Significance
Nucleosomal modifications have been implicated in fundamental epigenetic regulation, whereas the roles of nucleosome binding in shaping changes through evolution remain to be addressed. Here we performed a comparative study to clarify the roles of nucleosome occupancy in exon origination. By profiling a high-resolution, cross-species mononucleosome landscape for mammalian tissues, we found nucleosome occupancy profiles are conserved across tissues and species. Further, through a phylogenetic approach, we found that the feature of differential nucleosome occupancy appears prior to the origination of new exons and, presumably, facilitates the origin of new exons by increasing the splice strength of the ancestral nonexonic regions through driving a local difference in GC content, which suggests the function of nucleosome binding in exonization.
Keywords: nucleosome occupancy, exonization, comparative genomics, exon origination, primate evolution
Abstract
Nucleosomal modifications have been implicated in fundamental epigenetic regulation, but the roles of nucleosome occupancy in shaping changes through evolution remain to be addressed. Here we present high-resolution nucleosome occupancy profiles for multiple tissues derived from human, macaque, tree shrew, mouse, and pig. Genome-wide comparison reveals conserved nucleosome occupancy profiles across both different species and tissue types. Notably, we found significantly higher levels of nucleosome occupancy in exons than in introns, a pattern correlated with the different exon–intron GC content. We then determined whether this biased occupancy may play roles in the origination of new exons through evolution, rather than being a downstream effect of exonization, through a comparative approach to sequentially trace the order of the exonization and biased nucleosome binding. By identifying recently evolved exons in human but not in macaque using matched RNA sequencing, we found that higher exonic nucleosome occupancy also existed in macaque regions orthologous to these exons. Presumably, such biased nucleosome occupancy facilitates the origination of new exons by increasing the splice strength of the ancestral nonexonic regions through driving a local difference in GC content. These data thus support a model that sites bound by nucleosomes are more likely to evolve into exons, which we term the “nucleosome-first” model.
Eukaryotic genomes are packaged with nucleosomes that each consist of a histone core with ∼147 bp DNA wrapped around it (1, 2). Recent studies have linked modifications of nucleosome proteins to fundamental epigenetic regulation (3), whereas the roles of nucleosome occupancy (NOC) in shaping long-term changes through evolution remain to be addressed. Because nucleosome disassembly is required in transcription (4), it is plausible that nucleosome occupancy may regulate some cotranscriptional processes, such as RNA splicing (5, 6). Notably, previous studies in mammalian cell lines, or model organisms like Caenorhabditis elegans and Drosophila melanogaster, have found relatively higher levels of nucleosome occupancy at exonic regions than at intronic regions (7–11). Moreover, the average length of exons was found to be relatively constant among species and similar to the length of the nucleosome-binding DNA sequence (147 bp) (12). These findings thus suggest a role for nucleosome occupancy in shaping fundamental, long-term biological processes, such as the origination of new exons (exonization).
However, at least two fundamental issues should be addressed before mechanistically linking nucleosome occupancy regulation with exonization. First, previous studies typically investigate the profiles of nucleosome occupancy in mammalian cell lines or model organisms such as Drosophila and Caenorhabditis elegans (8–10). However, quantification of nucleosome occupancy profiles in tissues taken from both human and mammalian models that are closely related to human is a fundamental issue in need of immediate focus.
Second, previous studies have found relatively higher level of nucleosome occupancy at both exonic regions (7–11) and regions with high GC content (13, 14). Moreover, a biased fixation spectrum of DNA mutation to G or C was found in DNA regions with enriched nucleosome binding (15–17). Given the fact that exons typically show higher GC content than introns (9), it is difficult to clarify the relationship between the two interlinked processes—the preferential binding of nucleosomes in exons and the preferential binding of nucleosomes in GC-rich regions. Briefly, the patterns could be explained by two different evolutionary models of the exonization process. The “nucleosome-first” model posits that differential nucleosome occupancy occurs before the origination of new exons. Then the profile of varied nucleosome occupancy facilitates the exonization through mechanisms facilitating splicing-favorable features, such as a higher GC content in newly originated exonic regions. Alternatively, the “exon-first” model suggests that exon origination occurs first, and then the sequence feature of high GC content is gradually optimized by natural selection. Then the pattern of higher exonic nucleosome occupancy appears due to the preference of nucleosome binding in GC-rich regions. If this is true, then the feature of biased nucleosome occupancy might merely represent a downstream effect of differing exon–intron GC content found after the origination of exons. It is difficult for traditional approaches to distinguish between the two models because of the coexistence of relatively higher GC content with higher nucleosome occupancy in exons than in introns.
