5 A and B). State 2 is a meta-state, also shared by WT EGFR and all three insertion mutations (Fig. 5A). The K745–E762 distance in state 2 centers around 5 Å and the C-helix is in an “inward” conformation (Fig. 5B). State 3 represents a cluster of conformations in which the N terminus of the C-helix is unfolded/disordered (Fig. 5B). State 3 is sampled by N771_P772insN, but not by D770GY and D770_N771insG. In state 3, a truncated C-helix is formed only after residue K757, which makes the C-helix four residues shorter (Fig. 5B). Notably, such helix truncation is also observed in some of the recently solved EGFR crystal structures (PDB IDs: 4WRG and 4ZAU) (49), indicating that our simulations are sampling biologically relevant conformations. State 4 corresponds to a highly destabilized C-helix and a broken K745–E762 salt bridge, as observed in previous microsecond timescale MD simulations (11). In state 4, E762 in the C-helix makes a charged interaction with K860 from the activation loop, an interaction seen only in the inactive crystal structure of EGFR (Fig. 5B). None of the mutants sample state 4, suggesting that the conformational freedom of the activating mutants is restricted to the active state (state 1) or meta-states close to the active state (states 2–3), consistent with our cell-based assay in which these three insertion mutations are more active than WT (Figs. 2 and 5A). Interestingly, the conformation heterogeneity of the mutants in the active state is also correlated with kinase activity in that the conformationally flexible N771_P772insN mutant is less active compared with the conformationally restricted D770GY and D770_N771insG mutants (Figs. 2B and 5A).
Fig. 5.
MD simulation of EGFR and three activating insertion mutations in the active state. (A) The joint distribution of the K745/NZ–E762/(OE1,OE2) distance and the C-helix rmsd is shown in logarithmic scale. The plot is constructed using 100 different bins on both axes. The probability of each bin is estimated using the frequency of the 3-s MD snapshots that fall into each bin. The distribution plot is transformed into an energy unit by , where is set to 1.0 in generating the contour image. Four distinct state clusters are labeled. (B) Representative snapshots from each conformational state labeled in A are shown.
Activating insertion mutations occlude autoinhibitory interactions associated with the C-helix movement.
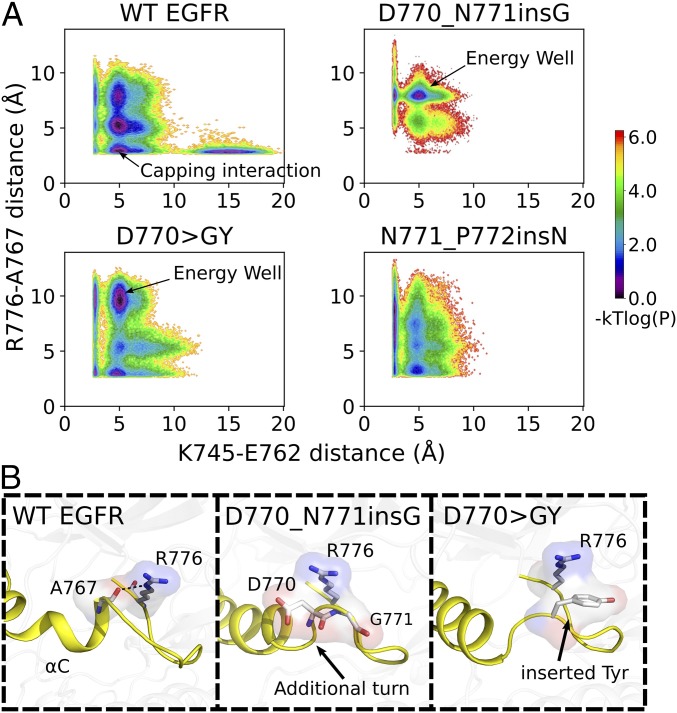
We previously reported that stabilization of the C-helix in the inactive state is correlated with an autoinhibitory capping interaction formed between R776/(NE,NH1,NH2) in the C–4 loop and A767/O in the C-helix (25). We hypothesize that activating insertion mutations in the C–4 loop could also modulate the C-helix movement by altering the C-helix capping interaction. To test this hypothesis, we analyzed our MD trajectories to correlate changes in the R776/(NE,NH1,NH2)–A767/O distance with the K745–E762 salt bridge, which serves as a proxy for kinase conformational transition (Fig. 6A). The Boltzmann weighted probability distribution plot of these two distance variables indicates an energy well at a K745–E762 distance of 5 Å and an R776–A767 distance of 2.5 Å for WT (Fig. 6A). However, for the mutants, the energy well is distributed across various R776–A767 distance values in the vertical axis (Fig. 6A). For the D770GY mutant, the energy well is maintained at 10 Å whereas for the D770_N771insG mutant, it is mostly populated at 7.5 Å (Fig. 6A), indicating that the capping interaction is not frequently sampled by these mutants. By visualizing the MD trajectories of WT and mutant EGFR, we identify key atomic interactions that provide plausible explanations for the observed differences in the conformational ensemble (Fig. 6B). In the D770GY mutant, for example, the inserted tyrosine residue orients in a way that prevents R776 from interacting with A767 (Fig. 6B). Likewise, in the D770_N771insG mutant, the inserted segment introduces an additional turn in the C-helix that prevents R776 from forming the canonical autoinhibitory capping interaction with A767 (Fig. 6B). Although no clear interactions are identified for N771_P772insN, the flat energy distribution observed for this mutant suggests that the R776–A767 interaction is destabilized relative to WT EGFR (Fig. 6A).
