Significance
A key goal in the study of influenza virus evolution is to forecast which viral strains will persist and which ones will die out. Here we experimentally measure the effects of all amino acid mutations to the hemagglutinin protein from a human H3N2 influenza strain on viral growth in cell culture. We show that these measurements have utility for distinguishing among viral strains that do and do not succeed in nature. Overall, our work suggests that new high-throughput experimental approaches may be useful for understanding virus evolution in nature.
Keywords: influenza virus, hemagglutinin, deep mutational scanning, epistasis, mutational shifts
Abstract
Human influenza virus rapidly accumulates mutations in its major surface protein hemagglutinin (HA). The evolutionary success of influenza virus lineages depends on how these mutations affect HA’s functionality and antigenicity. Here we experimentally measure the effects on viral growth in cell culture of all single amino acid mutations to the HA from a recent human H3N2 influenza virus strain. We show that mutations that are measured to be more favorable for viral growth are enriched in evolutionarily successful H3N2 viral lineages relative to mutations that are measured to be less favorable for viral growth. Therefore, despite the well-known caveats about cell-culture measurements of viral fitness, such measurements can still be informative for understanding evolution in nature. We also compare our measurements for H3 HA to similar data previously generated for a distantly related H1 HA and find substantial differences in which amino acids are preferred at many sites. For instance, the H3 HA has less disparity in mutational tolerance between the head and stalk domains than the H1 HA. Overall, our work suggests that experimental measurements of mutational effects can be leveraged to help understand the evolutionary fates of viral lineages in nature—but only when the measurements are made on a viral strain similar to the ones being studied in nature.
Seasonal H3N2 influenza virus evolves rapidly, fixing 3 to 4 amino acid mutations per year in its hemagglutinin (HA) surface protein (1, 2). Many of these mutations contribute to the rapid antigenic drift that necessitates frequent updates to the annual influenza vaccine (3). This evolution is further characterized by competition and turnover among groups of strains called clades bearing different complements of mutations (4–8). Clades vary widely in their evolutionary success, with some dying out soon after emergence and others going on to take over the virus population. Several lines of evidence indicate that successful clades have higher fitness than clades that remain at low frequency (4–6, 9). A key goal in the study of H3N2 evolution is to identify the features that enable certain clades to succeed as others die out.
Two main characteristics distinguish evolutionarily successful clades from their competitors: greater antigenic change, and efficient viral growth and transmission. In principle, experiments could be informative for identifying how mutations affect these features. Most work on influenza evolution to date has used experimental data to assess the antigenicity of circulating strains (11–16). However, the nonantigenic effects of mutations also play an important role (5, 7, 9, 17). Specifically, due to influenza virus’s high mutation rate (18–20) and lack of intrasegment recombination (21), deleterious mutations become linked to beneficial ones. The resulting accumulation of deleterious mutations can affect nonantigenic properties central to viral fitness (9). However, there are no large-scale quantitative characterizations of how mutations to H3N2 HA affect viral growth.
It is now possible to use deep mutational scanning (22) to measure the functional effects of all single amino acid mutations to viral proteins (10, 23–27). However, the only HA for which such large-scale measurements have previously been made is from the highly laboratory-adapted A/WSN/1933 (H1N1) strain (10, 23, 24). Here, we measure the effects on viral growth in cell culture of all mutations to the HA of a recent human H3N2 strain. We show that these experimental measurements can help discriminate evolutionarily successful mutations from those found in strains that quickly die out. However, the utility of the experiments for understanding natural evolution depends on the similarity between the experimental and natural strains: Measurements made on an H1 HA are less informative for understanding the evolutionary fate of H3 viral strains.
Results
Deep Mutational Scanning of HA from a Recent Strain of Human H3N2 Influenza Virus.
We performed a deep mutational scan to measure the effects of all amino acid mutations to HA from the A/Perth/16/2009 (H3N2) strain on viral growth in cell culture. This strain was the H3N2 component of the influenza vaccine from 2010–2012 (28, 29). Relative to the consensus sequence for this HA in GenBank, we used a variant with two mutations that enhanced viral growth in cell culture, G78D and T212I (see SI Appendix, Fig. S1 and Dataset S1). The G78D mutation occurs at low frequency in natural H3N2 sequences, and T212 is a site where a mutation to Ala rose to fixation in human influenza in 2011.
We mutagenized the entire HA coding sequence at the codon level to create mutant plasmid libraries harboring an average of 1.4 codon mutations per clone (see SI Appendix, Fig. S2). We then generated mutant virus libraries from the mutant plasmids using a helper-virus system that enables efficient generation of complex influenza virus libraries (10) (Fig. 1A). These mutant viruses derived all their non-HA genes from the laboratory-adapted A/WSN/1933 strain. Using WSN/1933 for the non-HA genes reduces biosafety concerns and also helped increase viral titers. To further increase viral titers, we used MDCK-SIAT1 cells (Madin–Darby canine kidney cells overexpressing 2,6-sialyltransferase) (30) that we engineered to constitutively express TMPRSS2 (Transmembrane Protease, Serine 2), which cleaves the HA precursor to activate it for membrane fusion (31, 32).
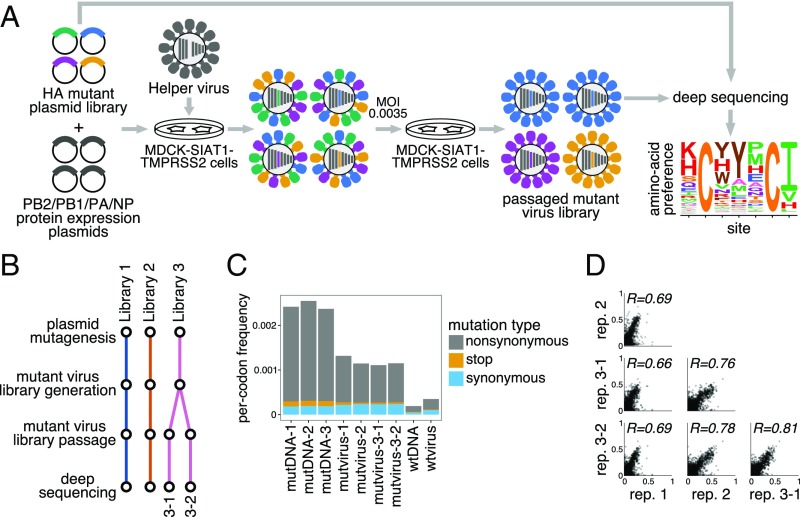
Fig. 1.
Deep mutational scanning of the Perth/2009 H3 HA. (A) We generated mutant virus libraries using a helper-virus approach (10) and passaged the libraries at low MOI to establish a genotype–phenotype linkage and to select for functional HA variants. Deep sequencing of the variants before and after selection allowed us to estimate each site’s amino acid preferences. (B) The experiments were performed in full biological triplicate. We also passaged and deep sequenced library 3 in duplicate. (C) Frequencies of nonsynonymous, stop, and synonymous mutations in the mutant plasmid DNA, the passaged mutant viruses, and wild-type DNA and virus controls. (D) The Pearson correlations among the amino acid preferences estimated in each replicate.
After generating the mutant virus libraries, we passaged them at low multiplicity of infection (MOI) in cell culture to create a genotype–phenotype link and select for functional HA variants (Fig. 1A). All experiments were completed in full biological triplicate (Fig. 1B). We also passaged and deep sequenced library 3 in duplicate (library 3-1 and 3-2) to gauge experimental noise within a single biological replicate. As a control to measure sequencing and mutational errors, we used the unmutated HA gene to generate and passage viruses carrying wild-type HA.
Deep sequencing of the initial plasmid mutant libraries and the passaged mutant viruses revealed selection for functional HA mutants. Specifically, stop codons were purged to 20% to 45% of their initial frequencies after correcting for error rates estimated by sequencing the wild-type controls (Fig. 1C). The incomplete purging of stop codons is likely because genetic complementation due to co-infection (33, 34) enabled the persistence of some virions with nonfunctional HAs. We also observed selection against many nonsynonymous mutations (Fig. 1C), with their frequencies falling to 30% to 40% of their initial values after error correction.
