Significance
N6-methyladenosine (m6A) has recently been found to regulate numerous aspects of RNA biology. Similar to methylation of cytosine residues in DNA, eukaryotic RNA is modified by enzymatic addition of methyl groups at adenosines. m6A modification of RNA affects a wide variety of RNA functions, including mRNA stability, translation, and in the case of viruses, viral replication and production. Our investigation revealed that the adenosine residues present in the known m6A consensus motif within the 5′ epsilon stem loop of pregenomic RNA and the 3′ ends of all the hepatitis B virus (HBV) transcripts are m6A modified. We demonstrate here that m6A modification differentially modulates HBV RNA stability and reverse transcription, thereby playing two distinct regulatory roles in the HBV life cycle.
Keywords: hepatitis B virus, RNA methylation, HBV reverse transcription, epsilon loop
Abstract
N6-methyladenosine (m6A) RNA methylation is the most abundant epitranscriptomic modification of eukaryotic messenger RNAs (mRNAs). Previous reports have found m6A on both cellular and viral transcripts and defined its role in regulating numerous biological processes, including viral infection. Here, we show that m6A and its associated machinery regulate the life cycle of hepatitis B virus (HBV). HBV is a DNA virus that completes its life cycle via an RNA intermediate, termed pregenomic RNA (pgRNA). Silencing of enzymes that catalyze the addition of m6A to RNA resulted in increased HBV protein expression, but overall reduced reverse transcription of the pgRNA. We mapped the m6A site in the HBV RNA and found that a conserved m6A consensus motif situated within the epsilon stem loop structure, is the site for m6A modification. The epsilon stem loop is located in the 3′ terminus of all HBV mRNAs and at both the 5′ and 3′ termini of the pgRNA. Mutational analysis of the identified m6A site in the 5′ epsilon stem loop of pgRNA revealed that m6A at this site is required for efficient reverse transcription of pgRNA, while m6A methylation of the 3′ epsilon stem loop results in destabilization of all HBV transcripts, suggesting that m6A has dual regulatory function for HBV RNA. Overall, this study reveals molecular insights into how m6A regulates HBV gene expression and reverse transcription, leading to an increased level of understanding of the HBV life cycle.
The N6-methyladenosine (m6A) modification of eukaryotic RNA is currently recognized as a cotranscriptional modification that can regulate RNA function. Many biological processes, including fertility, stem cell differentiation, circadian rhythm, stress response, and cancer are known to be regulated by m6A modification (1–4). While m6A was detected in viral transcripts, including those of influenza A virus, simian virus 40, Rous sarcoma virus, avian sarcoma virus, and adenovirus several decades ago (5–7), little was known about the effects of the m6A modification on viral replication and pathogenesis until recently. The recent discoveries of the enzymes that add and remove m6A modification, as well as technical advances in transcriptome-wide profiling of m6A through immunoprecipitation coupled with next generation sequencing, have permitted the elucidation of its effects on viral pathogenesis. We now know that m6A modification plays a role in regulating the life cycle of multiple viruses (8–12). For instance, m6A regulates HIV replication (8, 11, 13), potentially by regulating reverse transcription (13). In the case of Kaposi’s sarcoma-associated herpesvirus, m6A modification is required for splicing of the pre-mRNA encoding the replication transcription activator (RTA) and supports lytic replication of the virus (12, 14, 15). m6A is also found across the cytoplasmically replicating Flaviviridae members and negatively regulates the life cycle of hepatitis C virus and Zika virus (9, 10). Differences in the role of m6A among viruses may result in different outcomes in their viral life cycles but, taken together, these recent reports defining roles for m6A during viral infection reinforce the notion that this RNA modification likely plays an important role in viral pathogenesis.
Hepatitis B virus (HBV) infection is the leading cause of chronic hepatitis and carries the risk for the development of cirrhosis and hepatocellular carcinoma (16). It is estimated that about 350 million people are infected with HBV worldwide (16). After its entry into the hepatocytes via Na/taurocholate cotransporting polypeptide (NTCP) receptor (17), the small virion-associated, partially double-stranded relaxed circular DNA (rcDNA) genome (∼3.2 kb) undergoes several modifications in the nucleus to produce a covalently closed circular DNA (cccDNA). This cccDNA is organized into a minichromosome with histone and nonhistone proteins (16), and it is transcribed by the cellular polymerase II machinery to produce viral RNAs. HBV transcription begins from different transcription start sites on the HBV genome, but it ends at a common transcription termination signal. Thus, HBV transcripts differ in their 5′ terminus but share common 3′ terminal sequence. These HBV transcripts include RNAs of 2.4 and 2.1 kb that encode different forms of the hepatitis B surface antigen (HBsAg) proteins, a 0.7-kb HBx RNA that encodes HBx protein, and two RNAs longer than the genome length (3.5–3.6 kb), known as precore RNA (pcRNA) and pgRNA (18). The pcRNA starts ∼30 nucleotides (nt) upstream of the pgRNA start site and hence is longer than pgRNA. pcRNA is a message for e antigen (HBeAg) translation while pgRNA encodes for the polymerase (pol; a reverse transcriptase) and core (HBc) proteins. The majority of pgRNA serves as a template for translation of its encoded proteins, but a smaller fraction of pgRNA serves as a template for reverse transcription (18). This fraction of pgRNA associates with core and pol in the cytoplasm to become encapsidated (known as immature capsids), which then allows for reverse transcription by pol to produce a rcDNA and the mature capsids. These mature capsids either replenish the cccDNA pool or are secreted as infectious viral particles. The RNA regulatory mechanisms that regulate the functions of HBV transcripts in the viral life cycle are currently not well understood.