Fig. 6.
Modulation of autoinhibitory capping interactions [R776/(NE,NH1,NH2)–A767/O hydrogen bonds] by activating insertion mutations. (A) The joint distribution of K745–E762 distance and R776–A767 distance is shown in logarithmic scale. The same binning and log-transformation procedure as used in Fig. 5A was followed to generate the plot. (B) Representative snapshots from the energy wells (indicated by arrows in A).
Stabilized inactive state simulation.
We also performed MD simulations of WT and mutant forms in the inactive state. Unlike the active state, the overall thermal fluctuations of WT and mutant forms are similar, with only minor differences in the C terminus of the C-helix (SI Appendix, Fig. S7). This may be due to the overall global stabilization of the inactive state relative to the active state, as suggested by our free-energy landscape analysis described below.
Insertion Mutations Alter the Free-Energy Landscape of EGFR Conformational Transitions.
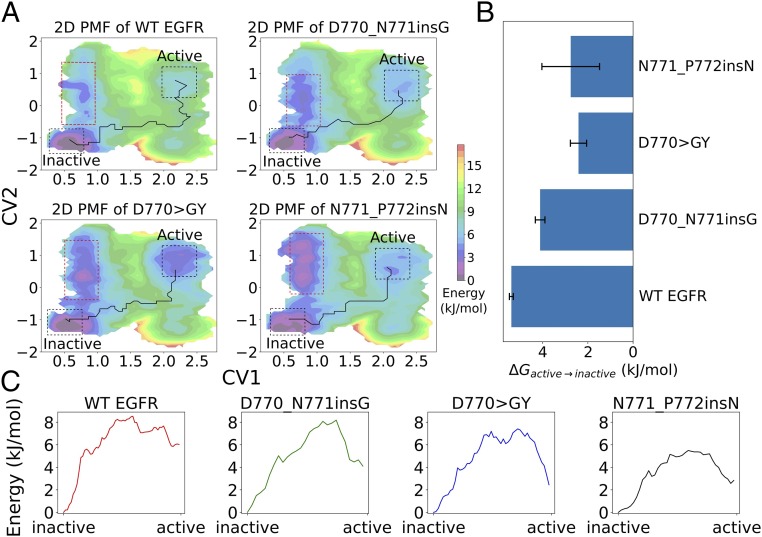
We next wanted to investigate how insertion mutations in the C–4 loop alter the free-energy landscape between active and inactive states. To this end, we used an umbrella sampling technique, which allows efficient sampling of the entire transition event by restraining the simulation at various intermediate states. We used two collective variables (CVs) to describe the transition between active and inactive states (SI Appendix, Fig. S9A). CV1 is the pseudodihedral angle formed by the C atom of the DFG motif and the DFG+1 residue (D855, F856, G857, L858). The pseudodihedral angle describes the conformation of the activation loop and serves as an important conformational feature for distinguishing kinase active and inactive conformations (50). CV2 is the difference in the distance between two salt bridge-forming pairs, i.e., K745–E762 () and K860–E762 () (Materials and Methods). The K745–E762 salt bridge is the catalytically important interaction formed in the active state of EGFR (9), whereas the K860–E762 salt bridge is observed only in the inactive state of EGFR (51). The difference between and (CV2) describes the C-helix movement during the conformational transition and is orthogonal to the DFG motif conformation captured by CV1. The initial transition pathway is generated by four independent steered molecular dynamics (SMD) simulations (SI Appendix, Fig. S9B). A total of 178 umbrella windows are chosen to cover the CV space explored by the SMD simulations. The 2D potential of mean force (PMF) profiles for WT and mutant EGFR are shown in (Fig. 7A). The lower left corner of the PMF profile corresponds to the inactive conformation of EGFR, whereas the upper right corner of the 2D PMF profile corresponds to the active state. Umbrella sampling identified the inactive state to be the global energy minimum for WT EGFR and all three insertion mutations, consistent with our unbiased MD simulations in which the inactive state was found to be more stable than the active state. Quantification of the free-energy difference between active and inactive states along the 2D PMF shows that insertion mutations lower the compared with WT (Fig. 7B and SI Appendix, Table S1). We also identified the lowest free-energy pathway (LFEP) connecting the active and inactive states in the free-energy landscape (Fig. 7A). The LFEP suggests stabilization of intermediate states along the transition pathway for the N771_P772insN and D770GY mutants (Fig. 7C). The three activating mutants also explore a unique low energy conformation in the 2D PMF (Fig. 7A, red dashed frame). Structurally, this state corresponds to a conformation in which the DFG-Phe (F856) is in an inward conformation, analogous to the inactive state of EGFR. However, the characteristic 3/10 helix right after the DFG motif is not formed (SI Appendix, Fig. S10). This inactive-like conformation appears to be readily accessed by the insertion mutations (D770_N771insG, D770GY, and N771_P772insN) due to the interaction between the extended C–4 loop and the DFG motif in the activation segment (SI Appendix, Fig. S10). Notably, this inactive-like conformation is less predominant in WT EGFR, suggesting that it is uniquely explored by the activating insertion mutations.