We next quantified the reproducibility of our deep mutational scanning across biological and technical replicates. We first used the deep sequencing data for each replicate to estimate the preference of each site in HA for all 20 amino acids as described in ref. 39. Because there are 566 residues in HA, there are distinct measurements [the 20 preferences at each site sum to 1 (39)]. The correlations of the amino acid preferences between pairs of replicates are shown in Fig. 1D. The biological replicates were well-correlated, with Pearson’s ranging from 0.69 to 0.78. Replicate 1 exhibited the weakest correlation with other replicates; this replicate also showed the weakest selection against stop and nonsynonymous mutations (Fig. 1C), perhaps indicating more experimental noise. The two technical replicates, 3-1 and 3-2, were only slightly more correlated than pairs of biological replicates, suggesting that bottlenecking of library diversity during viral passage contributes most of the experimental noise.
Our Measurements Are Consistent with Existing Knowledge About HA’s Evolution and Function.
How do the HA amino acid preferences measured in our experiments relate to the evolution of H3N2 influenza virus in nature? This question can be addressed by evaluating how well an experimentally informed codon substitution model (ExpCM) using our measurements describes H3N2 evolution compared with standard substitution models (35, 40). Table 1 shows that an ExpCM using the across-replicate average of our measurements greatly outperforms conventional substitution models. This result indicates that our experiments authentically capture some of the constraints on HA evolution. A substitution model in which the amino acid preferences were averaged across all sites (ExpCM, site average) performs no better than conventional substitution models, demonstrating that the reason that our measurements are informative is because they capture site-specific constraints. The relative rate of nonsynonymous to synonymous substitutions (dN/dS or ) is 1 for conventional substitution models (Table 1). However, the relative rate of nonsynonymous to synonymous substitutions after accounting for the amino acid preferences measured in our experiments ( for the ExpCM) is close to 1 (Table 1), indicating that most purifying selection against nonsynonymous substitutions is accounted for in the deep mutational scanning. The ExpCM stringency parameter (35) is 2.47 (Table 1), indicating that natural selection favors the same amino acids as the experiments but with greater stringency. Throughout the rest of this paper, we use the amino acid preferences rescaled (35, 40) by this stringency parameter. These rescaled preferences are shown in Fig. 2.
Table 1.
Substitution models informed by the experiments describe HA’s evolution better than traditional models
| Model | AIC | LnL | Stringency | |
| ExpCM | 0.0 | −8,441 | 2.47 | 0.91 |
| GY94 M5 | 2,094 | −9,482 | — | 0.36 (0.30, 0.84) |
| ExpCM, site average | 2,501 | −9,692 | 0.67 | 0.32 |
| GY94 M0 | 2,536 | −9,704 | — | 0.31 |
Shown are the maximum likelihood phylogenetic fits to an alignment of human H3N2 HAs using ExpCM (35), ExpCM in which the experimental measurements are averaged across sites (site average), and M0 and M5 versions of the Goldman–Yang (GY94) model (36). Models are compared by Akaike information criterion (AIC) (37) computed from the log likelihood (LnL) and number of model parameters. The ω parameter is dN/dS for the Goldman–Yang models and the relative dN/dS after accounting for the measurements for the ExpCM. For the M5 model, we give the mean followed by the shape and rate parameters of the gamma distribution over ω.
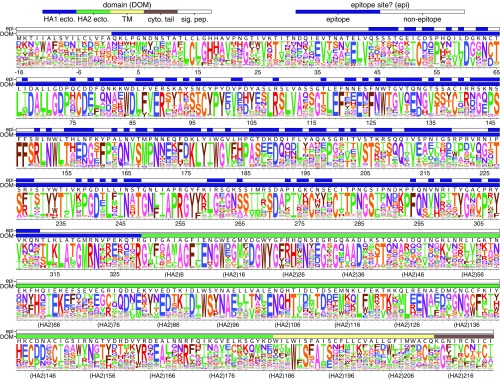
Fig. 2.
The site-specific amino acid preferences of the Perth/2009 HA measured in our experiments. The height of each letter is the preference for that amino acid, after taking the average over experimental replicates and rescaling (35) by the stringency parameter in Table 1. The sites are in H3 numbering. The top overlay bar indicates whether or not a site is in the set of epitope residues delineated in ref. 38. The bottom overlay bar indicates the HA domain (sig. pep., signal peptide; HA1 ecto., HA1 ectodomain; HA2 ecto., HA2 ectodomain; TM, transmembrane domain; cyto. tail, cytoplasmic tail). The letters directly above each logo stack indicate the wild-type amino acid at that site.
Examination of Fig. 2 reveals that the experimentally measured amino acid preferences generally agree with existing knowledge about HA’s structure and function. For instance, sites that form structurally important disulfide bridges (sites 52 and 277, 64 and 76, 97 and 139, 281 and 305, 14 and 137-HA2, 144-HA2 and 148-HA2) (43) strongly prefer cysteine. At residues involved in receptor binding, there are strong preferences for the amino acids that are known to be involved in binding sialic acid, such as Y98, D190, W153, and S228 (44–47). A positively charged amino acid at site 329 is important for cleavage of the HA0 precursor into the mature form (48), and this site strongly prefers arginine.
However, there are also some differences between the amino acid preferences measured in our experiments and amino acid frequencies in natural H3 HA sequences (see SI Appendix, Fig. S3). Most surprisingly, the start codon does not show a particularly strong preference for methionine (Fig. 2). We validated that a virus carrying a mutation at this site from methionine to lysine does in fact reach appreciable titers (see SI Appendix, Fig. S4), perhaps because of alternative translation-initiation at a downstream or upstream start site as has been described for other HAs (49). Our measurements also suggest mutational tolerance at some other sites that are relatively conserved among natural HAs, such as the N-linked glycosylation motifs near the beginning of HA1 and the transmembrane domain (Fig. 2). We validated that viruses with mutations to the glycosylation motifs at sites 22 or 38, or a site in the transmembrane domain, do in fact grow to high titers (SI Appendix, Figs. S5 and S6, respectively). The disparity between the relative conservation of these sites in nature and their mutational tolerance in our study could be because cell culture does not fully capture the constraints on HA function in nature or could be because these sites are not under strong immune pressure and so mutations at them are not positively selected in nature.
There Is Less Difference in Mutational Tolerance Between the HA Head and Stalk Domains for H3 than for H1.
Our experiments measure which amino acids are tolerated at each HA site under selection for viral growth. We can therefore use our experimentally measured amino acid preferences to calculate the inherent mutational tolerance of each site, which we quantify as the Shannon entropy of the rescaled preferences. In prior mutational studies of H1 HAs, the stalk domain was found to be substantially less mutationally tolerant than the globular head (10, 23, 24, 50).
We performed a similar analysis using our new data for the Perth/2009 H3 HA. Surprisingly, the head domain of the H3 HA is not more mutationally tolerant than the stalk domain (Fig. 3). Specifically, whereas solvent-exposed residues in the head domain are substantially more mutationally tolerant than those in the stalk domain for the WSN/1933 H1 HA, the trend is actually reversed for the Perth/2009 H3 HA (Fig. 3B). This difference between the relative mutational tolerances of the H1 and H3 HAs is robust to the cutoff used to define surface residues (see SI Appendix, Fig. S7). For instance, for the H3 HA, the short helix A in the stalk domain is as mutationally tolerant as many surface-exposed residues in the head domain—something that is not the case for the H1 HA. Helix A forms part of the epitope of many broadly neutralizing antistalk antibodies (51–53).
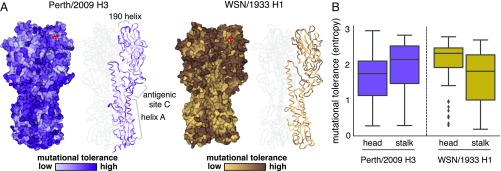
Fig. 3.
Mutational tolerance of each site in H3 and H1 HAs. (A) Mutational tolerance as measured in the current study is mapped onto the structure of the H3 trimer [Protein Data Bank (PDB) ID code 4O5N (41)]. Mutational tolerance of the WSN/1933 H1 HA as measured in ref. 10 is mapped onto the structure of the H1 trimer [PDB ID code 1RVX (42)]. Different color scales are used because measurements are comparable among sites within the same HA but not necessarily across HAs. Both trimers are shown in the same orientation. For each HA, the structure at Left shows a surface representation of the full trimer, while the structure at Right shows a ribbon representation of just one monomer. The sialic acid receptor is shown in red sticks. (B) The mutational tolerance of solvent-exposed residues in the head and stalk domains of the Perth/2009 H3 HA (purple) and WSN/1933 H1 HA (gold). Residues falling in between the two cysteines at sites 52 and 277 were defined as belonging to the head domain, while all other residues were defined as the stalk domain. A residue was classified as solvent exposed if its relative solvent accessibility was 0.2. The results are robust to the choice of solvent accessibility cutoff (see SI Appendix, Fig. S7). Note that the mutational tolerance values are not comparable between the two HAs but are comparable between domains of the same HA.