Results
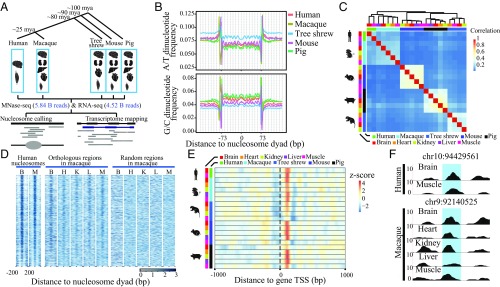
To study the possible regulatory roles of nucleosome occupancy in the origination of exons, we performed comparative studies in five mammalian species: human (Homo sapiens), rhesus macaque (Macaca mulatta), tree shrew (Tupaia belangeri), mouse (Mus musculus), and mini pig (Sus domesticus). We first profiled a comprehensive nucleosome occupancy map in tissue samples from these five species using micrococcal nuclease digestion of chromatin followed by high-throughput sequencing (MNase-seq) (18), in which mononucleosomal DNA fragments digested by MNase were subjected to deep sequencing to generate an average of 278 million paired-end reads for each sample (Fig. 1A; Table 1; and SI Appendix, Table S1). We further verified the specificity of this high-resolution nucleosome occupancy map with multiple known features of nucleosome-protected DNA sequences and MNase-seq profiles. These included an expected MNase-digested fragment size (18), predominant representations of ∼10-bp periodic repetitions of AA/AT/TA/TT dinucleotides, and out-of-phase ∼10-bp periodic repetitions of CC/CG/GC/GG dinucleotides in nucleosome-protected regions (19, 20), as well as a nucleosome-depleted region located upstream of the transcription start site (TSS), followed by one well-positioned nucleosome and a downstream nucleosome array (21) (Fig. 1 B and E and SI Appendix, Fig. S1). This is a high-quality, cross-species mononucleosome landscape for mammalian tissues.
Fig. 1.
Nucleosome occupancy profiles are conserved across species and tissues. (A) Overview of the experimental design. A tree showing the phylogenetic relationships between humans and four outgroup species, with estimated divergence times marked on the branches. The tissue samples used in the MNase-seq and RNA-seq are shown, along with the total number of sequencing reads. (B) Dinucleotide frequencies of A/T (the combined dinucleotide frequency of AA/AT/TA/TT) and G/C (the combined dinucleotide frequency of CC/CG/GC/GG) in nucleosome-protected regions in brain samples of the five species are shown. Nucleosome dyad, the midpoint position of the DNA bound by the nucleosome core. (C) Hierarchical clustering chart showing the correlations of nucleosome occupancy profiles in different tissue samples of the five species. (D) Heat map of nucleosome occupancy profiles for human samples and nucleosome occupancy profiles at macaque orthologous regions, as well as at properly aligned, randomly selected macaque genomic regions. The nucleosome occupancy profiles for all human nucleosome-occupied regions detected in both of the two human samples are shown. B, brain; H, heart; K, kidney; L, liver; M, muscle. (E) For all of the orthologous genes across the five species, the nucleosome occupancy profile near the TSS is shown in heat map, with focal TSS indicated by a dotted line. (F) The nucleosome occupancy profiles of two human tissues in one human chromosome region, together with the profiles of multiple macaque tissues in the orthologous regions are shown. In each profile, a 147-bp window, shaded in blue, indicates a region occupied by one nucleosome with the coordinate of its midpoint shown above each panel.
Table 1.
Summary of MNase-seq and RNA-seq in multiple tissues of the five species
| Species | Tissue | MNase-seq total reads, M | RNA-seq total reads, M |
| Human | Brain, muscle | 503.7 | 302.6 |
| Macaque | Brain, heart, kidney, liver, muscle | 1,305.2 | 2,475.8 |
| Tree shrew | Brain, heart, kidney, muscle | 760.2 | 557.3 |
| Mouse | Brain, heart, kidney, liver, muscle | 1,938.5 | 638.6 |
| Pig | Brain, heart, kidney, liver, muscle | 1,332.4 | 549.8 |
Next, we compared the global patterns of these nucleosome occupancy profiles to explore the conservation level among species and tissues. According to the clustering chart we generated based on the similarity of the nucleosome occupancy profiles, we found that different tissues from the same species were clustered together (Fig. 1C), indicating cross-tissue conservation of the nucleosome occupancy profile in the same species, presumably mediated by cis-regulatory elements. Despite the generally larger cross-species difference of the nucleosome occupancy profile in contrast to the cross-tissue variations of the same species, the profiles were generally consistent between closely related primate species. In contrast to randomly selected regions, orthologous regions of human nucleosome-occupied regions in macaque showed nucleosome occupancy profile similar to that in human (Fig. 1D and SI Appendix, Fig. S2). Moreover, when comparing the nucleosome occupancy of orthologous genes in the five species, we found a similar pattern of nucleosome binding nearby the TSS regions (Fig. 1E). Several demonstration cases in randomly selected chromosome regions were shown for the cross-tissue and cross-species conservation of the nucleosome occupancy profiles (Fig. 1F and SI Appendix, Fig. S3). Overall, although epigenetic regulators are typically dynamic, it seems that this nucleosome occupancy profile is largely stable across both different tissues and closely related species.
