Significance
We describe a class of protein–protein interactions mediated by the HORMA (Hop1, Rev7, Mad2) dimerization interface of Rev7, a multitasking scaffolding protein involved in translesion synthesis (TLS), repair of double-strand breaks, and mitosis. Biochemical and structural analyses of Rev7 dimerization reveal an unexpected architecture of the Rev1/Polζ TLS complex, which plays a central role in replication of damaged DNA, and describe the mechanism of Rev7 interactions with HORMA proteins from other pathways. Assays in Rev7−/− cells complemented with mutant Rev7 provide evidence that protein–protein interactions mediated by the Rev7 HORMA interface are important for the DNA damage response. These results contribute to the structural biology of DNA replication and repair and to understanding of the important class of HORMA proteins.
Keywords: DNA damage tolerance, translesion synthesis, protein–protein interactions, protein structure, HORMA domain proteins
Abstract
The translesion synthesis (TLS) polymerases Polζ and Rev1 form a complex that enables replication of damaged DNA. The Rev7 subunit of Polζ, which is a multifaceted HORMA (Hop1, Rev7, Mad2) protein with roles in TLS, DNA repair, and cell-cycle control, facilitates assembly of this complex by binding Rev1 and the catalytic subunit of Polζ, Rev3. Rev7 interacts with Rev3 by a mechanism conserved among HORMA proteins, whereby an open-to-closed transition locks the ligand underneath the “safety belt” loop. Dimerization of HORMA proteins promotes binding and release of this ligand, as exemplified by the Rev7 homolog, Mad2. Here, we investigate the dimerization of Rev7 when bound to the two Rev7-binding motifs (RBMs) in Rev3 by combining in vitro analyses of Rev7 structure and interactions with a functional assay in a Rev7−/− cell line. We demonstrate that Rev7 uses the conventional HORMA dimerization interface both to form a homodimer when tethered by the two RBMs in Rev3 and to heterodimerize with other HORMA domains, Mad2 and p31comet. Structurally, the Rev7 dimer can bind only one copy of Rev1, revealing an unexpected Rev1/Polζ architecture. In cells, mutation of the Rev7 dimer interface increases sensitivity to DNA damage. These results provide insights into the structure of the Rev1/Polζ TLS assembly and highlight the function of Rev7 homo- and heterodimerization.
DNA damage creates replication blocks leading to fork collapse, double-strand breaks, and genomic rearrangements (1, 2). To avert this scenario, specialized DNA polymerases (Y-family Rev1, Polη, Polι, and Polκ and B-family Polζ) help human cells tolerate DNA damage by replicating opposite the lesions or filling single-stranded gaps left after replication in a process called “translesion synthesis” (TLS) (3–6). Rev1/Polζ-dependent TLS occurs through a two-step mechanism in which one polymerase (typically Polη, Polι, or Polκ) inserts a nucleotide opposite the lesion, while another polymerase (typically Polζ) extends the distorted primer terminus (7–10). During this process, TLS DNA polymerases assemble into a multiprotein complex on the monoubiquitinated sliding clamp, proliferating cell nuclear antigen (PCNA) (11), with the aid of a scaffold protein, Rev1 (4, 5).
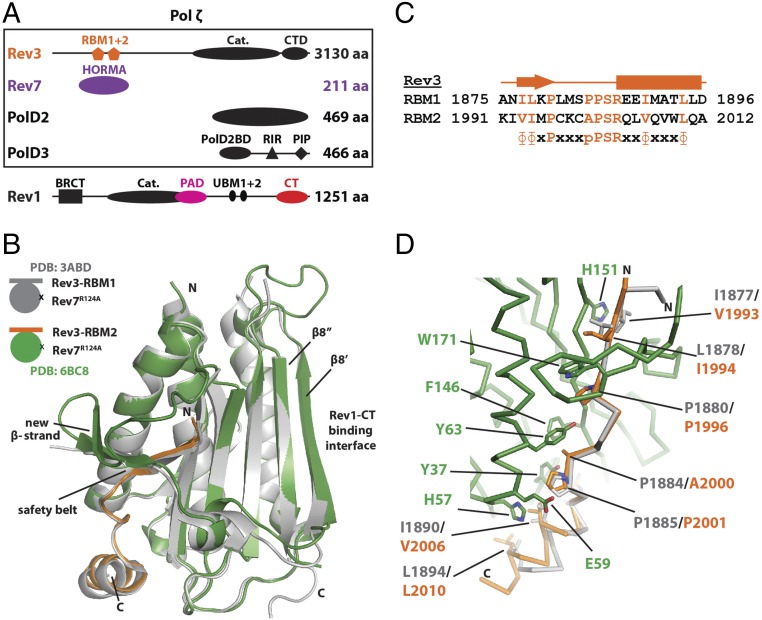
Polζ acts as the “extender” TLS DNA polymerase due to its proficiency in mismatched primer extension (7, 8), although it can insert nucleotides across certain lesions (12, 13). The catalytic subunit of Polζ, Rev3, forms a complex with Rev7 (called “Polζ2”), although Polζ is now known to function as a four-subunit complex (called “Polζ4”) composed of Rev3, Rev7, PolD2, and PolD3 (Fig. 1A) (14–18). PolD2 and PolD3 are subunits of the replicative DNA polymerase Polδ (19) but also enhance the efficiency of Polζ4 relative to Polζ2 (14–18). Polζ4 is assembled by protein–protein interactions, including those between the Rev7-binding motifs (RBMs) of Rev3 and Rev7 (20–22), between the C-terminal domain of Rev3 and PolD2 (16), and between PolD2 and PolD3 (23). The activity of Polζ4 is coordinated with other TLS polymerases through interactions of Rev7 and PolD3 with the Rev1 C-terminal (Rev1-CT) and Rev1 polymerase-associated (Rev1-PAD) domains (Fig. 1A) (24–31).
Fig. 1.
Components of the Rev1/Polζ4 complex. (A) The four subunits of TLS DNA polymerase Polζ (Upper, in box) and the TLS DNA polymerase Rev1 (Lower). (B) Structure comparison of Rev7R124A complexes with the two RBMs of Rev3: Rev3–RBM1 (gray; PDB ID code 3ABD) (21) and Rev3–RBM2 (green/orange, PDB ID code 6BC8) (this work). (C) Sequence alignment of the two Rev3–RBM motifs (20, 22). (D) Close-up of the Rev3–RBM1 (gray) and Rev3–RBM2 (orange) interaction interfaces with Rev7R124A (green).
Besides TLS, Polζ participates in the repair of DNA interstrand cross-links (32) and replication of “fragile-site” regions and non-B DNA structures (33, 34), while the individual subunits also act in other pathways. Rev7 (MAD2L2) regulates the metaphase-to-anaphase transition by sequestering CDH1, thus preventing premature activation of the anaphase-promoting complex/cyclostome (APC/C) (35). In addition, Rev7’s in vitro interactions with the spindle assembly checkpoint (SAC) protein Mad2 (36) and its interactions and colocalization with Ras-related nuclear GTPase (37) are consistent with a role in cell-cycle regulation. Rev7 also contributes to pathway choice for the repair of double-strand breaks (38, 39).
Rev7 belongs to the HORMA (Hop1, Rev7, Mad2) domain family (21, 40) whose members act as interaction modules in several cellular pathways (41). Structurally, HORMA domain proteins consist of a β-sheet flanked by three α-helices and a “safety-belt” region that can adopt two distinct conformations (open and closed) (42–45). The interaction between Rev7 and the Rev31847–1898 peptide (below called “Rev3–RBM1”) (21) occurs by a mechanism conserved among HORMA domains in which the safety-belt loop closes around a partner protein (Fig. 1B). This mechanism is best characterized (46) for the interaction of a related HORMA protein, Mad2, with a peptide motif from the SAC proteins Mad1 or Cdc20 (42–44), whereby, upon binding, Mad2 converts from the open to the closed state (42–44). In the case of Mad2, this conformational change is induced by dimerization (46). Notably, although two copies of Mad2 are held in immediate proximity by Mad1, the Mad2 homodimer can form only between open and closed monomers (47) or between the two closed apo-monomers (48) but not between two ligand-bound closed Mad2 monomers even if they are tethered together (46–48). In turn, disassembly of the Mad2/Mad1 complex involves active opening of Mad2 by the AAA+ ATPase TRIP13 aided by Mad2 heterodimerization with another HORMA protein, p31comet (MAD2L1BP) (49–53). Taken together, these studies suggest that homo- and heterodimerization mediates the formation and disassembly of HORMA domain complexes by the safety-belt mechanism. Accordingly, Rev7 also forms a homodimer (20) as well as a heterodimer with Mad2 (36), although these structures are not available. Instead, most studies on Rev7 used a dimer-breaking mutation, R124A, to induce a monomeric state (20, 21, 27–29).
The role of Rev7 dimerization remained unexplored until its relevance was underscored by a study that identified a second RBM on Rev3 (within residues 1974–2025, referred to as “RBM2”), which is in proximity to Rev3–RBM1 (Rev31847–1898) (Fig. 1 A and C) (22). This finding poses questions about the consequences of Rev7 dimerization for the assembly and function of the TLS machinery, given the role of Rev7 as an interaction module. (i) How many copies of Rev7 are present in human Polζ? (ii) If both Rev3–RBMs can bind Rev7 simultaneously, can the two copies of Rev7 form a dimer within Polζ [considering that the two bound closed Mad2 do not form a dimer (46)]? (iii) If the two Rev3-bound Rev7s can form a dimer, is it still competent to interact with the known Rev7 partners Rev1-CT and Rev1-PAD? (iv) If the Rev7 dimer can interact with these domains from Rev1, how many copies of Rev1 can bind Polζ, considering that Rev1 also functions as a scaffold? (v) What is the role of Rev7 dimerization in the response to DNA damage?
To answer these questions, we have taken an in vitro biophysical and biochemical approach combined with a functional assay in a Rev7−/− cell line. This study provides evidence that Rev7 can form a Rev3-tethered homodimer within Polζ while retaining interaction with Rev1 as well as heterodimers with other HORMA domains through the conserved homodimerization interface. Our functional assay in a Rev7−/− cell line demonstrates that Rev7 homo- or heterodimerization is necessary for the function of Rev7 that promotes DNA damage resistance. Overall, this work provides insights into the interactions that assemble the TLS machinery and highlights the role of Rev7 dimerization in mediating the response to DNA damage.
Results
Crystal Structure of Rev7R124A/Rev3–RBM2 Confirms a Second Rev7-Binding Site on Rev3.