In this report, we sought to define the role of m6A on HBV RNA during HBV infection. We found that HBV transcripts analyzed from both HBV-expressing cells and liver tissues of chronic HBV patients contain m6A. Furthermore, by silencing the m6A methyltransferase, we found that m6A affects the stability of the HBV transcripts and also regulates the reverse transcription of pgRNA. We identified that an m6A consensus motif within the epsilon stem loop of HBV RNAs as the site of m6A modification. Importantly, mutation of the adenosine residue in this m6A consensus motif to cytosine revealed that m6A positively regulates reverse transcription but also negatively regulates the stability of HBV mRNAs, resulting in decreased expression of viral proteins. To confirm that these results were not due to altered stem loop structures made by the cytosine mutation, we made compensatory mutations that restored full base pairing in the stem loop of 5′ epsilon. In these cases, we found that reverse transcription and HBV RNA stability were still affected, reinforcing the notion that m6A regulates these processes in HBV RNA. Taken together, our results reveal two roles for m6A-modified HBV RNA, regulating both RNA stability and reverse transcription. Overall, these functions of m6A in HBV RNA are essential to regulate the viral life cycle and likely play a role in the liver disease pathogenesis associated with HBV infection.
Results
HBV-Encoded Transcripts Are m6A Modified and Are Bound by YTHDF Proteins.
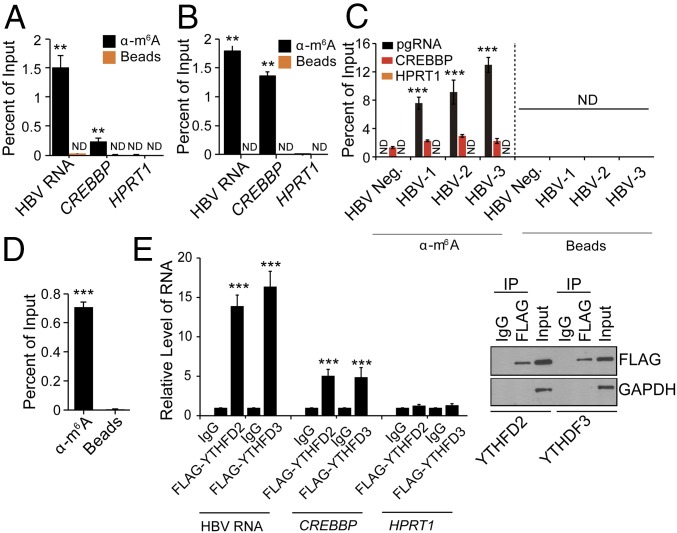
m6A has been identified on both cellular and viral RNAs (8–10, 19) but its function on HBV RNA has not been studied. We first determined that HBV transcripts do contain the m6A modification using a methylated RNA immunoprecipitation (MeRIP) assay with an m6A-specific antibody, as described previously (8–10) in the HepAD38 cell line in which the stably integrated HBV genome is under the control of an inducible tetracycline promoter (20). After inducing HBV expression by removing tetracycline from the culture medium, mRNAs were isolated from cellular lysates and subjected to MeRIP followed by RT-qPCR using primers that recognize a shared 3′ UTR sequence presented in all HBV transcripts, including pgRNA. The results presented in Fig. 1A and SI Appendix, Fig. S1A, indicate that HBV RNA contains m6A (Fig. 1A). Next, we validated these results using HepG2 cells transfected with the HBV genome expression vector (HBV 1.3mer) and found that HBV RNAs are also m6A modified in this system (Fig. 1B). In the MeRIP–RT-qPCR assay (Fig. 1 A and B and SI Appendix, Fig. S1A), CREBBP, a cellular RNA known to contain m6A and HPRT1, a cellular RNA that does not contain m6A, were used as positive and negative controls, respectively (21). Importantly, we found that HBV transcripts isolated from the liver tissues of patients with chronic hepatitis B also contain m6A, similar to the positive control CREBBP and different from the negative control HPRT1 (Fig. 1C), while beads alone did not pull down any HBV RNA. In HBV-expressing cells, while the majority of pgRNA is used to translate the corresponding encoded proteins, a fraction of pgRNA is encapsidated into core particles destined for reverse transcription. To test whether the core-associated pgRNA is also m6A modified, we isolated core particles from HBV 1.3mer-expressing HepG2 cells, extracted the core-associated RNA (22), and then performed MeRIP–RT-qPCR analysis using pgRNA-specific primers. The results show that the core-associated pgRNA is also m6A modified (Fig. 1D).
Fig. 1.
HBV transcripts contain the m6A RNA modification. MeRIP–RT-qPCR of m6A-modified HBV transcripts from total RNA extracted from (A) HepAD38 cells stably expressing HBV and (B) HepG2 cells transfected with the HBV 1.3mer genome using primers specific for the shared sequence in the 3′ UTR of all HBV RNAs. CREBBP and HPRT1 serve as positive and negative controls, respectively. (C) MeRIP–RT-qPCR analysis of total RNA from liver biopsy samples from a healthy individual (n = 1) and HBV patients (n = 3) using primers specific to pgRNA. (D) MeRIP–RT-qPCR analysis of core-associated RNA. (E) RNA immunoprecipitation (IP) from FLAG-YTHDF2 and -YTHDF3 HepAD38-HBV expression cells using an anti-FLAG antibody or IgG, with RT-qPCR analysis of HBV RNA, CREBBP, and HPRT1 were quantified as the percent of input and graphed as fold enrichment relative to IgG control. Immunoblot analysis of FLAG-YTHDF2/3 in the input and IP is shown on the Right. For A–D, the fraction of m6A-modified RNA was calculated as the percent of the level present in the eluate compared with the total input RNA. The data for this figure are from three independent experiments and the bars represent the mean ± SD. **P ≤ 0.01 and ***P ≤ 0.001. ND, not detected.