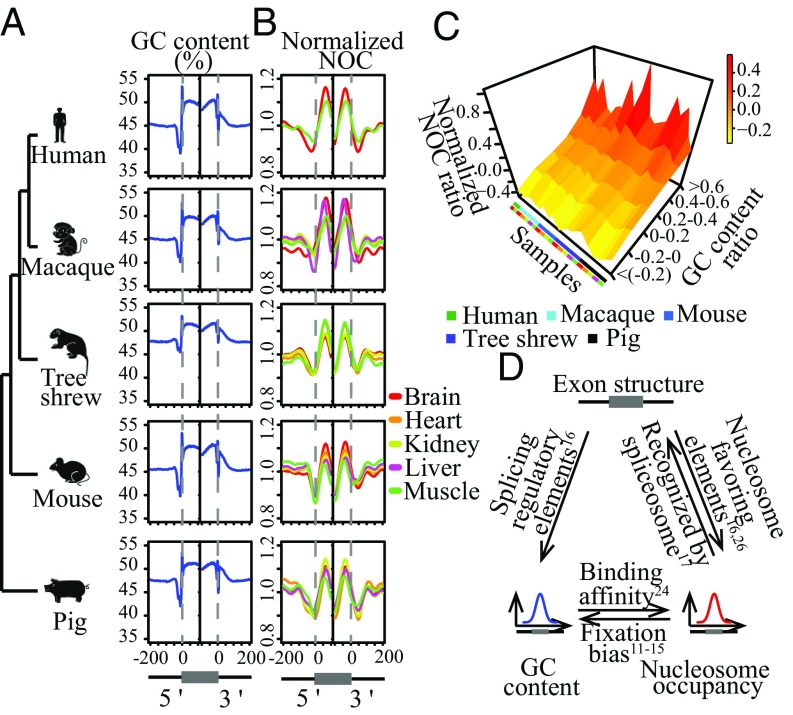
With cross-species nucleosome occupancy profiles identified, we then investigated whether the biased binding of nucleosomes on exonic regions or GC-rich regions, as previously detected in mammalian cell lines, could also be detected in real tissues. As expected, we found that the GC content in exons was markedly higher than in the flanking introns for all species (Fig. 2A; Wilcoxon rank-sum test, P < 2.2e-16). Notably, in different tissues derived from all of the tested species, we consistently found significantly higher nucleosome occupancy in exonic regions than in their flanking introns, as indicated by a sharp peak of nucleosome density at the boundaries of the exonic regions (Fig. 2B; Wilcoxon rank-sum test, P < 2.2e-16). To quantitatively investigate correlations between the two levels, we used NOC ratios (binary logarithm of the average nucleosome occupancy of exon to that of the upstream 150-bp intronic region) and GC content ratios (binary logarithm of the GC content of exon to that of the upstream 150-bp intronic region) to quantify levels of exon–intron differences in nucleosome occupancy and GC content, respectively (10). Notably, for comparisons with different tissues derived from different species, NOC ratios consistently showed a significant positive correlation with GC content ratios (Fig. 2C and SI Appendix, Fig. S4), indicating that increased nucleosome occupancy and GC content in exonic regions are correlated features conserved in multiple mammalian species. Because exonic regions are generally GC-rich and nucleosomes preferentially bind to GC-rich regions, challenges remain for traditional approaches to determine whether biased nucleosome occupancy may play some roles in the origination of new exons (Fig. 2D).
Fig. 2.
Positive correlation of exon–intron differences in nucleosome occupancy with GC content. The (A) GC contents and (B) normalized nucleosome occupancies of nearby exonic regions in five species. The original scores for nucleosome occupancy were normalized with the mean score of the plotted region. Exonic regions are indicated by gray dashed lines. (C) Exonic regions of each tissue type for each species were assigned to one of six bins based on the GC content ratios (y axis). Average NOC ratios for each bin were then calculated (z axis). (D) Schematic of the relationships among exon–intron structure, GC content, and nucleosome occupancy. Each relationship was labeled with reference studies that provide evidence supporting that relationship.
