Significance
Alternative pre-mRNA splicing (AS) is a critical gene regulatory mechanism to produce diverse, tissue-specific, and functionally distinct protein profiles in eukaryotes to maintain normal cellular functions. Aberrant AS patterns are constantly associated with many human diseases, including cancer. The exceptional complexity of AS imposes a major challenge to analyzing AS across various tissues and cell types. Here we present a computational algorithm to profile and quantitate tissue-specific AS profiles from RNA-sequencing data without any prior knowledge of the host transcriptome. The junction usage model shows consistent superior performance in both specificity and sensitivity compared with other currently available AS analysis methods, and can be readily applied to a wide range of RNA samples from different organisms for accurate and comprehensive analyses of AS.
Keywords: alternative pre-mRNA splicing, annotation-free, RNA-seq
Abstract
Alternative pre-mRNA splicing (AS) greatly diversifies metazoan transcriptomes and proteomes and is crucial for gene regulation. Current computational analysis methods of AS from Illumina RNA-sequencing data rely on preannotated libraries of known spliced transcripts, which hinders AS analysis with poorly annotated genomes and can further mask unknown AS patterns. To address this critical bioinformatics problem, we developed a method called the junction usage model (JUM) that uses a bottom-up approach to identify, analyze, and quantitate global AS profiles without any prior transcriptome annotations. JUM accurately reports global AS changes in terms of the five conventional AS patterns and an additional “composite” category composed of inseparable combinations of conventional patterns. JUM stringently classifies the difficult and disease-relevant pattern of intron retention (IR), reducing the false positive rate of IR detection commonly seen in other annotation-based methods to near-negligible rates. When analyzing AS in RNA samples derived from Drosophila heads, human tumors, and human cell lines bearing cancer-associated splicing factor mutations, JUM consistently identified approximately twice the number of novel AS events missed by other methods. Computational simulations showed JUM exhibits a 1.2 to 4.8 times higher true positive rate at a fixed cutoff of 5% false discovery rate. In summary, JUM provides a framework and improved method that removes the necessity for transcriptome annotations and enables the detection, analysis, and quantification of AS patterns in complex metazoan transcriptomes with superior accuracy.
Alternative pre-mRNA splicing (AS) is a major gene regulatory mechanism that greatly expands proteomic diversity and serves as a crucial determinant of cell fate and identity. More than 95% of human gene transcripts undergo AS that enables one single gene locus to produce multiple, and usually functionally distinct, pre-mRNA and protein isoforms (1, 2). AS is regulated by a large constellation of RNA-binding proteins that interact with cis-acting RNA elements embedded in nuclear pre-mRNA sequences (3, 4). Distinct cellular states or tissue types are associated with different AS profiles that affect almost every aspect of cellular function, including proliferation, differentiation, apoptosis, and migration (1, 5, 6). Furthermore, mutations that result in aberrant AS patterns are a major source for human diseases such as cancer as well as immune and neurological disorders (7–9). Thus, a thorough and comprehensive evaluation of global AS profiles in different tissues, cells, and disease states will be critical to understanding the role of AS in gene regulation and facilitating the development of screening and therapeutic strategies to diagnose, treat, and prevent many diseases linked to defects in AS. However, due to the exceptionally diverse and dynamic features of AS patterns, systematic quantification and analysis of cellular AS profiles among a complex array of tissues or cell types remain major unsolved challenges in the bioinformatics of gene expression.
Recent technical advances in short-read high-throughput Illumina transcriptome sequencing (RNA sequencing; RNA-seq) provide powerful tools to investigate AS at the genome-wide scale, but at the same time present a formidable computational challenge to accurately classify and quantitate global AS changes from raw RNA-seq data. Previously, a number of computational software tools and algorithms have been developed for this purpose (10–22), but most use a top-down approach that relies on preannotation of known AS events or an incomplete, preannotated transcriptome to draft the general picture of global AS patterns for quantification and analysis. As complete dependency on annotation (10) restricts AS analysis to only previously observed AS events, recent methods generally use two approaches to extend the analysis to unannotated splicing events: (i) Supplement the preannotated AS event library with novel splice junction-implicated AS events identified from the sample under analysis (15, 18, 22); or (ii) provide a de novo transcriptome annotation through ab initio transcriptome assembly from RNA-seq data using probabilistic models (12, 23–26). For the first approach, the library of preannotated AS events is still the primary source for calling AS events and can either mask or misclassify novel AS events in the specific RNA-seq sample. For the second approach, a precise and deterministic ab initio assembly of transcriptomes from shotgun RNA sequencing is still a big computational challenge for the field, especially for genes that produce multiple transcripts with complex AS patterns. Thus, the difficulties in transcriptome assembly will directly affect the quality of downstream AS analysis. Considering the caveats described above, there is an urgent need for development of computational tools that can perform accurate, comprehensive, and tissue-specific global AS analysis with a different approach.