We also see high mutational tolerance in many of the known antigenic regions of H3 HA (54). For instance, antigenic region B is an immunodominant area, and many recent major antigenic drift mutations have occurred in this region (14, 15, 55). We find that the most distal portion of the globular head near the 190-helix, which is part of antigenic region B, is highly tolerant of mutations (Fig. 3A). Antigenic region C is also notably mutationally tolerant.
Many residues inside HA’s receptor binding pocket are known to be highly functionally constrained (45, 56), and our data indicate that these sites are relatively mutationally intolerant in both H3 and H1 HAs (Fig. 3A). In contrast, the residues surrounding the receptor binding pocket are fairly mutationally tolerant, which may contribute to the rapidity of influenza’s antigenic evolution, since mutations at these sites can have large effects on antigenicity (14, 54).
Our Measurements Can Help Distinguish Between Mutations That Reach Low and High Frequencies in Nature.
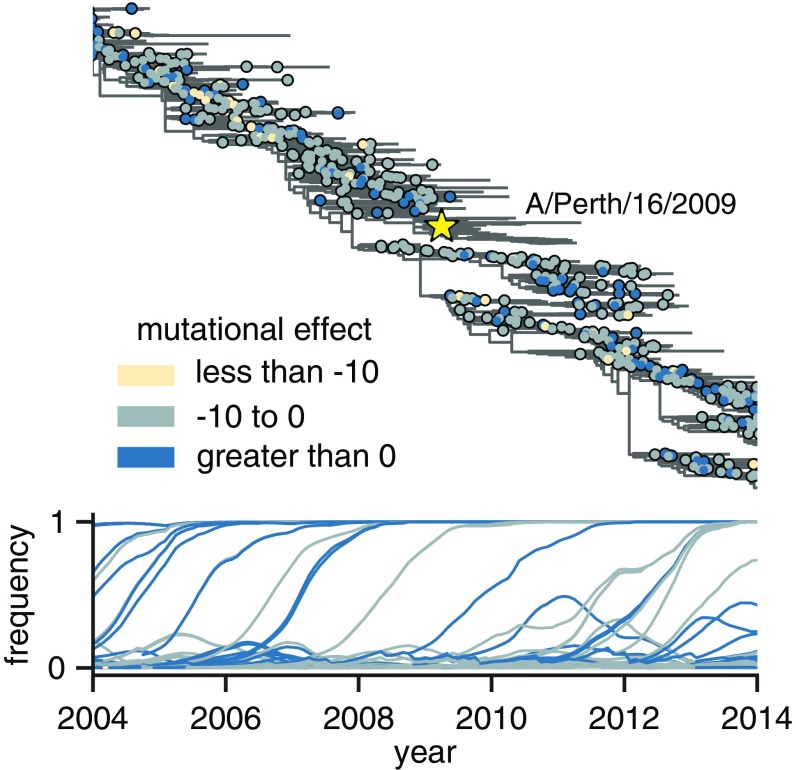
Mutations occurring in the H3N2 virus population experience widely varying evolutionary fates (Fig. 4). Some mutations appear, spread, and fix in the population, while others briefly circulate before disappearing. We take the maximum frequency reached by a mutation as a coarse indicator of its effect on fitness, since favorable mutations generally reach higher frequencies than unfavorable ones (57). Here, we follow the population genetic definition of “mutation” and track the outcome of each individual mutation event; for example, although R142G occurs multiple times on the phylogeny, we track each of these mutations occurring on different backgrounds separately. As such, each mutation is shown as a separate circle on a separate branch in Fig. 4. However, because multiple mutations on the same phylogeny branch cannot be disentangled, when multiple mutations occurred on a single branch, we assigned a single mutational effect based on the sum of effects of each mutation.
Fig. 4.
Frequency trajectories of individual mutations and their relation to the experimentally measured effects of these mutations. Top shows the subset of the full H3N2 HA tree in SI Appendix, Fig. S8 from 2004 to 2014. Circles indicate individual amino acid mutations and are colored according to the mutational effect measured in our deep mutational scanning (negative values indicate mutations measured to be deleterious to viral growth). The Perth/2009 strain is labeled with a star, and nodes in the clade containing the Perth/2009 strain were excluded from our analyses. Bottom shows the frequency trajectory of each mutation, with trajectories colored according to the mutational effects as in Top. It is clear that most mutations that reach high frequency are measured to be relatively favorable in our experiments. SI Appendix, Fig. S11 shows a similar layout but colors mutations by whether they are in HA’s head or stalk domain.
After annotating mutations and their frequencies on the phylogeny in this way (Fig. 4), it is visually obvious that there are relatively few circulating mutations that we measure to be strongly deleterious—and that such deleterious mutations rarely reach high frequency when they do occur.
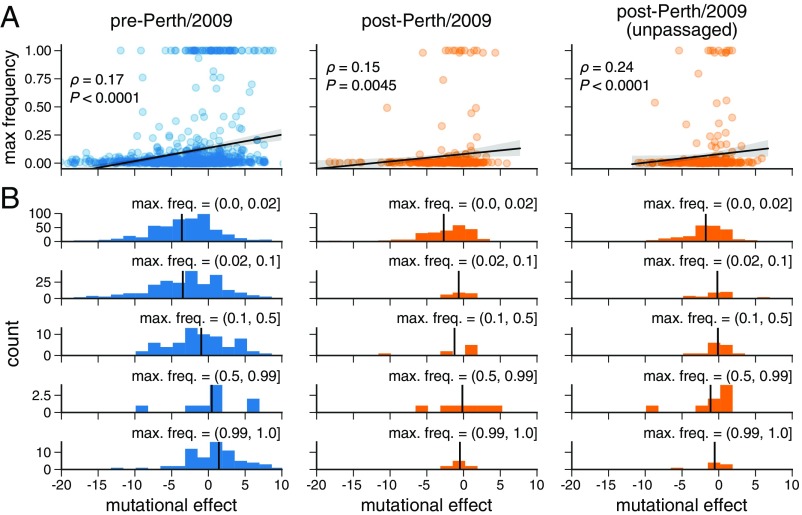
We next sought to quantify the correlation between a mutation’s experimentally measured effect and the maximum frequency it attained during natural evolution. To calculate a given mutation’s effect, we simply took the logarithm of the ratio of the preferences for the mutant and wild-type amino acids at that site. To minimize effects related to the genetic background of the strain used in the experiment, we excluded mutations closely related to the experimental strain itself and partitioned the remaining mutations into 1,022 mutations predating and 299 mutations postdating the Perth/2009 strain (see SI Appendix, Fig. S8). We additionally excluded mutations from the post-Perth partition that were sampled in 2014 or after, since these mutations have not had enough time for their evolutionary fates to be fully resolved. We used these pre-Perth and post-Perth partitions to test the utility of our measurements for both post hoc and prospective analyses, respectively. We quantified the relationship between mutational effects and maximum mutation frequencies in the H3N2 phylogeny via Spearman rank correlation (Fig. 5A). In both pre-Perth and post-Perth time periods, we found a modest but statistically significant relationship between mutational effect and maximum mutation frequency (pre-Perth , post-Perth ). The similar effect sizes for both the pre- and post-Perth partitions show that our experimental measurements can help explain the evolutionary fates of mutations in strains that postdate the experimentally studied strain as well as to retrospectively analyze mutations that precede the experimental strain.
Fig. 5.
Experimental measurements are informative about the evolutionary fate of viral mutations. (A) Correlation between the effects of mutations as measured in our deep mutational scanning of the Perth/2009 HA and the maximum frequency reached by these mutations in nature. The plots show Spearman and an empirical value representing the proportion of 10,000 permutations of the experimental measurements for which the permuted was greater than or equal to the observed . (B) The distribution of mutational effects partitioned by maximum mutation frequency. The vertical black line shows the mean mutation effect for each category. The analysis is performed separately for pre-Perth/2009, post-Perth/2009, and unpassaged isolates from the post-Perth/2009 partitions of the tree (see SI Appendix, Fig. S8).