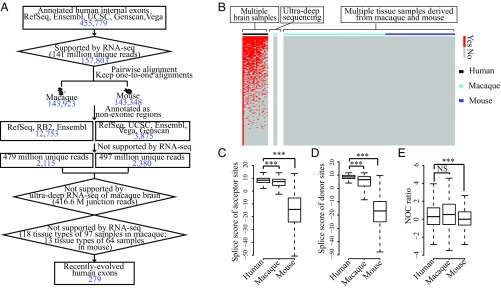
Alternatively, a comparative approach with both profiles of exon splicing and nucleosome binding in multiple mammalian species may provide a sequential order of the appearance of exons and biased nucleosome occupancy, providing a practical way to distinguish between the nucleosome-first and exon-first models and to clarify the potential role of nucleosome occupancy in exonization. Therefore, we performed strand-specific RNA sequencing (RNA-seq) in matched tissues to define the splicing events in the five species (Figs. 1A and 3A and SI Appendix, Fig. S5). Overall, an average of 126 million RNA-seq reads were generated for each sample, and a total of 157,803 human internal exons were identified. We then identified recently evolved human exons using macaque and mouse as outgroup species because their genomes and gene structures are well studied (22, 23). Notably, 2,115 and 2,380 human exons could not be found in the orthologous regions of macaque and mouse, respectively, based on gene annotations and the RNA-seq signals. Among these exons, 664 could not be found in both species.
Fig. 3.
Differential nucleosome occupancy appears before the origination of exons. (A) Flowchart identifying recently evolved human exons based on RNA-seq data and gene annotations. The number of human exons in each step of the flowchart is indicated in blue. Ensembl, gene annotations from Ensembl; Genscan, gene predictions from Genscan program; RB2, gene annotations from RhesusBase (version 2); RefSeq, gene annotations from NCBI Reference Sequence Database; UCSC, gene annotations from University of California, Santa Cruz genome browser; Vega, gene annotations from the Vertebrate Genome Annotation database. (B) Heat map showing the expression of recently evolved human exons in brain samples of different individuals, as well as the expression of the orthologous regions in macaque and mouse animals. Horizontal red bars indicate the existence of exon expression. Splice site scores of (C) acceptor sites and (D) donor sites and (E) NOC ratios are shown in boxplots for recently evolved exons in human and orthologous regions in macaque and mouse. NS, not significant; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Notably, confounding factors such as the limited detection sensitivity in outgroup species and cross-sample variations could introduce false-positives in the definition of recently evolved human exons. To further control for the potential false-positives, we first performed ultradeep RNA-seq on the macaque brain sample to increase the detection sensitivity of macaque exons. A total of 1.4 billion RNA-seq reads (with 416.6 million junction reads) were generated and uniquely mapped to the macaque genome. The sequencing depth is 12.5-fold higher than the initial sequencing of the same macaque sample or 10-fold higher than that of the human brain sample (SI Appendix, Table S1). Second, we further integrated more public RNA-seq data of rhesus macaque and mouse to control for the false-positives introduced by the variability among populations and tissue types. A total of 97 RNA-seq datasets from 18 types of macaque tissues, as well as 64 RNA-seq datasets from 13 types of mouse tissues were integrated. On the basis of these RNA-seq data from outgroup species, we ultimately identified 279 exons exclusively detected in human (Dataset S1), presumably representing recently evolved human exons that were not detected in any of the 172 samples from macaque and mouse (Fig. 3 A and B and SI Appendix, Tables S1 and S2).
To account for the variability among populations, we also used public RNA-seq data from brain samples of multiple human individuals to verify the specificity of these candidate young exons (24) (SI Appendix, Table S2). Overall, 84.59% of these exons were detected in at least two human individuals (Fig. 3B). The remaining undetected exons in samples from other human individuals were partially due to the relatively lower expression levels of these exons and to the relatively low sequencing depth of the public data (SI Appendix, Fig. S6). Therefore, population-level variability did not have a strong effect in defining recently evolved human exons.
On the basis of the 279 recently evolved human exons, we compared the levels of the splice site scores and nucleosome occupancy of the young exon-encoded regions in human and orthologous regions in macaque and mouse. As expected, the splice site scores of these orthologous regions were significantly lower in macaque and mouse, a finding consistent with the fact that these exons are specific to human (Fig. 3 C and D). As for the nucleosome occupancy, the NOC ratios for these recently evolved human exons are comparable with those of all human annotated exons (SI Appendix, Fig. S7). Notably, although the NOC ratios in mouse orthologous regions were significantly lower, we found human-comparable NOC ratios in macaque orthologous regions, although these exons are not encoded in macaque (Fig. 3E).
Considering the poor genome annotations of tree shrew and pig, we only used macaque and mouse as outgroup species to identify recently evolved human exons. When using the RNA-seq data for all five species to repeat the analyses, a smaller number of young human exons (41 exons) were identified (Dataset S2), largely due to difficulties in identifying orthologous regions in tree shrew and pig, given their relatively poor genome assembly and annotations. When we used this list of young human exons to investigate the levels of nucleosome occupancy in the orthologous regions of the four outgroup species, we found similar patterns of human-comparable NOC ratios in macaque (SI Appendix, Fig. S5).