Following previous reports identifying a second RBM in the Rev3 subunit of Polζ (Fig. 1 A and C) (22) and showing Rev7 and other HORMA domain proteins tend to form dimers (20, 36, 47–49, 54), we set out to determine the consequences of Rev7 dimerization on the structure and function of the Rev1/Polζ complex. First, given its significance for this study, we confirmed the second RBM in Rev3 (RBM2) (22) by solving a crystal structure of the Rev7R124A/Rev3–RBM2 complex [Protein Data Bank (PDB) ID code 6BD8] (Fig. 1B, green/orange and SI Appendix, Table S1). Similar to previous studies, we used the Rev7R124A mutation that prevents homodimerization (20). Importantly, although a previous structure of Rev7R124A/Rev3–RBM1 [PDB ID code 3ABD (21)] was used for molecular replacement, Rev3–RBM1 was omitted. Still, the resulting map showed well-defined electron density corresponding to the Rev3–RBM2 peptide bound to Rev7 (SI Appendix, Fig. S1A).
Our structure shows Rev7R124A in the closed conformation with Rev3–RBM2 bound underneath the safety-belt loop of Rev7 (Fig. 1B, green/orange). It has a 1.28-Å backbone rmsd against Rev7R124A/Rev3–RBM1 [PDB ID code 3ABD (21)] over residues 13–205 (Fig. 1B, gray) but with one difference. In the safety-belt region on Rev7 at residues 163–166, our structure shows a β-strand leading into a β-turn (Fig. 1B, green) whereas previous structures were either missing density (Fig. 1B, gray) (21) or modeled the density as an α-helix (28, 29). With respect to Rev3, despite variation in sequence (Fig. 1C), Rev3–RBM1 and Rev3–RBM2 adopt nearly identical conformations when bound to Rev7 (Fig. 1D). Overall, our analysis verifies that Rev3–RBM2 (22) is a bona fide Rev7 interaction motif.
Rev7 Uses a Conventional HORMA Interface for Homodimerization.
Previously, Hara et al. (20) showed that wild-type Rev7 (Rev7WT) undergoes dimerization by the analysis of sedimentation equilibrium data. We confirmed this by dilution isothermal titration calorimetry (ITC) measurements resulting in a Kd of 1.9 µM for the Rev7WT dimer (SI Appendix, Fig. S2A). However, Rev7WT/Rev3–RBM1 failed to crystallize, unlike Rev7R124A/Rev3–RBM1 which harbors a dimer-breaking mutation (20) and displays no heat change attributed to dimerization in the dilution ITC experiment (SI Appendix, Fig. S2A). We attempted to determine the structure of the Rev7 dimer but only obtained crystals of Rev7WT/Rev3–RBM2 that diffracted to 2.80 Å under high-salt conditions where the protein crystallized as a monomer (SI Appendix, Table S1, PDB ID code 6BI7). This is not unexpected, considering that the R124A mutation that abolishes electrostatic interactions of the arginine side-chain prevents the formation of the dimer (20). Our structure of Rev7WT/Rev3–RBM2 shows no substantial differences from structures of Rev7R124A/Rev3–RBM1 (21) or Rev7R124A/Rev3–RBM2 (SI Appendix, Fig. S1B).
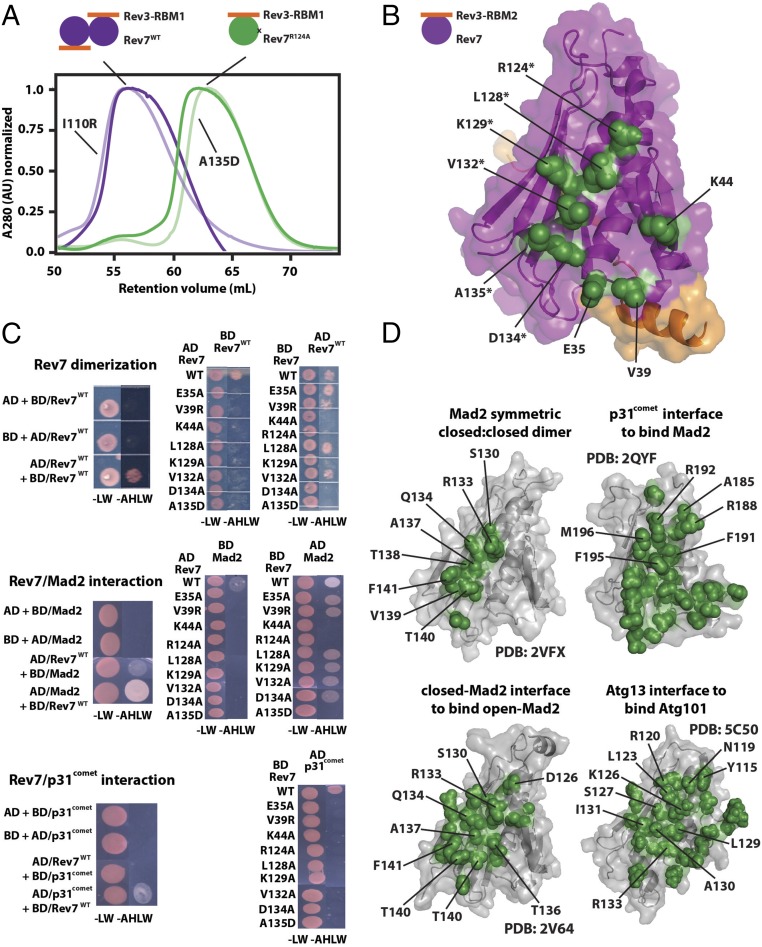
To map the Rev7 dimerization interface, we introduced 32 single mutations to solvent-exposed residues and first determined the oligomeric state using gel filtration chromatography (SI Appendix, Table S2). The Rev7/Rev3–RBM1 complexes were loaded on a gel filtration column so the concentration of eluted protein was about 0.3–0.4 mM. Under these conditions, Rev7WT/Rev3–RBM1 and Rev7R124A/Rev3–RBM1 elute as distinct peaks corresponding to the monomer and the dimer (Fig. 2A) (20). Using this approach, we identified eight mutations (in addition to R124A) that disrupt the Rev7 dimer: E35A, V39R, K44A, L128A, K129A, V132A, D134A, and A135D. When mapped onto Rev7R124A/Rev3–RBM2, these residues form a continuous surface centered on helix αC (Fig. 2B).
Fig. 2.
Mutational mapping of the Rev7 homo- and heterodimerization interface. (A) An example of gel filtration profiles of the monomeric Rev7R124A/Rev3–RBM1 (green) and dimeric Rev7WT/Rev3–RBM1 (purple) complexes; gel filtration profiles for additional Rev7 mutants which had no effect (I110R) or broke the dimer (A135D) are pictured in lighter shades. (B) Rev7 residues whose mutation abolishes dimerization (green) mapped on our structure of Rev7R124A/Rev3–RBM2, outlining the Rev7 dimerization interface. Residues on helix αC are marked with an asterisk. (C) Yeast two-hybrid studies of Rev7 homodimerization (Top) and its heterodimerization with Mad2 (Middle) or p31comet (Bottom): growth on −LW and −AHLW plates of the PJ69-4A strain of yeast transformed with fusions to the GAL4-BD or GAL4-AD as indicated. (D) Homo- and heterodimerization interface of other HORMA domains (47–49, 54). All structures appear in the same orientation as in C, and residues on helix αC are labeled.
Next, we cross-validated the mutations with a yeast two-hybrid assay using fusions of Rev7 with the activation domain (AD) and DNA-binding domain (BD) of the GAL4 transcription factor (55). As expected, transformation of yeast strain PJ69-4A with plasmids encoding AD- and BD-fused Rev7WT resulted in growth on medium lacking adenine, histidine, leucine, and tryptophan (−AHLW plates) (Fig. 2C, Top and SI Appendix, Fig. S2B), indicating that this assay is sensitive to formation of the Rev7 dimer. In contrast, yeast did not grow on −AHLW plates when cells were transformed with AD-fused Rev7mutant and BD-fused Rev7WT despite the presence of viable transformants on plates lacking leucine and tryptophan (−LW plates), suggesting that all mutations abolished Rev7 dimerization (Fig. 2C, Top and SI Appendix, Fig. S2B). In the reverse orientation (AD-fused Rev7WT, BD-fused Rev7mutant), the mutations K44A, R124A, K129A, D134A, and A135D abolished the interaction, while the mutations E35A, V39R, L128A, and V132A permitted growth on −AHLW plates (Fig. 2C, Top). A possible explanation for this discrepancy is the difference in the number of mutations per Rev7 dimer. During the gel filtration analysis, both copies of Rev7 harbored a dimer-breaking mutation, whereas during the yeast two-hybrid assay only one protomer contained the mutation.
Overall, our analyses show that Rev7 homodimerization is mediated by the interface centered around helix αC. This is the canonical interface responsible for homo- and heterodimerization of other HORMA proteins (Fig. 2D) (47–49, 54), providing us with confidence in the identification of this region.
Rev7 Interacts with Mad2 and p31comet Through the Dimerization Interface.
Capitalizing on our analysis of Rev7 homodimerization, we used the yeast two-hybrid assay to probe heterodimerization of Rev7 with two other HORMA domains, Mad2 and p31comet. Consistent with the previous report of a Rev7/Mad2 interaction (36), growth on −LW and −AHLW plates was observed in both orientations when yeast strain PJ69-4A was transformed with Mad2 and Rev7WT (Fig. 2C, Middle and SI Appendix, Fig. S2C). We then tested our dimer-breaking mutations (SI Appendix, Table S2) to determine whether the homodimerization interface in Rev7 mediates this interaction. In one orientation (AD-fused Rev7mutant, BD-fused Mad2), all mutations broke the interaction. In the reverse orientation (AD-fused Mad2, BD-fused Rev7mutant), K44A, R124A, and A135D broke the interaction, while the remaining mutations still grew on −AHLW plates (Fig. 2C, Middle and SI Appendix, Fig. S2C). The overlap in mutations that broke the Rev7 homodimer (Fig. 2C, Top) and Rev7/Mad2 heterodimer (Fig. 2C, Middle) in this orientation is likely not a coincidence but instead reports on the relative contributions of these residues to the binding energy.
To date, no interaction has been reported between p31comet and Rev7, although a crystal structure of the p31comet/Mad2 HORMA heterodimer is available (49). Considering the tendency of HORMA domain proteins for heterodimerization, we investigated whether Rev7 binds p31comet and observed an interaction between AD-fused p31comet and BD-fused Rev7WT (Fig. 2C, Bottom and SI Appendix, Fig. S2D). The growth on −AHLW plates for Rev7WT/p31comet transformants appears to be less robust than for Rev7WT/Mad2 or Rev7WT dimer, indicating a weaker interaction. We then tested the mutations to the Rev7 dimer interface (SI Appendix, Table S2) and found that all mutations broke the interaction between AD-fused p31comet and BD-fused Rev7mutant (Fig. 2C, Bottom and SI Appendix, Fig. S2D).