Many of the HAs in sequence databases are from viral isolates that were passaged in cell culture or eggs, which can cause laboratory-adaptation mutations that confound evolutionary analyses (58). To check that our results were robust to such laboratory-adaptation mutations, we repeated our analysis using only HA sequences derived from viruses that had not been passaged in the laboratory. Because sequencing of unpassaged primary isolates has only recently become commonplace, we could only perform this analysis for the post-Perth partition of the phylogenetic tree. Fig. 5A shows that the correlation between our measured mutational effects and the maximum frequency was even stronger for mutations from unpassaged viral isolates ().
The trends in Fig. 5A are most strongly driven by the behavior of substantially deleterious mutations. We investigated this further by partitioning mutations into those that reach low, medium, and high frequencies and those that fix in the population (Fig. 5B). The mutations that reach higher frequencies have a more favorable mean effect. Mutations measured to be substantially deleterious almost never reach high frequency. Overall, these results demonstrate that measurements of how mutations affect viral growth in cell culture are informative for understanding the fates of these mutations in nature: In particular, if a mutation is measurably deleterious to viral growth, that mutation is unlikely to prosper in nature.
Measurements Made on an H1 HA Are Less Informative for Understanding the Evolution of H3 Influenza.
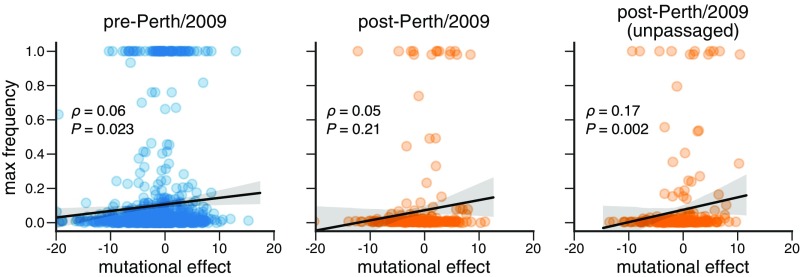
To determine how broadly experimental measurements can be generalized across HAs, we repeated the foregoing analysis of H3N2 mutation frequencies but using mutational effects measured in our prior deep mutational scanning of the WSN/1933 H1 HA (10) (see SI Appendix, Fig. S9), which is highly diverged from the Perth/2009 H3 HA (the two HAs only have 42% protein sequence identity). Fig. 6 shows that the correlations between the H1 experimental measurements and the maximum frequency that mutations reach during H3N2 viral evolution are consistently weaker than those using H3 experimental measurements (compare Fig. 6 to Fig. 5A). Therefore, the utility of an experiment for understanding natural evolution degrades as the experimental sequence becomes more diverged from the natural sequences that are being studied.
Fig. 6.
Experimental measurements on an H1 HA are less informative about the evolutionary fate of H3N2 mutations. This figure repeats the analysis of the H3N2 mutation frequencies in Fig. 5A but uses the deep mutational scanning data for an H1 HA as measured in ref. 10. SI Appendix, Fig. S10 shows the histograms comparable to those in Fig. 5B. The empirical value represents the result of 1,000 permutations.
There Are Large Differences Between H3 and H1 HAs in the Amino Acid Preferences of Many Sites.
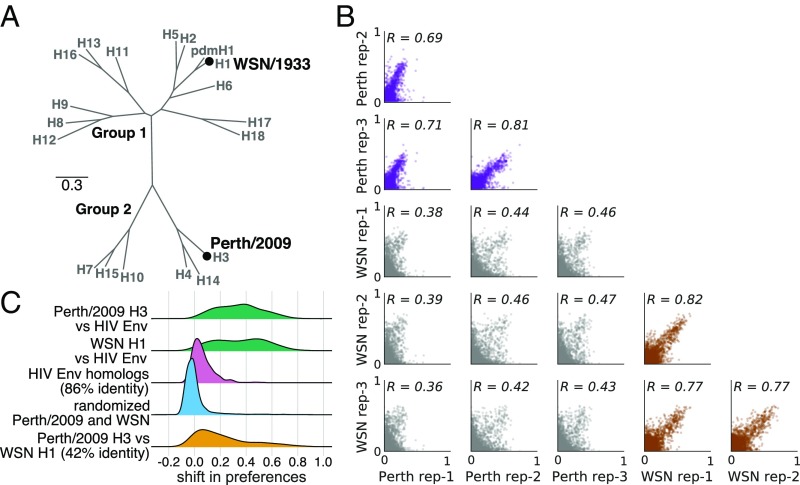
An obvious hypothesis for why the H1 deep mutational scanning is less useful for understanding the evolution of H3N2 influenza viruses is that the effect of the same mutation is often different between these two HA subtypes. To determine if this is the case, we examined how much the amino acid preferences of homologous sites have shifted between H3 and H1 HAs. Prior experiments have found only modest shifts in amino acid preferences between two variants of influenza nucleoprotein with 94% amino acid identity (59) and variants of HIV envelope (Env) with 86% amino acid identity (27). However, the H1 and H3 HAs are far more diverged, with only 42% amino acid identity (Fig. 7A). One simple way to investigate the extent of shifts in amino acid preferences is to correlate measurements from independent deep mutational scanning replicates on H1 and H3 HAs. Fig. 7B shows that replicate measurements on the same HA variant are more correlated than those on different HA variants.
Fig. 7.
There are large shifts in the effects of mutations between H1 and H3 HAs. (A) Phylogenetic tree of HA subtypes, with the WSN/1933 H1 and Perth/2009 H3 HAs labeled. (B) All pairwise correlations of the amino acid preferences measured in the three individual deep mutational scanning replicates in the current study and the three replicates in prior deep mutational scanning of the H1 HA (10). Comparisons between H3 replicates are in purple, those between H1 replicates are in brown, and those across H1 and H3 replicates are in gray. indicates the Pearson correlation coefficient. (C) We calculated the shift in amino acid preferences at each site between H3 and H1 HAs using the method in ref. 27 and plotted the distribution of shifts for all sites. The shifts between H3 and H1 (yellow) are much larger than the null distribution (blue) expected if all differences are due to experimental noise. The shifts are also much larger than those previously observed between two variants of HIV Env that share 86% amino acid identity (pink). However, the shifts between H3 and H1 are less than the differences between HA and HIV Env (green).
To more rigorously quantify shifts in amino acid preferences after correcting for experimental noise, we used the statistical approach in refs. 27 and 59. Fig. 7C shows the distribution of shifts in amino acid preferences between H3 and H1 HAs after correcting for experimental noise. Although some sites have small shifts near zero, many sites have large shifts. These shifts between H3 and H1 are much larger than expected from the null distribution that would be observed purely from experimental noise. They are also much larger than the shifts previously observed between two HIV Envs with 86% amino acid identity (27). However, the typical shift between H3 and H1 is still smaller than that observed when comparing HA to the nonhomologous HIV Env protein. Therefore, there are very substantial shifts in mutational effects between highly diverged HA homologs, although the effects of mutations remain more similar than for nonhomologous proteins.
Properties Associated with the Shifts in Amino Acid Preferences Between H3 and H1 HAs.
What features distinguish the sites with shifted amino acid preferences between H3 and H1 HAs? The sites of large shifts do not obviously localize to one specific region of HA’s structure (Fig. 8A). However, at the domain level, sites in HA’s stalk tend to have smaller shifts than sites in HA’s globular head (Fig. 8B). The HA stalk domain is also more conserved in sequence (60), suggesting that conservation of amino acid sequence is correlated with conservation of amino acid preferences. Consistent with this idea, sites that are absolutely conserved across all 18 HA subtypes are significantly less shifted than sites that are variable across HA subtypes (Fig. 8B). Presumably these sites are under consistent functional constraint across all HAs.
Fig. 8.
Sites with strongly shifted amino acid preferences between H3 and H1 HAs. (A) The shift in amino acid preferences between the H3 and H1 HA at each site as calculated in Fig. 7C is mapped onto the structure of the H3 HA. (B) Amino acid preferences of sites in the stalk domain are less shifted than those in the head domain. Sites absolutely conserved in all 18 HA subtypes are less shifted than other sites. Sites with one amino acid identity in the clade containing H1, H2, H5, and H6 and another identity in the clade containing H3, H4, and H14 are more shifted than other sites. (C) Sites 107 and 75(HA2) help determine the different orientation of the globular head domain in H1 versus H3 HAs. These sites are shown in spheres on the structure of H1 and H3 and colored as in A, and the experimentally measured amino acid preferences in the H1 and H3 HAs are shown. One monomer is in dark gray, while the HA1 domain of the neighboring monomer is in lighter gray.