In the above analyses, the recently evolved human exons were defined solely based on the deeply sequenced RNA-seq data in outgroup species. Because these human exons could not be detected in samples from both macaque and mouse, they presumably represent newly originated human exons after the divergence of human and rhesus macaque according to the parsimony role. However, despite the use of an extensive dataset of 172 deeply sequenced RNA-seq data in outgroup species (including an ultradeep RNA-seq on macaque brain), false-positives may still exist due to the detection sensitivity in outgroup species. Notably, although AG–GY boundaries do not necessarily constitute sufficient prerequisite for regions undergoing splicing, regions without the boundaries are less likely to be spliced by the spliceosome (25–27). Among the 279 recently evolved human exons defined based on the RNA-seq data, we found 39 candidates without AG–GY boundaries in the orthologous regions of rhesus macaque, crab-eating macaque, baboon, squirrel monkey, tarsier, mouse lemur, bush baby, and mouse, indicating that these exons are more likely to represent exons newly originated in human after the divergence of human and rhesus macaque. We then repeated the analysis using the 39 exons and found similar patterns of human–macaque comparable NOC ratios (SI Appendix, Fig. S8).
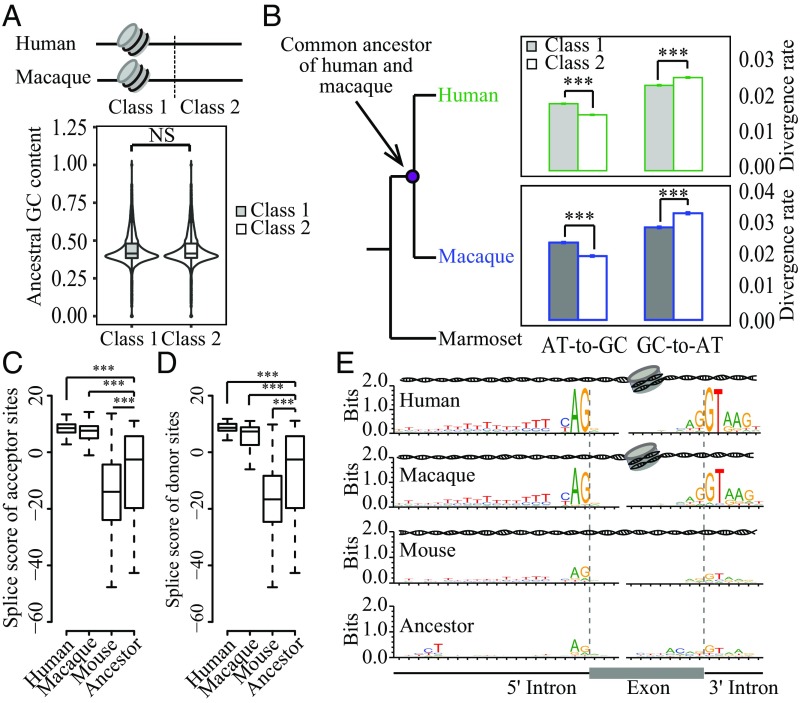
The above findings suggest a nucleosome-first model because differential nucleosome occupancy appears before the origination of exons. Previous studies have shown a biased fixation of DNA mutation spectrum for DNA regions protected by nucleosomes (15–17), although the biased spectrum was partially explained by intrinsic differences in the spectra of genomic regions with different GC content (SI Appendix, Fig. S9). Promoted by this clue, we then investigated whether the profile of varied nucleosome occupancy could facilitate exon origination by modulating the GC content in ancestral nonexonic regions. As a comparative study, we designated two classes of paired orthologous regions in human and macaque based on the levels of nucleosome occupancy: class 1, with an average NOC > 2 in both species, and class 2, with an average NOC < 0.5 in both species. To further circumvent the influence of GC content, these datasets were selected with comparable ancestral GC content (Fig. 4A). When counting the divergence sites in both human and macaque lineages after the divergence of the two species, we found a significantly higher AT-to-GC and lower GC-to-AT divergence rate for class 1 regions than for class 2 regions in human and macaque, a pattern consistent with previous reports (Fig. 4B; Wilcoxon rank-sum test, P < 2.2e-16) (15–17). This finding thus indicates that the binding of nucleosomes could increase the GC content of their occupied regions by influencing the fixation of different types of DNA mutations.
Fig. 4.
