Significance
Ion channels are pore-forming proteins spanning biological membranes. Transient receptor potential ion channels are a subclass of ion channel proteins, characterized by nonselective permeability to cations such as sodium, calcium, magnesium, and zinc, and little voltage sensitivity; their gating is still an area of active investigation. TRPM6 and TRPM7 are ubiquitously expressed with prominent roles in early embryonic development. Uniquely, these channels also include an active kinase domain. The functions of TRPM6 and TRPM7 are correlated with proteolytic cleavage of the kinase domain, which is then translocated to the nucleus to phosphorylate histones and regulate gene expression. Here we describe the structure of the TRPM7 transmembrane regions and compare its features to other ion channels.
Keywords: ion channel, chanzyme, TRP channel, kinase, zinc
Abstract
The transient receptor potential ion channel subfamily M, member 7 (TRPM7), is a ubiquitously expressed protein that is required for mouse embryonic development. TRPM7 contains both an ion channel and an α-kinase. The channel domain comprises a nonselective cation channel with notable permeability to Mg2+ and Zn2+. Here, we report the closed state structures of the mouse TRPM7 channel domain in three different ionic conditions to overall resolutions of 3.3, 3.7, and 4.1 Å. The structures reveal key residues for an ion binding site in the selectivity filter, with proposed partially hydrated Mg2+ ions occupying the center of the conduction pore. In high [Mg2+], a prominent external disulfide bond is found in the pore helix, which is essential for ion channel function. Our results provide a structural framework for understanding the TRPM1/3/6/7 subfamily and extend the knowledge base upon which to study the diversity and evolution of TRP channels.
Among the more than 300 mammalian ion channels are two unique members, TRPM6 and TRPM7, that are ion channels and protein kinases (chanzymes) (1–3). TRPM6 and TRPM7 are ubiquitously expressed with prominent roles in early embryonic development (4–6). Tissue-specific knockout mice indicate that TRPM7 is required for normal organ development of the heart (7) and kidneys (4).
Similar to other TRPs, TRPM6 and TRPM7 comprise tetrameric ion channels with each subunit containing six-transmembrane (6-TM) segments. However, unlike most TRP channels, they are permeable to several divalent cations, including Zn2+, Mg2+, and Ca2+, as well as common monovalent cations such as Na+ and K+ (8–11). As chanzymes, the functions of TRPM6 and TRPM7 are correlated with proteolytic cleavage of the kinase domain, which is then translocated to the nucleus to phosphorylate histones and regulate gene expression (12–14). The majority of native TRPM6 and TRPM7 cellular protein is in unique intracellular vesicles (15–17). However, since these vesicles are inaccessible to patch clamp recording, all recordings are from channels in the plasma membrane. In addition to relative ion selectivities and biophysical properties, electrophysiological recordings have demonstrated that TRPM7 inactivation/deactivation is enhanced by phosphatidylinositol 4,5 bisphosphate [PI(4.5)P2] hydrolysis (18), intracellular Mg2+ (9), and intracellular acidification (19). Activators of a TRPM7 divalent cation current are unknown. However, removal of external divalent cations (9), acidification of the external environment (20), or addition of glutathione (17) potentiate TRPM7 currents by enhancing monovalent cation permeability.
Kuriyan and coworkers crystallized and solved the structure of TRPM7’s C-terminal isolated kinase domain (21). The kinase’s NT is similar to classical protein kinases, but its C-terminal lobe resembles ATP-grasp fold enzymes. The aspartic acid and glutamine residues important for catalysis are separated by one residue on a continuous kinked β-strand, with a critical lysine forming hydrogen bonds with the α-phosphate group and the adenine ring of ATP. A Zn2+ appears to be a resident stabilizing cation in the hydrophobic core of the C-terminal lobe. TRPM7’s isolated C-terminal coiled-coil domain was proposed to participate in tetrameric chanzyme assembly by bundling the membrane-proximal C-terminal ∼50 amino acids (22).
Based on sequence homology, the TRPMs can be divided into two subfamilies as TRPM2/4/5/8 and TRPM1/3/6/7 (23). This familial divide is significant because TRPM6/7 are permeable to divalent cations (including Zn2+ and Mg2+) whereas TRPM4/5 are monovalent cation-selective channels. Cryo-EM structures of human TRPM4, bird TRPM8, and sea anemone TRPM2 have been solved recently (24–27); however, none are available for the TRPM1/3/6/7 subfamily. In this study, we report cryo-EM structures of TRPM7 in EDTA (TRPM7-EDTA, to remove most free divalent cations), in high external Mg2+ (TRPM7-Mg2+), and in buffer without added external Mg2+ (TRPM7-divalent-free: DVF), to overall resolutions of 3.3, 3.7, and 4.1 Å, respectively.
Overall Structure of the Mouse TRPM7 Tetrameric Ion Channel
We screened orthologs of TRPM7 and determined that mouse TRPM7 was a promising candidate for our structural studies. To improve the stability and homogeneity of mouse TRPM7, residues C-terminal to natural cleavage sites (12) (almost always 1281–1863) were removed, including the previously characterized kinase domain (21) (almost always 1580–1863). Maltose binding protein (MBP) was fused to the N terminus of TRPM7, and the fusion protein was expressed in the BacMam expression system (Methods). The MBP tag was not removed before structural determination, since its cleavage significantly reduced protein yield and stability. Purified and detergent-solubilized protein was exchanged with the amphipol, PMAL-C8 for structure determination (SI Appendix, Fig. S1 A and B).
To evaluate the functional properties of our tagged and truncated TRPM7 construct without endogenous TRPM6/7 contamination [TRPM6 can heteromultimerize with TRPM7 (10)], we patch-clamped transiently transfected HEK293T cells in which TRPM6 and TRPM7 were both deleted (13). The resulting current-voltage (I-V) relationship elicited from a 200-mV ramp protocol (8, 9, 11, 28) displayed several TRPM7 characteristics, including a small and linear inward current, reversal near 0 mV, and a prominent outwardly rectifying current (SI Appendix, Fig. S1 C and D). Whole-cell currents also exhibited dialysis-induced growth (SI Appendix, Fig. S1 E and F), with a mean peak time (113 ± 21 s at +100 mV; n = 7) within the range of full-length TRPM7 (9). Moreover, as with full-length TRPM7 (8), currents activated rapidly and did not inactivate over a 500-ms test pulse; however, some current decay was observed at voltages positive to +80 mV (SI Appendix, Fig. S1G). Divalent cations are proposed to permeate TRPM7 through an anomalous mole fraction mechanism in which they also act as pore blockers (9). We therefore determined whether our truncated construct was blocked by high concentrations of external Mg2+ and observed a substantial reduction of outward current with minimal change to inward current (SI Appendix, Fig. S1 H and I), analogous to full-length TRPM7 (9, 28). As our truncated construct is suitable for evaluating the Mg2+ regulatory/permeability properties of TRPM7, we solved structures of TRPM7-EDTA, TRPM7-Mg2+, and TRPM7-DVF using cryo-EM. Resolutions were estimated using the gold-standard Fourier shell correlation cutoff of 0.143 (SI Appendix, Fig. S2 and Table S1). We will primarily discuss TRPM7-Mg2+, which represents the main features of all three structures.