Fig. 7.
Free-energy landscape of EGFR and insertion mutations from umbrella sampling. (A) The 2D PMF of CV1 and CV2. The LFEP connecting the active and inactive states is highlighted. Red dashed frames show the CV space that is stabilized by insertion mutations. (B) The between active and inactive states. The error bars are estimated from three independent umbrella sampling simulations. (C) The free energy associated with the LFEP identified from 2D-PMF from A of various mutants.
Conclusion and Discussion
Using a combination of computational and experimental approaches we find that patient-derived insertion mutations in the EGFR C–4 loop impact kinase autophosphorylation and drug sensitivity to varying degrees. We identify three gain-of-function mutations (D770_N771insG, D770GY, and N771_P772insN) that activate EGFR in a ligand-independent manner. Interestingly, these activating mutations rely more on the N-lobe asymmetric dimer interface than the C-lobe interface for kinase activation. Consistent with our findings, a previous study focusing on the dimerization dependence of the D770_N771insNPG mutant found that the N-lobe dimerization-deficient mutation (L704N) had a more dramatic effect on the transforming activity of EGFR compared with the C-lobe dimerization-deficient mutation (I941R) (23). These findings, however, cannot be reconciled based on the only available low-resolution crystal structure of an exon 20 insertion mutation (D770_N771insNPG mutation; PDB ID: 4LRM) in which the C lobe of the donor kinase docks to the N lobe of the acceptor in a manner similar to WT EGFR (15). Thus, based on the asymmetric dimer model, it is unclear how activating insertion mutations in the C–4 loop can maintain EGFR activity in the presence of the C-lobe dimerization-deficient mutant. Based on our cell-based mutagenesis experiment, we conclude that the asymmetric dimer is important for the full activation of EGFR insertion mutations. However, with a compromised C-lobe dimer interface, the mutants can still achieve activation through alternative configurations (Fig. 3B).
The C–4 loop is a conserved regulatory segment present in all eukaryotic protein kinases (ePKs) (39). Through quantitative comparisons of diverse eukaryotic protein kinases and distantly related small molecule kinases, we previously proposed that the HxN motif in the C–4 loop (NPH motif in EGFR) serves as a hinge point for C-helix and interlobe movement (39). While the length of the C–4 loop is typically seven to eight residues long in most protein kinases, some kinases such as CK2, VRK1, VRK2, and SRPK1 (52–54) naturally contain an extended C–4 loop due to sequence insertions. Interestingly, the C-helix in these kinases is stabilized in an active conformation (52–54), although the precise mechanisms by which such stabilization occurs are unclear. Our studies suggest that modulation of key autoinhibitory interactions associated with the C-helix movement, such as the R776–A767 capping interaction in EGFR, is one possible mechanism by which sequence and conformational variation in the C–4 loop may impact C-helix conformational freedom and, consequently, kinase activity. The variable impact on EGFR autophosphorylation activity based on the nature, location, and size of the inserted segment further supports the hypothesis that the C–4 loop is a rheostat that can be fine-tuned to regulate kinase activity (55). For example, the two insertion mutations (N771_P772insN and N771_P772insH) that differ only by a single residue at the NPH-Asn position have a dramatically different effect on kinase autophosphorylation. N771_P772insH abolishes Y1197 autophosphorylation, whereas N771_P772insN is constitutively active (Fig. 2). Thus, subtle sequence and conformational variations in the C–4 loop may have long-range allosteric effects on ATP binding and activation loop conformation as proposed previously for other tyrosine kinases (37, 38).
Our drug response assays indicate that activating insertion mutations are more sensitive to osimertinib compared with WT EGFR. This is consistent with a recently established high-throughput functional evaluation assay in which the relative viability of Ba/F3 cells expressing N771_P772insN is reported to be sensitive to osimertinib treatment (19). Another recently published study showed that D770GY is partially responsive to dacomitinib compared with other insertion mutations in the C–4 loop (16). This is in agreement with our experimental result in that the other two activation insertion mutations (D771_N772insG and N771_P772insN) show a similar inhibition profile compared with WT EGFR. Our finding also suggests that nonreversible EGFR inhibitors, originally designed to overcome the drug-resistant T790M mutant, may also be repurposed to treat patients with activating C–4 loop insertion mutations. Moreover, establishing a comprehensive drug response profile for C–4 loop insertion mutations to irreversible EGFR inhibitors is critical to guide therapeutic strategies for patient treatment.