Overall, our data corroborate the interaction between Rev7 and Mad2 (36), which we mapped to the Rev7 homodimerization interface. In addition, we identified an interaction between Rev7 and p31comet through the same interface. These results demonstrate that Rev7 uses the dimerization interface for interaction with other HORMA domain proteins.
Two Copies of Rev7 Can Bind Adjacent Sites on Rev3 and Form a Tethered Dimer.
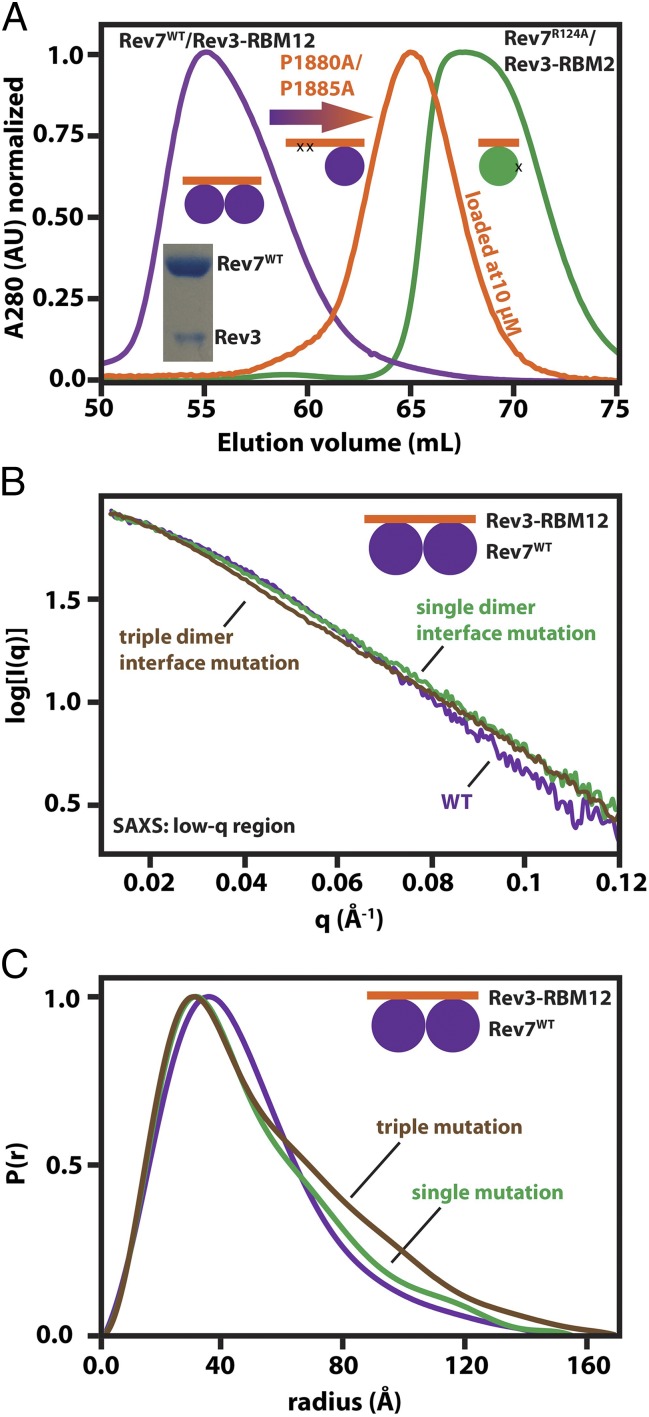
Because Rev7 is an interaction module, the number of Rev7s in Polζ and its oligomeric state will have implications for assembly of the TLS machinery. Therefore, we set out to determine if both Rev7-binding sites on Rev3 can be simultaneously occupied and, if so, to test whether these tethered Rev7s form a dimer. Specifically, we used a fragment (Rev3–RBM12, residues 1871–2014) that includes both RBMs, which likely mimics the interaction with full-length Rev3 because both RBMs are located within a disordered region ∼200 residues away from the nearest structured domain. Since RBM1 and RBM2 are separated by ∼90 residues, one might assume there are no constraints preventing Rev3 from binding a Rev7 dimer. On the other hand, the Rev7 homolog Mad2 does not form a symmetric homodimer between two protomers that are similarly bound by two sites on Mad1 (46–48). Instead, Mad1-bound closed Mad2 interacts with an open apo-Mad2, promoting the open-to-closed transition and interaction with Mad1, followed by dissociation of the dimer (46). With this in mind, we investigated the stoichiometry of the Rev3/Rev7 interaction and asked whether Rev7 exhibits behavior similar to that of Mad2.
Rev7WT/Rev3–RBM12 elutes as a single peak from a gel filtration column at a volume consistent with a 2:1 stoichiometry (Fig. 3A, purple), and an SDS gel shows two proteins of the expected size in the peak fraction (Fig. 3A, Inset). We then introduced a double mutation, P1880A/P1885A, to knock out Rev3–RBM1, which shifted the elution volume (Fig. 3A, orange) to nearly the volume of monomeric Rev7R124A/Rev3–RBM2 (Fig. 3A, green) when loaded at low concentration to preclude intermolecular Rev7 dimerization (SI Appendix, Fig. S3A). Still, an SDS gel of the mutant shows two bands corresponding to Rev7 and Rev3–RBM12P1880A/P1885A coeluting.
Fig. 3.
Two copies of Rev7 bind Rev3 and form a tethered dimer. (A) Gel filtration profiles of Rev7WT/Rev3–RBM12 (purple), Rev7WT/Rev3–RBM12P1880A/P1885A (orange), and Rev7R124A/Rev3–RBM2 (green) suggesting a 2:1 stoichiometry for the Rev7:Rev3 interaction. Each curve was normalized to its maximum absorbance. (Inset) An SDS gel of the Rev7WT/Rev3–RBM12 peak fraction with the two bands corresponding to Rev7 (Upper) and Rev3–RBM12 (Lower). (B) SAXS scattering curves of Rev7WT/Rev3–RBM12 (purple), Rev7R124A/Rev3–RBM12 (green), and Rev7K44A,R124A,A135D/Rev3–RBM12 (brown) at low q. The first points from the scattering intensities were aligned to highlight the effect of the mutations on the shape of the curve. (C) P(r) distributions indicating that the dimer-breaking mutations elongate the complex.
Furthermore, we collected small-angle X-ray scattering (SAXS) measurements (SI Appendix, Fig. S3B and Table S3) on Rev7WT/Rev3–RBM12 and calculated a mass consistent with a 2:1 stoichiometry (68.8 ± 0.6 kDa vs. theoretical 68.4 kDa) (SI Appendix, Fig. S3C, Left). We also collected SAXS measurements on Rev7R124A/Rev3–RBM12, which is unable to form an intermolecular Rev7 dimer, resulting in a mass of 72.5 ± 4.2 kDa (SI Appendix, Fig. S3C, Right). This eliminates the possibility of a 2:2 complex held together by Rev7 dimerization. Taken together, the data indicate both Rev7-binding sites on Rev3 can be occupied simultaneously.
To address whether the two Rev7s tethered by Rev3 form a dimer, we collected SAXS measurements on Rev7WT/Rev3–RBM12 and two complexes harboring one or three dimer-breaking mutations (Rev7R124A/ or Rev7K44A,R124A,A135D/Rev3–RBM12) (SI Appendix, Figs. S3 and S4). Because SAXS can report on molecular shape, we reasoned that if Rev7 forms a tethered dimer, then mutating the dimer interface will alter the shape of the complex, and the inverse. Consistent with a Rev7 dimer, the mutations altered the scattering intensity at low q where the data are sensitive to larger-scale structural perturbations (Fig. 3B and SI Appendix, Fig. S3D). As expected, the pair-distance distribution functions [P(r)] show the effect of the mutations was to expand the complex (Fig. 3C and SI Appendix, Fig. S4 B–D). Interestingly, the P(r) distributions point to a more extended conformation for Rev7K44A,R124A,A135D than for Rev7R124A when in complex with Rev3–RBM12 (Fig. 3C), suggesting the two Rev7 protomers are more accessible to each other when tethered and may retain residual interaction resulting in a fraction of Rev7R124A/Rev3–RBM12 sampling a dimeric state. In agreement with a mutation-induced increase in flexibility, the density of the complex with Rev3–RBM12 decreased from 1.03 g/cm3 for Rev7WT to 0.92 g/cm3 for Rev7R124A and 0.85 g/cm3 for Rev7K44A,R124A,A135D (SI Appendix, Table S3) (56).
To illustrate that neither the mutations themselves nor protomer-level conformational changes explain the mutation-induced variations in the SAXS data (Fig. 3B), we determined a crystal structure of Rev7K44A,R124A,A135D/Rev3–RBM2 (PDB ID code 6BCD) (SI Appendix, Fig. S5A and Table S1) and simulated SAXS data for this structure and the structures of Rev7R124A/Rev3–RBM2 and Rev7WT/Rev3–RBM2, which resulted in identical scattering intensity profiles at low q (SI Appendix, Fig. S5 B and C vs. Fig. 3B). Furthermore, we collected 1H-15N heteronuclear single-quantum correlation (HSQC) NMR spectra of Rev7K44A,R124A,A135D/Rev3–RBM2 and Rev7R124A/Rev3–RBM2, which confirmed the lack of mutation-induced conformational changes (SI Appendix, Fig. S5D).
Finally, to cross-validate the formation of a tethered dimer, we collected 1H-15N HSQC spectra on Rev7WT/ and Rev7R124A/Rev3–RBM12 (mass 68 kDa) (SI Appendix, Fig. S6). The Rev7WT/Rev3–RBM12 spectrum displays little to no peak intensity for structured protein residues, consistent with slow tumbling of the Rev7 dimer that behaves as a single entity. When the R124A mutation is introduced to the dimer interface, which should loosen the tethered dimer, the spectrum improves, reflecting faster tumbling of independent Rev7 protomers connected by a flexible linker.
Overall, the data indicate that two copies of Rev7WT can bind Rev3, where they form a tethered dimer.
Tethered Rev7 Dimer Retains Interaction with Rev1-CT but Does Not Bind Rev1-PAD.