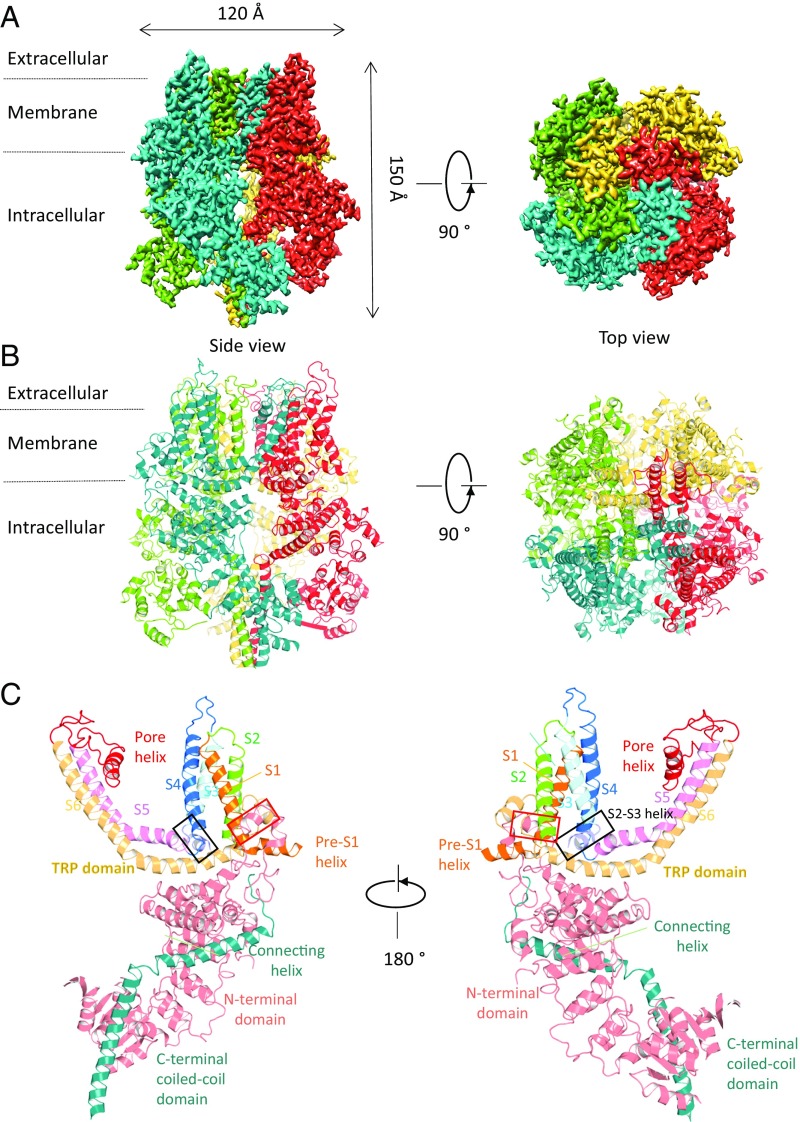
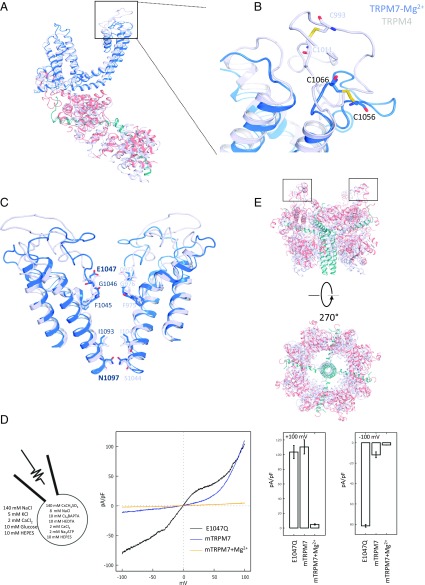
The tetrameric structure of TRPM7-Mg2+ surrounds an ion channel pore which empties into a ∼50 Å × 50 Å cytoplasmic space created by an ∼100-Å-tall, ∼120-Å-wide intracellular skirt (Fig. 1 A and B). Our density map was sufficient to enable tracing of the complete polypeptide chain, guided by strong densities for the bulky side chains in Arg, Lys, His, Trp, Tyr, and Phe (SI Appendix, Fig. S3). Despite <20% protein sequence identity between TRPM7 and other solved TRP channels [TRPVs (29, 30), polycystin-2 (31), TRPML1 (32–34) and TRPA1 (35)], the TRPM7 structure exhibits the fourfold symmetry and domain-swapping features of these and other 6-TM channels, such as the well-studied voltage-gated K+ channels. The orientation and position of helices in the 6-TM bundles of TRPM7 are quite similar to other TRP channel structures (SI Appendix, Fig. S4). A short helix between S2 and S3 (“S2−S3 helix”) lies parallel to the intracellular lipid bilayer located close to the amphipathic TRP domain. This resembles that of other TRPM structures, but is shorter by one helical turn compared with TRPM2 and TRPM4 (black boxes in Fig. 1C and SI Appendix, Fig. S5 A and B). We speculate that these partially membrane-embedded orthogonal helices may affect gating of the channel upon changes in bilayer composition and thickness.
Fig. 1.
Overall structure of TRPM7-Mg2+. (A) Cryo-EM reconstruction density map of TRPM7-Mg2+ at 3.7-Å overall resolution. Each channel subunit is color-coded. (B) Ribbon diagram of the mouse TRPM7 model. (C) Ribbon diagrams depicting two side views of a single subunit. Red boxes: hydrophobic helix (almost alawys 759–769) anchored to inner leaflet of the plasma membrane. Black boxes: S2–S3 helix.
Compared with other TRPM channels, TRPM7’s long N terminus is ∼30% conserved by sequence homology; however, the overall architecture of this region is surprisingly similar, including the N-terminal domain (NT), the ankyrin-repeat domain, and the linker-helical domain (SI Appendix, Fig. S5D). A hydrophobic helix, formed by almost always 759–769 and anchored to the inner leaflet of the plasma membrane (red boxes of Fig. 1C and SI Appendix, Fig. S5 A and C), is similar to that observed in available TRPM structures, but is not present in other TRP channel structures. A long “connecting helix” (25 residues), present in TRPC3 (36), TRPC6 (37), TRPM2 (26), TRPM4 (24, 27), and TRPM8 (25), links the transmembrane domain and coiled-coil domain (Fig. 1C). Sequence alignments indicate that this connecting helix is conserved across TRPM family members (SI Appendix, Figs. S5E and S6 A and B).