Here, we present a computational method called the junction usage model (JUM) that uses a bottom-up approach to profile, analyze, and quantitate tissue-specific global AS patterns without any prior knowledge of the transcriptome. JUM exclusively uses sequence reads spanning splice junctions to faithfully assemble complete AS patterns in the RNA-seq samples based on their unique topological features and to quantify splicing changes. We applied JUM to analyze AS patterns in RNA samples from Drosophila heads, mouse embryonic neurons, human cancer tumor samples, as well as human cell lines bearing cancer-associated splicing factor mutations. We demonstrate that JUM consistently identified numerous novel, previously not observed, true tissue-specific AS events that were missed or misclassified when analyzed using annotation-based methods. Furthermore, computational simulations showed that JUM exhibits superior performance in terms of both specificity and sensitivity compared with several popular annotation-based methods. Thus, JUM provides a new framework and improved analytical approach to studying the extraordinarily diverse global cellular AS transcriptome profiles and the dynamic regulation of AS without the necessity of transcriptome annotation. JUM can be readily applied to a wide range of RNA samples from different organisms for accurate and quantitative analysis of differential AS patterns.
Results
JUM Utilizes Sequence Reads Spanning Splice Junctions to Construct AS Structures as the Basic Quantitation Unit for AS Analysis.
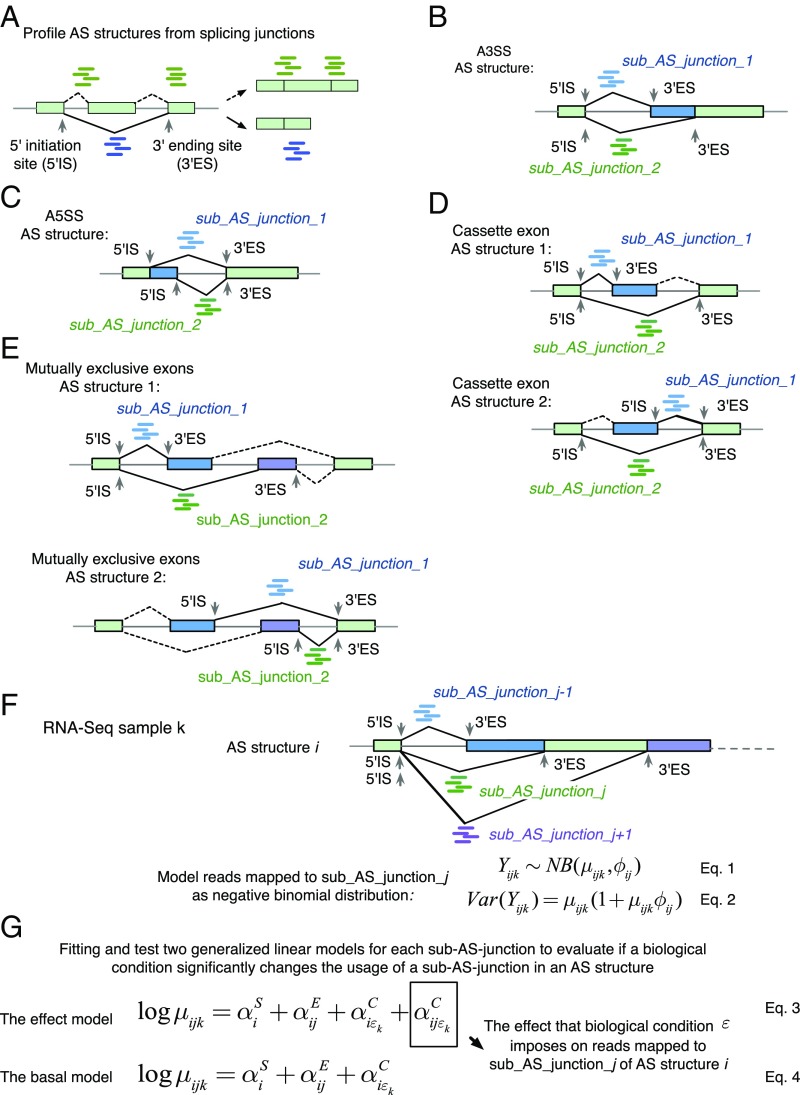
JUM exclusively uses sequence reads that map over splice junctions to detect and quantitate splicing events (Fig. 1A), as these reads provide the most direct evidence for the splicing of the corresponding intron and quantitatively reflect the level of splicing. These splice junction reads can be inferred through the mapping of the shotgun sequencing reads to the genome as reads that cannot be completely mapped to one location in the genome but instead map as “split” reads. From there, JUM defines the AS structure as the basic quantitation unit for AS analysis. An AS structure is a set of splice junctions that share the same start site or the same ending site, with each splice junction in an AS structure defined as a sub-AS-junction (Fig. 1 A and B). JUM uses AS structures for AS analysis because not only are AS structures the basic graphical nodes that compose the conventionally recognized AS patterns (alternative 5′ splice site, A5SS; alternative 3′ splice site, A3SS; skipped cassette exon, SE; mutually exclusive exon, MXE; intron retention, IR) but also the relative levels of sub-AS-junctions within an AS structure directly reflect the level of alternative splicing, greatly facilitating AS quantification. As a result, an A5SS or A3SS event is composed of one AS structure with two sub-AS-junctions (Fig. 1 B and C); an SE event is composed of two AS structures, each with two sub-AS-junctions (Fig. 1D); and an MXE event with two mutually exclusive exons is composed of two AS structures, each with two sub-AS-junctions (Fig. 1E).