After identifying the second Rev7 interaction motif on Rev3, Tomida et al. (22) proposed a model in which Rev7 mediates a bivalent interaction with Rev1 by binding Rev1-CT (27–29) and Rev1-PAD (31). We tested this model and determined whether the tethered Rev7 dimer retains the ability to bind known Rev7 interaction partners. Because Rev7 interacts with Rev1-CT, which is also known to bind Polη, Polι, Polκ, and PolD3 through a second interface (25–30), the number of Rev1s attached to Polζ through the Rev7/Rev1-CT interaction will have implications for assembly of the TLS machinery. With respect to Rev1-PAD, a previous report using a pull-down assay showed that yeast Rev7 can bind Rev1-PAD (31), although this has not been confirmed in higher eukaryotes.
First, we tested the interaction of human Rev1-PAD with Rev7R124A/Rev3–RBM2 by NMR, which can detect weak binding. We collected 1H-15N HSQC and 1H-13C HMQC spectra of 15N- or 15N/ILV-13CH3–labeled monomeric Rev7R124A/Rev3–RBM2 with unlabeled Rev1-PAD added to molar excess; however, chemical shift perturbations characteristic of binding were not observed (SI Appendix, Fig. S7A). We next considered the possibility that Rev1-PAD can interact only with tethered dimeric Rev7 and tested the interaction of Rev1-PAD with Rev7WT/Rev3–RBM12 using gel filtration chromatography; however, the proteins eluted as separate peaks despite concentrations of 750 μM and 150 μM, respectively, in the elution fractions, again suggesting a lack of interaction (SI Appendix, Fig. S7B). Overall, the data indicate human Rev1-PAD does not interact with Rev3-bound Rev7.
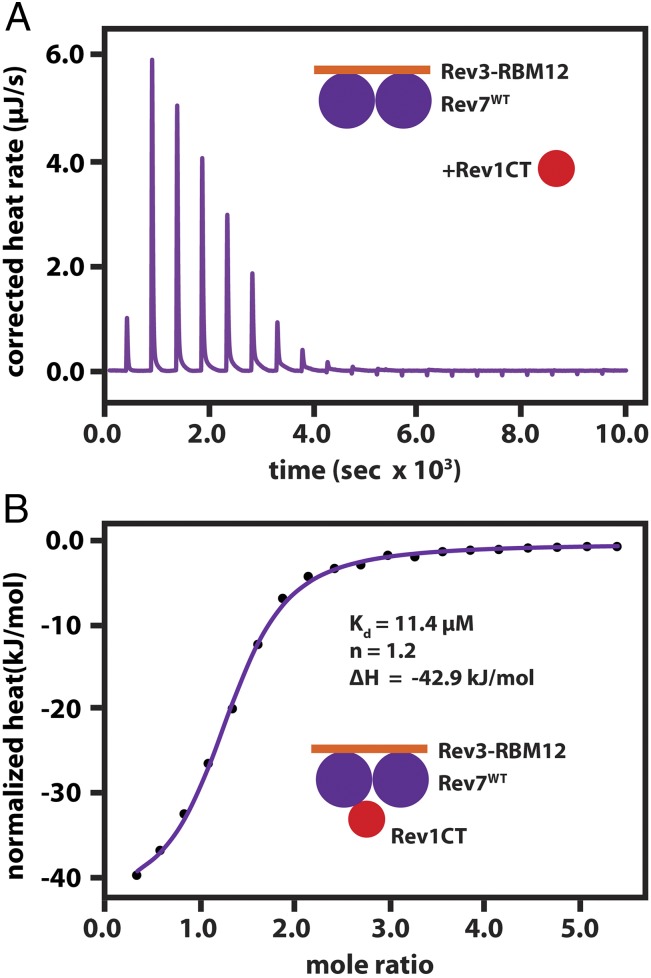
The interaction of human Rev1-CT with Rev7R124A/Rev3–RBM1 has been shown by NMR titrations and ITC from our laboratory and subsequent crystal structures, which revealed a 1:1 stoichiometry for the Rev7R124A/Rev1-CT complex (27–29). Here, we found Rev7WT/Rev3–RBM12 still interacts with Rev1-CT, as the complex coelutes from a gel filtration column (SI Appendix, Fig. S7C). We further validated this interaction by ITC (Fig. 4), which, surprisingly, indicates that only one copy of Rev1-CT can bind Rev7WT/Rev3–RBM12 despite the presence of two Rev7s (stoichiometry parameter n = 1.2). Notably, the Kd of 11.4 µM and association enthalpy (ΔH) of −42.9 kJ/mol for this complex are consistent with our previous ITC data for the interaction of Rev7R124A/Rev3–RBM1 with Rev1-CT (Kd = 1.3 µM, ΔH = −39.2 kJ/mol) (27), indicating that the same heat is released when Rev1-CT binds to the Rev7R124A monomer or the Rev7WT dimer.
Fig. 4.
ITC measurements suggest only one copy of Rev1-CT can bind the Rev7WT dimer when tethered by Rev3–RBM12. (A) Raw ITC data and (B) integrated heat changes obtained during titration of Rev1-CT into Rev7WT/Rev3–RBM12. The best fit to the ITC data (solid line) results in a Kd of 11.4 ± 1.9 µM, ΔH of −42.9 ± 1.4 kJ/mol, and stoichiometry parameter (n) of 1.24 ± 0.02.
Model of the Rev7 Dimer Suggests 2:1 Stoichiometry for the Rev7:Rev1-CT Interaction.
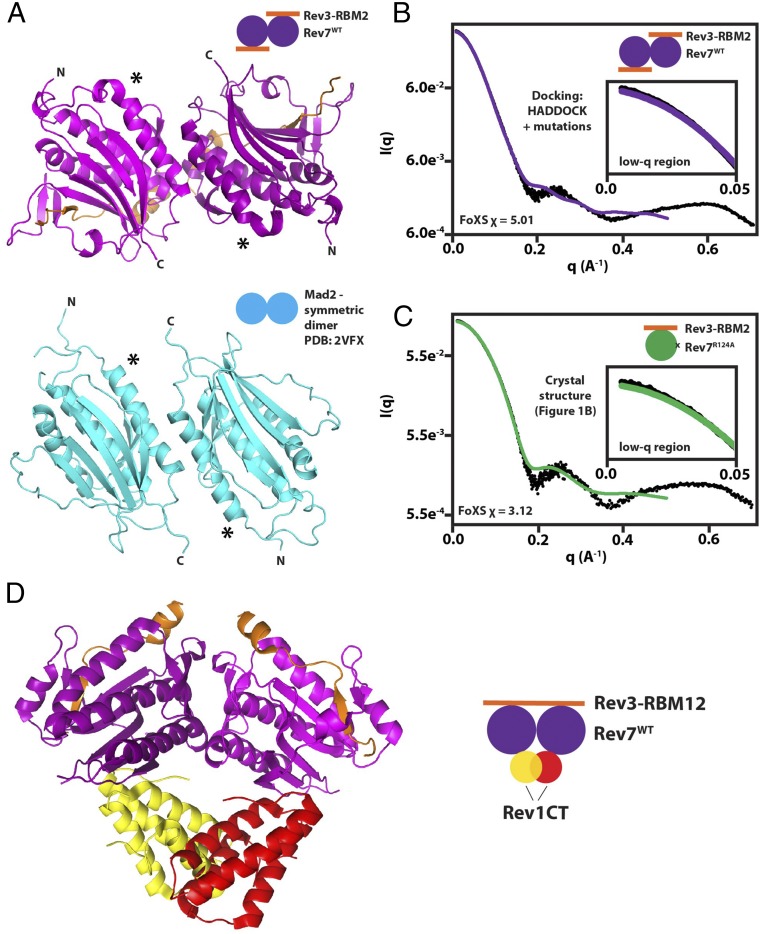
To confirm and understand the 2:1 stoichiometry of the Rev7:Rev1 interaction when Rev7 is forming a tethered dimer in Polζ, we modeled the structure of the Rev7WT/Rev3–RBM2 dimer using HADDOCK (57) with the dimer-breaking mutations described above as input (Fig. 2B and SI Appendix, Table S2). The resulting 200 models of the Rev7WT/Rev3–RBM2 dimer were all grouped into a single cluster by HADDOCK and show the two Rev7 protomers in an antiparallel orientation with the C terminus of helix αC forming the core of the interface (Fig. 5A, Upper). Encouragingly, the lowest-energy structure of the Rev7WT/Rev3–RBM2 dimer exhibits remarkable similarity to the symmetric apo-Mad2 dimer (Fig. 5A, Lower) (48), providing us with confidence in our model.
Fig. 5.
Structural modeling of the Rev7 dimer and its complex with Rev1-CT. (A) Model of Rev7WT/Rev3–RBM2 (purple) obtained using HADDOCK (57) based on mutational mapping of the Rev7 dimerization interface and its comparison with the structure of the symmetric apo-Mad2 dimer (PDB ID code 3VFX) (cyan) (48). Asterisks denote the N terminus of helix αC to demonstrate alignment. (B) Agreement between SAXS/WAXS scattering data (black dots) and scattering intensities calculated from the model of Rev7WT/Rev3–RBM2 dimer in A using FoXS (68). (C) Comparison of experimental SAXS/WAXS data with scattering data predicted from our crystal structure of the Rev7R124A/Rev3–RBM2 complex using FoXS (68). Insets in B and C show low-q regions in plots. (D) Structure of Rev7R124A (purple)/Rev3–RBM1 (orange)/Rev1-CT (yellow/red) (29) superimposed on our model of the Rev7 dimer generated using HADDOCK (57) shown in A. The Rev7 dimer is unable to bind a second Rev1-CT due to a steric clash.
To cross-validate our model, we collected SAXS/WAXS (wide-angle X-ray scattering) data on the dimeric Rev7WT/Rev3–RBM2 construct and compared the experimental scattering with our model of the dimer (Fig. 5B and SI Appendix, Fig. S8). Agreement was observed up to q = 0.16–0.18⋅Å−1, suggesting our model captures the shape of the complex. To rationalize the discrepancy at higher q, we also collected SAXS/WAXS data on monomeric Rev7R124A/Rev3–RBM2 and predicted data based on our crystal structure (Fig. 1B, green/orange), resulting in the same level of agreement up to q = 0.16–0.18⋅Å−1 (Fig. 5C). This discrepancy at high q may be caused by dynamics of Rev7/Rev3 in solution, as a 1H-15N HSQC spectrum of Rev7R124A/Rev3–RBM2 contains ∼25% fewer peaks than expected, presumably due to microsecond–millisecond exchange line-broadening (SI Appendix, Fig. S5D). Another possible explanation is sensitivity of SAXS curves for Rev7WT/Rev3–RBM2 to the presence of a small fraction of Rev7 monomers; however, modeling the volume fraction of dimers using the program Oligomer (58) and molecular weight calculations (SI Appendix, Table S3 and Fig. S8) suggest an almost homogeneous solution of Rev7WT/Rev3–RBM2 dimers.