Ion Conduction Pore and Magnesium Binding Site
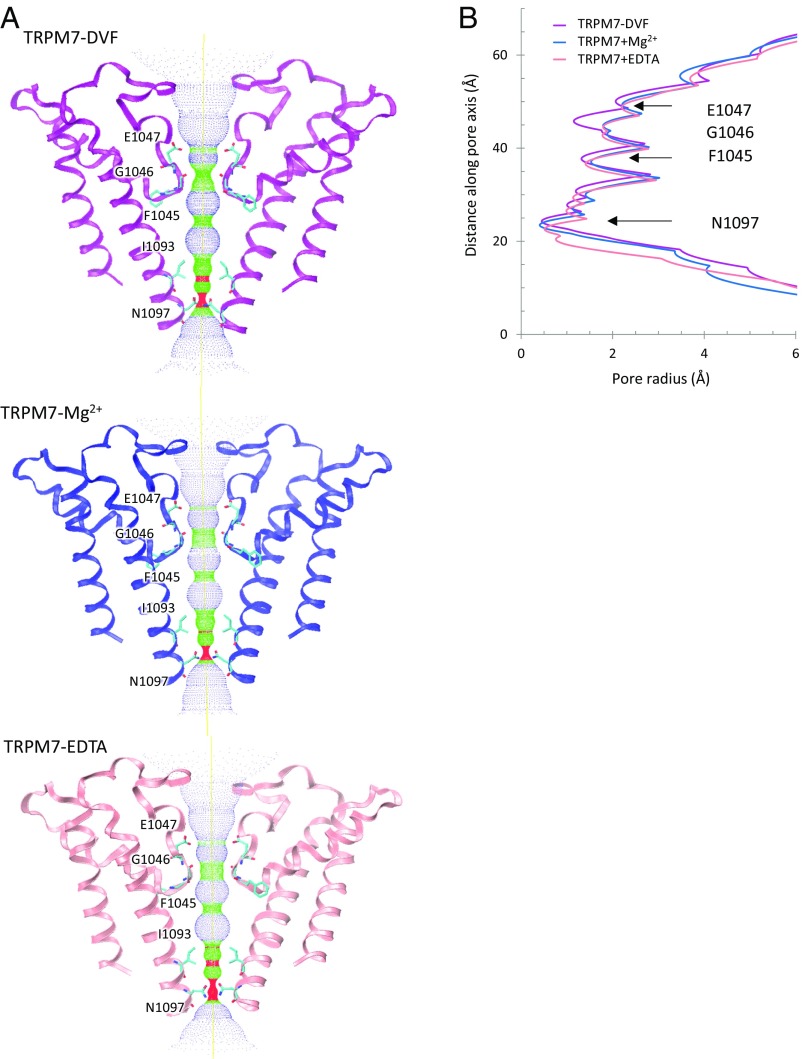
TRPM7’s selectivity filter is formed by the backbone carbonyls of Phe1045/Gly1046 and Glu1047s side chain. The narrowest opening is 6.9 Å across the diagonally opposed Gly1046 residues (SI Appendix, Fig. S7A). Along the pore, Ile1093 (S6 cytoplasmic end) and Asn1097 (before the TRP domain) form another restriction site, defined as the lower gate. The most constricted point is Asn1097, where S6 helices from each subunit form a hydrophobic seal (SI Appendix, Fig. S7A). The distances between diagonal (4.1 Å) and neighboring (2.9 Å) side chains of Asn1097 are narrower than corresponding sites in the TRPA and TRPV structures. For comparison, the most restricted opening in TRPV1 is in the outer pore region, measuring 7.6 Å diagonally between opposed Gly643 residues (29). In TRPA1, the narrowest point (6.4 Å) is the lower gate (Val961) (35).
Regardless of condition (with EDTA, DVF, or with high [Mg2+]), the ion conduction pathway is essentially unchanged (Fig. 2A). The pore radius in all three structures differs by ∼0.5 Å to 1 Å (Fig. 2B), which is most likely due to differences in resolution rather than structural modification.
Fig. 2.
Ion conduction pathway. (A) Ion conduction pathway along the pore of TRPM7-DVF (Top), TRPM7-Mg2+ (Middle), and TRPM7-EDTA (Bottom), shown as dots and mapped using HOLE. Three continuous residues (Glu1047, Gly1046, and Phe1045) in the pore helix form the funnel-shaped selectivity filter (1045FGE1047). The lower gate, Asn1097, is located before the TRP domain. (B) Pore radius along the central axis (calculated with HOLE). Glu1047-Phe1045 and Asn1097 form narrow constrictions at the selectivity filter and lower gate, respectively.
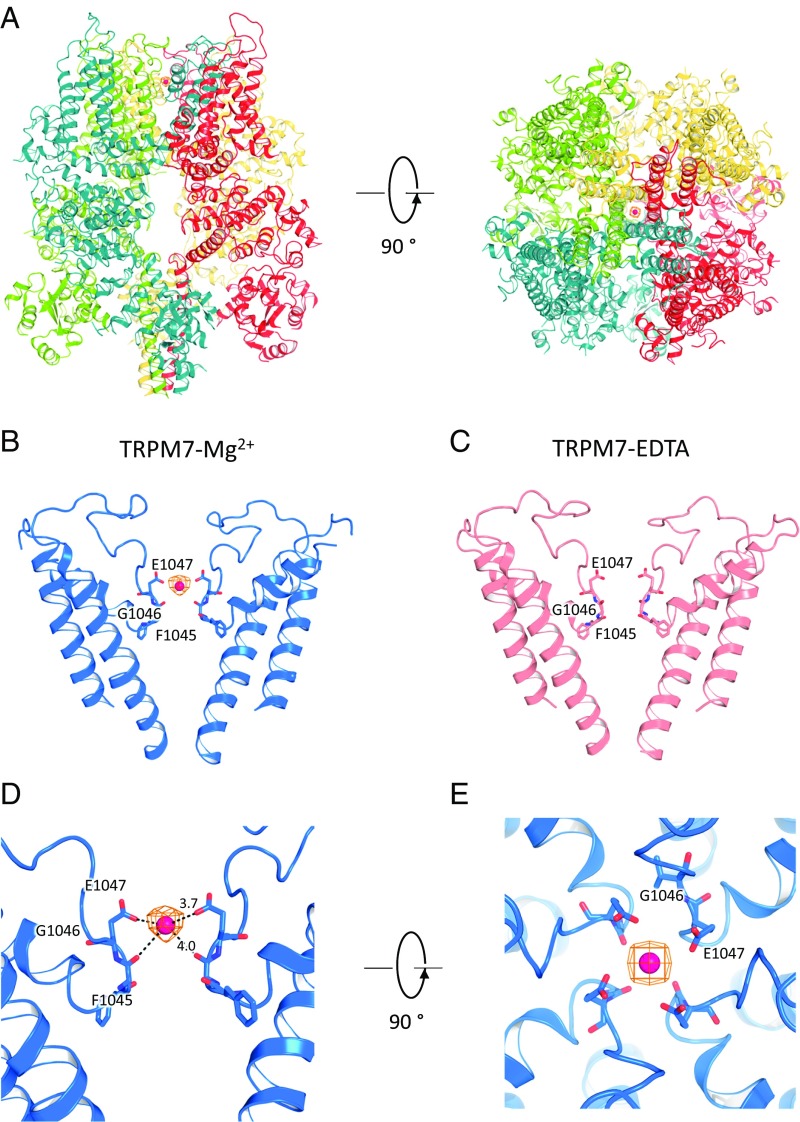
A strong nonprotein density with a high signal-to-noise ratio is present in the ion permeation pathway of the TRPM7-Mg2+ structure (Fig. 3 A and B). In contrast, spherical densities are not found in the TRPM7-EDTA structure (Fig. 3C) at various contour levels (SI Appendix, Fig. S8). We interpret this density as a probable Mg2+ ion located at the most restricted site of the selectivity filter (Fig. 3 D and E). Moreover, the distance between Mg2+ and Glu1047s side chain (3.7 Å) and the backbone carbonyl of Gly1046 (4.0 Å; Fig. 3D) is large enough to accommodate a partially hydrated Mg2+ ion.
Fig. 3.
Mg2+ binding site. (A) Top and side views of the tetrameric TRPM7-Mg2+ structure with bound Mg2+. (B) Side view of the TRPM7-Mg2+ pore, with residues important for cation binding shown in stick representation. (C) Side view of the TRPM7-EDTA pore, with residues in the selectivity filter shown in stick representation. (D) Enlarged side view of the TRPM7-Mg2+ pore. Residues that contribute to cation-binding sites are shown as sticks, and the putative Mg2+ is the red sphere. (E) Top view of the TRPM7-Mg2+ pore.