Despite their high sequence divergence, H1 and H3 adopt very similar protein folds (61, 62). However, there are differences in the rotation and upward translation of the globular head subdomains relative to the central stalk domain among different HA subtypes (61, 62). Previous work has defined clades of structurally related HA subtypes (61, 62). One such clade includes H1, H2, H5, and H6, whereas another clade includes H3, H4, and H14 HAs (Fig. 7A). Sites that are conserved at different amino acid identities in these two clades tend to have exceptionally large shifts in amino acid preferences (Fig. 8B). The clade containing H1 has an upward translation of the globular head relative to the clade containing H3. This structural shift has been attributed largely to the interaction between sites 107 and 75(HA2) (61, 62). Specifically, the clade containing H1 has a taller turn in the interhelical loop connecting helix A and helix B in the stalk domain, and this tall turn is stabilized by a hydrogen bond between Glu-107 and Lys-75(HA2) (Fig. 8C). In deep mutational scanning of the H1 HA, site 107 has a high preference for Glu and 75(HA2) strongly prefers positively charged Lys and Arg. In contrast, the interhelical loop in H3 HA makes a sharper and shorter turn that is facilitated by a Gly at 75(HA2). In the deep mutational scanning of the Perth/2009 H3 HA, site 75(HA2) prefers Gly and to a lesser extent Val, while site 107 is fairly tolerant of mutations. Therefore, some of the shifts in HA amino acid preferences can be rationalized in terms of changes in HA structure.
Discussion
We have measured the effects of all possible single amino acid mutations to the Perth/2009 H3 HA on viral growth in cell culture and demonstrated that these measurements have some value for understanding the evolutionary fate of these mutations in nature. Specifically, mutations measured to be more beneficial for viral growth tend to reach higher frequencies in nature than mutations measured to be more deleterious for viral growth. The fact that our experiments can help identify evolutionarily successful mutations suggests that they might inform evolutionary forecasting. In their landmark paper introducing predictive viral fitness models that accounted for both antigenic and nonantigenic mutations, Łuksza and Lässig (9) noted that the models could in principle be improved by integrating “diverse genotypic and phenotypic data” that more realistically represented the effects of specific mutations. Our work suggests that deep mutational scanning may be able to provide such data.
It is important to emphasize that measurements of viral growth in cell culture do not represent true fitness in nature. Indeed, a vast amount of work in virology has chronicled the ways in which experiments can select for laboratory artifacts or fail to capture important pressures that are relevant in nature (63–66). As an example, although we identified G78D as favorable for viral growth in cell culture, this mutation never fixes in nature. Mutations in viral genes other than HA are also important in determining strain success (67, 68). Given these caveats, it might seem surprising that measuring viral growth in cell culture can be informative about the success of viral strains in nature. However, before our work, there were no comprehensive studies of the functional effects of mutations to H3 HA on any property that even resembled viral fitness in nature, and modeling work has either omitted the nonantigenic effects of mutations (11–13) or assumed that all nonepitope mutations had equivalent deleterious effects (9). The strength of our measurements are not that they perfectly capture fitness in nature but that they are systematic and quantitative—and so represent an improvement over no information at all. We suspect that performing similar experiments using more realistic and complex selections (e.g., ferrets or primary human airway cultures) might further improve their utility and possibly their generalizability to more divergent strains.
We measured the effects of all single amino acid mutations to a specific HA and then generalized these measurements to other H3N2 HAs from a 50-y timespan. These generalizations will only be valid to the extent that the effects of mutations are conserved during HA’s evolution. Extensive prior work has shown that epistasis can shift the effects of mutations as proteins evolve (69–75). Our work suggests that measurements on a HA from a single human H3N2 viral strain can be usefully generalized to at least some extent across the entire evolutionary history of human H3N2 HA. On the other hand, when we compared our measurements for an H3 HA to prior measurements on H1 HA, we found substantial shifts at many sites—much greater than those observed in prior protein-wide comparisons of more closely related homologs (27, 59). Further investigation of how mutational effects shift as proteins diverge will be important for determining how broadly any given experiment can be generalized when attempting to make evolutionary forecasts.
Our work did not characterize the antigenic effects of mutations, which also play an important role in determining strain success in nature (13, 14). However, our basic selection and deep-sequencing approach can be harnessed to completely map how mutations affect antibody recognition (76, 77). But so far, experiments using this approach have not examined antibodies or sera that are relevant to driving the evolution of H3N2 influenza (76, 77) or have used relevant sera but examined a noncomprehensive set of mutations (16). Future experiments that completely map how HA mutations affect recognition by human sera seem likely to be especially fruitful for informing viral forecasting.
Materials and Methods
Data and Computer Code.
Deep sequencing data are available from the Sequence Read Archive under BioSample accession nos. SAMN08102609 and SAMN08102610. Computer code used to analyze the data are at https://github.com/jbloomlab/Perth2009-DMS-Manuscript.
HA Numbering.
Sites are in H3 numbering, with the signal peptide in negative numbers, HA1 in plain numbers, and HA2 denoted with “(HA2).” Sequential 1, 2, ... numbering of the Perth/2009 HA can be converted to H3 numbering by subtracting 16 for the HA1 subunit and subtracting 345 for the HA2 subunit.
Creation of MDCK–SIAT1–TMPRSS2 Cells.
When growing influenza virus in cell culture, trypsin is normally added to cleave HA into its mature form. To obviate the need for trypsin, we engineered MDCK–SIAT1 cells and MDCK–SIAT1–CMV–PB1 (78) cells to constitutively express the TMPRSS2 protease, which cleaves and activates HA in the human airways (31, 32). The human TMPRSS2 cDNA ORF was ordered from OriGene (NM_005656) and cloned into a pHAGE2 lentiviral vector under an EF1-Int promoter followed by an IRES driving expression of mCherry to create plasmid pHAGE2–EF1aInt–TMPRSS2–IRES–mCherry-W. We used the lentiviral vector to transduce MDCK–SIAT1 or MDCK–SIAT1–CMV–PB1 cells and sorted an intermediate mCherry-positive population by flow cytometry. We refer to the sorted bulk population as MDCK–SIAT1–TMPRSS2 cells or MDCK–SIAT1–CMV–PB1–TMPRSS2 cells. There is no selectable marker for the TMPRSS2; however, we maintain the cells at low passage number and have seen no indication that they lose their ability to support the growth of viruses with H3 HAs in the absence of exogenous trypsin.
Generation of HA Codon Mutant Plasmid Libraries.
HA and NA genes for the Perth/2009 viral strain were cloned from recombinant virus obtained from BEI Resources (NR-41803) into the pHW2000 (79) influenza reverse-genetics plasmids to create pHW-Perth09-HA and pHW-Perth09-NA.
We initially created a virus with the HA and NA from Perth/2009 and internal genes from WSN/1933 and passaged it in cell culture to test its genetic stability. To generate this virus, we transfected a coculture of 293T and MDCK–SIAT1–TMPRSS2 in D10 media (DMEM, supplemented with 10% heat-inactivated FBS, or fetal bovine serum, 2 mM L-glutamine, 100 U of penicillin per milliliter, and 100 g of streptomycin per milliliter) with equal amounts of pHW-Perth09-HA, pHW-Perth09-NA, the pHW18 series of plasmids (79) for all non-HA/NA viral genes, and pHAGE2–EF1aInt–TMPRSS2–IRES-mCherry-W. The next day, we changed the media to influenza growth media (IGM, consisting of Opti-MEM supplemented with 0.01% heat-inactivated FBS, 0.3% BSA, 100 U of penicillin per milliliter, 100 g of streptomycin per milliliter, and 100 g of calcium chloride per milliliter; no trypsin was added since there was TMPRSS2) and then collected the viral supernatant at 72 h posttransfection. This viral supernatant was blind passaged in MDCK–SIAT1–TMPRSS2 a total of six additional times. We isolated viral RNA from these passaged viruses and sequenced the HA gene. The passaged HA had two mutations, G78D and T212I, which enhanced viral growth as shown in SI Appendix, Fig. S1. The HA with these two mutations was cloned into pHW2000 (79) and pICR2 (80) to create pHW-Perth09-HA-G78D-T212I and pICR2-Perth09-HA-G78D-T212I. For all subsequent experiments, we used viruses with the HA containing these two mutations to improve titers and viral genetic stability, and this is the HA that we refer to as Perth/2009. We used all non-HA genes (including NA) from WSN/1933 to help increase titers and reduce biosafety concerns.