Fig. 1.
JUM exclusively uses sequence reads mapped to splice junctions and defines AS structures as the basic quantitation unit for differential AS analysis. (A) JUM uses RNA-seq reads mapped to splice junctions for AS quantification. Green rectangles indicate exons, and lines indicate introns. Green and blue short lines represent reads that mapped to splice junctions connecting exons, which are the most direct evidence for the existence and quantitative assessment of a given splice junction. JUM defines the start coordinate of a splice junction as the 5′ initiation site (5′IS) and the end coordinate of a splice junction as the 3′ ending site (3′ES). An “AS structure” is defined as a set of junctions that share the same 5′IS or the same 3′ES. Each splice junction in an AS structure is defined as a sub-AS-junction. (B–E) AS structures are the basic element that comprises all conventionally recognized AS patterns (B) for A3SS; (C) for A5SS; (D) for SE; (E) for MXE. (F) JUM models the sequence reads that map to a sub-AS-junction as a negative binomial distribution to quantify the usage of each sub-AS-junction in an AS structure under one biological condition. (G) JUM fits two generalized linear models to evaluate the influence of a given biological condition on the usage of a specific sub-AS-junction in an AS structure.
After the profiling of all AS structures, JUM counts sequence reads that are mapped to each sub-AS-junction in every AS structure under a biological condition and defines the read count as the “usage” of a sub-AS-junction relative to other sub-AS-junctions in the same AS structure under that condition. To quantify AS changes, JUM compares the usage of every profiled sub-AS-junction in the AS structure between conditions and profiles for AS structures that contain sub-AS-junctions with differential usage (Fig. 1F). To do this, JUM models the total number of reads that map to a sub-AS-junction as a negative binomial distribution (Fig. 1F, Eq. 1). Negative binomial distributions have been widely applied in high-throughput sequencing data analysis to model read counts, as these models nicely depict the overdispersion phenomenon observed in next-generation RNA-sequencing experiments (11, 27–30). In negative binomial distributions, the variance among biological replicates is dependent on the mean through a parameter that describes dispersion (Fig. 1F, Eq. 2). To infer the dispersion parameter, JUM applies a similar empirical Bayesian approach as described (28–31). JUM first estimates a dispersion parameter for each sub-AS-junction with Cox–Reid–adjusted maximum likelihood. JUM then fits a mean-variance function for all sub-AS-junctions from all AS structures on their average normalized count values. Finally, JUM shrinks the dispersion parameter for each individual sub-AS-junction toward the fitted value depending on how close the real dispersion tends to be to the fitted value and the replicate sample size (28–31). To evaluate if a biological condition significantly changes the usage of a sub-AS-junction in the AS structure, JUM adapts a generalized linear model (GLM) approach as described (11, 30, 32), so that two GLMs are fitted and tested for each sub-AS-junction in the AS structure (11) (Fig. 1G). The basal model evaluates the effect from the following three elements on the usage of the sub-AS-junction: the basal expression level of the AS structure of the corresponding gene (; Fig. 1G, Eq. 4), the fraction of sequence reads that mapped to each sub-AS-junction from the total number of reads mapped to the AS structure (; Fig. 1G, Eq. 4), as well as the overall change of basal expression of the AS structure upon a biological condition (; Fig. 1G, Eq. 4). On the other hand, the effect model evaluates an additional influence imposed on the usage of a sub-AS-junction by a biological condition (; Fig. 1G, Eq. 3). The fitting of the effect and basal model are compared and a likelihood-ratio test is performed (11) so as to test if a biological condition causes significant differential usage of a sub-AS-junction in the AS structure.
JUM Profiles a Tissue-Specific Global AS Atlas by Faithfully Assembling AS Structures into Conventionally Recognized AS Patterns Without Any Prior Knowledge of the Transcriptome Annotation.