Assembly and Domain Interactions
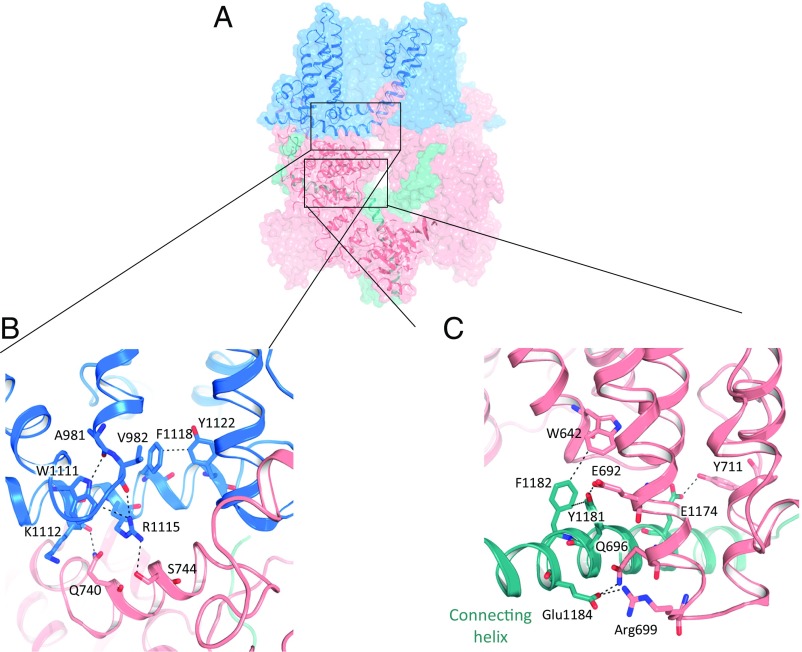
TRPM7’s “TRP domain” is the α-helical structure bending sharply toward the membrane immediately adjacent to the near-perpendicular S6 transmembrane helix (Fig. 1C). This TRP domain is a common feature of other TRP family members and is proposed to participate in subunit assembly or allosteric modulation of channel gating (38–41). In our structures, unlike those for TRPA1 and TRPV1, the pre-S1 helix does not interact with the TRP domain. Instead, Arg1115 and Lys1112 in the TRP domain (almost always 1099–1123) interact with the N-terminal Ser744 and Gln740, which are located on an elbow helix immediately after the pre-S1 helix (Fig. 4 A and B). In addition, Trp1111 and Arg1115 form hydrogen bonds with Ala981 and Val982 on the S4−S5 linker, respectively (Fig. 4 A and B). A π−π stacking interaction (Phe1118/Tyr1122) and a cation−π interaction (Arg1115/Trp1111) are also found on the TRP domain (Fig. 4 A and B). These interactions are consistent with the TRP domain’s potential importance in gating. This hypothesis is supported by prior mutagenic studies in which Lys1112Gln and Arg1115Gln generated nonfunctional channels (42). Based on these and other structure−function studies of tetrameric ion channels (43–45), we hypothesize that the TRP domain and S4−S5 linker couple the movement of the “voltage sensor-like” S1 to S4 domains to the channel gate.
Fig. 4.
Assembly and domain interactions. (A) One TRPM7 subunit from the plane of the membrane with regions of interest depicted by boxes. (B) Expanded view of A showing interactions between the N terminus (pink) and the TRP domain (blue). (C) Expanded view of A showing interactions between N (pink) and C (green) termini. Residues at domain interfaces are labeled, with potential hydrogen bonds and electrostatic interactions shown as dashed lines.
The transmembrane domains of TRPM7 are overlapped with other known core TRP structures in SI Appendix, Fig. S4, exhibiting remarkably similarity. The largest differences between TRP channels lie in the N and C termini. TRPV1’s ankyrin repeats 3 and 4 interact with the C-terminal domain via three-stranded antiparallel β-sheets (46). In TRPA1, a β-strand links a short, poorly resolved α-helix, buried in the inner leaflet of the membrane, to a TRP-like domain/C-terminal coiled-coil domain connecting loop (35). In contrast, TRPM7’s N/C-terminal interaction is through a well-resolved connecting helix, encoded by sequences that are relatively conserved among TRPM subfamily members (SI Appendix, Fig. S6 A and B). The center of these interactions is Tyr1181 and Phe1182, which form a π−π interaction and further interact with residues Trp642 and Glu692 on the NT (Fig. 4 A and C). A hydrogen bond and salt bridge involving Glu1184 and Glu1174 in the connecting helix with Arg699 and Try711 in the NT stabilize these domain interactions (Fig. 4 A and C).
Major Structural Differences Between TRPM7 and TRPM4
We next compared the structure of TRPM7-Mg2+ with the structure of TRPM4, a monovalent cation-selective channel and member of TRPM2/4/5/8 subfamily (26). First, the conserved disulfide bond near the pore loop in TRPM7 (formed by Cys1056 and Cys1066; SI Appendix, Fig. S6 C and D) is closer to the S5−S6 helices than that of TRPM4 (Fig. 5 A and B). Second, the TRPM7 (1045FGE1047) and TRPM4 (975FGQ977) selectivity filters differ by one amino acid (Fig. 5C). This disparity is essential to TRPM7 divalent cation permeation, as mutating Glu1047 to Gln (the equivalent TRPM4 residue) abolished Ca2+ and Mg2+ permeability in full-length TRPM7 (28). We tested this mutation (Glu1047Gln) in our truncated TRPM7 protein and observed a similar result, with inward currents becoming inwardly rectifying in the mutant (6.5-fold greater at −100 mV; Fig. 5D), while outward rectification was unchanged above 0 mV. Moreover, in the Nematostella vectensis (NvTRPM2) structure, the corresponding outermost selectivity filter residue is also a glutamate. However, in Homo sapiens (HsTRPM2), the equivalent residue is a glutamine (by sequence analysis). Electrophysiologically, this difference was correlated with an ∼70-fold greater PCa/PNa in NvTRPM2 compared with HsTRPM2 (26). Thus, a negatively charged glutamate in the outermost selectivity filter position likely facilitates divalent cation permeability in TRPM channels (SI Appendix, Fig. S7 B and C). Third, the residues of the lower gate, which create the most constricted region of both pores, differ between TRPM7 (Asn1097) and TRPM4 (Ser1044). Here, asparagine−asparagine side chain separations in TRPM7 are smaller than those between serines in TRPM4 (Fig. 5C and SI Appendix, Fig. S7 B and C). However, the isoleucine and asparagine in the lower gate are well conserved between TRPM7, NvTRPM2, and hTRPM2 (SI Appendix, Fig. S7 B and C). Finally, the cytosolic domains are distinctly organized. In TRPM7, hydrophobic loops and two helices belonging to the NT are anchored to the inner leaflet of the plasma membrane (black boxes, Fig. 5E). This is more complex than the loop and short helix in TRPM4. The movement of this region may produce additional conformational changes in the cytosolic domains, perhaps leading to large vertical and rotational motions of the NT of the channel (Fig. 5E).
Fig. 5.
Structural comparison of TRPM7 and TRPM4. (A) Overlaid monomer of TRPM7 (blue, pink, and cyan) and TRPM4 (gray). (B) Enlarged view of key pore loop disulfide bond between Cys1056 and Cys1066. (C) Aligned TRPM7 (blue) and TRPM4 (gray) dimer pores. Residues of interest are labeled and shown as sticks. (D) Whole-cell I-V relationships of HEK293T TRPM6/7 double KO cells expressing MBP-tagged truncated mouse TRPM7 (blue), with 4 mM Mg2+(gold), mTRPM7 in 4 mM Mg2+, or truncated mouse E1047Q-TRPM7 (black). Bar charts represent the mean currents recorded at +100 mV and −100 mV; E1047Q (n = 18, 103 10 pA/pF and −82 1 pA/pF), mTRPM7 (n = 10, 111 10 pA/pF and −12 0.3 pA/pF), and mTRPM7+4 mM Mg2+ (mTRPM7+Mg2+, n = 10, 5 1 pA/pF and −2 0.1 pA/pF). (E) Side and bottom views of superimposed cytosolic domains of TRPM7 (blue and pink) and TRPM4 (gray); boxes highlight extracellular regions containing cysteines.