Differential nucleosome occupancy facilitates the exonization. (A) Schematic of two classes of paired orthologous regions based on nucleosome occupancy in human and macaque. The two regions show different NOC values, with class 1 showing NOC > 2 in both human and macaque and class 2 showing NOC < 0.5 in both species (Upper). The two classes show similar ancestral GC content (Lower). (B) Divergence rates of different substitution types of the mutations accumulated in the lineages of human (Upper) and macaque (Lower) after the two species diverged. AT-to-GC, A:T to G:C and A:T to C:G substitutions; GC-to-AT, G:C to A:T and G:C to T:A substitutions. The strength of splice (C) acceptor and (D) donor sites are shown in boxplots for young human exons (Human); the orthologous regions in macaque (Macaque) and mouse (Mouse); and regions of the common ancestor of human, macaque, and mouse (Ancestor). (E) Sequence motifs flanking splice sites of the young human exons (Human); the orthologous regions in macaque (Macaque) and mouse (Mouse); and the regions of the common ancestor of human, macaque, and mouse (Ancestor). The nucleosome occupancy status in each species is schematically indicated above the sequence motif. NS, not significant; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Given that intronic regions are typically AT-rich, whereas exonic regions are GC-rich, we speculated that this biased GC content, mediated by varied nucleosome occupancy, may facilitate the origination of exons by increasing the splice strength of the ancestral nonexonic regions. Subsequently, we compared the splice strength of the 279 recently evolved human exons, their orthologous regions in macaque and mouse, and their ancestral sequences. Notably, the ancestral sequences of these species show relatively weaker splice strength and sequence motif for the splice sites (Fig. 4 C–E). Compared with the ancestral sequences, the splice site scores were significantly lower in mouse with low NOC ratios (Wilcoxon rank-sum test, P = 1.01e-10 for splice donor sites and P = 1.92e-5 for splice acceptor sites), whereas both human and macaque with higher NOC ratios showed significantly higher splice strength compared with the ancestral sequences (Fig. 4 C and D) (Wilcoxon rank-sum test, P < 2.2e-16), a pattern consistent with the sequence motif analyses (Fig. 4E). These findings thus suggest that the binding of nucleosomes at ancestral nonexonic regions may increase the splice strength and facilitate the origination of the new exons (Fig. 4 C–E).
Discussion
Although previous studies have suggested a regulatory role of nucleosome occupancy in RNA splicing, the roles of nucleosome occupancy in exon origination remain to be addressed (9–11, 13). Considering both the preference of nucleosome binding in GC-rich DNA regions and the fact that exonic regions are generally GC-rich, a challenge remains to determine whether biased nucleosome occupancy may play some roles in the origination of new exons through evolution (nucleosome-first model) or whether it merely represents a downstream effect of differing exon–intron GC content after the origination of exons (exon-first model). Here we generated high-resolution nucleosome occupancy profiles and matched transcriptome profiles in multiple tissue samples derived from five mammalian species and verified cross-tissue/species conservation of nucleosome occupancy profiles. By using a phylogenetic approach to sequentially trace the order of the origination of exons and the appearance of biased nucleosome occupancy, our findings support a nucleosome-first model of potential roles of nucleosome occupancy in exonization. In this model, nucleosomes initially bind nonexonic regions and mark them for biased evolutionary changes (Fig. 5). Relatively higher nucleosome occupancy at an ancestral nonexonic region than at its flanking regions tends to drive a local difference in GC content, presumably facilitating the origination of a new exon by increasing the splice strength (Fig. 5). It is plausible that a role in exonization is a previously underestimated function of nucleosome binding.
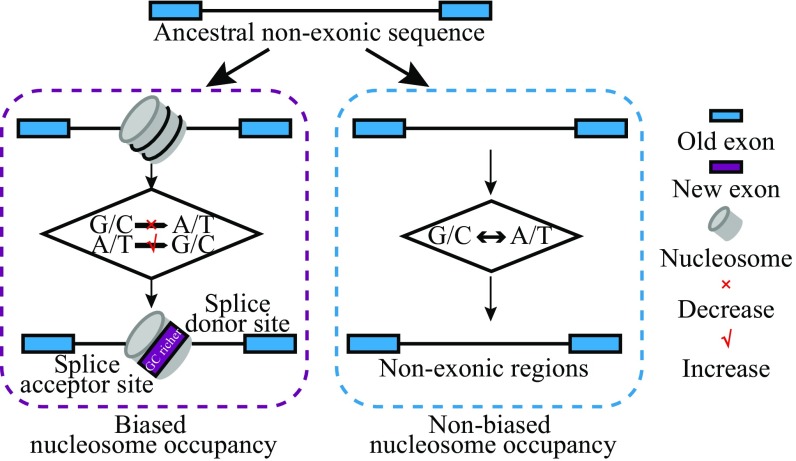
Fig. 5.
Nucleosome-first model for the origination of exons. Potential evolutionary outcomes of an ancestral nonexonic region toward exonization (at the top) yield two possible results. Biased nucleosome occupancy (purple dashed box) facilitates the formation of a local difference of GC content through biased fixation of DNA mutations. This, in turn, presumably facilitates the origination of a new exon by increasing the splice strength. In contrast, without the existence of biased nucleosome occupancy, most of these regions remain to be nonexonic similar to their ancestral states (blue dashed box).