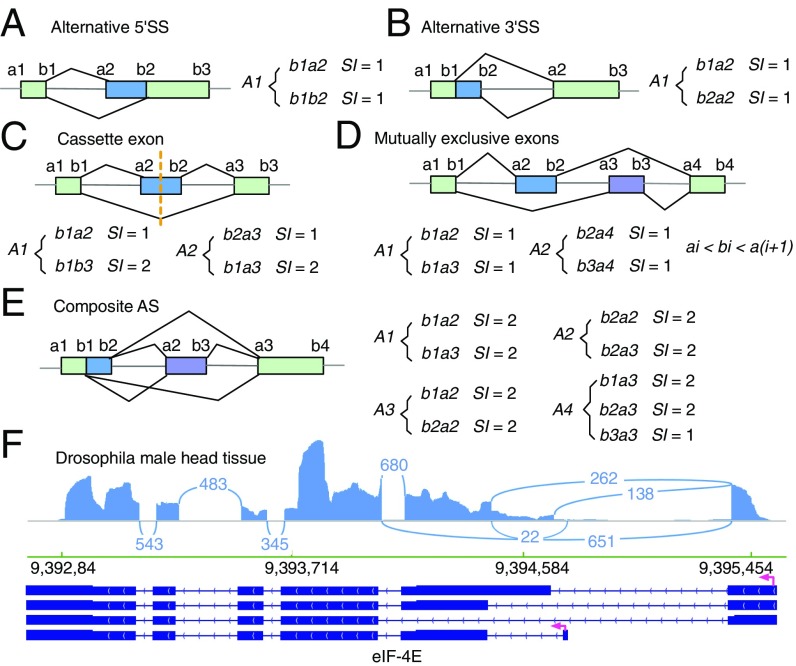
After differential AS analysis using AS structures, JUM assembles profiled AS structures into conventionally recognized categories of AS patterns using graph theory based on the unique topological feature of each pattern. To do this, JUM first converts each AS pattern into a graph by converting exons into nodes and splice junctions as arcs that connect exon nodes. JUM then defines a frequency parameter SI for each sub-AS-junction as the number of AS structures that share the specific sub-AS-junction. Because of the definition of AS structures, it can be proven that a given sub-AS-junction can only be included in up to two AS structures (i.e., SI can only be 1 or 2). For the A5SS or A3SS patterns, the representative graphs are asymmetric and are composed of one AS structure with SI value equal to 1 for all sub-AS-junctions (Fig. 2 A and B). For the SE pattern, the representative graphs are symmetric, composed of two AS structures, each containing two sub-AS-junctions with SI values equal to 1 and 2, respectively (Fig. 2C). For SE, JUM utilizes extra quarantine steps here, including tiled sequence reads that support the coverage over the entire cassette exon region to avoid false positive calls. For MXE with mutually exclusive exons, the representative graph is composed of one pair of AS structures that each has n sub-AS-junctions with SI values all equal to 1 (Fig. 2D). For the MXE pattern, JUM utilizes extra quality control steps, including that coordinates of MXEs meet the condition ai < bi < a(i + 1), where i = 1, …, n (Fig. 2D), and tiled sequence reads that support coverage over the entire regions of all mutually exclusive exons. Based on the unique topological features of each AS pattern described above, JUM searches for sets of AS structures that match the composition of each AS pattern and bundles them together as one AS event under the corresponding AS pattern category.
Fig. 2.
JUM profiles the AS atlas specific to the sample by assembling AS structures into conventionally recognized categories of AS patterns based on the unique topological features of each AS pattern type. (A–D) The topological features of AS patterns A5SS (A), A3SS (B), SE (C), and MXE (D) represented by the splicing graphs that are composed of a unique set of AS structures and the values of frequency parameter SI of each sub-AS-junction in the AS structures. (E) JUM defines an additional, previously unclassified AS pattern category—the composite AS, which is a complex combination of several conventionally recognized AS patterns. (F) An example for such a composite AS pattern is shown for the eIF-4E gene transcripts found in Drosophila male head tissue RNA-seq samples (49). Arcs represent splice junctions that connect different exons.
Additionally and uniquely, JUM also recognizes and defines an additional AS pattern called “composite,” which describes an AS event that is an inseparable combination of several conventionally recognized AS patterns (Fig. 2 E and F). Such composite AS patterns are found extensively in Drosophila, rodent, and human tissues and cell line RNA samples (Fig. 2F), further illustrating the complexity and diversity of AS in complex tissues. For example, JUM identifies an AS event in the transcripts from the eIF-4E gene in Drosophila male heads that is an intertwined combination of SE, A5SS, and A3SS and cannot be deconvoluted into any of the three individual patterns separately (Fig. 2F). Although such complex AS events have been reported before (33), they are mostly divided into the conventionally recognized AS patterns by the currently available AS analysis tools. This approach overlooks the fact that such composite AS events are usually an intertwined combination of the basic AS patterns and a forced separation of these events into the basic patterns can lead to inaccurate quantification of the AS changes. JUM thus classifies these complex AS events in their own category. For composite AS events in JUM, the representative graph includes an interconnected combination of AS structures corresponding to each AS pattern that composes the composite AS pattern. For example, a composite AS pattern event that is a combination of an A5SS and SE is composed of four AS structures, three of which have sub-AS-junctions with SI values all equal to 2 and one that has one sub-AS-junction with an SI value equal to 1 and the rest with SI value equal to 2 (Fig. 2E).
JUM Applies Stringent Criteria for the Analysis of Intron Retention Events.