Pore Loop Cysteines
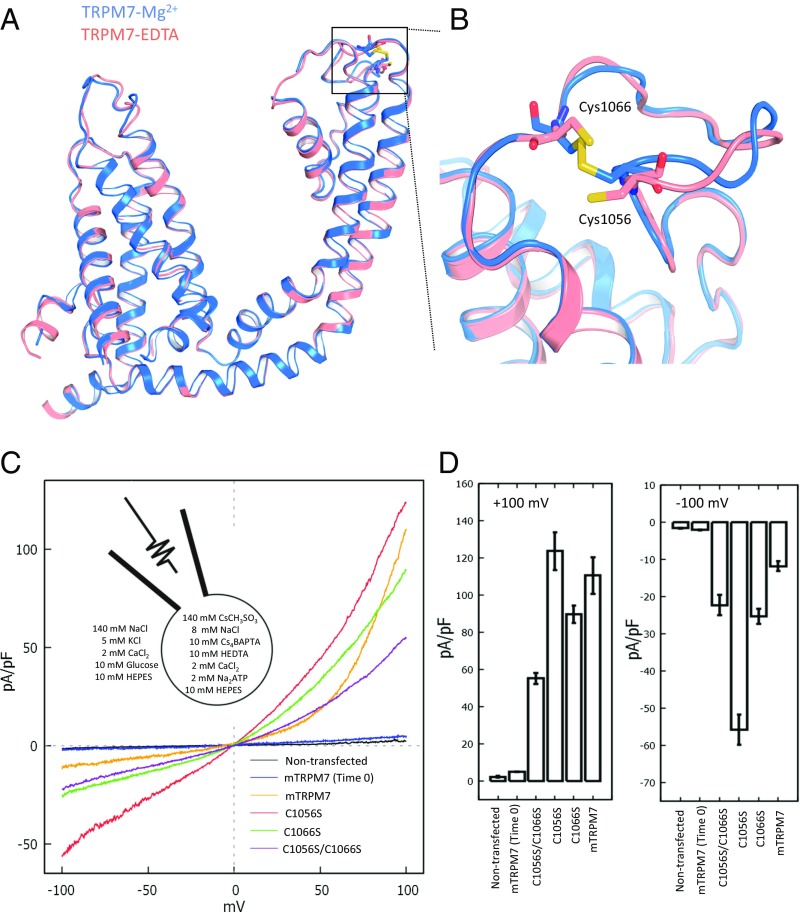
The movement and organization of the extracellular pore loop may be important to TRPM7 channel function. Conformational changes of the neighboring pore helices, S5 and S6, usually affect key channel properties. In the TRPM7-Mg2+ structure, we observed a pore loop disulfide bond between Cys1056 and Cys1066 (Fig. 6 A and B), which is conserved across the TRPM family (SI Appendix, Fig. S6 C and D). If this configuration is not due to protection from radiation damage (47), we hypothesize that divalent cations, such as Mg2+, may stabilize this disulfide bond, since it is only observed in the TRPM7-Mg2+ structure. Moreover, external application of the reducing agent glutathione potentiated TRPM7 currents by enhancing monovalent cation permeability (17), implicating an externally located disulfide bond. We tested whether the Cys1056/Cys1066 disulfide bond is essential for divalent permeability by mutagenesis and patch-clamp electrophysiology. When either or both cysteines are mutated, the resulting currents (I-V relationships) are more linear (Fig. 6 C and D), in contrast to the inward rectification noted in the prior glutathione results (17). Thus, mutation of these cysteines globally disrupts TRPM7 function by mechanisms which will require more investigation.
Fig. 6.
Comparison of TRPM7-Mg2+ and TRPM7-EDTA structures. (A) Superimposition of the transmembrane module from TRPM7-Mg2+ (blue) and TRPM7-EDTA structures (pink). (B) Site of the broken disulfide bond between Cys1056 and Cys1066 in the pore domain. Key discernible residues, Cys1056 and Cys1066, are labeled. Shown is superimposition of the disulfide bond between S5 and S6 in the TRPM7-Mg2+ (blue) and TRPM7-EDTA (pink) structures. (C) Whole-cell I-V relationship of HEK293T TRPM6/7 double KO cells expressing MBP-tagged truncated mouse TRPM7 (mTRPM7; orange), C1056S (red), C1065S (green), and C1056S/C1066S (violet). (D) Mean currents of mTRPM7 and mTRPM7 cysteine(s) mutant measured at +100 mV and −100 mV; nontransfected (n = 10, 2.3 0.5 and −1.6 0.1), mTRPM7 (Time0) (n = 12, 5.1 0.1 and −2.1 0.1 pA/pF), C1056S/C1066S (n = 12, 55 3 and −22 3 pA/pF), C1056S (n = 12, 124 5 and −56 2 pA/pF), C1066S (n = 10, 90 and −25 4 pA/pF), and mTRPM7 (n = 10, 111 10 and −12 1 pA/pF).
Discussion
Mg2+ is the second most abundant cellular divalent cation. However, relatively little is known about the molecular components and mechanisms that regulate Mg2+ compared with Ca2+ homeostasis. In this study, we presented cryo-EM structures of the Mg2+-permeable TRPM7 in three ionic conditions: TRPM7-DVF, TRPM7-Mg2+, and TRPM7-EDTA. There are three conclusions from the work. First, in the TRPM7-Mg2+ structure, a nonprotein density we presume is Mg2+ is found within the selectivity filter. Mg2+ has well-defined first and second hydration shells; the dehydration energy of the first hydration shell is 476 kcal/mol (48), at least 4 times that of Na+ and K+, making the passage of fully dehydrated Mg2+ energetically unfavorable. In contrast, the second hydration shell of Mg2+ is similar to bulk water (49). Thus, we propose that the backbone carbonyl of Gly1046 and the carboxyl group of Glu1047 replace the waters of the second hydration shell and compensate for the energetic cost of dehydration when hydrated Mg2+ enters the pore. Importantly, the key negatively charged selectivity filter Glu1047 differs from the orthologous Gln in TRPM4/5’s monovalent cation selective filter. Second, the channel’s lower gate has of two constriction sites (Ile1093 and Asn1097), which is identical to that of NvTRPM2. Interestingly, Asn1097 occupies a similar position in TRPM1, TRPM3, TRPM6, and TRPM7, but not in the monovalent-selective TRPM4/5. As the only polar amino acid in the lower gate, the side chains of Asn1097 from each monomer form a polar ring at the cytoplasmic pore entrance. In addition, asparagine may also bind the first hydration shell of Mg2+ (50). Third and finally, the pore helix’s disulfide bond is required for proper channel assembly/function and is a general feature of the TRPM family.
In summary, TRPM7’s structure sheds light on many questions raised by previous electrophysiological and physiological studies. The next steps will be to define open, desensitized, and blocked conformations states, perhaps requiring X-ray crystallography, or extensive subparticle classification (51).
Methods
Protein Expression and Purification of MBP-TRPM7.