The codon-mutant libraries were generated using the approach in ref. 81 with the modifications in ref. 82. See SI Appendix, Supplementary Text for full details.
Generation and Passaging of Mutant Viruses.
The mutant virus libraries were generated using the helper-virus approach described in ref. 10 with several modifications, most notably the cell line used. Briefly, we transfected MDCK–SIAT1–TMPRSS2 cells in suspension with 937.5 ng each of four protein expression plasmids encoding the ribonucleoprotein complex (HDM–Nan95–PA, HDM–Nan95–PB1, HDM–Nan95–PB2, and HDM–Aichi68–NP) (71) and 1,250 ng of one of the three pICR2-mutant-HA libraries (or the wild-type control) using Lipofectamine 3000 (ThermoFisher L3000008). We allowed the transfected cells to adhere in six-well plates and 4 h later changed the media to D10 media. Eighteen hours after transfection, we infected the cells with the WSN/1933 HA-deficient helper virus (10) by preparing an inoculum of 500 per microliter of helper virus (as computed on HA-expressing cells) in IGM, aspirating the D10 media from the cells, and adding 2 mL of the helper-virus inoculum to each well. After 3 h, we changed the media to fresh IGM. At 24 h after helper-virus infection, we harvested the viral supernatants for each replicate, froze aliquots at –C, and titered them in MDCK–SIAT1–TMPRSS2 cells. The titers were 92, 536, 536, and 734 per microliter for the three library replicates and the wild-type control, respectively.
We passaged of the transfection supernatants at an MOI of 0.0035 per cell. To do this, we plated MDCK–SIAT1–TMPRSS2 cells per dish in fifteen 15-cm dishes in D10 media and allowed the cells to grow for 24 h, at which time they were at cells per dish. We replaced the media in each dish with 25 mL of an inoculum of 2.5 of virus per microliter in IGM. Three hours postinfection, we replaced the inoculum with fresh IGM for replicates 1, 2, and 3-2. We did not perform a media change for replicate 3-1. As can be seen in Fig. 1D, the media change does not appear to have a substantial effect, as replicate 3-1 looks comparable to the other replicates. We collected viral supernatant for sequencing 48 h postinfection.
Barcoded Subamplicon Sequencing.
To extract viral RNA from the three replicate HA virus libraries and the wild-type HA virus, we clarified the viral supernatant by centrifuging at 2,000 × g for 5 min, then ultracentrifuged 24 mL of the clarified supernatant at 22,000 rpm for 1.5 h at C in a Beckman Coulter SW28 rotor, and extracted RNA using the Qiagen RNeasy Mini Kit by resuspending the viral pellet in 400 L of buffer RLT supplemented with -mercaptoethanol, pipetting 30 times, transferring the liquid to a microcentrifuge tube, adding 600 L 70% ethanol, and proceeding according to the manufacturer’s instructions. The HA gene was reverse-transcribed with AccuScript Reverse Transcriptase (Agilent 200820) using primers P09-HA-For (5′-AGCAAAAGCAGGGGATAATTCTATTAATC-3′) and P09-HA-Rev (5′-AGTAGAAACAAGGGTGTTTTTAATTACTAATACAC-3′). The barcoded-subamplicon library prep and deep sequencing were then performed as in ref. 10. See SI Appendix, Supplementary Text for full details.
Analysis of Deep Sequencing Data.
We used the dms_tools2 software package (39) (https://github.com/jbloomlab/dms_tools2, version 2.2.5) to analyze the deep sequencing data. The amino acid preferences are provided in Dataset S3. Computer code and detailed plots about read depth and other quality control metrics are at https://github.com/jbloomlab/Perth2009-DMS-Manuscript/blob/master/analysis_code/analysis_notebook.ipynb.
Phylogenetic Model Comparison and Stringency Parameter.
Phylogenetic model comparisons and fitting of a stringency parameter were performed using phydms as described in ref. 35. See SI Appendix, Supplementary Text for full details.
Shannon Entropy and Relative Solvent Accessibility.
We calculated Shannon entropy for site as , where is the preference for amino acid at site .
We quantified the absolute solvent accessibility of each site of the H3 HA [PDB ID code 4O5N (41)] or the H1 HA [PDB ID code 1RVX (42)] structure using DSSP (Define Secondary Structure of Proteins) (83). We then normalized to a relative solvent accessibility using the absolute accessibilities in ref. 84.
Quantification of Mutational Effects.
The effect of mutating site from amino acid to was quantified as
| [1] |
where and are the rescaled preferences for amino acids or at site as shown in Fig. 2. The WSN/1933 H1 HA amino acid preferences are the replicate-average values reported in ref. 10, rescaled by a stringency parameter of 2.05 (see https://github.com/jbloomlab/dms_tools2/blob/master/examples/Doud2016/analysis_notebook.ipynb).
H3N2 Phylogenetic Tree and Maximum Mutation Frequencies.
The pylogenetic tree was generated using Nextstrain’s augur pipeline (85), and ancestral state reconstruction and branch length timing were performed with TreeTime (86). Frequency trajectories of mutations were estimated following Nextstrain’s augur pipeline as first implemented in Nextflu (87). See SI Appendix, Supplementary Text for full details.
Analysis of Mutational Shifts.
We compared the preferences for the Perth/2009 H3 and WSN/1933 H1 HAs using the approach in ref. 27. See SI Appendix, Supplementary Text for full details.
Validation of Individual Point Mutants.
To validate the viral growth of Perth/2009 HA point mutants M(-16)K, C52A, C52C, T24F, T40V, S287A, and C199(HA2)K, we examined the supernatant titers of each of these variants after reverse-genetics generation in the context of PB1flank-GFP viruses (78, 88). See SI Appendix, Supplementary Text for full details.
Supplementary Material
Acknowledgments
We thank Sarah Hilton, Hugh Haddox, and Sidney Bell for helpful discussions about data analysis and Richard Neher for sharing analysis code and providing helpful comments on the manuscript. We thank the Fred Hutch Genomics Core for performing the Illumina deep sequencing. This work was supported by NIH National Institute of Allergy and Infectious Diseases (NIAID) Grants R01 AI127893 (to J.D.B. and T.B.) and U19 AI117891 (to T.B.). J.M.L. was supported in part by the Center for Inference and Dynamics of Infectious Diseases (CIDID), which is funded by NIH National Institute of General Medical Sciences (NIGMS) Grant U54GM111274. The research of J.D.B. is supported in part by a Faculty Scholar grant from the Howard Hughes Medical Institute and the Simons Foundation and a Burroughs Wellcome Young Investigator in the Pathogenesis of Infectious Diseases grant. T.B. is a Pew Biomedical Scholar and is supported by NIH Grant R35 GM119774.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Deep sequencing data are available from the Sequence Read Archive under BioSample accessions nos. SAMN08102609 and SAMN08102610. Computer code used to analyze the data and produce the results in the paper are on GitHub at https://github.com/jbloomlab/Perth2009-DMS-Manuscript.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1806133115/-/DCSupplemental.