The “reader proteins” [YTH domain containing protein family (YTHDF)] are cellular m6A RNA-binding proteins that regulate the stability and translation of m6A-modified RNAs (21, 23). We next determined whether HBV transcripts are recognized by the YTHDF proteins. We transfected HepAD38 cells with plasmids encoding FLAG-YTHDF2 and FLAG-YTHDF3 and immunoprecipitated lysates using an anti-FLAG antibody. RT-qPCR analysis showed that pgRNA was enriched in both YTHDF2 and YTHDF3 immunoprecipitates relative to the IgG control, confirming that pgRNA is bound by the YTHDF2 and YTHDF3 proteins (Fig. 1E). Cellular RNAs, CREBBP and HPRT1, were analyzed as m6A-positive and m6A negative controls, respectively (Fig. 1E). Taken together, these results (Fig. 1) reveal that HBV transcripts, including the core-associated pgRNA that is destined for reverse transcription into the DNA genome, are m6A modified.
m6A RNA Methylation Modulates HBV Gene Expression and Reverse Transcription.
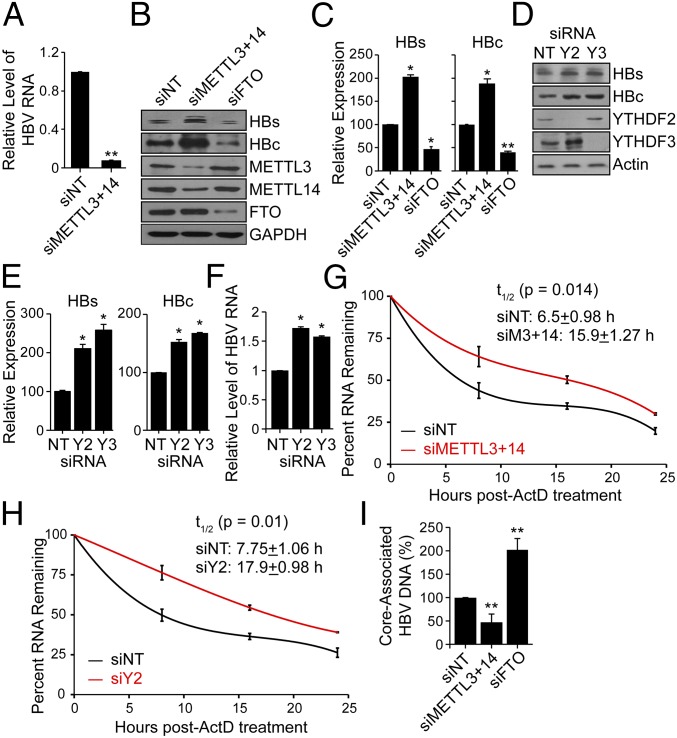
m6A is added to mRNA by a complex of proteins that includes the m6A methyltransferases METTL3 and METTL14 (24). To determine whether the cellular m6A machinery affects HBV replication, we depleted both METTL3 and METTL14 in HBV-expressing cells using siRNAs and confirmed that the levels of m6A were reduced on pgRNA by using MeRIP–qPCR (Fig. 2A). We then analyzed HBV protein expression in METTL3- and METTL14-depleted cells and found that loss of these proteins resulted in increased expression of the HBc and HBs proteins compared with control siRNA-treated cells, as revealed by immunoblotting and quantification of these immunoblots (Fig. 2 B and C). Conversely, depletion of the putative m6A demethylases, fat mass and obesity-associated protein (FTO) (25) and ALKBH5 (26), decreased the expression of these HBV proteins relative to control siRNA-treated cells (Fig. 2 B and C and SI Appendix, Fig. S1 B and C). These results suggest that m6A methylation of HBV transcripts negatively regulates HBV protein expression.
Fig. 2.
Effect of depletion of methyltransferases (METTL3/14) and demethylases (FTO) on HBV protein expression, HBV RNA stability, and reverse transcription. (A) MeRIP–RT-qPCR analysis of HBV RNA harvested from HBV-induced HepAD38 cells following siRNA depletion of METTL3 and METTL14 or nontargeting control (NT). RNA was immunoprecipitated with an anti-m6A antibody and eluted RNA was quantified as a percent of input and graphed as fraction relative to the m6A level in siNT. (B) Immunoblot analysis of HBV proteins (surface antigen, HBs) and (core, HBc) from extracts of HepG2 cells expressing the HBV 1.3mer plasmid following siRNA depletion of METTL3+METTL14, and FTO, or NT at 5 d post-HBV transfection. (C) HBV proteins levels relative to the housekeeping gene GAPDH from three independent experiments, as in B, were quantified using ImageJ. (D) Immunoblot analysis of HBs and HBc from extracts of HepG2 cells expressing the HBV 1.3mer plasmid following siRNA depletion of YTHDF2 or YTHDF3, or NT at 5 d post-HBV transfection. (E) HBV protein levels relative to the housekeeping gene Actin from three independent experiments, as in D, were quantified using ImageJ. (F) RT-qPCR analysis of HBV RNA relative to GAPDH in HepG2 cells expressing the HBV 1.3mer plasmid. At 2 d post-HBV transfection, siYTHDF2 or siYTHDF3 were added and RNA was harvested at 5 d post-HBV transfection. (G) RT-qPCR analysis of HBV RNA relative to GAPDH in the HBV 1.3mer-expressing HepG2 cells. The HBV 1.3mer-transfected HepG2 cells were depleted for METTL3 and METTL14 by siRNA, following actinomycin D treatment at 24 h post-siRNA transfection. RNA was harvested at 0, 8, 16, and 24 h post actinomycin D treatment and relative levels of remaining HBV transcript were analyzed. (H) RT-qPCR analysis of HBV RNA relative to GAPDH in the HBV 1.3mer-expressing HepG2 cells. The HBV 1.3mer-transfected HepG2 cells were depleted for YTHDF2 by specific siRNA, following actinomycin D treatment at 24 h post-siRNA transfection. RNA was harvested at 0, 8, 16, and 24 h post actinomycin D treatment and relative levels of remaining HBV transcript were analyzed. (I) Following siRNA treatment for depletion of METTL3 and METTL14, FTO, or NT in HBV-expressing HepG2 cells, the core particles were isolated (Materials and Methods). The core-associated HBV DNA was then purified and quantified by qPCR assay. The values are graphed as percent relative to the siNT, which was set at 100%. All experiments were performed in triplicate. Immunoblots shown are representative of three independent experiments, and the graph bars represent the mean ± SD of these three independent experiments. *P ≤ 0.05 and **P ≤ 0.01 by unpaired Student’s t test.