The mouse TRPM7 construct (amino acids 1 to 1280) was cloned into the pEG BacMam vector (52). An MBP tag was added to TRPM7’s N terminus via the linker, NSSSNNNNNNNNNNLGIELEVLFQ/GP, containing the HRV 3C cleavage site (underlined). P3 baculovirus was produced in the Bac-to-Bac Baculovirus Expression System (Invitrogen) using Spodoptera frugiperda (Sf9) cells transduced at a density of 2 to 3 × 106 cells/mL. For protein expression, HEK293S GnTI− cells (lacking N-acetylglucosaminyltransferase I) were grown in Freestyle 293 Expression medium (Life Technologies) supplemented with 2% FBS and 1% penicillin−streptomycin (37 °C, 8% CO2), and transduced with P3 baculovirus once cells reached a density of 2 to 3 × 106 cells/mL. After 12 h to 24 h, 10 mM sodium butyrate was added, and the temperature was reduced to 30 °C. Cells were harvested 72 h after transduction and frozen at −80 °C. Before solubilization, cells were resuspended for 30 min in a DVF buffer containing 30 mM Hepes, 150 mM NaCl, 1 mM dithiothreotol (DTT), pH 7.5 with EDTA-free protease inhibitor mixture (Roche). MBP-TRPM7 membranes were mechanically homogenized and solubilized for 2 h to 3 h in a buffer containing 1.0% (wt/vol) N-dodecyl-beta-D-maltopyranoside (DDM; Affymetrix), 0.1% (wt/vol) cholesteryl hemisuccinate (CHS; Sigma), 25 mM Hepes, 150 mM NaCl, 1 mM DTT, pH 7.5 with EDTA-free protease inhibitor mixture (Roche). The supernatant was isolated by centrifugation at 100,000 × g for 60 min, followed by incubation in amylose resin (New England BioLabs) at 4 °C overnight. The resin was washed with 20 column volumes of 25 mM Hepes, 150 mM NaCl, 0.05% (wt/vol) DDM, 0.01% (wt/vol) CHS, 1 mM DTT, pH 7.5 with EDTA-free protease inhibitor mixture (Roche). The protein was eluted with four column volumes of 25 mM Hepes, 150 mM NaCl, 0.05% (wt/vol) DDM, 0.01% (wt/vol) CHS, 1 mM DTT, pH 7.5 with EDTA-free protease inhibitor mixture (Roche) and 40 mM maltose. The protein was then concentrated and mixed with poly (maleic anhydride-alt-1-decene) substituted with 3-(dimethylamino) propylamine (PMAL-C8, anatrace) at 1:4 (wt/wt) with gentle agitation for 4 h. Bio-Beads SM-2 (50 mg/mL reconstitution mixture; Bio-Rad) were added to remove additional detergent, and a disposable Poly-Prep column was used to remove Bio-Beads. The protein was then concentrated to 0.5 mL with a 100-kDa molecular weight cutoff concentrator (Millipore) before further purification on a Superose 6 column in a buffer composed of 25 mM Hepes, 150 mM NaCl, 1 mM DTT, pH 7.5. The peak, corresponding to tetrameric C-terminally truncated TRPM7, was collected and concentrated to 4 mg/mL to 5 mg/mL for electron cryomicroscopy.
For the high Mg2+ structure, 300 mM MgCl2 was added to the DVF buffer for every step of extraction and purification. For the TRPM7-EDTA structure, 2 mM EDTA (pH 8.2) was added to the protein extraction DVF buffer, and 5 mM EDTA was added to purified protein before freezing. In our DVF buffer, we determined that NaCl (Fisher Scientific) contributed the most contaminating Ca2+ (0.002%; 3 µM) and Mg2+ (<0.001%; <1.5 µM). Using MaxChelator software (https://web.stanford.edu/∼cpatton/webmaxcS.htm), the free Ca2+ and Mg2+ in the purified TRPM7-EDTA protein were estimated as 3.8 pM and 895 pM, respectively. We note that these values may change under cryoconditions, as MaxChelator estimates are limited to a minimum temperature of 0 °C.
Electron Microscopy Data Collection.
For the TRPM7-DVF, TRPM7-Mg2+ and TRPM7-EDTA structures, 3.5 µL of purified TRPM7 in PMAL-C8 at 4 mg/mL to 4.5 mg/mL was applied onto a glow-discharged, 400 mesh copper Quantifoil R1.2/1.3 holey carbon grid (Quantifoil). Grids were blotted for 7 s at 100% humidity and flash frozen in liquid nitrogen-cooled liquid ethane using an FEI Vitrobot Mark I (FEI). The grid was loaded onto an FEI TF30 Polara electron microscope operating at 300 kV accelerating voltage. TRPM7 image stacks were recorded on Gatan K2 Summit (Gatan) direct detector set in superresolution counting mode using SerialEM (53), with a defocus range between 1.5 μm and 3.0 μm. The electron dose was set to 8 e per physical pixel per second; subframe time of 200 ms and total exposure time of 10 s resulted in 50 subframes per image stack. The total electron dose was 52.8 e− per square angstrom (∼1.1 e− per square angstrom per subframe).
Image Processing and 3D Reconstruction.
Similar strategies were used for the image processing and 3D reconstruction of TRPM7-DVF, TRPM7-Mg2+, and TRPM7-EDTA structures. Image stacks were gain-normalized and binned by 2× to a pixel size of 1.23 Å before drift and local movement correction using motionCor2 (54). The images from the sum of all frames with dose weighting were subjected to visual inspection, and poor images were removed before particle picking. Particle picking and subsequent bad particle elimination through 2D classification was performed using Python scripts/programs developed by Maofu Liao (55) with minor modifications in the 8× binned images. The selected, good 2D class averages were used to build an initial map using the common lines approach implemented in SPIDER (56) through Maofu Liao’s Python scripts (55), which was needed in later 3D refinement and classification in RELION (57). The contrast transfer function (CTF) parameters were estimated using CTFFIND4 (58) with the sum of all frames without dose weighting. Quality particle images were then boxed out from the dose-weighted sum of all 50 frames and subjected to RELION 3D classification. RELION 3D refinements were then performed on selected classes. The resolution for the final map was further improved by using subframes 1 to 14 from 3.8 Å to 3.7 Å for the TRPM7-Mg2+ structure, 3.3 Å to 3.3 Å for the TRPM7-EDTA structure, and 4.2 Å to 4.1 Å for the TRPM7-DVF structure.
Model Building, Refinement, and Validation.
At 3.3-Å resolution for the TRPM7-EDTA structure, the cryo-EM density map was of sufficient quality for de novo atomic model building, despite the lower density of the N terminus. For the full-length (1 to 1280) protein, a polyalanine model was first built in COOT (59). Taking advantage of the defined geometry of helices and clear bumps for Cα atoms in the transmembrane domains, amino acid assignment was subsequently achieved based primarily on the clearly defined side chain densities of bulky residues such as Phe, Tyr, and Trp, as well as some Arg and Lys residues. Resolution of the first part of NT (1 to 150) were insufficient for backbone tracing, and the polyalanine model was used for that region. The refined atomic model was further visualized in COOT. A few residues with side chains moving out of the density during the refinement were fixed manually, followed by further refinement. The TRPM7 model was then subjected to global refinement and minimization in real space using the PHENIX (60) module “phenix.real_space_refine” (61), and geometries of the model were assessed using MolProbity (62) in the comprehensive model validation section of PHENIX. The final model of TRPM7-EDTA structure exhibited good geometry as indicated by the Ramachandran plot (preferred region, 97.8%; allowed region, 2.1%; outliers, 0.1%). The pore radius was calculated using HOLE (63). For the TRPM7-DVF and TRPM7-Mg2+ structures, the structure of TRPM7-EDTA at 3.3 Å was docked into the cryo-EM maps, followed by manual adjustment in COOT. They were then subjected to the same global refinement and minimization process described for TRPM7-EDTA, and geometries of the models were also assessed using MolProbity (62). The final models of TRPM7-DVF and TRPM7-Mg2+ structures exhibited good geometry as indicated by the Ramachandran plot.
Electrophysiology.