References
- 1.Fitch WM, Bush RM, Bender CA, Cox NJ. Long term trends in the evolution of H(3) HA1 human influenza type A. Proc Natl Acad Sci USA. 1997;94:7712–7718. doi: 10.1073/pnas.94.15.7712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhatt S, Holmes EC, Pybus OG. The genomic rate of molecular adaptation of the human influenza A virus. Mol Biol Evol. 2011;28:2443–2451. doi: 10.1093/molbev/msr044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith DJ, et al. Mapping the antigenic and genetic evolution of influenza virus. Science. 2004;305:371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- 4.Bedford T, Cobey S, Pascual M. Strength and tempo of selection revealed in viral gene genealogies. BMC Evol Biol. 2011;11:220. doi: 10.1186/1471-2148-11-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strelkowa N, Lässig M. Clonal interference in the evolution of influenza. Genetics. 2012;192:671–682. doi: 10.1534/genetics.112.143396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neher RA, Russell CA, Shraiman BI. Predicting evolution from the shape of genealogical trees. eLife. 2014;3:e03568. doi: 10.7554/eLife.03568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koelle K, Rasmussen DA. The effects of a deleterious mutation load on patterns of influenza A/H3N2’s antigenic evolution in humans. eLife. 2015;4:e07361. doi: 10.7554/eLife.07361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bedford T, et al. Global circulation patterns of seasonal influenza viruses vary with antigenic drift. Nature. 2015;523:217–220. doi: 10.1038/nature14460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Łuksza M, Lässig M. A predictive fitness model for influenza. Nature. 2014;507:57–61. doi: 10.1038/nature13087. [DOI] [PubMed] [Google Scholar]
- 10.Doud MB, Bloom JD. Accurate measurement of the effects of all amino-acid mutations to influenza hemagglutinin. Viruses. 2016;8:155. doi: 10.3390/v8060155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun H, et al. Using sequence data to infer the antigenicity of influenza virus. MBio. 2013;4:e00230–13. doi: 10.1128/mBio.00230-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harvey WT, et al. Identification of low- and high-impact hemagglutinin amino acid substitutions that drive antigenic drift of influenza A (H1N1) viruses. PLoS Pathog. 2016;12:e1005526. doi: 10.1371/journal.ppat.1005526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neher RA, Bedford T, Daniels RS, Russell CA, Shraiman BI. Prediction, dynamics, and visualization of antigenic phenotypes of seasonal influenza viruses. Proc Natl Acad Sci USA. 2016;113:E1701–E1709. doi: 10.1073/pnas.1525578113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koel BF, et al. Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science. 2013;342:976–979. doi: 10.1126/science.1244730. [DOI] [PubMed] [Google Scholar]
- 15.Chambers BS, Parkhouse K, Ross TM, Alby K, Hensley SE. Identification of hemagglutinin residues responsible for H3N2 antigenic drift during the 2014–2015 influenza season. Cell Rep. 2015;12:1–6. doi: 10.1016/j.celrep.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li C, et al. Selection of antigenically advanced variants of seasonal influenza viruses. Nat Microbiol. 2016;1:16058. doi: 10.1038/nmicrobiol.2016.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pybus OG, et al. Phylogenetic evidence for deleterious mutation load in RNA viruses and its contribution to viral evolution. Mol Biol Evol. 2007;24:845–852. doi: 10.1093/molbev/msm001. [DOI] [PubMed] [Google Scholar]
- 18.Holland J, et al. Rapid evolution of RNA genomes. Science. 1982;215:1577–1585. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- 19.Steinhauer D, Holland J. Rapid evolution of RNA viruses. Annu Rev Microbiol. 1987;41:409–431. doi: 10.1146/annurev.mi.41.100187.002205. [DOI] [PubMed] [Google Scholar]
- 20.Lauring AS, Andino R. Quasispecies theory and the behavior of RNA viruses. PLoS Pathog. 2010;6:e1001005. doi: 10.1371/journal.ppat.1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boni MF, Zhou Y, Taubenberger JK, Holmes EC. Homologous recombination is very rare or absent in human influenza A virus. J Virol. 2008;82:4807–4811. doi: 10.1128/JVI.02683-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fowler DM, Fields S. Deep mutational scanning: A new style of protein science. Nat Methods. 2014;11:801–807. doi: 10.1038/nmeth.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thyagarajan B, Bloom JD. The inherent mutational tolerance and antigenic evolvability of influenza hemagglutinin. eLife. 2014;3:e03300. doi: 10.7554/eLife.03300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu NC, et al. High-throughput profiling of influenza A virus hemagglutinin gene at single-nucleotide resolution. Sci Rep. 2014;4:4942. doi: 10.1038/srep04942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haddox HK, Dingens AS, Bloom JD. Experimental estimation of the effects of all amino-acid mutations to HIV’s envelope protein on viral replication in cell culture. PLoS Pathog. 2016;12:e1006114. doi: 10.1371/journal.ppat.1006114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qi H, Wu NC, Du Y, Wu TT, Sun R. High-resolution genetic profile of viral genomes: Why it matters. Curr Opin Virol. 2015;14:62–70. doi: 10.1016/j.coviro.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 27.Haddox HK, Dingens AS, Hilton SK, Overbaugh J, Bloom JD. Mapping mutational effects along the evolutionary landscape of HIV envelope. eLife. 2018;7:e34420. doi: 10.7554/eLife.34420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.WHO 2010 Recommended viruses for influenza vaccines for use in the 2010-2011 northern hemisphere influenza season. www.who.int/influenza/vaccines/virus/recommendations/201002_Recommendation.pdf?ua=1. Accessed April 9, 2018.
- 29.WHO 2011 Recommended composition of influenza virus vaccines for use in the 2011-2012 northern hemisphere influenza season. www.who.int/influenza/vaccines/2011_02_recommendation.pdf?ua=1. Accessed April 9, 2018.
- 30.Matrosovich M, Matrosovich T, Carr J, Roberts NA, Klenk HD. Overexpression of the -2, 6-sialyltransferase in mdck cells increases influenza virus sensitivity to neuraminidase inhibitors. J Virol. 2003;77:8418–8425. doi: 10.1128/JVI.77.15.8418-8425.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Böttcher E, et al. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J Virol. 2006;80:9896–9898. doi: 10.1128/JVI.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Böttcher-Friebertshäuser E, et al. Cleavage of influenza virus hemagglutinin by airway proteases TMPRSS2 and HAT differs in subcellular localization and susceptibility to protease inhibitors. J Virol. 2010;11:5605–5614. doi: 10.1128/JVI.00140-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marshall N, Priyamvada L, Ende Z, Steel J, Lowen AC. Influenza virus reassortment occurs with high frequency in the absence of segment mismatch. PLoS Pathog. 2013;9:e1003421. doi: 10.1371/journal.ppat.1003421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brooke CB, et al. Most influenza a virions fail to express at least one essential viral protein. J Virol. 2013;87:3155–3162. doi: 10.1128/JVI.02284-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hilton SK, Doud MB, Bloom JD. phydms: Software for phylogenetic analyses informed by deep mutational scanning. PeerJ. 2017;5:e3657. doi: 10.7717/peerj.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Z, Nielsen R, Goldman N, Pedersen AMK. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Posada D, Buckley TR. Model selection and model averaging in phylogenetics: Advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst Biol. 2004;53:793–808. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- 38.Wolf Y, Viboud C, Holmes E, Koonin E, Lipman D. Long intervals of stasis punctuated by bursts of positive selection in the seasonal evolution of influenza A virus. Biol Direct. 2006;1:34. doi: 10.1186/1745-6150-1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bloom JD. Software for the analysis and visualization of deep mutational scanning data. BMC Bioinformatics. 2015;16:168. doi: 10.1186/s12859-015-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bloom JD. Identification of positive selection in genes is greatly improved by using experimentally informed site-specific models. Biol Direct. 2017;12:1. doi: 10.1186/s13062-016-0172-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee PS, et al. Receptor mimicry by antibody F045-092 facilitates universal binding to the H3 subtype of influenza virus. Nat Commun. 2014;5:3614. doi: 10.1038/ncomms4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gamblin S, et al. The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science. 2004;303:1838–1842. doi: 10.1126/science.1093155. [DOI] [PubMed] [Google Scholar]
- 43.Waterfield M, Scrace G, Skehel J. Disulphide bonds of haemagglutinin of Asian influenza virus. Nature. 1981;289:422–424. doi: 10.1038/289422a0. [DOI] [PubMed] [Google Scholar]
- 44.Weis W, et al. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature. 1988;333:426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- 45.Martin J, et al. Studies of the binding properties of influenza hemagglutinin receptor-site mutants. Virology. 1998;241:101–111. doi: 10.1006/viro.1997.8958. [DOI] [PubMed] [Google Scholar]