The C–4 loop has recently been appreciated as an important region for Hsp90 association and cochaperone cdc37 binding (40). While EGFR is not a typical client protein for Hsp90, insertion mutations in the C–4 loop could alter this preference (56). A previous study has shown that D770_N771insNPG and D770_N771insNPH mutants are sensitive to Hsp90 inhibitors, opening another therapeutic window for clinical treatment (56). In addition, our umbrella sampling analysis of WT and mutant EGFR identified alternative inactive states that are uniquely sampled by C–4 loop insertion mutants (Fig. 7 and SI Appendix, Fig. S10). Thus, selectively targeting these inactive states can be beneficial for treating patients harboring C–4 loop insertion mutations. Finally, by mechanistically characterizing oncogenic mutations in the C–4 loop, our study provides a framework for investigating the underappreciated role of the C–4 loop in kinase structure, function, regulation, and disease.
Materials and Methods
Materials and Reagents.
Q5 site-directed mutagenesis kits for generating mutant EGFR and DH5 competent cells were purchased from New England Biolabs. Qiaprep spin miniprep/midiprep kits were obtained from Qiagen. The 1 Dulbecco’s modified Eagle’s medium (DMEM), 1 Ham’s F12, and PBS were purchased from Mediatech. Lipofectamine-2000 was obtained from Invitrogen. Anti–pY1197-EGFR, anti–pY869-EGFR, anti-GFP, anti-STAT3, anti–pY705-STAT3, anti-Akt, anti–pS473-Akt, and horseradish peroxidase (HRP)-conjugated mouse monoclonal and rabbit polyclonal antibodies were obtained from Cell Signaling. Anti-FLAG human recombinant EGF and dacomitinib were bought from Sigma-Aldrich. BSA was purchased from Rockland Immunochemicals. Pierce ECL Western blotting substrate was bought from Thermo Scientific. Gefitinib, erlotinib, and lapatinib were kind gifts from Kojo Mensa-Wilmot, University of Georgia Department of Cellular Biology. Osimertinib was purchased from LC Laboratories.
The pEGFP-N1-EGFR plasmid from our previous studies was used for mutagenesis (25, 26). The PCR primers for site-directed mutagenesis are designed using the NEBaseChanger v1.2.6 web server from New England Biolabs. All mutated plasmids were confirmed via Sanger sequencing through Eurofins Genomics.
Cell Culture, Transfection, and Starvation.
Chinese hamster ovary (CHO) cells were cultured in DMEM with 10% FBS. DNA transfection was performed using Lipofectamine-2000 by following the manufacturer’s protocol. After 12–16 h, the transfection efficacy was inspected under the microscope for GFP signal. Transfected CHO cells were washed with PBS and starved in Ham’s F12 media for more than 12 h before EGF stimulation/drug treatment.
EGF Stimulation, Cell Lysis, and Immunoblotting.
EGF stimulation was carried out by incubating serum-starved CHO cells in 50 human EGF for 5 min. Cells were washed and immediately lysed with lysis buffer (50 mM TrisHCl, pH 7.4, 150 mM NaCl, 10% glycerol, 1 mM EDTA, 10% Triton X-100, 1 mM PMSF, and 1× protease inhibitor mixture Set V, EDTA-free). The total cell lysate was spun at 15,000 rpm at 4 °C for 5 min. Samples for SDS/PAGE gel were prepared in 4 Laemmli buffer. Proteins were resolved on 8% or 5% SDS/PAGE and transferred onto a polyvinylidenedifluoride (PVDF) membrane using the Trans-Blot Turbo Transfer System (Bio-Rad). Western blotting was done using target-specific primary antibodies, followed by the rabbit/mouse secondary antibodies. Protein luminescence was detected using Pierce Western blotting ECL substrate.
Drug Inhibition Assay.
Cells were transfected and then incubated with drugs at five different concentrations (0.001 M, 0.01 M, 0.1 M, 1.0 M, and 10.0 M) in Ham’s F12 media without serum. After 2 h drug treatment, the cells were processed as described above.
Image Digitization and Densitometry Analysis.
The exposed Western blotting films were scanned using an EPSON perfection 4490 photo scanner at 300 dpi resolution and saved in portable document format (PDF). The resulting images were converted to black and white using Inkscape software without any additional means of postprocessing. Densitometry analysis was performed using ImageJ software.
Structural Modeling.
The PDB structures 2GS6 (residues 696–1019) and 3W32 (residues 701–1017) were used to model the active and inactive states of the EGFR kinase domain, respectively (9, 51). All crystallographic ligands, ions, and water molecules were removed from the original structures. The two missing loops of 2GS6 (C loop and part of the C-terminal tail) were fixed using the automodel class of MODELLER (version 9.12) (57). We then introduced the C–4 loop insertions in the WT EGFR structure using the same protocol. The insertion loop of various mutants was then refined by the robotics-inspired kinematic closure (KIC) algorithm implemented in Rosetta, in which 1,000 loop structures were generated (58). We clustered the structure ensemble of the insertion mutations based on the rms similarity of the modeled C–4 loop. The analysis revealed two clusters for D770_N771insG and three clusters for D770GY and N771_P772insN in the active state and four clusters for D770_N771insG and five clusters for D770GY and N771_P772insN in the inactive state. The cluster center of each cluster was selected for subsequent MD simulations. The number of structures and Rosetta energy distribution for each cluster are shown in SI Appendix, Figs. S4 and S5.