All electrophysiological experiments were performed in HEK293T TRPM6/7 double KO cells to eliminate endogenous TRPM6 and TRPM7 currents (13). The cells were cultured in DMEM (ATCC 30-2002) with 10% FBS, 1% Pen/Strep, 10 mM MgCl2, 3 μg/mL of blasticidin and 0.5 μg/mL of puromycin. Cells were transiently transfected (48 h) with the MBP-tagged truncated mouse TRPM7 construct used for structural investigation or various mutants (C1056S, C1066S, C1056S/C1066S). E1047Q mouse TRPM7 was kindly provided by Lixia Yue, University of Connecticut Health Center, Farmington, and we truncated this construct to mimic the mouse TRPM7 channel used for structural investigation. Cells were cotransfected with EGFP at 10% of total DNA for visualization and cell selection.
Cells were perfused at a rate of 1 mL/min to 2 mL/min in a modified Tyrode’s solution containing (millimolars): 140 NaCl, 5 KCl, 2 CaCl2, 10 glucose, and 10 Hepes; pH 7.4. Where indicated, cells were perfused with a high Mg2+ solution containing (millimolars): 110 MgCl2, 10 Hepes; pH 7.4. Patch pipettes of 2 MΩ to 5 MΩ contained (millimolars): 120 CsCH3SO3, 8 NaCl, 10 Cs4BAPTA, 2 CaCl2, 2 Na2ATP, 10 Hepes; pH 7.4. For experiments utilizing the high Mg2+ external solution, a modified pipette solution contained (millimolars): 120 CsCH3SO3, 8 NaCl, 10 mM HEDTA, 5 Cs4BAPTA, 1 CaCl2, 2 Na2ATP, 10 Hepes; pH 7.4. All solutions were 300 ± 5 mOsm.
Whole-cell currents were recorded at room temperature using an Axopatch 200B patch-clamp amplifier controlled via a Digidata 1440A (Molecular Devices) and pClamp 10.3 software. Data were digitized at 10 kHz and low-pass-filtered at 2 kHz. In most recordings, cells were held at 0 mV, and 200-ms ramps from −100 mV to 100 mV were applied at 0.5 Hz. Where indicated, cells were held at 0 mV and stepped from −100 mV to +100 mV in +10-mV increments (2 Hz per step). Before recordings, currents were corrected for pipette (fast) capacitance, whole-cell capacitance, and series resistance compensated to 80%. Currents were offline-corrected for liquid junction potential using previously described methods (64).
Data and Software Availability.
Cryo-EM electron density maps of the mouse TRPM7-EDTA, TRPM7-Mg2+, and TRPM7-DVF have been deposited in the Electron Microscopy Data Bank under accession code EMD-6975, EMD-7297 and EMD-7298, respectively. Atomic coordinates of the mouse TRPM7-EDTA, TRPM7-Mg2+ and TRPM7-DVF have been deposited in the Protein Data Bank under accession code 5ZX5, 6BWD and 6BWF, respectively.
Supplementary Material
Acknowledgments
We thank Dr. Shu-Hsien Sheu [Howard Hughes Medical Institute (HHMI)] and Dr. Lihua Sun (Shanghai Institute of Applied Physics, Chinese Academy of Sciences) for providing help with the HOLE program; Dr. Steve Harrison and the Cryo-EM Facility (Harvard Medical School) for use of their microscopes; Dr. Maofu Liao for providing the Python scripts and help in image processing; Dr. Lixia Yue (University of Connecticut Health Center) for kindly provided the E1047Q construct; Dr. Nikolaus Grigorieff (HHMI) for reviewing the manuscript and offering insightful and helpful comments; and members of the D.E.C. laboratory for productive discussion. This work was supported by funds from HHMI (to D.E.C.). J.Z. was supported by the Thousand Young Talents program of China and National Natural Science Foundation of China (Grant 31770795).
Footnotes
The authors declare no conflict of interest.
Data deposition: Cryo-EM electron density maps of the mouse TRPM7-EDTA, TRPM7-Mg2+, and TRPM7-DVF have been deposited in the Electron Microscopy Data Bank under accession codes EMD-6975, EMD-7297, and EMD-7298, respectively. Atomic coordinates of the mouse TRPM7-EDTA, TRPM7-Mg2+ and TRPM7-DVF have been deposited in the Protein Data Bank, www.wwpdb.org, under accession codes 5ZX5, 6BWD, and 6BWF, respectively.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810719115/-/DCSupplemental.
References
- 1.Clapham DE. TRP channels as cellular sensors. Nature. 2003;426:517–524. doi: 10.1038/nature02196. [DOI] [PubMed] [Google Scholar]
- 2.Ramsey IS, Delling M, Clapham DE. An introduction to TRP channels. Annu Rev Physiol. 2006;68:619–647. doi: 10.1146/annurev.physiol.68.040204.100431. [DOI] [PubMed] [Google Scholar]
- 3.Runnels LW. TRPM6 and TRPM7: A Mul-TRP-PLIK-cation of channel functions. Curr Pharm Biotechnol. 2011;12:42–53. doi: 10.2174/138920111793937880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin J, et al. The channel kinase, TRPM7, is required for early embryonic development. Proc Natl Acad Sci USA. 2012;109:E225–E233. doi: 10.1073/pnas.1120033109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin J, et al. Deletion of Trpm7 disrupts embryonic development and thymopoiesis without altering Mg2+ homeostasis. Science. 2008;322:756–760. doi: 10.1126/science.1163493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Komiya Y, Runnels LW. TRPM channels and magnesium in early embryonic development. Int J Dev Biol. 2015;59:281–288. doi: 10.1387/ijdb.150196lr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sah R, et al. Ion channel-kinase TRPM7 is required for maintaining cardiac automaticity. Proc Natl Acad Sci USA. 2013;110:E3037–E3046. doi: 10.1073/pnas.1311865110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Runnels LW, Yue L, Clapham DE. TRP-PLIK, a bifunctional protein with kinase and ion channel activities. Science. 2001;291:1043–1047. doi: 10.1126/science.1058519. [DOI] [PubMed] [Google Scholar]
- 9.Nadler MJS, et al. LTRPC7 is a Mg.ATP-regulated divalent cation channel required for cell viability. Nature. 2001;411:590–595. doi: 10.1038/35079092. [DOI] [PubMed] [Google Scholar]
- 10.Chubanov V, et al. Disruption of TRPM6/TRPM7 complex formation by a mutation in the TRPM6 gene causes hypomagnesemia with secondary hypocalcemia. Proc Natl Acad Sci USA. 2004;101:2894–2899. doi: 10.1073/pnas.0305252101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li M, Jiang J, Yue L. Functional characterization of homo- and heteromeric channel kinases TRPM6 and TRPM7. J Gen Physiol. 2006;127:525–537. doi: 10.1085/jgp.200609502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krapivinsky G, Krapivinsky L, Manasian Y, Clapham DE. The TRPM7 chanzyme is cleaved to release a chromatin-modifying kinase. Cell. 2014;157:1061–1072. doi: 10.1016/j.cell.2014.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krapivinsky G, et al. Histone phosphorylation by TRPM6’s cleaved kinase attenuates adjacent arginine methylation to regulate gene expression. Proc Natl Acad Sci USA. 2017;114:E7092–E7100. doi: 10.1073/pnas.1708427114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desai BN, et al. Cleavage of TRPM7 releases the kinase domain from the ion channel and regulates its participation in Fas-induced apoptosis. Dev Cell. 2012;22:1149–1162. doi: 10.1016/j.devcel.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brauchi S, Krapivinsky G, Krapivinsky L, Clapham DE. TRPM7 facilitates cholinergic vesicle fusion with the plasma membrane. Proc Natl Acad Sci USA. 2008;105:8304–8308. doi: 10.1073/pnas.0800881105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krapivinsky G, Mochida S, Krapivinsky L, Cibulsky SM, Clapham DE. The TRPM7 ion channel functions in cholinergic synaptic vesicles and affects transmitter release. Neuron. 2006;52:485–496. doi: 10.1016/j.neuron.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 17.Abiria SA, et al. TRPM7 senses oxidative stress to release Zn2+ from unique intracellular vesicles. Proc Natl Acad Sci USA. 2017;114:E6079–E6088. doi: 10.1073/pnas.1707380114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Runnels LW, Yue L, Clapham DE. The TRPM7 channel is inactivated by PIP(2) hydrolysis. Nat Cell Biol. 2002;4:329–336. doi: 10.1038/ncb781. [DOI] [PubMed] [Google Scholar]