Besides this hypothesis that neutral nucleosome occupancy facilitates the origination of new exons by improving the splicing motif, biased nucleosome occupancy may represent a preadapted feature of alternative mechanisms involved in the origination of new exons. First, previous studies found that nucleosomes can behave as a barrier resulting in suppressed RNA polymerase II elongation or even polymerase II pausing (28–30). These processes may likely increase recognition of the splice motif by spliceosomes because of the extended action time for spliceosome assembly (31). In line with this hypothesis, a recent study by Merkin et al. (27) found that mouse-specific exons with upstream intron deletions displayed increased nucleosome occupancy difference and enhanced RNA polymerase II pausing. This may serve as an indication that the increased nucleosome occupancy may enhance the inclusion of new exons in part by slowing polymerase elongation and extending action time for spliceosome assembly. Second, higher nucleosome occupancy also provides a molecular basis for histone modifications, which may promote the recognition of exons and substantially increase the flexibility of specific DNA regions involved in precise temporospatial regulation (7, 32).
The nucleosome-first model presented by this study focused on recently originated exons in human and the regulatory role of nucleosome occupancy in the recent evolutionary process of exonization. When considering more ancient events over evolutionary time, such as the order of the formation of the exon recognition signals and nucleosome binding motifs, additional possibilities actually exist. Briefly, although prokaryotes lack both nucleosome structure and splicing regulation, eukaryotes possess both, with underlying sequences similar in the GC-rich property and the average length (12). Two models may exist for the consistent features of the two regulatory levels: A coincidence model suggests that the exon recognition and nucleosome binding signals were coincidentally similar at the initial stage after the divergence of prokaryotes and eukaryotes. Alternatively, a nucleosome-first, spliceosome optimization model highlighted the roles of nucleosome binding in exonization, which suggests that after the binding of the nucleosomes, mechanisms such as rapid linker-biased insertion of transposable elements may create precursor exons with a nucleosome in length (33), and the differential nucleosome occupancy may introduce a local difference of GC content. Then the spliceosome may be expected to evolve to recognize these features over time, from which new exons with these features tend to be better recognized and more efficiently spliced. Under such circumstance, the signal of exon recognition gradually becomes similar to that of nucleosome binding. In this study, although we could not further differentiate the two models, the consistent features of the two regulatory levels and the finding that a new exon tends to evolve from nucleosome wraps highlight the significance to account for nucleosome occupancy and modification in the investigation of the exonization and alternative splicing.
Materials and Methods
Tissue samples used in this study were obtained from the animal facility of the Institute of Molecular Medicine in Peking University (accredited by the Association for Assessment and Accreditation of Laboratory Animal Care). The human participants provided informed consent. The study was approved by the Institutional Animal Care and Use Committee of Peking University, and the Medical Ethics Committee of Peking University Third Hospital. MNase-seq and strand-specific, poly(A)-positive RNA-seq were performed and analyzed to profile the nucleosome occupancy and transcriptome in multiple tissues derived from human, macaque, tree shrew, mouse, and pig. The similarities of the nucleosome occupancy profiles across both tissues and species were then evaluated. On the basis of the identification of recently evolved human exons, we further compared the nucleosome occupancy of the young exon-encoded regions in human and orthologous regions in macaque and mouse to sequentially trace the order of exonization and biased nucleosome binding. The sequencing data in this study are available in the Gene Expression Omnibus under accession no. GSE106580. Additional details are provided in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Heping Cheng at Peking University; Dr. Yong E. Zhang at Institute of Zoology, Chinese Academy of Science; Dr. Tim Liu at the National Institutes of Health (United States); and Dr. Qing Sunny Shen and Wanqiu Ding at Peking University for insightful suggestions. We acknowledge Dr. I. C. Bruce and Dr. Bertrand Chin-Ming Tan for critical reading of the manuscript. This work was supported by grants from the National Natural Science Foundation of China (31522032, 31471240, and 31521062) and the National Young Top-Notch Talent Support Program of China.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The MNase-seq and RNA-seq data in this study are available in the Gene Expression Omnibus (accession no. GSE106580).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1802561115/-/DCSupplemental.