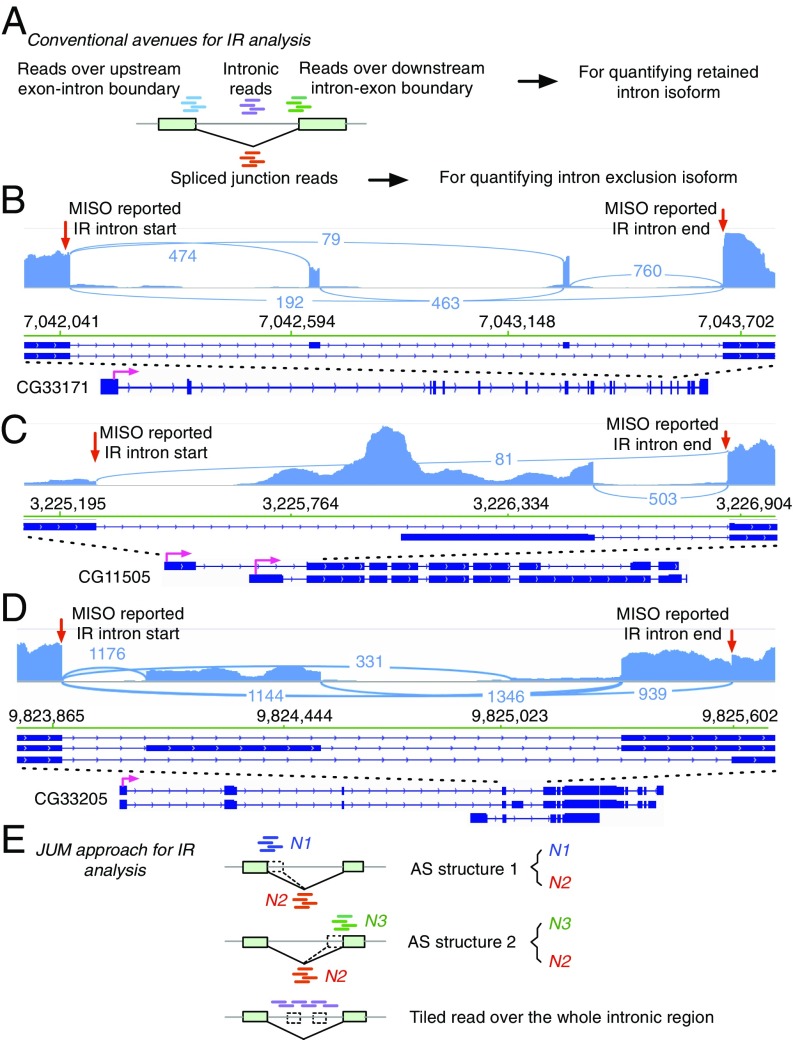
Intron retention has been a relatively understudied category compared with other AS patterns, but nevertheless a key AS pattern. IR events have been reported to be frequently found in mammals and have been shown to play crucial roles for the normal functioning of the organism and in disease in eukaryotes (34, 35). For example, tissue-specific IR of the Drosophila P-element transposase pre-mRNA underlies the restriction of transposon activity to germ-line tissues (36, 37). Recently, a bioinformatics study reported that widespread retained introns were associated with various cancer types compared with matched normal tissues (38). Increased IR has also been shown to be associated with the pluripotent state of embryonic stem cells (39). However, intron retention is an intricate AS pattern that can be easily misclassified. The most common approaches to quantifying retained intron-containing isoforms in currently available AS analysis tools are either to use the sum of sequence reads mapped to the upstream exon–intron boundary and the downstream intron–exon boundary, or to use any reads mapped to the intronic region or just the center of the intron (Fig. 3A). A major caveat of these approaches is that other AS patterns can be mistaken as IR, especially when alternative SEs or MXEs reside within an intron (Fig. 3 B–D) or an A5SS/A3SS event resides at the edge of the intron (Fig. 3D). In such scenarios, sequence reads from the intronic region can in fact come from the SEs or MXEs and reads mapped to exon–intron or intron–exon boundaries can come from A5SS or A3SS events, but with the conventionally available methods these reads can be mistakenly interpreted as support for intron-retained isoforms.
Fig. 3.
JUM applies stringent criteria for detection, quantitation, and analysis of IR events. (A) The conventional avenues for IR analysis in the current available AS analysis tools. RNA-seq reads spanning intron–exon or exon–intron boundaries are represented by short green or blue lines, respectively. Short purple lines represent sequence reads mapped to intronic regions. Short red lines represent sequence reads mapped to the splice junction for the corresponding intron. (B–D) The commonly used strategies in other AS analysis software can misclassify other AS patterns as IR. Three MISO-reported (49) significantly changed IR events were shown that were actually an MXE (B), alternative promoter event (C), and SE mixed with A3SS (D) from Drosophila male head tissue in a comparison of a control wild-type fly strain and a transgenic fly strain that expresses the truncated PSI protein. The start and end points of the retained intron events reported by MISO are denoted by red arrows. (E) The approach that JUM uses to analyze IR. Short blue and green lines represent reads mapped to the exon–intron or intron–exon boundaries, respectively. Short red lines represent sequence reads mapped to the splice junctions. Two AS structures are constructed to analyze the level of retained intron isoform versus spliced intron isoform. Short purple reads represent sequence reads mapped to the intronic regions and are required to be approximately uniformly distributed across the entire intronic region of the retained intron.