- 19.Chokshi R, Matsushita M, Kozak JA. Detailed examination of Mg2+ and pH sensitivity of human TRPM7 channels. Am J Physiol Cell Physiol. 2012;302:C1004–C1011. doi: 10.1152/ajpcell.00422.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang J, Li M, Yue L. Potentiation of TRPM7 inward currents by protons. Gen Physiol. 2005;126:137–150. doi: 10.1085/jgp.200409185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamaguchi H, Matsushita M, Nairn AC, Kuriyan J. Crystal structure of the atypical protein kinase domain of a TRP channel with phosphotransferase activity. Mol Cell. 2001;7:1047–1057. doi: 10.1016/s1097-2765(01)00256-8. [DOI] [PubMed] [Google Scholar]
- 22.Fujiwara Y, Minor DL., Jr X-ray crystal structure of a TRPM assembly domain reveals an antiparallel four-stranded coiled-coil. J Mol Biol. 2008;383:854–870. doi: 10.1016/j.jmb.2008.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clapham DE, Runnels LW, Strübing C. The TRP ion channel family. Nat Rev Neurosci. 2001;2:387–396. doi: 10.1038/35077544. [DOI] [PubMed] [Google Scholar]
- 24.Autzen HE, et al. Structure of the human TRPM4 ion channel in a lipid nanodisc. Science. 2018;359:228–232. doi: 10.1126/science.aar4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yin Y, et al. Structure of the cold- and menthol-sensing ion channel TRPM8. Science. 2018;359:237–241. doi: 10.1126/science.aan4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Z, Toth B, Szollosi A, Chen J, Csanady L. Structure of a TRPM2 channel in complex with Ca(2+) explains unique gating regulation. eLife. 2018;7:e36409. doi: 10.7554/eLife.36409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duan J, et al. Structure of full-length human TRPM4. Proc Natl Acad Sci USA. 2018;115:2377–2382. doi: 10.1073/pnas.1722038115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li M, et al. Molecular determinants of Mg2+ and Ca2+ permeability and pH sensitivity in TRPM6 and TRPM7. J Biol Chem. 2007;282:25817–25830. doi: 10.1074/jbc.M608972200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao E, Liao M, Cheng Y, Julius D. TRPV1 structures in distinct conformations reveal activation mechanisms. Nature. 2013;504:113–118. doi: 10.1038/nature12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao Y, Cao E, Julius D, Cheng Y. TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature. 2016;534:347–351. doi: 10.1038/nature17964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen PS, et al. The structure of the polycystic kidney disease channel PKD2 in lipid nanodiscs. Cell. 2016;167:763–773.e711. doi: 10.1016/j.cell.2016.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Q, et al. Structure of mammalian endolysosomal TRPML1 channel in nanodiscs. Nature. 2017;550:415–418. doi: 10.1038/nature24035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmiege P, Fine M, Blobel G, Li X. Human TRPML1 channel structures in open and closed conformations. Nature. 2017;550:366–370. doi: 10.1038/nature24036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirschi M, et al. Cryo-electron microscopy structure of the lysosomal calcium-permeable channel TRPML3. Nature. 2017;550:411–414. doi: 10.1038/nature24055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paulsen CE, Armache JP, Gao Y, Cheng Y, Julius D. Structure of the TRPA1 ion channel suggests regulatory mechanisms. Nature. 2015;520:511–517. doi: 10.1038/nature14367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fan C, Choi W, Sun W, Du J, Lu W. Structure of the human lipid-gated cation channel TRPC3. eLife. 2018;7:7. doi: 10.7554/eLife.36852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang Q, et al. Structure of the receptor-activated human TRPC6 and TRPC3 ion channels. Cell Res. 2018;28:746–755. doi: 10.1038/s41422-018-0038-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Minke B, Cook B. TRP channel proteins and signal transduction. Physiol Rev. 2002;82:429–472. doi: 10.1152/physrev.00001.2002. [DOI] [PubMed] [Google Scholar]
- 39.Rohács T, Lopes CMB, Michailidis I, Logothetis DE. PI(4,5)P2 regulates the activation and desensitization of TRPM8 channels through the TRP domain. Nat Neurosci. 2005;8:626–634. doi: 10.1038/nn1451. [DOI] [PubMed] [Google Scholar]
- 40.Wang T, Jiao Y, Montell C. Dissecting independent channel and scaffolding roles of the Drosophila transient receptor potential channel. J Cell Biol. 2005;171:685–694. doi: 10.1083/jcb.200508030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.García-Sanz N, et al. A role of the transient receptor potential domain of vanilloid receptor I in channel gating. J Neurosci. 2007;27:11641–11650. doi: 10.1523/JNEUROSCI.2457-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xie J, et al. Phosphatidylinositol 4,5-bisphosphate (PIP(2)) controls magnesium gatekeeper TRPM6 activity. Sci Rep. 2011;1:146. doi: 10.1038/srep00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Catterall WA. ion channel voltage sensors: Structure, function, and pathophysiology. Neuron. 2010;67:915–928. doi: 10.1016/j.neuron.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swartz KJ. Sensing voltage across lipid membranes. Nature. 2008;456:891–897. doi: 10.1038/nature07620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gouaux E, Mackinnon R. Principles of selective ion transport in channels and pumps. Science. 2005;310:1461–1465. doi: 10.1126/science.1113666. [DOI] [PubMed] [Google Scholar]
- 46.Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504:107–112. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hattne J, et al. Analysis of global and site-specific radiation damage in cryo-EM. Structure. 2018;26:759–766.e754. doi: 10.1016/j.str.2018.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sperelakis N. Cell Physiology Source Book: Essentials of Membrane Biophysics. Academic; San Diego: 2011. [Google Scholar]
- 49.Markham GD, Glusker JP, Bock CL, Trachtman M, Bock CW. Hydration energies of divalent beryllium and magnesium ions: An ab initio molecular orbital study. J Phys Chem. 1996;100:3488–3497. [Google Scholar]
- 50.Guskov A, et al. Structural insights into the mechanisms of Mg2+ uptake, transport, and gating by CorA. Proc Natl Acad Sci USA. 2012;109:18459–18464. doi: 10.1073/pnas.1210076109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hite RK, MacKinnon R. Structural titration of Slo2.2, a Na+-dependent K+ channel. Cell. 2017;168:390–399.e311. doi: 10.1016/j.cell.2016.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goehring A, et al. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat Protoc. 2014;9:2574–2585. doi: 10.1038/nprot.2014.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 54.Zheng SQ, et al. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ru H, et al. Molecular mechanism of V(D)J recombination from synaptic RAG1-RAG2 complex structures. Cell. 2015;163:1138–1152. doi: 10.1016/j.cell.2015.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Frank J, et al. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 57.Scheres SH. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Afonine PV, Headd JJ, Terwilliger TC, Adams PD. New tool: Phenix.real_space_refine. Comput Crystallogr Newsl. 2013;4:43–44. [Google Scholar]
- 62.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MS. HOLE: A program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph. 1996;14:354–360, 376. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
- 64.Neher E. Correction for liquid junction potentials in patch clamp experiments. Methods Enzymol. 1992;207:123–131. doi: 10.1016/0076-6879(92)07008-c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.