After determining that the m6A machinery regulates expression of HBV proteins, we next tested whether the m6A reader proteins (YTHDFs) could similarly affect HBV protein expression. We found that depletion of either YTHDF2 or YTHDF3 significantly increased the expression of the HBV proteins HBs and HBc (Fig. 2 D and E), suggesting that the YTHDF proteins also negatively regulate HBV protein expression. The enhanced expression of HBV proteins in YTHDF- and METTL3/14-depleted cells could be due to either elevated protein translation or increased abundance of HBV transcripts. As the YTHDF proteins are known to regulate RNA turnover and degradation of m6A-modified transcripts, including viral RNAs (15), we next tested whether this was the case with HBV RNAs. To determine the role of YTHDF proteins in the turnover of HBV transcripts, we quantified the amount of pgRNA (as a representative HBV transcript) in YTHDF-depleted cells and found that its overall level was increased in YTHDF-depleted cells (Fig. 2F). We then determined the half-life of pgRNA in cells depleted of m6A writers (METTL3/14) following actinomycin D treatment. The results show that the depletion of METTL3/14 significantly enhances the half-life of pgRNA, from about 6.5 h to 15.9 h (Fig. 2G). Similarly, the depletion of YTHDF2 in HBV-expressing HepG2 cells, also increased the half-life of HBV transcripts from 7.75 h to 17.9 h (Fig. 2H). Thus, m6A modification of HBV RNAs negatively regulates their abundance (Fig. 2 F and G) and thereby the levels of the proteins they encode (Fig. 2 B and C). In addition to assessing the role of m6A in HBV RNA stability and protein expression, we also analyzed its role in regulating HBV reverse transcription. Following depletion of the m6A machinery, we measured the synthesis of core-associated DNA following reverse transcription in the isolated core particles. We found that reverse transcription was reduced in METTL3/14-depleted cells but increased in FTO-depleted cells (Fig. 2I). Together, these results suggest that m6A modification lowers the stability of HBV RNA transcripts, and the expression of HBV proteins (Fig. 2 F and G), but positively regulates the reverse transcription step of the viral life cycle (Fig. 2I).
The DRACH Motif Within the HBV Epsilon Loop Is m6A Modified.
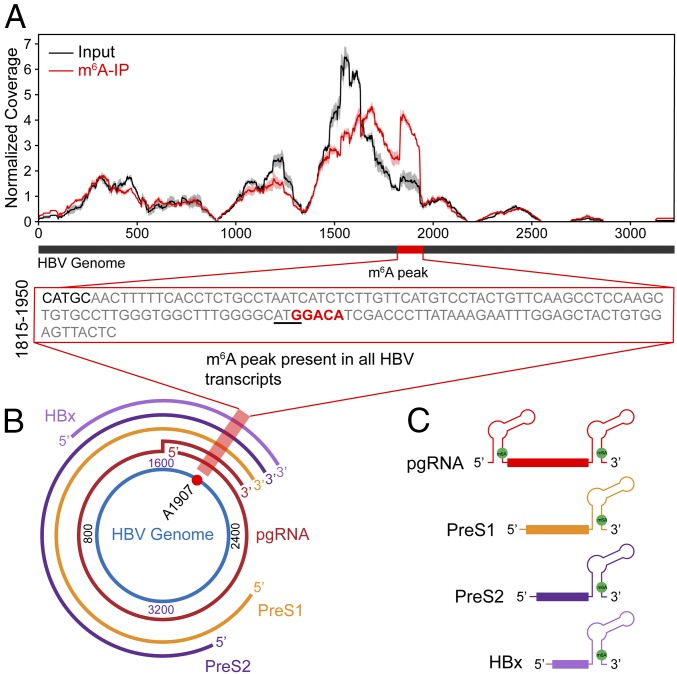
m6A modification occurs within a distinct consensus motif composed of 5′RA*C3′ (where R = G or A, and A* denotes methylated adenosine on viral and cellular transcripts) (19, 27, 28). Transcriptome-wide m6A mapping studies further identified important residues flanking RAC motifs that suggest a broader consensus motif of DRACH (where D = A, G, or U and H = A, C, or U) (29). Sequence analysis of the HBV genome using the sequence-based m6A modification site predictor (SRAMP) (30) revealed a high frequency of DRACH clusters in the HBV genome. However, as not all DRACH motifs undergo m6A modification, we experimentally determined the locations of m6A sites in HBV RNA using MeRIP-seq (19). PolyA-enriched RNAs from control and HBV-expressing hepatocytes were fragmented into 60–200 nucleotides. A fraction of this RNA was immunoprecipitated using an m6A-specific antibody (IP) and the associated RNA was eluted and then sequenced, along with the input RNA. Reads were mapped to HBV (ayw) reference and coverage was compared to identify regions enriched in m6A. In this study, we focused on the identification of m6A on the viral RNAs. We identified a distinct m6A peak in the region of HBV genome spanning from position 1815–1950 (relative to the unique single EcoRI site) (Fig. 3A). This region contains only one DRACH motif located at position 1905–1909 (GGACA) (Fig. 3A, Inset), suggesting that A1907 is the site of m6A methylation. During transcription of the HBV genome, this m6A site is encoded in the lower stem of the epsilon stem loop present in all HBV transcripts and is conserved across all known HBV genotypes (Fig. 3 B and C and SI Appendix, Fig. S2 A and B).