MD Simulations.
Unbiased MD simulation.
Full-atom unbiased MD simulations were performed for each insertion mutation using GROMACS version 5.0.14 patched with plumed version 2.3.3 (59, 60). Representative structural models from each cluster center were used as the starting conformation for MD. All hydrogen atoms were converted to virtual sites to remove the fastest vibrational freedom. The protein was parameterized with the amber99sb-ildn force field and solvated with the TIP3P water model in a dodecahedron box that was 1 nm larger than the protein in all directions (61). Sodium and chloride ions were added to the system to neutralize the charge of the protein. The Verlet cutoff scheme was used to maintain the neighbor list for calculating nonbonded interactions (62). Long-range electrostatics were calculated using particle mesh Ewald (PME) algorithms. Energy minimization was performed with the steepest-descent algorithm followed by the conjugate-gradient algorithm until the was less than 100 kJ. The canonical ensemble was carried out by heating the system from 0 K to 310 K, using velocity rescaling for 200 ps (63). The isothermal–isobaric ensemble (P = 1 bar, T = 310 K) was carried out using the Berendsen barostat for 200 ps (64). Position restraint was applied to nonhydrogen atoms of the protein during the equilibration steps. The unrestrained MD productions were collected using a time step of 5 fs after the isothermal–isobaric ensemble. The detailed simulation information can be found in SI Appendix, Table S2. The trajectories were processed and analyzed using the built-in tools of Gromacs and plumed. Structural visualization was performed using PyMOL (65).
SMD simulation.
SMD is a technique to transform the system from an initial configuration to a target configuration by applying an external potential biased toward the target state. In our simulation, the external potential takes the form of a harmonic function of the instantaneous atom coordinates () to the corresponding target atoms coordinates (): , where is the external potential being added to the system Hamiltonian at each time step and is the spring constant controlling the strength of the pulling force. Because the crystal structure of the active state of EGFR is several residues longer than that of the inactive state, we trimmed the active model of EGFR to match the inactive model so that the two structures contain identical sequences (residues 701–1017). The system preparation protocol for SMD simulation is similar to the MD simulation described in the previous subsection. In the production run, the plumed engine was enabled to calculate the CVs and bias the system potential (60). A carefully chosen CV is important to obtain the reliable intermediate transition states connecting the two conformations (35). In this study, we used two different CVs to perform SMD simulation, i.e., (i) the C atoms rmsd of the whole protein (residues 701–1017) and (ii) the C atoms rmsd of the C-helix and activation loop (residues 756–769 and 855–863). A total of four independent SMD simulations were run by performing bidirectional transformations (active to inactive or inactive to active). The detailed information for each SMD simulation can be found in SI Appendix, Table S3. The convergence of conformational transition was determined by monitoring the endpoint rmsd value to be less than 0.05 nm to the target state.
Umbrella sampling.
Umbrella sampling (US) is a widely used enhanced sampling technique to obtain the PMF of certain CVs. We used two CVs to describe the conformational transition of the EGFR kinase domain. CV1 is defined as the pseudodihedral angle formed by four atoms in the DFG motif, C(D855)–C(F856) –C(G857) –C(L858), and CV2 is defined as the distance between two salt bridge-forming pairs , where is K745/NZ–E762/(OE1,OE2) distance and is K860/NZ–E762/(OE1,OE2) distance (SI Appendix, Fig. S9A). A total of 178 US centers were selected to cover the 2D CV space explored by the SMD simulations (SI Appendix, Fig. S9B). A 5-ns US simulation on each of the selected US centers was carried out by applying a restraining potential with the form , where and are the current US centers. and are the instantaneous CV values evaluated at time . and are the spring constants controlling the width of US exploration at each chosen CV center. In our simulation, spring constants for CV1 and CV2 were 500 kJ and 750 kJ, respectively. A sufficient overlap in the CV space between each US simulation and the rest of the simulations was checked before subsequent analysis. To reconstruct the 2D PMF, the time-series data of the last 4-ns simulation were analyzed by the weighted histogram analysis method (WHAM) implemented in the PyEMMA package (66, 67). The convergence of US sampling was ensured by block analysis. Specifically, we split the last 4 ns of each US simulation into four equal-length blocks (1 ns each). WHAM calculation was done by using the time-series data of each block. The average difference of the 2D PMF profile between adjacent blocks was less than 1.0 kJ for our US sampling, indicating PMF convergence. The statistical error was estimated by running three independent US simulations for WT and the three activating insertion mutations. Localization of the LFEP along the 2D-PMF connecting two different states is estimated using the algorithm described in a previous study (68).