To avoid false positive calls of IR as described above, JUM applies a stringent three-criterion strategy to profile and analyze IR patterns (Fig. 3E). First, JUM profiles for splice junctions that do not overlap with any other splice junctions from the RNA-seq data. Second, for each of the resulting splice junctions and the corresponding intron, JUM counts the number of sequence reads mapped to the upstream exon–intron boundary (N1), reads spanning across the splice junction (N2), and reads mapped to the downstream intron–exon boundary (N3) (Fig. 3E). JUM then defines two AS structures for each candidate intron: N1 versus N2, and N3 versus N2. Both AS structures must be differentially “used” with the same trend (N1 and N3 both significantly more used than N2 or both significantly less used than N2) in order for the candidate to be classified as a potential IR event. These two criteria are set to avoid A5SS or A3SS events from being mistaken as IR (Fig. 3D). Finally, JUM requires evidence from mapped sequence reads that are approximately evenly distributed across every base of the intron, to confirm a real IR event (Fig. 3E). This criterion aims at preventing SE or MXE events from being misclassified as IR, as reads from SEs or MXEs residing in the intron will present a much higher, “spikey” distribution pattern compared with other regions of the intron.
JUM Demonstrates Superior Performance in Both Specificity and Sensitivity Compared with Other Methods in Computationally Simulated RNA-Seq Experiments.
To fully assess the performance of JUM in differential global AS analysis, we performed computational simulations of RNA-seq datasets with varying degrees of alternative splicing in a prefixed set of genes (SI Appendix, Table S1) and used the simulated datasets to test the ability of JUM to profile true differentially spliced AS events. We compared JUM with five other commonly used annotation-based tools—MISO (10), Whippet (21), Cufflinks (40), MAJIQ (18), and rMATS (15) (SI Appendix, Table S2). The five tools were chosen to represent the three most commonly used strategies in annotation-based AS analysis methods: MISO is completely dependent on a developer-provided preannotation of AS events and cannot detect novel AS events outside of the provided database (10, 21); Whippet also analyzes known AS events from a user-provided transcriptome annotation, but in addition can detect a subclass of novel splicing events that utilize various combinations of annotated 5′ or 3′ splice sites that are present in the provided annotation database (21); Cufflinks first performs the challenging de novo transcriptome assembly from shotgun sequencing and then quantifies AS changes based on the annotation of the assembled transcriptome (40); MAJIQ and rMATS both use a preannotation of the transcriptome to guide the AS analysis but add in novel splicing junctions detected in the specific RNA-seq sample (15, 18). The latter four methods can extend analysis to novel splicing events in the sample.
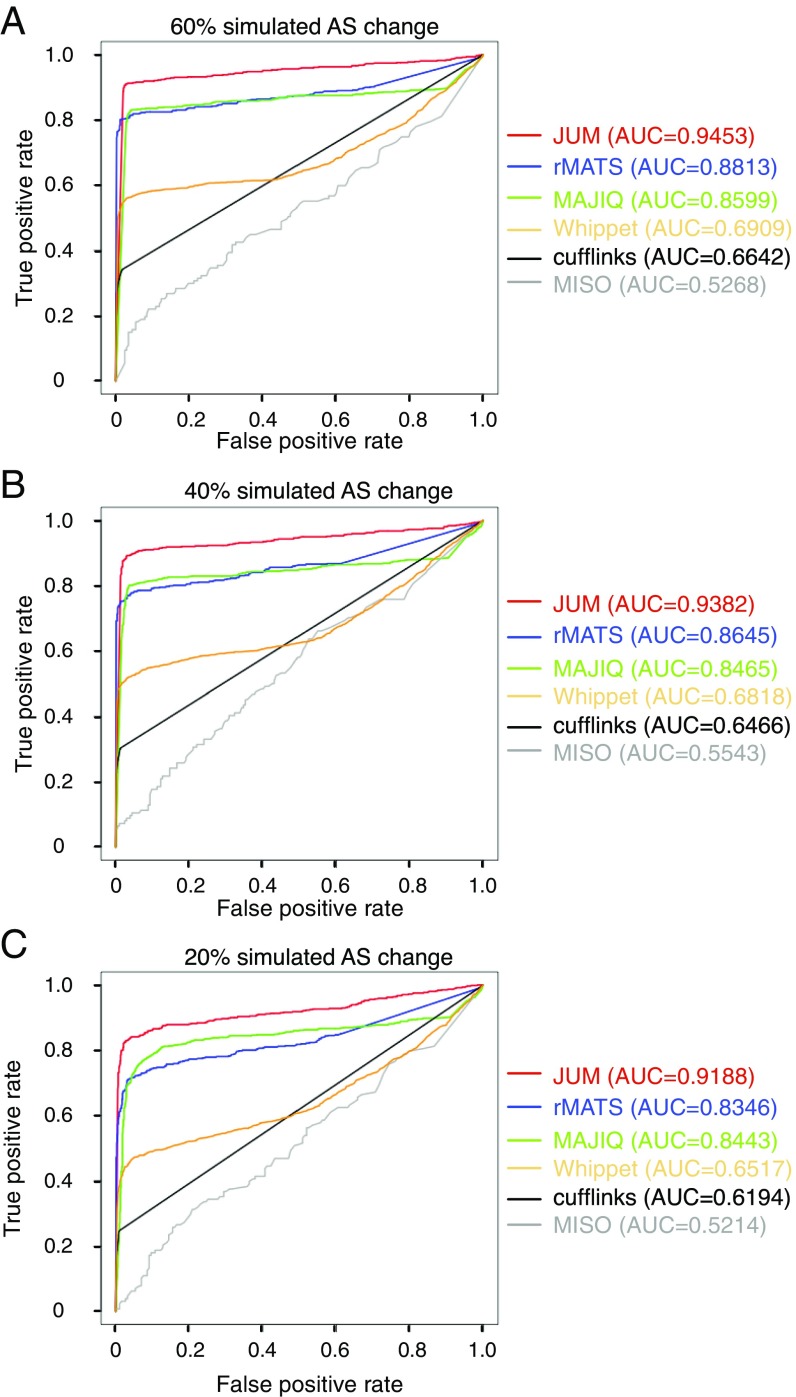
The test datasets are simulated based on real experimental RNA-seq data in Drosophila Schneider-2 cell lines comparing AS changes brought about by the RNA interference (RNAi) knockdown of a splicing factor called PSI (41) and a nontargeting, control RNAi knockdown. The simulation follows a method that has been previously described (42). Specifically, a randomly chosen 2,000 genes from expressed genes in the Drosophila Schneider-2 cell line are set to be alternatively spliced, serving as the “true” AS genes, with AS patterns covering all patterns. Triplicates of ∼80 million total 100-bp RNA-seq reads were simulated for both the PSI knockdown and control knockdown samples. Three independent simulations were performed, with 20, 40, and 60% differential splicing changes in the true AS genes between the control and knockdown conditions. The performance of each AS analysis software under comparison was evaluated in terms of the receiver operating characteristic (ROC) curves and the area under the curve (AUC) metric. The ROC curve depicts the true positive rate (sensitivity) against the false positive rates (1-specificity) for each threshold setting to call an AS event. AUC is a numerical metric that determines how well a method can distinguish between the true AS events and non-AS events. AUC scores range between 0.5 and 1, and a higher AUC score indicates a method with better discrimination between AS and non-AS events.