With respect to the Rev7:Rev1 stoichiometry, superposition of the crystal structure of Rev7R124A/Rev3–RBM1/Rev1-CT (29) onto our model of the Rev7 dimer reveals a steric clash between the two Rev1-CT domains (Fig. 5D), providing an explanation for the 2:1 Rev7:Rev1 stoichiometry observed by ITC: Once the first Rev1-CT domain binds the Rev7 dimer, binding of the second is occluded by the first. Furthermore, superposition of the Rev7R124A/Rev3–RBM1/Rev1-CT structure (29) onto the dimer structures formed by other HORMA proteins, including the symmetric apo-Mad2 homodimer (48), p31comet/Mad2 heterodimer (49), open/closed Mad2 dimer (47), and Atg13/Atg101 heterodimer (54), resulted in steric clash between the two Rev1-CT in all cases (SI Appendix, Fig. S9).
Taken together, our ITC data for the Rev7WT/Rev3–RBM12–Rev1-CT interaction (Fig. 4) and structural model for the Rev7WT/Rev3–RBM2 dimer (Fig. 5) provide evidence that the Rev7 dimer can bind only one copy of Rev1-CT.
The Rev7 Dimerization Mutant Is Unable to Restore Cisplatin Resistance of Rev7−/− Cells.
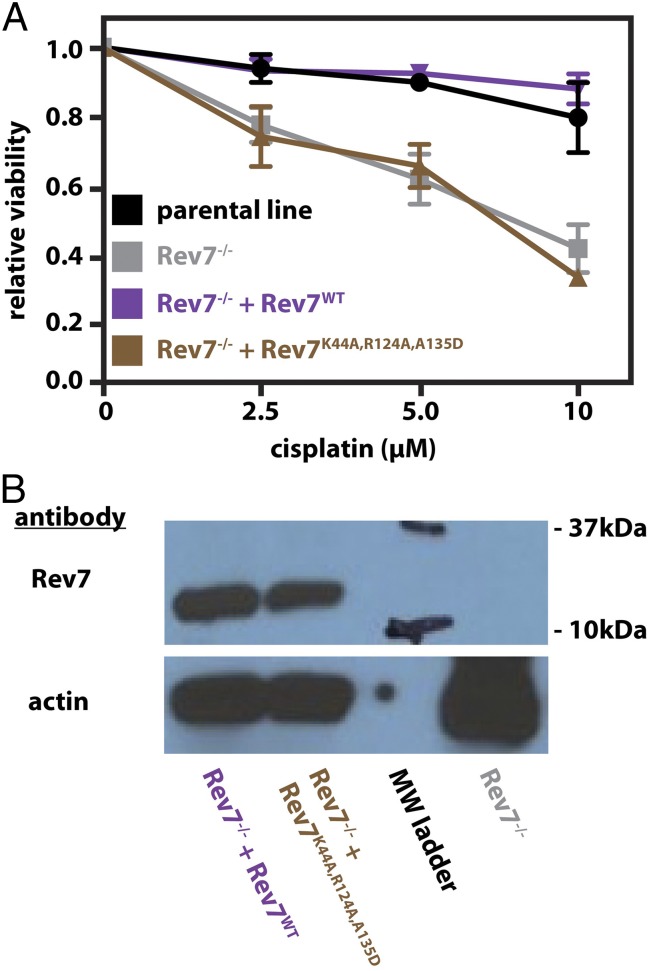
Given the role of Rev7-mediated interactions in the assembly and function of Polζ (7–10), we were interested in whether Rev7 dimerization has a functional significance in DNA damage tolerance. To address this, we used a Rev7-knockout cell line developed by F.-M.V. (59). This cell line was generated using the CRISPR/Cas9 system in the KrasG12D/+;Trp53−/− (KP) lung adenocarcinoma cell line background (60), which is a murine model for human nonsmall cell lung cancer that is intrinsically resistant to front-line chemotherapeutics such as cisplatin (61). The resulting Rev7−/− cells were more sensitive to the DNA cross-linking agent cisplatin and showed reduced viability compared with the parental cell line (Fig. 6, gray vs. black). To test whether the sensitivity to cisplatin is contingent upon Rev7 dimerization, the Rev7−/− cells were complemented with either Rev7WT or the triple dimer interface mutant Rev7K44A,R124A,A135D. While complementation with Rev7WT restored resistance to cisplatin, complementation with Rev7K44A,R124A,A135D was unable to rescue the sensitized phenotype (Fig. 6A, purple vs. brown) despite the appearance of stably expressed protein by Western blot (Fig. 6B, brown). Overall, the data indicate that interactions mediated by the Rev7 HORMA dimerization interface are required for cell viability after treatment with cisplatin.
Fig. 6.
Interactions of the Rev7 dimer interface are required for resistance to cisplatin. (A) Relative viability of the parental KrasG12D;p53−/− cells (black), Rev7−/− cells (gray), Rev7−/− cells complemented with Rev7WT (purple), and dimerization-deficient Rev7K44A,R124A,A135D cells (brown) after treatment with cisplatin for 48 h. (B) Western blot showing knockout of Rev7 (gray) and expression of Rev7WT (labeled in purple) and Rev7K44A,R124A,A135D (labeled in brown) during complementation. In the lane marked “MW ladder,” the black marker bands were traced by hand with marker.
Discussion
The TLS DNA polymerases Rev1, Polη, Polι, Polκ, and Polζ are recruited to replication-blocking DNA lesions and assemble into a multiprotein complex that enables DNA synthesis (7–10). The B-family polymerase Polζ participates in TLS by extending from the aberrant primer–template junction after another TLS polymerase has inserted a nucleotide opposite the lesion (7, 8, 14). The subunit composition of Polζ, which had been known as a complex of the catalytic Rev3 and accessory Rev7 subunits (12), was recently revised after several groups discovered that Polζ contains two additional subunits, PolD2 and PolD3 in humans or Pol31 and Pol32 in yeast, that are bound through interaction with the C-terminal domain of Rev3 (14–18). The latter two subunits are known subunits of the replicative DNA polymerase Polδ (19), suggesting that Polζ has an architecture typical for B-family polymerases. On the other hand, Rev7, which is a HORMA domain protein (21), is a unique component of Polζ with no analogs in other DNA polymerases.
One function of Rev7 is to bridge Polζ with other TLS polymerases through interaction with the scaffold protein Rev1 (4, 5, 25–30). With this in mind, a recent study from Tomida et al. (22) that revealed a second Rev7 interaction motif on human Rev3 (RBM2) in proximity to the previously described Rev3–RBM1 (20) raised several questions about the assembly and stoichiometry of the TLS machinery. The authors proposed a model in which both Rev7 interaction sites can be occupied simultaneously by two copies of Rev7, whose function is to mediate a bivalent interaction with two binding modules on Rev1 (Rev1-PAD and Rev1-CT). Noting that, like other HORMA domain proteins (47–49, 54), Rev7 is prone to dimerization (20), we tested this model and addressed whether Rev7 can bind to both sites on Rev3 and form a dimer in the context of Polζ and whether the presence of two copies of Rev7 affects the interaction of Polζ with Rev1.
We confirmed the second RBM on Rev3 and showed that both Rev3–RBMs can be occupied by two Rev7s at the same time, suggesting a 2:1 Rev7:Rev3 stoichiometry. Thus, human Polζ includes two copies of Rev7, resulting in a five-subunit complex. When tethered together, these two Rev7s form a homodimer through the canonical HORMA interface (47–49, 54). However, we were unable to detect an interaction between human Rev7 and Rev1-PAD that was shown in yeast by pull-down (31). Instead, our binding studies and structural modeling revealed that the tethered Rev7 dimer is still able to interact with Rev1-CT but with only a single copy. This suggests a 1:1 stoichiometry for assembly of the human Rev1:Polζ complex. Beyond studying the role of Rev7 dimerization in Rev1/Polζ-dependent TLS, we also established that Rev7 uses its dimerization interface for interaction with other HORMA domains, Mad2 and p31comet. Importantly, we also demonstrated that the intact Rev7 dimerization interface is functionally significant in vivo, as it is required for cell viability after cisplatin treatment.
Our study raises questions about the mechanistic role of human Rev7 dimerization. One consequence of tethering two Rev7s by the high-affinity interaction sites on Rev3 is strengthening of the Rev7 dimer interaction relative to that of unbound Rev7. This will reduce the access of other Rev7 interaction partners that use the dimerization interface in a bimolecular interaction, such as other HORMA proteins, which often form heterodimers through the homodimerization interface (36, 47–49, 54). Given the multiple functions of Rev7 (14, 35–39, 41), the formation of a tight Rev3-tethered Rev7 dimer in the context of Polζ may act as a mechanism to separate the functions of Rev7 by hiding the Rev7 dimerization interface from its interactors from other pathways. In line with this thinking, Rev7 is three orders of magnitude more abundant than Rev3 in human (293T) cells (22) and thus is mostly bound to other proteins. Here, we confirmed the interaction of Rev7 with Mad2 (36) and showed that it occurs through the canonical HORMA dimerization interface. Considering the role for Rev7 in the metaphase-to-anaphase transition (35) and the role of Mad2 as the spindle checkpoint (62), based on our model, a mitotically relevant Rev7/Mad2 interaction would be disfavored through tethering of the two subunits when Rev7 is participating in TLS. Thus, homodimerization of Rev7 may serve to protect the functional Rev1/Polζ TLS complex.
We have also demonstrated that Rev7 uses its dimerization interface to bind another HORMA protein, p31comet, which plays a role in the disassembly of the mitotic checkpoint complex by the AAA+ ATPase TRIP13. In this process, p31comet recognizes a target HORMA protein (Mad2) and brings it in contact with TRIP13, which catalyzes the closed-to-open conversion (50–53). Given the similarity between Mad2 and Rev7 (MAD2L2) (36) and that both Mad2 and Rev7 interact with p31comet through a conserved HORMA dimerization interface (49), one might hypothesize that p31comet and TRIP13 also participate in the active opening of Rev7 and its dissociation from Rev3. In this case, Rev3-mediated Rev7 dimerization would interfere with a Rev7/p31comet interaction and thus would prevent premature unloading of Rev7 and deactivation of Polζ. In this regard, one should note that Rev7 and Mad2 exhibit differing behavior: The two Rev3-bound closed Rev7s interact to form a dimer, whereas Mad1-bound closed Mad2 interacts with ligand-free open Mad2 to facilitate ligand uptake accompanied by the conformational change and dissociation of the dimer (46–48).