Fig. 3.
Consensus m6A site within the HBV genome. (A) Map of m6A-binding sites in the HBV ayw genome by MeRIP-seq of polyA-RNA isolated from HBV expressing HepG2 cells at 5 d postinfection. Read coverage, normalized to the total number of reads mapping to the viral genome for each experiment, is in red for MeRIP-seq and in gray for input RNA-seq. Means and SDs across replicates are shown for each position in the genome. One m6A peak was identified after normalizing for coverage, indicated by the red bar within the black bar that depicts a linear representation of the HBV genome. The Inset presents nt 1815–1950 of the HBV genome, with the m6A site highlighted by red text and the ATG of core ORF underlined. (B) The location of the m6A site (A1907) in the pictorial representation of transcripts within the HBV genome is indicated by red shading. (C) Schematic showing the position of the m6A site (A1907), indicated by the green filled circle in all of the HBV transcripts. Note that it is present at both the 5′ and 3′ ends of pgRNA and only at 3′ ends of the other HBV transcripts.
m6A Modification Exerts Dual Regulatory Role on pgRNA.
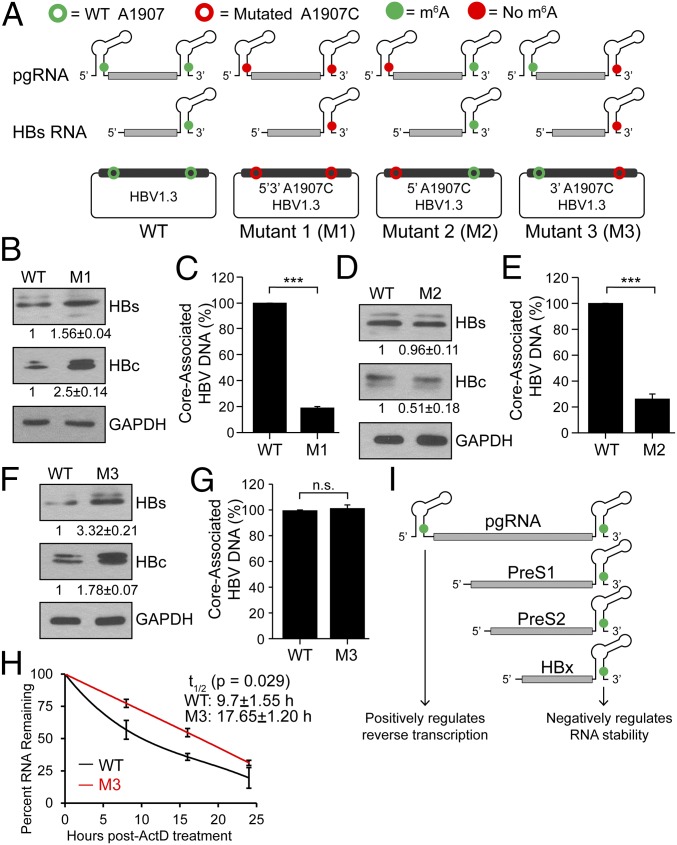
The m6A site that we identified is encoded at the 3′ epsilon stem loop of all HBV transcripts, but pgRNA acquires this site in both the 5′ and 3′ epsilon stem loops. To characterize the effect of m6A in either of these stem loops on the HBV life cycle, we generated an A1907C mutation to disrupt the identified m6A site in either of the stem loops at both the termini (HBV-M1), only the pgRNA 5′ stem loop (HBV-M2), or only the 3′ stem loop (HBV-M3) (SI Appendix, Fig. S3 A and B) in the context of the HBV1.3mer plasmid, which contains additional genomic sequence to permit the acquisition of terminal redundancy for pgRNA synthesis (SI Appendix, Fig. S3A). Possible patterns of m6A modification of the viral RNAs transcribed by these mutants are depicted in Fig. 4A. We used these mutants (Fig. 4A) to test the effect of m6A on HBV RNA stability and associated viral protein expression, as well as viral DNA synthesis by measuring core-associated RT activity. We found that when transfected into HepG2 cells, the HBV-M1 construct (loss of 5′ and 3′ m6A) resulted in increased levels of the HBc and HBs proteins compared with the WT HBV-expressing cells (Fig. 4B). However, this construct resulted in decreased levels of DNA synthesis from isolated core particles, indicating reduced reverse transcriptase activity (Fig. 4C). These results (Fig. 4 B and C) mirrored those observed in the METTl3/14-depleted cells (Fig. 2 B and H). In contrast to HBV-M1 (Fig. 4B), the HBV-M2 construct displayed no significant increase in HBV protein expression (Fig. 4D). However, the level of core-associated DNA was reduced in the HBV-M2–expressing cells (Fig. 4E), similar to HBV-M1 (Fig. 4C). On the other hand, the HBV-M3 construct had increased HBV protein expression (Fig. 4F), similar to HBV-M1 (Fig. 4B), but no significant change in the levels of core-associated DNA (Fig. 4G). Taken together, these results support a model in which m6A modification at both termini of the HBV RNAs (5′ and 3′ epsilon stem loop) exerts different effects. To test whether the increased HBV protein expression following loss of the 3′ m6A site was due to alterations in HBV RNA stability, we determined the half-life of HBV RNA from the HBV-M3 construct compared with WT following actinomycin D treatment. The results show that HBV RNAs from HBV-M3 have an increased half-life, from about 9.7 h to 17.7 h (Fig. 4H). Based on these results, we conclude that the m6A at the 5′ stem loop plays a positive role for reverse transcription of pgRNA while the m6A at the 3′ stem loop, present in all HBV transcripts, negatively regulates HBV RNA stability (Fig. 4I).
Fig. 4.