Importantly, JUM received the best AUC score in all three simulated RNA-seq experiments among the six methods tested, indicating its superior performance in both sensitivity and specificity (Fig. 4). The AUC scores for JUM in all three simulations ranged from 0.92 to 0.95, indicating that JUM is a superior method for accurate differential AS analysis by the AUC standard (AUC value between 0.9 and 1). rMATS and MAJIQ show comparable performance, with AUC values ranging between 0.83 and 0.88; Whippet and Cufflinks perform worse than both rMATS and MAJIQ, with AUC values ranging between 0.62 and 0.69, but still better than the completely annotation-dependent method MISO, which has AUC values of 0.52 to 0.55 (Fig. 4). The poor performance of MISO is to some extent understandable, because the true AS genes in the test set used here are randomly chosen, which is vastly different from the developer-provided MISO annotation. The results from these simulations further demonstrate the importance for an AS analysis method to account for sample-specific, novel AS patterns rather than using a preannotated library of AS events to achieve better performance in AS analysis.
Fig. 4.
Comparison of JUM with five other of the most widely used computational methods (rMATS, MAJIQ, Whippet, Cufflinks, and MISO) for AS analysis using computationally simulated RNA-seq experiments. Receiver operating characteristic curves are shown for each method to illustrate their sensitivity and specificity in identifying true differentially spliced AS events. The y axis of the ROC figure shows the true positive rate and the x axis shows the false positive rate. The metric area under a ROC curve is listed (Right). Three independent simulations were done by varying the alternative splicing changes at levels of 60 (A), 40 (B), and 20% (C).
The computation times of the six software tools in analyzing the simulated datasets are summarized in SI Appendix, Fig. S5.
Following the simulation, we applied JUM to multiple experimental RNA-seq datasets from two different organisms to evaluate the performance of JUM in real RNA-seq datasets.
JUM Revealed Remarkable Heterogeneity of IR Splicing in Colon Cancer Patient Tumor Samples and Significantly Reduced the False Positive Rate of IR Detection.
To assess the performance of JUM in IR analysis, we used JUM to analyze differential splicing of IR between the tumor and the matched normal tissue samples from colon cancer patients in The Cancer Genome Atlas (TCGA) database. A previous study (38) used MISO and the MISO built-in preannotation of human AS events to compare the AS profiles between patient tumor and matched normal tissues and reported that extensive intron retention is a prevalent feature and was a highly elevated splicing pattern observed in cancer, with colon cancer among the cancer types where this phenomenon was the most obvious (38). However, MISO restricts cancer IR analysis to a fixed set of only 6,895 IR events in its annotation library (10), which may only be partially present in various cancer cells or tissues and also includes numerous false positive events called by MISO (Fig. 3 B–D). Moreover, cancer cells are well-known to display diverse splicing patterns that are novel and cancer-specific (6, 43–45). To explore the features and functions of IR splicing in cancer, we conducted a detailed IR analysis on the colon cancer patient RNA-seq datasets from TCGA using JUM (Datasets S1–S34).