Finally, this study demonstrates that the Rev7 dimerization interface is required for cell viability after treatment with cisplatin, although the mechanism is unresolved. Presumably, the loss of Rev7 dimerization affects Polζ activity and sensitizes cells to cisplatin because Polζ participates in TLS across cisplatin DNA adducts (18) and repair of cisplatin DNA interstrand cross-links (32). However, the sensitization cannot be deductively attributed to a loss of Rev7 dimerization in Polζ, as Rev7 contributes to pathway choice for the repair of double-strand breaks (38, 39) and potentially functions in mitosis (35). A global loss of the Rev7 dimerization interface may also operate in other contexts, including those with Mad2 or p31comet. For example, one might envision a scenario in which the dimerization-deficient Rev7 mutants cannot be unloaded from their partner proteins by TRIP13 through interaction with p31comet, resulting in a decrease in Rev7 available for interaction with Rev3.
In summary, this study yields insights into the role of Rev7 dimerization in mediating the assembly of the TLS machinery and the interactions of Rev7 with HORMA proteins from other cellular pathways. How and whether dimerization affects other functions of Rev7 in cell-cycle control (35) or the repair of double-strand breaks (38, 39) and any cross-talk between these functions and TLS remain to be determined.
Methods
Subcloning and Mutagenesis.
Subcloning and mutagenesis were carried out using standard molecular biology techniques. The pETDuet-1 (Novagen) based construct for coexpression of human Rev7R124A with the Rev3–RBM1 fragment (the first RBM, Rev31847–1898) (20) was used as a template to design corresponding constructs for coexpression of Rev7R124A with other Rev3 fragments, including Rev3–RBM2 (the second RBM, Rev31988–2014) (22) and Rev3–RBM12 (a fragment containing two consecutive RBMs, Rev31871–2014). New fragments were introduced by PCR amplifying codon-optimized Rev3 from a custom-ordered gBlock Gene Fragment (Integrated DNA Technologies) using Q5 DNA polymerase (New England Biolabs), followed by ligating the digested product into the NdeI/XhoI restriction enzyme (Thermo Fisher) sites of pETDuet-1 with T4 DNA ligase (New England Biolabs).
Mutations in the Rev7 and Rev3 genes were introduced using the modified inverse PCR procedure (63) without DPN1 digestion. After generating linear double-stranded DNA by amplification of the plasmid with Q5 DNA polymerase (New England Biolabs) using an extension time of 4 min, 2 μL of the PCR mixture was phosphorylated in a 25-μL reaction with T4 polynucleotide kinase (New England Biolabs) in T4 DNA ligase buffer. After heat inactivation, 3.5 μL of the phosphorylation mixture was ligated at room temperature for 30 min using T4 DNA ligase (New England Biolabs) in a 20-μL reaction and transformed into Top10 cells (Thermo Fisher). The correct nucleotide sequence was confirmed in all cases by sequencing (Genewiz).
Protein Expression and Purification.
All Rev7 complexes with Rev3–RBM fragments were expressed from pETDuet-1 (Novagen) constructs described above (20). Rev1-CT and Rev1-PAD domains were expressed from pET28b+ (Novagen) constructs described elsewhere (26). All proteins demonstrated excellent stability and were expressed and purified following a standard protocol. In brief, Escherichia coli BL21(DE3) cells were transformed with the plasmid encoding the protein(s) of interest, and bacteria were grown to midlog phase and then induced overnight with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at 20 °C. When necessary for NMR, 15N-labeled proteins were produced by growing bacteria in M9 minimal medium containing 1 g/L 15NH4Cl. Ile, Val, and Leu ILV-3CH3 labeling was achieved by adding 70 mg/L α-ketobutyrate and 120 mg/L α-ketoisovalerate precursors 1 h before induction. The following morning, cells were spun down in an F10S-6 × 500y rotor for 10 min at 6,000 rpm. The cell pellet was resuspended in the lysis buffer consisting of 50 mM sodium phosphate, 300 mM NaCl, and 10 mM imidazole at pH 8 and was lysed by sonication. The lysate was then centrifuged in an SS-34 rotor for 45 min at 18,500 rpm, and the supernatant was filtered through a 0.45-μM PVDF membrane directly into TALON cobalt resin that was equilibrated with lysis buffer for purification using the His6 affinity tag. The column was run by gravity flow and was washed extensively with lysis buffer, and the protein was eluted using the elution buffer consisting of 50 mM sodium phosphate, 300 mM NaCl, and 300 mM imidazole at pH 8. The proteins were then purified by gel filtration chromatography on a HiLoad 16/600 Superdex75 pg column (GE Healthcare). During this step, the proteins were exchanged into their final buffers as noted. Proteins were concentrated using Amicon Ultra Centrifugal Filters (Millipore) when necessary.
The interaction of Rev7 with Rev3–RBM2 and Rev3–RBM12 was assessed by coexpressing the two proteins using pETDuet-1 and coeluting the complex on a gel filtration column. After SDS gels of the peak fractions showed that two proteins corresponding to Rev7 and either Rev3–RBM2 or Rev3–RBM12 had eluted together, the identity of Rev3–RBM2 and Rev3–RBM12 was further confirmed by in-gel trypsin digestion and LC-MS/MS.
Protein Crystallization, X-Ray Data Collection, and Structure Determination.
Rev7R124A/Rev3–RBM2, Rev7K44A,R124A,A135D/Rev3–RBM2, and Rev7WT/Rev3–RBM2 were exchanged into 5 mM Hepes, 100 mM NaCl, and 10 mM DTT at pH 7.4 (20) by gel filtration and concentrated to 45 mg/mL, 45 mg/mL, and 60 mg/mL, respectively. In all cases, diffraction-quality crystals were obtained by vapor diffusion in hanging-drop format at 16 °C when protein solution was mixed at a 1:1 ratio with reservoir solution in 4-μL drops. For Rev7R124A/Rev3–RBM2, the reservoir solution consisted of 100 mM sodium acetate, 200 mM NaCl, and 1.4 M ammonium sulfate at pH 5.0, and crystals were flash frozen in the reservoir solution containing 20% (vol/vol) PEG400. For Rev7K44A,R124A,A135D/Rev3–RBM2, the reservoir solution contained 100 mM citrate and 1.6 M ammonium sulfate at pH 5.25, and crystals were frozen in the reservoir solution containing 20% (vol/vol) sucrose. For Rev7WT/Rev3–RBM2, the reservoir solution contained 100 mM citrate, 1 M LiCl, and 7.5% (wt/vol) PEG6000 at pH 4.75, and crystals were frozen in the reservoir solution containing 20% (wt/vol) sucrose.
X-ray diffraction data were collected at the Cornell High Energy Synchrotron Source (CHESS) F1 beamline using a Pilatus 6M detector at a wavelength of 0.976 Å. For all structures, 360 frames were collected in 0.5° wedges with a collection time of 3–6 s per image over a total range of 180°. Data were processed using the HKL-2000 package (64). The structures were solved by molecular replacement using the previously reported structure of Rev7R124A/Rev3–RBM1 (21) with the program Phaser (65) and were refined by iterative cycles of model building and refinement with Coot and Refmac (as part of the CCP4i2 package) (66).
The details of data collection and structure refinement statistics are summarized in SI Appendix, Table S1. Structures were deposited in the PDB with ID codes 6BC8, 6BCD, and 6BI7.
SAXS.
SAXS measurements were collected on Rev7R124A/Rev3–RBM2, Rev7WT/Rev3–RBM12, Rev7R124A/Rev3–RBM12, and Rev7K44A,R124A,A135D/Rev3–RBM12 complexes in buffer containing 50 mM Tris, 150 mM NaCl, 10 mM DTT, 1 mM EDTA, and 5% glycerol at pH 8.4. Measurements for Rev7WT/Rev3–RBM2 were carried out in 20 mM Hepes, 10 mM DTT, and 5% glycerol at pH 8.0. Buffers were matched by loading the proteins on a gel filtration column at a high enough concentration to ensure the peak fractions would be sufficient to take for measurement without concentrating. The concentration series for each construct is listed in SI Appendix, Table S3. The matched buffer was taken from buffer that had passed through the column.
SAXS data collected for the above samples are summarized in SI Appendix, Table S3. Ten 1-s exposures were collected with the sample oscillating in the capillary flow-cell at the CHESS G1 station. The data were integrated, averaged, buffer subtracted, and subjected to Guinier analysis to determine the gyration radius (Rg) and forward scattering [I(0)] using the software RAW (67). To calculate the molecular weight of the complexes using the forward scattering, human PCNA was used as a standard. For all samples, the Rg and molecular weight were consistent across concentrations, indicating concentration-dependent effects were not present (SI Appendix, Table S3). The P(r) function was calculated using the software GNOM as part of the ATSAS suite (58). Real-space values for Rg and I(0) derived from the P(r) analysis are in agreement with the values derived from the Guinier analysis and also show no concentration dependence except, as expected, in the case of Rev7WT/Rev3–RBM2 (SI Appendix, Table S3).
To simulate SAXS curves from the Rev7/Rev3 coordinate files, the FoXS server was used with default parameters (68). Similar results were obtained with AXES (69). When simulating the SAXS data, residues at the N and C termini of Rev7 that were missing in the crystal structures were built in an extended conformation.
NMR Spectroscopy.
1H-15N HSQC spectra of 15N-labeled Rev7WT/Rev3–RBM12 and Rev7R124A/Rev3–RBM12 (SI Appendix, Fig. S6A) were collected at 30 °C on an Agilent VNMRS 800 MHz (1H) spectrometer equipped with a cold probe on protein samples dissolved in 20 mM Tris, 100 mM NaCl, and 10 mM DTT, pH 8.4, at a Rev7 monomer concentration of 240 μM. Spectra of 15N-labeled Rev7R124A/Rev3–RBM2 and Rev7K44A,R124A,A135D/Rev3–RBM2 (SI Appendix, Fig. S5D) were collected at 30 °C in 20 mM Hepes, 100 mM NaCl, and 10 mM DTT, pH 7.4. To probe the Rev7/Rev1-PAD interaction, unlabeled Rev1-PAD was added to 15N-labeled Rev7R124A/Rev3–RBM2 to molar excess (440 μM Rev1-PAD, 260 μM Rev7R124A/Rev3–RBM2) in 20 mM Hepes, 100 mM NaCl, and 10 mM DTT at pH 7.4; however, chemical shift perturbations indicating binding were not observed in the 1H-15N HSQC spectra (SI Appendix, Fig. S6A). Consistent with the lack of binding, chemical shift changes were not observed in the 1H-13C HMQC spectra of ILV-13CH3–labeled Rev7R124A/Rev3–RBM2 titrated with unlabeled Rev1-PAD (SI Appendix, Fig. S6A, Inset). All spectra were processed with NMRPipe (70) and analyzed using the software CCPNmr Analysis (71).