Mutations of 5′ and 3′ DRACH motifs and their effect on HBV protein expression and DNA synthesis. (A) Schematics indicate the location of the A1907C mutations of 5′ and 3′ m6A sites in HBV RNAs. Open circles represent WT (green) and A1907C mutations (red) in HBV plasmid DNA. Green solid circles indicate m6A modification while red solid circles represent lack of m6A (due to A1907C mutations) in encoded RNAs. HBV-M1 having the A1907C mutation at both termini, HBV-M2 only at the 5′ end, and HBV-M3 only at the 3′ end. (B–G) Immunoblot analysis of HBV proteins following transfection of indicated HBV constructs in HepG2 cells with relative quantifications below the respective blots [(B) M1, (D) M2, (F) M3] and core-associated DNA isolated from core particles and quantified by qPCR using equal amounts of purified DNA in the input. Relative levels of core-associated DNA from the various constructs [(C) M1, (E) M2, (G) M3] graphed as percent relative to HBV-WT. (H) RT-qPCR analysis of relative levels of remaining HBV transcript relative to GAPDH isolated at indicated times following 24 h of actinomycin D treatment from HBV 1.3mer (WT or M3)-expressing HepG2 cells. (I) Model for how m6A at the 5′ or 3′ epsilon stem loops of HBV transcripts differentially regulates their stability and reverse transcription. Data here are presented from three independent experiments and the bars represent mean ± SD. ***P ≤ 0.001 by unpaired Student’s t test, and n.s., not significant.
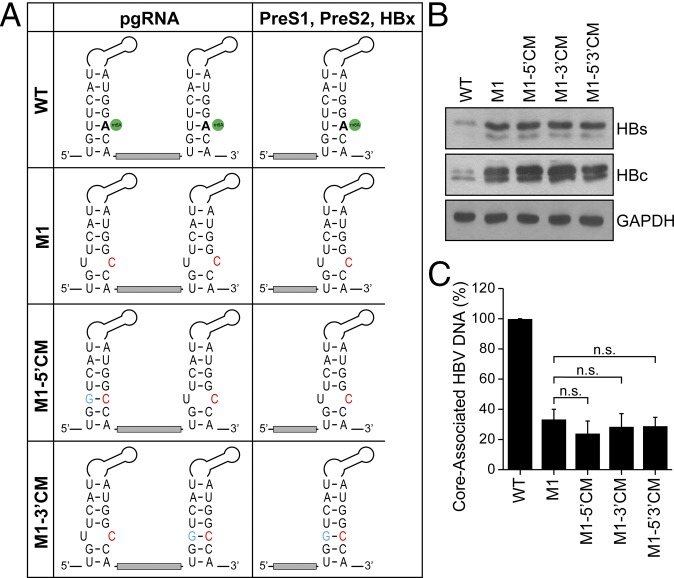
The A1907C mutation in the HBV-M1, -M2 and -M3 constructs produces a base pair mismatch in the lower stem of the epsilon structure (Fig. 5A). As this base pair mismatch slightly alters the secondary structure of the lower stem of the epsilon loop, it is possible that our observed effects of the A1907C substitution on protein expression and reverse transcription seen in Fig. 4 could be due to these structural alterations. We therefore restored the base pairing in the stem loop by generating the compensatory guanine mutation (CM) U1851G in the HBV-M1 construct, either at the 5′ end to make HBV-M1-5′CM or at the 3′ end to make HBV-M1-3′CM, or at both ends to make HBV-M1-5′3′ CM (Fig. 5A). Importantly, when expressed in HepG2 cells, each of these constructs had no difference in HBV protein expression and reverse transcription relative to parental HBV-M1 mutant, and they all still showed increased HBV protein expression and decreased RT activity compared with wild-type HBV-expressing cells (Fig. 5 B and C). Therefore, these results reveal that the loss of m6A, and not any structural alterations to the epsilon stem loop, is regulating the effects on HBV RNA stability and reverse transcription that we observed above (Fig. 4 B–G).
Fig. 5.
Compensatory mutations in the epsilon structure in the HBV 1.3-expressing plasmid do not restore reverse transcription. (A) The epsilon loop within HBV RNAs is depicted, with the m6A site indicated in green, to highlight the HBV base pairing (nt 1849–1861 with nt 1897–1909) in the lower stem and to show that the A1907C substitution in HBV-M1 (red) is predicted to create a bubble. To restore the base pairing with C1907 the compensatory U1851G mutation (blue) was introduced either at the 5′ end (HBV-M1-5′ CM), the 3′ end (HBV-M1-3′ CM), or both (HBV-M1-5′3′ CM). (B and C) Using these constructs, HBV protein expression (B) and core-associated DNA (C) were analyzed, as done previously. The data are the presentation of three independent experiments. The bars represent mean ± SD. n.s., not significant by unpaired Student’s t test.
Discussion
The modification of RNA by m6A has been shown to regulate the function of both cellular and viral RNAs (2, 3, 10, 14, 21, 23, 31). Since HBV contains only five major transcripts, we hypothesized that HBV RNA might be chemically modified as a way of regulating the function of these RNAs within the viral life cycle. We found that HBV transcripts, isolated from HBV-expressing cells in culture as well as from the livers of HBV-positive patients, contain m6A, and that the cellular m6A machinery regulates HBV infection. We used MeRIP-seq to map the m6A site on HBV RNAs and found that the m6A consensus motif situated within the epsilon stem loop is the site for m6A modification (Fig. 3) and that this m6A consensus motif is evolutionarily conserved. By making HBV mutants that lack m6A either in the 5′ or 3′ epsilon structures (or both), we were able to define unique positional effects of m6A on specific HBV RNA functions, including increased stability of HBV transcripts and reduced reverse transcription from pgRNA. Importantly, depletion of the m6A methylatransferase complex (METTL3/14) also resulted in similar alterations to HBV RNA function (Fig. 2 B, C, and G) to those obtained by terminus-specific inactivation of m6A in the epsilon stem loop (Fig. 4 B, D, and H). Based on these results, we conclude that m6A on HBV RNAs dually regulates HBV transcripts by modulating their stability and thereby protein expression, as well as by promoting reverse transcription of pgRNA.