- 46.Nobusawa E, Ishihara H, Morishita T, Sato K, Nakajima K. Change in receptor-binding specificity of recent human influenza A viruses (H3N2): A single amino acid change in hemagglutinin altered its recognition of sialyloligosaccharides. Virology. 2000;278:587–596. doi: 10.1006/viro.2000.0679. [DOI] [PubMed] [Google Scholar]
- 47.Yang H, et al. Structure and receptor binding preferences of recombinant human A (H3N2) virus hemagglutinins. Virology. 2015;477:18–31. doi: 10.1016/j.virol.2014.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stech J, Garn H, Wegmann M, Wagner R, Klenk H. A new approach to an influenza live vaccine: Modification of the cleavage site of hemagglutinin. Nat Med. 2005;11:683–689. doi: 10.1038/nm1256. [DOI] [PubMed] [Google Scholar]
- 49.Girard G, Gultyaev A, Olsthoorn R. Upstream start codon in segment 4 of North American H2 avian influenza A viruses. Infect Genet Evol. 2011;11:489–495. doi: 10.1016/j.meegid.2010.12.014. [DOI] [PubMed] [Google Scholar]
- 50.Heaton NS, Sachs D, Chen CJ, Hai R, Palese P. Genome-wide mutagenesis of influenza virus reveals unique plasticity of the hemagglutinin and NS1 proteins. Proc Natl Acad Sci USA. 2013;110:20248–20253. doi: 10.1073/pnas.1320524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mallajosyula VV, et al. Influenza hemagglutinin stem-fragment immunogen elicits broadly neutralizing antibodies and confers heterologous protection. Proc Natl Acad Sci USA. 2014;111:E2514–E2523. doi: 10.1073/pnas.1402766111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laursen NS, Wilson IA. Broadly neutralizing antibodies against influenza viruses. Antiviral Res. 2013;98:476–483. doi: 10.1016/j.antiviral.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chai N, et al. Two escape mechanisms of influenza A virus to a broadly neutralizing stalk-binding antibody. PLoS Pathog. 2016;12:e1005702. doi: 10.1371/journal.ppat.1005702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wiley D, Wilson I, Skehel J. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 1981;289:373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- 55.Popova L, et al. Immunodominance of antigenic site B over site A of hemagglutinin of recent H3N2 influenza viruses. PLoS One. 2012;7:e41895. doi: 10.1371/journal.pone.0041895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilson I, Skehel J, Wiley D. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 57.Ewens WJ. Mathematical Population Genetics 1: Theoretical Introduction. Springer Science & Business Media; New York: 2012. [Google Scholar]
- 58.McWhite C, Meyer A, Wilke C. Sequence amplification via cell passaging creates spurious signals of positive adaptation in influenza virus H3N2 hemagglutinin. Virus Evol. 2016;2:vew026. doi: 10.1093/ve/vew026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Doud MB, Ashenberg O, Bloom JD. Site-specific amino acid preferences are mostly conserved in two closely related protein homologs. Mol Biol Evol. 2015;32:2944–2960. doi: 10.1093/molbev/msv167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nobusawa E, et al. Comparison of complete amino acid sequences and receptor-binding properties among 13 serotypes of hemagglutinins of influenza A viruses. Virology. 1991;182:475–485. doi: 10.1016/0042-6822(91)90588-3. [DOI] [PubMed] [Google Scholar]
- 61.Ha Y, Stevens DJ, Skehel JJ, Wiley DC. H5 avian and H9 swine influenza virus haemagglutinin structures: Possible origin of influenza subtypes. EMBO J. 2002;21:865–875. doi: 10.1093/emboj/21.5.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Russell R, et al. H1 and H7 influenza haemagglutinin structures extend a structural classification of haemagglutinin subtypes. Virology. 2004;325:287–296. doi: 10.1016/j.virol.2004.04.040. [DOI] [PubMed] [Google Scholar]
- 63.Daniels R, et al. Fusion mutants of the influenza virus hemagglutinin glycoprotein. Cell. 1985;40:431–439. doi: 10.1016/0092-8674(85)90157-6. [DOI] [PubMed] [Google Scholar]
- 64.Sun X, Longping VT, Ferguson AD, Whittaker GR. Modifications to the hemagglutinin cleavage site control the virulence of a neurotropic H1N1 influenza virus. J Virol. 2010;84:8683–8690. doi: 10.1128/JVI.00797-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee HK, et al. Comparison of mutation patterns in full-genome A/H3N2 influenza sequences obtained directly from clinical samples and the same samples after a single MDCK passage. PLoS One. 2013;8:e79252. doi: 10.1371/journal.pone.0079252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu N, et al. A structural explanation for the low effectiveness of the seasonal influenza H3N2 vaccine. PLoS Pathog. 2017;13:e1006682. doi: 10.1371/journal.ppat.1006682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Memoli MJ, et al. Recent human influenza A/H3N2 virus evolution driven by novel selection factors in addition to antigenic drift. J Infect Dis. 2009;200:1232–1241. doi: 10.1086/605893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Raghwani J, Thompson RN, Koelle K. Selection on non-antigenic gene segments of seasonal influenza A virus and its impact on adaptive evolution. Virus Evol. 2017;3:vex034. doi: 10.1093/ve/vex034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pollock DD, Thiltgen G, Goldstein RA. Amino acid coevolution induces an evolutionary Stokes shift. Proc Natl Acad Sci USA. 2012;109:E1352–E1359. doi: 10.1073/pnas.1120084109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shah P, McCandlish DM, Plotkin JB. Contingency and entrenchment in protein evolution under purifying selection. Proc Natl Acad Sci USA. 2015;112:E3226–E3235. doi: 10.1073/pnas.1412933112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gong LI, Suchard MA, Bloom JD. Stability-mediated epistasis constrains the evolution of an influenza protein. eLife. 2013;2:e00631. doi: 10.7554/eLife.00631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Natarajan C, et al. Epistasis among adaptive mutations in deer mouse hemoglobin. Science. 2013;340:1324–1327. doi: 10.1126/science.1236862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Harms MJ, Thornton JW. Historical contingency and its biophysical basis in glucocorticoid receptor evolution. Nature. 2014;512:203–207. doi: 10.1038/nature13410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Starr TN, Thornton JW. Epistasis in protein evolution. Protein Sci. 2016;25:1204–1218. doi: 10.1002/pro.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Starr TN, Picton LK, Thornton JW. Alternative evolutionary histories in the sequence space of an ancient protein. Nature. 2017;549:409–413. doi: 10.1038/nature23902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Doud MB, Hensley SE, Bloom JD. Complete mapping of viral escape from neutralizing antibodies. PLoS Pathog. 2017;13:e1006271. doi: 10.1371/journal.ppat.1006271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Doud MB, Lee JM, Bloom JD. How single mutations affect viral escape from broad and narrow antibodies to H1 influenza hemagglutinin. Nat Commun. 2018;9:1386. doi: 10.1038/s41467-018-03665-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bloom JD, Gong LI, Baltimore D. Permissive secondary mutations enable the evolution of influenza oseltamivir resistance. Science. 2010;328:1272–1275. doi: 10.1126/science.1187816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci USA. 2000;97:6108–6113. doi: 10.1073/pnas.100133697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ashenberg O, Padmakumar J, Doud MB, Bloom JD. Deep mutational scanning identifies sites in influenza nucleoprotein that affect viral inhibition by MxA. PLoS Pathog. 2017;13:e1006288. doi: 10.1371/journal.ppat.1006288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bloom JD. An experimentally determined evolutionary model dramatically improves phylogenetic fit. Mol Biol Evol. 2014;31:1956–1978. doi: 10.1093/molbev/msu173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dingens AS, Haddox HK, Overbaugh J, Bloom JD. Comprehensive mapping of HIV-1 escape from a broadly neutralizing antibody. Cell Host Microbe. 2017;21:777–787.e4. doi: 10.1016/j.chom.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kabsch W, Sander C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 84.Tien M, Meyer AG, Spielman SJ, Wilke CO. Maximum allowed solvent accessibilites of residues in proteins. PLoS One. 2013;8:e80635. doi: 10.1371/journal.pone.0080635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hadfield J, et al. Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics. 2018 doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sagulenko P, Puller V, Neher RA. TreeTime: Maximum-likelihood phylodynamic analysis. Virus Evol. 2018;4:vex042. doi: 10.1093/ve/vex042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Neher RA, Bedford T. nextflu: Real-time tracking of seasonal influenza virus evolution in humans. Bioinformatics. 2015;31:3546–3548. doi: 10.1093/bioinformatics/btv381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hooper KA, Bloom JD. A mutant influenza virus that uses an N1 neuraminidase as the receptor-binding protein. J Virol. 2013;87:12531–12540. doi: 10.1128/JVI.01889-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.