Mutational Analysis of the Rev7 Dimerization Interface.
A series of 32 site-directed mutations (SI Appendix, Table S2) were introduced to solvent-exposed residues covering the entire surface of Rev7. Rev7WT/Rev3–RBM2 or Rev7mutant/Rev3–RBM2 was loaded onto a gel filtration column at ∼1.5–1.7 mM, resulting in a peak concentration of ∼300–400 µM (average of peak) after elution. At these concentrations, Rev7WT/Rev3–RBM2 and the previously described dimer-breaking Rev7R124A/Rev3–RBM2 mutant (20) elute as separate peaks at volumes corresponding to the dimer and the monomer. The oligomeric state of Rev7WT/Rev3–RBM2 and Rev7R124A/Rev3–RBM2 was cross-validated by SAXS measurements (SI Appendix, Fig. S8) here and by analytical ultracentrifugation previously (20). All mutants identified in this study as breaking the Rev7 dimer (Fig. 2B) were also cross-validated with a yeast two-hybrid assay (Fig. 2C and SI Appendix, Fig. S2B).
Yeast Two-Hybrid Assay.
Studies of Rev7 dimerization using the yeast two-hybrid assay were carried out in yeast strain PJ69-4A (55). DNA encoding for Rev7 (wild type and/or mutants) was subcloned as fusions of the GAL4 AD and GAL4 BD in pGAD-C1 and pGBD-C1 plasmids marked with leucine and tryptophan, respectively. Yeast harboring the two plasmids, one encoding wild-type and another mutant Rev7, were grown at 30 °C for 2 d in a 3-mL culture lacking leucine and tryptophan and then were spotted on −LW plates to confirm the presence of viable transformants and on −AHLW plates to score the interaction (Fig. 2C and SI Appendix, Fig. S2B). This yeast two-hybrid protocol was also used to probe interactions between the wild-type or mutant Rev7 with the wild-type Mad2 and p31comet (Fig. 2C and SI Appendix, Fig. S2 C and D).
Docking.
Two PDB files of Rev7WT/Rev3–RBM2 were used as input for the software HADDOCK (57) in combination with R124A and our eight experimentally determined dimer-breaking mutations to guide the docking (SI Appendix, Table S2). For all other parameters, the default setting was used. The structure used for docking was our 1.68-Å structure of Rev7R124A/Rev3–RBM2 with the sequence mutated back to wild type using PyMOL (72). This was done because our structure of Rev7WT/Rev3-RMB2, while similar to that of the R124A mutant, is of lower quality, has residues missing, and lacks side-chain density in loop regions (SI Appendix, Fig. S1). All 200 lowest energy structures of the dimer calculated by HADDOCK were grouped into a single final cluster. To assess a possible steric clash that may arise when Rev1-CT binds to the Rev7WT/Rev3–RBM2 dimer, we used the “align” function in PyMOL (72) to superimpose two copies of Rev7R124A/Rev3–RBM1/Rev1-CT [PDB ID code 3VU7 (29)] onto our structure of the Rev7WT/Rev3–RBM2 dimer predicted by HADDOCK or other HORMA dimers as noted (Fig. 5D and SI Appendix, Fig. S9). The model of the (Rev7WT/Rev3-RBM2)2 dimer was deposited to PDB-dev with ID PDBDEV_00000009.
ITC.
ITC measurements were collected on a Nano ITC calorimeter (TA Instruments) at the University of Connecticut Biophysics Core. To study binding of the Rev7WT/Rev3–RBM12 dimer and the Rev1-CT domain, 50 μL of 2 mM Rev1-CT was titrated in 2.5-μL aliquots into a 170-μL solution of 250 μM Rev7 (125 μM Rev7WT2/Rev3–RBM12) in buffer (50 mM sodium phosphate, 100 mM NaCl, pH 8.2) at 25 °C. The data were fit with NanoAnalyze software to extract the Kd for the complex, ΔH, and stoichiometry parameter (n) (Fig. 4). ITC dilution experiments for Rev7WT/Rev3–RBM2 and Rev7R124A/Rev3–RBM2 were performed by titrating 2.5-μL aliquots of 400 μM protein into 170 μL of buffer (20 mM Hepes, 100 mM NaCl, pH 7.4) at 20 °C. The data were analyzed to extract the Kd and ΔH for the Rev7 dimer (SI Appendix, Fig. S2A).
DNA Damage Sensitivity Assay in the Rev7−/− Cell Line.
The Rev7−/−;KrasG12D/+;Trp53−/− cells used in this study were generated by F.-M.V. (59) using the CRISPR/Cas9 system from the KrasG12D/+; Trp53−/− murine lung adenocarcinoma cell line kindly provided by Tyler Jack’s laboratory (MIT, Cambridge, MA) (60). All cell lines were cultured in standard DMEM/10% FBS medium.
To assess cell viability following cisplatin-induced DNA damage, cells were seeded in triplicate (8 × 103) in 96-well plates and treated with cisplatin as indicated. After 48-h treatment, cell viability was assessed using Cell Titer-Glo (Promega) on an Applied Biosystems microplate luminometer.
Supplementary Material
Acknowledgments
We thank the faculty at CHESS, including Richard Gillilan and Jesse Hopkins for SAXS and Irina Kriksunov and David Schuller for crystallography. CHESS is supported by the National Science Foundation (NSF) and the NIH/National Institute of General Medical Science (NIGMS) via NSF Award DMR-1332208, and the MacCHESS resource is supported by NIH/NIGMS Award GM103485. The plasmid for human Rev7R124A/Rev3–RBM1 in pETDuet-1 was provided by Dr. Hiroshi Hashimoto. Research in the D.M.K. laboratory is supported by NSF/Division of Molecular and Cellular Biology Grant 1615866 and NIH/NIGMS Grant R01-GM123239. Work in the G.C.W. laboratory is supported by NIH/National Institute of Environmental Health Science Grants R01-ES015818 and R35-ES028303 and Grant P30-ES002109 to the MIT Center of Environmental Health Sciences. G.C.W. is an American Cancer Society Professor. B.H. is supported by NIH/NIGMS Grant R01-GM099948 and Connecticut Regenerative Medicine Research Fund Grant 15-RMA-UCHC-03.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank (PDB) database, www.wwpdb.org (PDB ID codes 6BC8, 6BCD, and 6BI7), and in the PDB-dev database, pdb-dev.wwpdb.org (PDB-dev ID code PDBDEV_00000009).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1801149115/-/DCSupplemental.
References
- 1.Friedberg EC, Walker GC, Siede W, Wood RD. DNA Repair and Mutagenesis. American Society for Microbiology; Washington, DC: 2005. [Google Scholar]
- 2.Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol. 2014;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang W, Woodgate R. What a difference a decade makes: Insights into translesion DNA synthesis. Proc Natl Acad Sci USA. 2007;104:15591–15598. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waters LS, et al. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev. 2009;73:134–154. doi: 10.1128/MMBR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sale JE, Lehmann AR, Woodgate R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat Rev Mol Cell Biol. 2012;13:141–152. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ulrich HD. Timing and spacing of ubiquitin-dependent DNA damage bypass. FEBS Lett. 2011;585:2861–2867. doi: 10.1016/j.febslet.2011.05.028. [DOI] [PubMed] [Google Scholar]
- 7.Livneh Z, Ziv O, Shachar S. Multiple two-polymerase mechanisms in mammalian translesion DNA synthesis. Cell Cycle. 2010;9:729–735. doi: 10.4161/cc.9.4.10727. [DOI] [PubMed] [Google Scholar]
- 8.Shachar S, et al. Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. EMBO J. 2009;28:383–393. doi: 10.1038/emboj.2008.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawrence CW. Cellular functions of DNA polymerase ζ and Rev1 protein. Adv Protein Chem. 2004;69:167–203. doi: 10.1016/S0065-3233(04)69006-1. [DOI] [PubMed] [Google Scholar]
- 10.Prakash S, Prakash L. Translesion DNA synthesis in eukaryotes: A one- or two-polymerase affair. Genes Dev. 2002;16:1872–1883. doi: 10.1101/gad.1009802. [DOI] [PubMed] [Google Scholar]
- 11.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 12.Nelson JR, Lawrence CW, Hinkle DC. Thymine-thymine dimer bypass by yeast DNA polymerase ζ. Science. 1996;272:1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- 13.Lin Y-C, et al. Error-prone replication bypass of the primary aflatoxin B1 DNA adduct, AFB1-N7-Gua. J Biol Chem. 2014;289:18497–18506. doi: 10.1074/jbc.M114.561563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Makarova AV, Burgers PM. Eukaryotic DNA polymerase ζ. DNA Repair (Amst) 2015;29:47–55. doi: 10.1016/j.dnarep.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Makarova AV, Stodola JL, Burgers PM. A four-subunit DNA polymerase ζ complex containing Pol δ accessory subunits is essential for PCNA-mediated mutagenesis. Nucleic Acids Res. 2012;40:11618–11626. doi: 10.1093/nar/gks948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baranovskiy AG, et al. DNA polymerase δ and ζ switch by sharing accessory subunits of DNA polymerase δ. J Biol Chem. 2012;287:17281–17287. doi: 10.1074/jbc.M112.351122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson RE, Prakash L, Prakash S. Pol31 and Pol32 subunits of yeast DNA polymerase δ are also essential subunits of DNA polymerase ζ. Proc Natl Acad Sci USA. 2012;109:12455–12460. doi: 10.1073/pnas.1206052109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee Y-S, Gregory MT, Yang W. Human Pol ζ purified with accessory subunits is active in translesion DNA synthesis and complements Pol η in cisplatin bypass. Proc Natl Acad Sci USA. 2014;111:2954–2959. doi: 10.1073/pnas.1324001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johansson E, Macneill SA. The eukaryotic replicative DNA polymerases take shape. Trends Biochem Sci. 2010;35:339–347. doi: 10.1016/j.tibs.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 20.Hara K, et al. Purification, crystallization and initial X-ray diffraction study of human REV7 in complex with a REV3 fragment. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009;65:1302–1305. doi: 10.1107/S1744309109046181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hara K, et al. Crystal structure of human REV7 in complex with a human REV3 fragment and structural implication of the interaction between DNA polymerase ζ and REV1. J Biol Chem. 2010;285:12299–12307. doi: 10.1074/jbc.M109.092403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomida J, et al. REV7 is essential for DNA damage tolerance via two REV3L binding sites in mammalian DNA polymerase ζ. Nucleic Acids Res. 2015;43:1000–1011. doi: 10.1093/nar/gku1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baranovskiy AG, et al. X-ray structure of the complex of regulatory subunits of human DNA polymerase delta. Cell Cycle. 2008;7:3026–3036. doi: 10.4161/cc.7.19.6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelson JR, Gibbs PE, Nowicka AM, Hinkle DC, Lawrence CW. Evidence for a second function for Saccharomyces cerevisiae Rev1p. Mol Microbiol. 2000;37:549–554. doi: 10.1046/j.1365-2958.2000.01997.x. [DOI] [PubMed] [Google Scholar]