Supplementary Material
Acknowledgments
We acknowledge the constructive feedback from Dr. Robert J. Woods, Dr. Eileen J. Kennedy, and Dr. Shaying Zhao. We thank members of the N.K. laboratory for helpful comments and suggestions on the manuscript. Funding for N.K. from National Institutes of Health Grant 5RO1GM114409 is acknowledged. Z.R. is the recipient of a 2017 Innovative and Interdisciplinary Research Grant for Doctoral Students at the University of Georgia. This study was supported in part by resources and technical expertise from the Georgia Advanced Computing Resource Center, a partnership between the University of Georgia’s Office of the Vice President for Research and Office of the Vice President for Information Technology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. Y.S. is a guest editor invited by the Editorial Board.
Data deposition: The MD snapshots, CV dataset obtained from US sampling, and the python code for generating the figures are available in github repository (https://github.com/esbg/PNAS_Ruan_2018).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1803152115/-/DCSupplemental.
References
- 1.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 2.Frattini V, et al. The integrated landscape of driver genomic alterations in glioblastoma. Nat Genet. 2013;45:1141–1149. doi: 10.1038/ng.2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Day KC, et al. Her2 and EGFR overexpression support metastatic progression of prostate cancer to bone. Cancer Res. 2017;77:74–85. doi: 10.1158/0008-5472.CAN-16-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Masuda H, et al. Role of epidermal growth factor receptor in breast cancer. Breast Cancer Res Treat. 2012;136:331–345. doi: 10.1007/s10549-012-2289-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gazdar AF. Activating and resistance mutations of EGFR in non-small-cell lung cancer: Role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene. 2009;28:S24–S31. doi: 10.1038/onc.2009.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arcila ME, et al. EGFR exon 20 insertion mutations in lung adenocarcinomas: Prevalence, molecular heterogeneity, and clinicopathologic characteristics. Mol Cancer Ther. 2013;12:220–229. doi: 10.1158/1535-7163.MCT-12-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackman DM, et al. Exon 19 deletion mutations of epidermal growth factor receptor are associated with prolonged survival in non-small cell lung cancer patients treated with gefitinib or erlotinib. Clin Cancer Res. 2006;12:3908–3914. doi: 10.1158/1078-0432.CCR-06-0462. [DOI] [PubMed] [Google Scholar]
- 8.Tsai CJ, Nussinov R. The molecular basis of targeting protein kinases in cancer therapeutics. Semin Cancer Biol. 2013;23:235–242. doi: 10.1016/j.semcancer.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 10.Yun CH, et al. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: Mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11:217–227. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shan Y, et al. Oncogenic mutations counteract intrinsic disorder in the EGFR kinase and promote receptor dimerization. Cell. 2012;149:860–870. doi: 10.1016/j.cell.2012.02.063. [DOI] [PubMed] [Google Scholar]
- 12.Yun CH, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci USA. 2008;105:2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster SA, et al. Activation mechanism of oncogenic deletion mutations in BRAF, EGFR, and HER2. Cancer Cell. 2016;29:477–493. doi: 10.1016/j.ccell.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 14.Yasuda H, Kobayashi S, Costa DB. EGFR exon 20 insertion mutations in non-small-cell lung cancer: Preclinical data and clinical implications. Lancet Oncol. 2012;13:e23–e31. doi: 10.1016/S1470-2045(11)70129-2. [DOI] [PubMed] [Google Scholar]
- 15.Yasuda H, et al. Structural, biochemical, and clinical characterization of epidermal growth factor receptor (EGFR) exon 20 insertion mutations in lung cancer. Sci Transl Med. 2013;5:216ra177. doi: 10.1126/scitranslmed.3007205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kosaka T, et al. Response heterogeneity of EGFR and HER2 exon 20 insertions to covalent EGFR and HER2 inhibitors. Cancer Res. 2017;77:2712–2721. doi: 10.1158/0008-5472.CAN-16-3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He M, et al. EGFR exon 19 insertions: A new family of sensitizing EGFR mutations in lung adenocarcinoma. Clin Cancer Res. 2012;18:1790–1797. doi: 10.1158/1078-0432.CCR-11-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naidoo J, et al. Epidermal growth factor receptor exon 20 insertions in advanced lung adenocarcinomas: Clinical outcomes and response to erlotinib. Cancer. 2015;121:3212–3220. doi: 10.1002/cncr.29493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohsaka S, et al. A method of high-throughput functional evaluation of EGFR gene variants of unknown significance in cancer. Sci Transl Med. 2017;9:eaan6566. doi: 10.1126/scitranslmed.aan6566. [DOI] [PubMed] [Google Scholar]