References
- 1.Kornberg RD. Chromatin structure: A repeating unit of histones and DNA. Science. 1974;184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- 2.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 3.Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17:487–500. doi: 10.1038/nrg.2016.59. [DOI] [PubMed] [Google Scholar]
- 4.Ehrenhofer-Murray AE. Chromatin dynamics at DNA replication, transcription and repair. Eur J Biochem. 2004;271:2335–2349. doi: 10.1111/j.1432-1033.2004.04162.x. [DOI] [PubMed] [Google Scholar]
- 5.Beyer AL, Bouton AH, Miller OL., Jr Correlation of hnRNP structure and nascent transcript cleavage. Cell. 1981;26:155–165. doi: 10.1016/0092-8674(81)90299-3. [DOI] [PubMed] [Google Scholar]
- 6.Beyer AL, Osheim YN. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988;2:754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- 7.Andersson R, Enroth S, Rada-Iglesias A, Wadelius C, Komorowski J. Nucleosomes are well positioned in exons and carry characteristic histone modifications. Genome Res. 2009;19:1732–1741. doi: 10.1101/gr.092353.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nahkuri S, Taft RJ, Mattick JS. Nucleosomes are preferentially positioned at exons in somatic and sperm cells. Cell Cycle. 2009;8:3420–3424. doi: 10.4161/cc.8.20.9916. [DOI] [PubMed] [Google Scholar]
- 9.Schwartz S, Meshorer E, Ast G. Chromatin organization marks exon-intron structure. Nat Struct Mol Biol. 2009;16:990–995. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- 10.Tilgner H, et al. Nucleosome positioning as a determinant of exon recognition. Nat Struct Mol Biol. 2009;16:996–1001. doi: 10.1038/nsmb.1658. [DOI] [PubMed] [Google Scholar]
- 11.Amit M, et al. Differential GC content between exons and introns establishes distinct strategies of splice-site recognition. Cell Rep. 2012;1:543–556. doi: 10.1016/j.celrep.2012.03.013. [DOI] [PubMed] [Google Scholar]
- 12.Schwartz S, Ast G. Chromatin density and splicing destiny: On the cross-talk between chromatin structure and splicing. EMBO J. 2010;29:1629–1636. doi: 10.1038/emboj.2010.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohanim AB, Haran TE. The coexistence of the nucleosome positioning code with the genetic code on eukaryotic genomes. Nucleic Acids Res. 2009;37:6466–6476. doi: 10.1093/nar/gkp689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaplan N, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sasaki S, et al. Chromatin-associated periodicity in genetic variation downstream of transcriptional start sites. Science. 2009;323:401–404. doi: 10.1126/science.1163183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langley SA, Karpen GH, Langley CH. Nucleosomes shape DNA polymorphism and divergence. PLoS Genet. 2014;10:e1004457. doi: 10.1371/journal.pgen.1004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prendergast JG, Semple CA. Widespread signatures of recent selection linked to nucleosome positioning in the human lineage. Genome Res. 2011;21:1777–1787. doi: 10.1101/gr.122275.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui K, Zhao K. Genome-wide approaches to determining nucleosome occupancy in metazoans using MNase-seq. Methods Mol Biol. 2012;833:413–419. doi: 10.1007/978-1-61779-477-3_24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Satchwell SC, Drew HR, Travers AA. Sequence periodicities in chicken nucleosome core DNA. J Mol Biol. 1986;191:659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- 20.Brogaard K, Xi L, Wang JP, Widom J. A map of nucleosome positions in yeast at base-pair resolution. Nature. 2012;486:496–501. doi: 10.1038/nature11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schones DE, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang SJ, et al. Evolutionary interrogation of human biology in well-annotated genomic framework of rhesus macaque. Mol Biol Evol. 2014;31:1309–1324. doi: 10.1093/molbev/msu084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blake JA, et al. the Mouse Genome Database Group Mouse Genome Database (MGD)-2017: Community knowledge resource for the laboratory mouse. Nucleic Acids Res. 2017;45:D723–D729. doi: 10.1093/nar/gkw1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lonsdale J, et al. GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burset M, Seledtsov IA, Solovyev VV. Analysis of canonical and non-canonical splice sites in mammalian genomes. Nucleic Acids Res. 2000;28:4364–4375. doi: 10.1093/nar/28.21.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parada GE, Munita R, Cerda CA, Gysling K. A comprehensive survey of non-canonical splice sites in the human transcriptome. Nucleic Acids Res. 2014;42:10564–10578. doi: 10.1093/nar/gku744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merkin JJ, Chen P, Alexis MS, Hautaniemi SK, Burge CB. Origins and impacts of new mammalian exons. Cell Rep. 2015;10:1992–2005. doi: 10.1016/j.celrep.2015.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bondarenko VA, et al. Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol Cell. 2006;24:469–479. doi: 10.1016/j.molcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 29.Bintu L, et al. The elongation rate of RNA polymerase determines the fate of transcribed nucleosomes. Nat Struct Mol Biol. 2011;18:1394–1399. doi: 10.1038/nsmb.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gilchrist DA, et al. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carrillo Oesterreich F, Bieberstein N, Neugebauer KM. Pause locally, splice globally. Trends Cell Biol. 2011;21:328–335. doi: 10.1016/j.tcb.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 32.Zhou HL, Luo G, Wise JA, Lou H. Regulation of alternative splicing by local histone modifications: Potential roles for RNA-guided mechanisms. Nucleic Acids Res. 2014;42:701–713. doi: 10.1093/nar/gkt875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huff JT, Zilberman D, Roy SW. Mechanism for DNA transposons to generate introns on genomic scales. Nature. 2016;538:533–536. doi: 10.1038/nature20110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.