- 25.Ohashi E, et al. Identification of a novel REV1-interacting motif necessary for DNA polymerase κ function. Genes Cells. 2009;14:101–111. doi: 10.1111/j.1365-2443.2008.01255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pozhidaeva A, et al. NMR structure and dynamics of the C-terminal domain from human Rev1 and its complex with Rev1 interacting region of DNA polymerase η. Biochemistry. 2012;51:5506–5520. doi: 10.1021/bi300566z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pustovalova Y, Bezsonova I, Korzhnev DM. The C-terminal domain of human Rev1 contains independent binding sites for DNA polymerase η and Rev7 subunit of polymerase ζ. FEBS Lett. 2012;586:3051–3056. doi: 10.1016/j.febslet.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wojtaszek J, et al. Structural basis of Rev1-mediated assembly of a quaternary vertebrate translesion polymerase complex consisting of Rev1, heterodimeric polymerase (Pol) ζ, and Pol κ. J Biol Chem. 2012;287:33836–33846. doi: 10.1074/jbc.M112.394841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kikuchi S, Hara K, Shimizu T, Sato M, Hashimoto H. Structural basis of recruitment of DNA polymerase ζ by interaction between REV1 and REV7 proteins. J Biol Chem. 2012;287:33847–33852. doi: 10.1074/jbc.M112.396838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pustovalova Y, et al. Interaction between the Rev1 C-terminal domain and the PolD3 subunit of Polζ suggests a mechanism of polymerase exchange upon Rev1/Polζ-dependent translesion synthesis. Biochemistry. 2016;55:2043–2053. doi: 10.1021/acs.biochem.5b01282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Acharya N, et al. Complex formation of yeast Rev1 and Rev7 proteins: A novel role for the polymerase-associated domain. Mol Cell Biol. 2005;25:9734–9740. doi: 10.1128/MCB.25.21.9734-9740.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Budzowska M, Graham TG, Sobeck A, Waga S, Walter JC. Regulation of the Rev1-pol ζ complex during bypass of a DNA interstrand cross-link. EMBO J. 2015;34:1971–1985. doi: 10.15252/embj.201490878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhat A, Andersen PL, Qin Z, Xiao W. Rev3, the catalytic subunit of Polζ, is required for maintaining fragile site stability in human cells. Nucleic Acids Res. 2013;41:2328–2339. doi: 10.1093/nar/gks1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Northam MR, et al. DNA polymerases ζ and Rev1 mediate error-prone bypass of non-B DNA structures. Nucleic Acids Res. 2014;42:290–306. doi: 10.1093/nar/gkt830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Listovsky T, Sale JE. Sequestration of CDH1 by MAD2L2 prevents premature APC/C activation prior to anaphase onset. J Cell Biol. 2013;203:87–100. doi: 10.1083/jcb.201302060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murakumo Y, et al. A human REV7 homolog that interacts with the polymerase ζ catalytic subunit hREV3 and the spindle assembly checkpoint protein hMAD2. J Biol Chem. 2000;275:4391–4397. doi: 10.1074/jbc.275.6.4391. [DOI] [PubMed] [Google Scholar]
- 37.Medendorp K, et al. The mitotic arrest deficient protein MAD2B interacts with the small GTPase RAN throughout the cell cycle. PLoS One. 2009;4:e7020. doi: 10.1371/journal.pone.0007020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boersma V, et al. MAD2L2 controls DNA repair at telomeres and DNA breaks by inhibiting 5′ end resection. Nature. 2015;521:537–540. doi: 10.1038/nature14216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu G, et al. REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature. 2015;521:541–544. doi: 10.1038/nature14328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aravind L, Koonin EV. The HORMA domain: A common structural denominator in mitotic checkpoints, chromosome synapsis and DNA repair. Trends Biochem Sci. 1998;23:284–286. doi: 10.1016/s0968-0004(98)01257-2. [DOI] [PubMed] [Google Scholar]
- 41.Rosenberg SC, Corbett KD. The multifaceted roles of the HORMA domain in cellular signaling. J Cell Biol. 2015;211:745–755. doi: 10.1083/jcb.201509076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo X, et al. Structure of the Mad2 spindle assembly checkpoint protein and its interaction with Cdc20. Nat Struct Biol. 2000;7:224–229. doi: 10.1038/73338. [DOI] [PubMed] [Google Scholar]
- 43.Luo X, Tang Z, Rizo J, Yu H. The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol Cell. 2002;9:59–71. doi: 10.1016/s1097-2765(01)00435-x. [DOI] [PubMed] [Google Scholar]
- 44.Sironi L, et al. Crystal structure of the tetrameric Mad1-Mad2 core complex: Implications of a ‘safety belt’ binding mechanism for the spindle checkpoint. EMBO J. 2002;21:2496–2506. doi: 10.1093/emboj/21.10.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luo X, et al. The Mad2 spindle checkpoint protein has two distinct natively folded states. Nat Struct Mol Biol. 2004;11:338–345. doi: 10.1038/nsmb748. [DOI] [PubMed] [Google Scholar]
- 46.Luo X, Yu H. Protein metamorphosis: The two-state behavior of Mad2. Structure. 2008;16:1616–1625. doi: 10.1016/j.str.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mapelli M, Massimiliano L, Santaguida S, Musacchio A. The Mad2 conformational dimer: Structure and implications for the spindle assembly checkpoint. Cell. 2007;131:730–743. doi: 10.1016/j.cell.2007.08.049. [DOI] [PubMed] [Google Scholar]
- 48.Yang M, et al. Insights into mad2 regulation in the spindle checkpoint revealed by the crystal structure of the symmetric mad2 dimer. PLoS Biol. 2008;6:e50. doi: 10.1371/journal.pbio.0060050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang M, et al. p31comet blocks Mad2 activation through structural mimicry. Cell. 2007;131:744–755. doi: 10.1016/j.cell.2007.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eytan E, et al. Disassembly of mitotic checkpoint complexes by the joint action of the AAA-ATPase TRIP13 and p31(comet) Proc Natl Acad Sci USA. 2014;111:12019–12024. doi: 10.1073/pnas.1412901111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang K, et al. Thyroid hormone receptor interacting protein 13 (TRIP13) AAA-ATPase is a novel mitotic checkpoint-silencing protein. J Biol Chem. 2014;289:23928–23937. doi: 10.1074/jbc.M114.585315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ye Q, et al. TRIP13 is a protein-remodeling AAA+ ATPase that catalyzes MAD2 conformation switching. eLife. 2015;4:e07367. doi: 10.7554/eLife.07367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miniowitz-Shemtov S, Eytan E, Kaisari S, Sitry-Shevah D, Hershko A. Mode of interaction of TRIP13 AAA-ATPase with the Mad2-binding protein p31comet and with mitotic checkpoint complexes. Proc Natl Acad Sci USA. 2015;112:11536–11540. doi: 10.1073/pnas.1515358112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qi S, Kim DJ, Stjepanovic G, Hurley JH. Structure of the human Atg13-Atg101 HORMA heterodimer: An interaction hub within the ULK1 complex. Structure. 2015;23:1848–1857. doi: 10.1016/j.str.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rambo RP, Tainer JA. Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers. 2011;95:559–571. doi: 10.1002/bip.21638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karaca E, Melquiond AS, de Vries SJ, Kastritis PL, Bonvin AM. Building macromolecular assemblies by information-driven docking: Introducing the HADDOCK multibody docking server. Mol Cell Proteomics. 2010;9:1784–1794. doi: 10.1074/mcp.M000051-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Franke D, et al. ATSAS 2.8: A comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J Appl Cryst. 2017;50:1212–1225. doi: 10.1107/S1600576717007786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vassel F-M. 2018. Rev7 is a Novel Regulator Chemotherapeutic Response in Drug-Resistant Lung Cancer. PhD thesis (MIT, Cambridge, MA)
- 60.Dimitrova N, et al. Stromal expression of miR-143/145 promotes neoangiogenesis in lung cancer development. Cancer Discov. 2016;6:188–201. doi: 10.1158/2159-8290.CD-15-0854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oliver TG, et al. Chronic cisplatin treatment promotes enhanced damage repair and tumor progression in a mouse model of lung cancer. Genes Dev. 2010;24:837–852. doi: 10.1101/gad.1897010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu H. Structural activation of Mad2 in the mitotic spindle checkpoint: The two-state Mad2 model versus the Mad2 template model. J Cell Biol. 2006;173:153–157. doi: 10.1083/jcb.200601172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Erster O, Liscovitch M. A modified inverse PCR procedure for insertion, deletion, or replacement of a DNA fragment in a target sequence and its application in the ligand interaction scan method for generation of ligand-regulated proteins. Methods Mol Biol. 2010;634:157–174. doi: 10.1007/978-1-60761-652-8_12. [DOI] [PubMed] [Google Scholar]
- 64.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 65.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nielsen S, et al. BioXTAS RAW, a software program for high-throughput automated small-angle X-ray scattering data reduction and preliminary analysis. J Appl Cryst. 2009;42:959–964. [Google Scholar]
- 68.Schneidman-Duhovny D, Hammel M, Tainer JA, Sali A. FoXS, FoXSDock and MultiFoXS: Single-state and multi-state structural modeling of proteins and their complexes based on SAXS profiles. Nucleic Acids Res. 2016;44:W424–W429. doi: 10.1093/nar/gkw389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Grishaev A, Guo L, Irving T, Bax A. Improved fitting of solution X-ray scattering data to macromolecular structures and structural ensembles by explicit water modeling. J Am Chem Soc. 2010;132:15484–15486. doi: 10.1021/ja106173n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 71.Vranken WF, et al. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 72. Schrödinger, LLC (2015) The PyMOL Molecular Graphics System (Schrödinger, LLC, New York), Version 1.8.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.