m6A regulates numerous aspects of cellular RNA biology (1–4). These effects of m6A often depend on its position (such as coding region, 5′ UTR or 3′ UTR) in the target RNA or the complement RNA-binding proteins that recognize this modification (21, 23). For instance, the presence of m6A in the 3′ UTR of cellular RNA enhances RNA decay mediated by interactions with YTHDF2 and the CCR4-Not complex (21, 32). We observed that m6A regulates HBV RNAs in more than one way, depending on its position in the RNA. First, we observed that m6A at HBV 3′ UTRs renders these RNAs less stable, ultimately affecting the expression of proteins encoded by these RNAs. We also found that HBV transcripts bind YTHDF proteins (Fig. 1E) and that depletion of YTHDF proteins increased HBV protein expression, likely through alterations in HBV transcript stability (Fig. 2 D–F). Importantly, mutational inactivation of the m6A site within the 3′ epsilon loop of all HBV transcripts also had similar effects: increased HBV RNA stability and subsequently protein expression (Fig. 4 F and G). While we could not analyze the expression of HBx or polymerase proteins, as reliable antibodies are currently not available, it is likely that their expression is also negatively regulated by m6A through the regulation of their mRNAs. This negative regulation of viral gene expression by m6A is congruent with the widely recognized role of m6A in RNA stability (15).
Second, we found that m6A at the 5′ epsilon loop, which is present only in pgRNA, positively regulates pgRNA reverse transcription. The exact mechanisms by which m6A regulates reverse transcription need to be explored further. Before pgRNA is reverse transcribed, it is associated with polymerase via its 5′ epsilon stem loop (33). This association serves as the signal for encapsidation, facilitating reverse transcription (33, 34). Then, initiation and priming occur at the 5′ epsilon stem loop followed by a jump of the polymerase–primer complex to the 3′ end and subsequent synthesis of minus strand synthesis of the HBV genome (35). Multiple distinct interactions modulated by m6A modification could affect the association of polymerase with pgRNA and exert an overall positive effect on minus strand DNA synthesis. In the case of duck hepatitis B virus, analogous to HBV, the polymerase–epsilon stem loop interaction is a dynamic and a multistep process, in which the initial RNA binding is followed by conformational changes in both polymerase and RNA (36). These changes are crucial to facilitate reverse transcription and require assistance by cellular chaperones (37). Since m6A can modulate both RNA structure and RNA–protein interactions (31), further studies are required to characterize the m6A-mediated remodeling of the 5′ epsilon loop of HBV transcript and how this structural switch contributes to the differential recruitment of cellular chaperones (possibly m6A reader proteins) to promote reverse transcription. NMR-based studies of the epsilon stem loop with or without m6A at A1907 may provide more structural insights into how this RNA modification regulates reverse transcription. In conclusion, our investigation has revealed a dual regulatory role for m6A modification in the HBV life cycle, based on its position within a viral transcript, which may bear relevance to chronic hepatitis and other associated pathological syndromes. This work also reveals that rather than exerting overall proviral or antiviral effects, m6A can be exploited by viruses for finer temporal control over events of their life cycle.
Materials and Methods
A detailed description of the methods used for site-directed mutagenesis, MeRIP-seq, and all other related experiments can be found in SI Appendix.
Cell Culture and Transfection.
HepG2 cells, transfected with whole HBV genome 1.3mer plasmid (Addgene) and HepAD38 (20) cells stably expressing HBV, were used in this study. Cells were maintained in DMEM supplemented with 10% FBS. For plasmid transfection, TransIT-LT1 reagent (Mirus) was used. Gene-specific siRNAs were derived from siRNA-SMARTpool, siGENOME (Dharmacon) and transfected using Lipofectamine RNAiMAX reagent (Thermo Fisher Scientific).
Quantitative PCR.
Quantitative PCR was carried out on the RNA extracted from the immunoprecipitated poly-A purified RNA from HBV-expressing HepG2 or HepAD38 cells using TRIzol reagent (Life Technologies). m6A RNA was immunoprecipitated according to the protocol described by Dominissini et al. (19).
Human Liver Biopsy Specimens.
Frozen human liver biopsy specimens (deidentified, n = 4) were collected in the Soonchunhyang University Hospital, Seoul, South Korea from anti–HBV-negative (n = 1) and anti–HBV-positive (n = 3) patients. Liver biopsy samples were collected according to institutional review board (IRB) protocols of the hospital. The IRB has approved this study as a whole.
Supplementary Material
Acknowledgments
We thank Dr. Christoph Seeger (Fox Chase Cancer Center) for providing HepAD38 cells and Dr. Tariq Rana (University of California, San Diego) for constant advice and encouragement. This work was supported in part by National Institutes of Health Grants AI125350, AI139234 (to A.S.), MH117406 (to C.E.M.), AI125416, and AI129851 (to S.M.H. and C.E.M.); the Burroughs Wellcome Fund (S.M.H.); the American Heart Association (N.S.G.); the Irma T. Hirschl and Monique Weill-Caulier Charitable Trusts; the Bert L. and N. Kuggie Vallee Foundation; the WorldQuant Foundation; the Pershing Square Sohn Cancer Research Alliance; NASA (NNX14AH50G and NNX17AB26G); the Bill and Melinda Gates Foundation (OPP1151054 to C.E.M.); and National Research Council of Science and Technology Grant CRC-16-01-KRICT from the Korean government (Ministry of Science, Institute of Chemical Technology and Future Planning) (to S.-J.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. G.R. is a guest editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE114486).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1808319115/-/DCSupplemental.