- 20.Dawson JP, et al. Epidermal growth factor receptor dimerization and activation require ligand-induced conformational changes in the dimer interface. Mol Cell. Biol. 2005;25:7734–7742. doi: 10.1128/MCB.25.17.7734-7742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang Y, et al. Molecular basis for multimerization in the activation of the epidermal growth factor receptor. Elife. 2016;5:e14107. doi: 10.7554/eLife.14107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z, et al. Mechanistic insights into the activation of oncogenic forms of EGF receptor. Nat Struct Mol Biol. 2011;18:1388–1393. doi: 10.1038/nsmb.2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho J, et al. Cetuximab response of lung cancer-derived EGF receptor mutants is associated with asymmetric dimerization. Cancer Res. 2013;73:6770–6779. doi: 10.1158/0008-5472.CAN-13-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cho J, et al. Colon cancer-derived oncogenic EGFR G724S mutant identified by whole genome sequence analysis is dependent on asymmetric dimerization and sensitive to cetuximab. Mol Cancer. 2014;13:141. doi: 10.1186/1476-4598-13-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruan Z, Kannan N. Mechanistic insights into R776H mediated activation of epidermal growth factor receptor kinase. Biochemistry. 2015;54:4216–4225. doi: 10.1021/acs.biochem.5b00444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruan Z, Katiyar S, Kannan N. Computational and experimental characterization of patient derived mutations reveal an unusual mode of regulatory spine assembly and drug sensitivity in EGFR kinase. Biochemistry. 2017;56:22–32. doi: 10.1021/acs.biochem.6b00572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li S, et al. Structural basis for inhibition of the epidermal growth factor receptor by cetuximab. Cancer Cell. 2005;7:301–311. doi: 10.1016/j.ccr.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 28.Fulton MD, et al. Conformationally constrained peptides target the allosteric kinase dimer interface and inhibit EGFR activation. Bioorg Med Chem. 2017;26:1167–1173. doi: 10.1016/j.bmc.2017.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shan Y, Arkhipov A, Kim ET, Pan AC, Shaw DE. Transitions to catalytically inactive conformations in EGFR kinase. Proc Natl Acad Sci USA. 2013;110:7270–7275. doi: 10.1073/pnas.1220843110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White KA, et al. Cancer-associated arginine-to-histidine mutations confer a gain in pH sensing to mutant proteins. Sci Signal. 2017;10:eaam9931. doi: 10.1126/scisignal.aam9931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang DD, Zhou W, Yan H, Wong M, Lee V. Personalized prediction of EGFR mutation-induced drug resistance in lung cancer. Sci Rep. 2013;3:2855. doi: 10.1038/srep02855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma L, et al. EGFR mutant structural database: Computationally predicted 3D structures and the corresponding binding free energies with gefitinib and erlotinib. BMC Bioinformatics. 2015;16:85. doi: 10.1186/s12859-015-0522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kannan S, et al. Hydration effects on the efficacy of the epidermal growth factor receptor kinase inhibitor afatinib. Sci Rep. 2017;7:1540. doi: 10.1038/s41598-017-01491-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Li X, Ma W, Dong Z. Conformational transition pathways of epidermal growth factor receptor kinase domain from multiple molecular dynamics simulations and Bayesian clustering. J Chem Theor Comput. 2014;10:3503–3511. doi: 10.1021/ct500162b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan AC, Weinreich TM, Shan Y, Scarpazza DP, Shaw DE. Assessing the accuracy of two enhanced sampling methods using EGFR kinase transition pathways: The influence of collective variable choice. J Chem Theor Comput. 2014;10:2860–2865. doi: 10.1021/ct500223p. [DOI] [PubMed] [Google Scholar]
- 36.Sutto L, Gervasio FL. Effects of oncogenic mutations on the conformational free-energy landscape of EGFR kinase. Proc Natl Acad Sci USA. 2013;110:10616–10621. doi: 10.1073/pnas.1221953110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen H, et al. A molecular brake in the kinase hinge region regulates the activity of receptor tyrosine kinases. Mol Cell. 2007;27:717–730. doi: 10.1016/j.molcel.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klein T, et al. Structural and dynamic insights into the energetics of activation loop rearrangement in FGFR1 kinase. Nat Commu. 2015;6:7877. doi: 10.1038/ncomms8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kannan N, Neuwald AF. Did protein kinase regulatory mechanisms evolve through elaboration of a simple structural component? J Mol Biol. 2005;351:956–972. doi: 10.1016/j.jmb.2005.06.057. [DOI] [PubMed] [Google Scholar]
- 40.Verba KA, et al. Atomic structure of Hsp90-Cdc37-Cdk4 reveals that Hsp90 traps and stabilizes an unfolded kinase. Science. 2016;352:1542–1547. doi: 10.1126/science.aaf5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu W, et al. Surface charge and hydrophobicity determine ErbB2 binding to the Hsp90 chaperone complex. Nat Struct Mol Biol. 2005;12:120–126. doi: 10.1038/nsmb885. [DOI] [PubMed] [Google Scholar]