References
- 1.Yue Y, Liu J, He C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2015;29:1343–1355. doi: 10.1101/gad.262766.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li S, Mason CE. The pivotal regulatory landscape of RNA modifications. Annu Rev Genomics Hum Genet. 2014;15:127–150. doi: 10.1146/annurev-genom-090413-025405. [DOI] [PubMed] [Google Scholar]
- 3.Saletore Y, et al. The birth of the epitranscriptome: Deciphering the function of RNA modifications. Genome Biol. 2012;13:175. doi: 10.1186/gb-2012-13-10-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou J, et al. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature. 2015;526:591–594. doi: 10.1038/nature15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dimock K, Stoltzfus CM. Sequence specificity of internal methylation in B77 avian sarcoma virus RNA subunits. Biochemistry. 1977;16:471–478. doi: 10.1021/bi00622a021. [DOI] [PubMed] [Google Scholar]
- 6.Kane SE, Beemon K. Precise localization of m6A in Rous sarcoma virus RNA reveals clustering of methylation sites: Implications for RNA processing. Mol Cell Biol. 1985;5:2298–2306. doi: 10.1128/mcb.5.9.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krug RM, Morgan MA, Shatkin AJ. Influenza viral mRNA contains internal N6-methyladenosine and 5′-terminal 7-methylguanosine in cap structures. J Virol. 1976;20:45–53. doi: 10.1128/jvi.20.1.45-53.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lichinchi G, et al. Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nat Microbiol. 2016;1:16011. doi: 10.1038/nmicrobiol.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lichinchi G, et al. Dynamics of human and viral RNA methylation during Zika virus infection. Cell Host Microbe. 2016;20:666–673. doi: 10.1016/j.chom.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gokhale NS, et al. N6-methyladenosine in Flaviviridae viral RNA genomes regulates infection. Cell Host Microbe. 2016;20:654–665. doi: 10.1016/j.chom.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kennedy EM, et al. Posttranscriptional m(6)A editing of HIV-1 mRNAs enhances viral gene expression. Cell Host Microbe. 2016;19:675–685. doi: 10.1016/j.chom.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye F, Chen ER, Nilsen TW. Kaposi’s sarcoma-associated herpesvirus utilizes and manipulates RNA N6-adenosine methylation to promote lytic replication. J Virol. 2017;91:e00466-17. doi: 10.1128/JVI.00466-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tirumuru N, et al. N(6)-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. eLife. 2016;5:e15528. doi: 10.7554/eLife.15528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hesser CR, Karijolich J, Dominissini D, He C, Glaunsinger BA. N6-methyladenosine modification and the YTHDF2 reader protein play cell type specific roles in lytic viral gene expression during Kaposi’s sarcoma-associated herpesvirus infection. PLoS Pathog. 2018;14:e1006995. doi: 10.1371/journal.ppat.1006995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan B, et al. Viral and cellular N6-methyladenosine and N6,2′-O-dimethyladenosine epitranscriptomes in the KSHV life cycle. Nat Microbiol. 2018;3:108–120. doi: 10.1038/s41564-017-0056-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levrero M, et al. Control of cccDNA function in hepatitis B virus infection. J Hepatol. 2009;51:581–592. doi: 10.1016/j.jhep.2009.05.022. [DOI] [PubMed] [Google Scholar]
- 17.Yan H, et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife. 2012;1:e00049. doi: 10.7554/eLife.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seeger C, Mason WS. Hepatitis B virus biology. Microbiol Mol Biol Rev. 2000;64:51–68. doi: 10.1128/mmbr.64.1.51-68.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dominissini D, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 20.Ladner SK, et al. Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: A novel system for screening potential inhibitors of HBV replication. Antimicrob Agents Chemother. 1997;41:1715–1720. doi: 10.1128/aac.41.8.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belloni L, et al. Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function. Proc Natl Acad Sci USA. 2009;106:19975–19979. doi: 10.1073/pnas.0908365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, He C. Reading RNA methylation codes through methyl-specific binding proteins. RNA Biol. 2014;11:669–672. doi: 10.4161/rna.28829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu J, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jia G, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng G, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyer KD, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwartz S, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 2014;8:284–296. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linder B, et al. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods. 2015;12:767–772. doi: 10.1038/nmeth.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Y, Zeng P, Li YH, Zhang Z, Cui Q. SRAMP: Prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res. 2016;44:e91. doi: 10.1093/nar/gkw104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu N, et al. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du H, et al. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016;7:12626. doi: 10.1038/ncomms12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bartenschlager R, Schaller H. Hepadnaviral assembly is initiated by polymerase binding to the encapsidation signal in the viral RNA genome. EMBO J. 1992;11:3413–3420. doi: 10.1002/j.1460-2075.1992.tb05420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pollack JR, Ganem D. Site-specific RNA binding by a hepatitis B virus reverse transcriptase initiates two distinct reactions: RNA packaging and DNA synthesis. J Virol. 1994;68:5579–5587. doi: 10.1128/jvi.68.9.5579-5587.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tavis JE, Perri S, Ganem D. Hepadnavirus reverse transcription initiates within the stem-loop of the RNA packaging signal and employs a novel strand transfer. J Virol. 1994;68:3536–3543. doi: 10.1128/jvi.68.6.3536-3543.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tavis JE, Ganem D. Evidence for activation of the hepatitis B virus polymerase by binding of its RNA template. J Virol. 1996;70:5741–5750. doi: 10.1128/jvi.70.9.5741-5750.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stahl M, Beck J, Nassal M. Chaperones activate hepadnavirus reverse transcriptase by transiently exposing a C-proximal region in the terminal protein domain that contributes to epsilon RNA binding. J Virol. 2007;81:13354–13364. doi: 10.1128/JVI.01196-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.