- 42.den Dunnen JT, Antonarakis SE. Mutation nomenclature extensions and suggestions to describe complex mutations: A discussion. Hum Mutat. 2000;15:7–12. doi: 10.1002/(SICI)1098-1004(200001)15:1<7::AID-HUMU4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 43.McSkimming DI, et al. Prokino: A unified resource for mining the cancer kinome. Hum Mutat. 2015;36:175–186. doi: 10.1002/humu.22726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Politi K, Lynch TJ. Two sides of the same coin: EGFR exon 19 deletions and insertions in lung cancer. Clin Cancer Res. 2012;18:1490–1492. doi: 10.1158/1078-0432.CCR-11-3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Red Brewer M, et al. Mechanism for activation of mutated epidermal growth factor receptors in lung cancer. Proc Natl Acad Sci USA. 2013;110:E3595–E3604. doi: 10.1073/pnas.1220050110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cross DAE, et al. AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov. 2014;4:1046–1061. doi: 10.1158/2159-8290.CD-14-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gonzales AJ, et al. Antitumor activity and pharmacokinetic properties of PF-00299804, a second-generation irreversible pan-erbB receptor tyrosine kinase inhibitor. Mol Cancer Ther. 2008;7:1880–1889. doi: 10.1158/1535-7163.MCT-07-2232. [DOI] [PubMed] [Google Scholar]
- 48.Minkovsky N, Berezov A. BIBW-2992, a dual receptor tyrosine kinase inhibitor for the treatment of solid tumors. Curr Opin Investig Drugs. 2008;9:1336–1346. [PubMed] [Google Scholar]
- 49.Yosaatmadja Y, et al. Binding mode of the breakthrough inhibitor AZD9291 to epidermal growth factor receptor revealed. J Struct Biol. 2015;192:539–544. doi: 10.1016/j.jsb.2015.10.018. [DOI] [PubMed] [Google Scholar]
- 50.McSkimming DI, Rasheed K, Kannan N. Classifying kinase conformations using a machine learning approach. BMC Bioinformatics. 2017;18:86. doi: 10.1186/s12859-017-1506-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kawakita Y, et al. Design and synthesis of novel pyrimido[4,5-b]azepine derivatives as HER2/EGFR dual inhibitors. Bioorg Med Chem. 2013;21:2250–2261. doi: 10.1016/j.bmc.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 52.Sajnaga E, Kubiński K, Szyszka R. Catalytic activity of mutants of yeast protein kinase CK2alpha. Acta Biochim Pol. 2008;55:767–776. [PubMed] [Google Scholar]
- 53.Scheeff ED, Eswaran J, Bunkoczi G, Knapp S, Manning G. Structure of the pseudokinase VRK3 reveals a degraded catalytic site, a highly conserved kinase fold, and a putative regulatory binding site. Structure. 2009;17:128–138. doi: 10.1016/j.str.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ngo JCK, et al. SR protein kinase 1 is resilient to inactivation. Structure. 2007;15:123–133. doi: 10.1016/j.str.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 55.Meinhardt S, Manley MWJ, Parente DJ, Swint-Kruse L. Rheostats and toggle switches for modulating protein function. PLoS One. 2013;8:e83502. doi: 10.1371/journal.pone.0083502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu W, et al. Sensitivity of epidermal growth factor receptor and ErbB2 exon 20 insertion mutants to Hsp90 inhibition. Br J Cancer. 2007;97:741–744. doi: 10.1038/sj.bjc.6603950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eswar N, et al. Comparative protein structure modeling using modeller. Curr Protoc Bioinformatics. 2006;Chapter 5:Unit-U5.6. doi: 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mandell DJ, Coutsias EA, Kortemme T. Sub-angstrom accuracy in protein loop reconstruction by robotics-inspired conformational sampling. Nat Methods. 2009;6:551–552. doi: 10.1038/nmeth0809-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abraham MJ, et al. Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1-2:19–25. [Google Scholar]
- 60.Bonomi M, et al. Plumed: A portable plugin for free-energy calculations with molecular dynamics. Comput Phys Commun. 2009;180:1961–1972. [Google Scholar]
- 61.Lindorff-Larsen K, et al. Improved side-chain torsion potentials for the Amber ff99sB protein force field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Páll S, Hess B. A flexible algorithm for calculating pair interactions on SIMD architectures. Comput Phys Commun. 2013;184:2641–2650. [Google Scholar]
- 63.Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. J Chem Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 64.Berendsen HJC, Postma JPM, WFv G, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 65.PyMOL 2015. The PyMOL Molecular Graphics System (Schrödinger, LLC, New York), Version 1.8.
- 66.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. The weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. J Comput Chem. 1992;13:1011–1021. [Google Scholar]
- 67.Scherer MK, et al. PyEMMA 2: A software package for estimation, validation, and analysis of Markov models. J Chem Theor Comput. 2015;11:5525–5542. doi: 10.1021/acs.jctc.5b00743. [DOI] [PubMed] [Google Scholar]
- 68.Ensing B, Laio A, Parrinello M, Klein ML. A recipe for the computation of the free energy barrier and the lowest free energy path of concerted reactions. J Phys Chem B. 2005;109:6676–6687. doi: 10.1021/jp045571i. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.