To avoid technical and sampling bias brought about by factors other than the cancerous state, we chose samples from two sets of colon cancer patients in TCGA database: (i) five male colon cancer patients that are of similar ages (60 to 68 y old), same colon tumor type (primary tumor), same vital states (alive), and with matched tumor and normal tissue samples sequenced using the same platform; and (ii) six female colon cancer patients chosen with similar parameters as described above except with a larger age span from 40 to 85 y old, as there were limited choices in age for female patient samples in TCGA database (SI Appendix, Table S4).
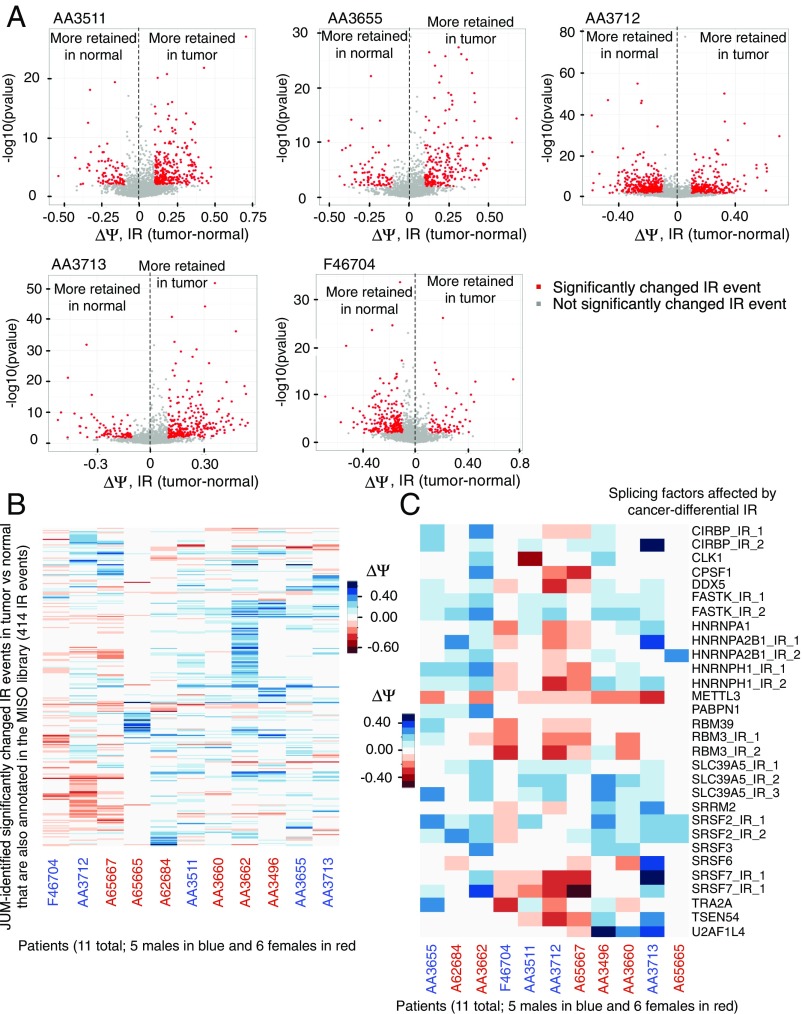
JUM identified 168 to 544 significantly different IR events in colon tumor versus matched normal tissues in each of the 11 patients (SI Appendix, Tables S5–S6). Among them, three patients (two males, AA3712 and F46704, and one female, A65667) displayed more retained introns in normal tissues compared with matched tumor tissue, while for the rest of the patients the tumor samples were associated with more retained introns (Fig. 5A and SI Appendix, Fig. S2 A and B). We also observed a wide range in the magnitude of retained intron isoform levels () and changes between tumor and matched normal tissues () across these patients’ samples (Fig. 5A and SI Appendix, Fig. S2A). To evaluate IR diversity among these patients and to compare our results with the previous study (38), we specifically profiled both the magnitude and direction of retained intron isoform-level changes (ΔΨ; IRtumor–normal) across all patients for a set of 414 IR events that were identified by JUM as significantly changed in at least one patient’s tumor versus matched normal tissue samples and also in the MISO annotation library (Fig. 5B and Dataset S36). Remarkably, we found that each patient has his or her own spectrum of significantly tumor- or normal tissue-differential IR events and the direction, as well as the extent of the retained intron isoform-level changes in these IR events, is heterogeneous across all patient samples analyzed (Fig. 5B). An overview of all JUM-identified significantly changed IR events from patients (including novel, tumor-specific, and known IR events) also showed a similar pattern (SI Appendix, Fig. S2C and Dataset S37). Moreover, each patient’s cancer-differential IR events affect distinct sets of genes in each patient’s tumor tissue (SI Appendix, Fig. S2D). A Gene Ontology (GO) analysis of the cancer-differential IR-affected genes in each patient revealed distinct functional enrichments that cover multiple aspects of carcinogenic metabolisms and activities (SI Appendix, Fig. S3A and Dataset S35), except for the GO category of RNA splicing, which is significantly enriched in most patients (8 out of 11) (SI Appendix, Fig. S3A). This result further highlights the association between aberrant splicing and cancerous cellular states, as previously recognized (6, 43–45). We profiled a total of 90 IR events influencing 44 splicing factors that are affected by cancer-differential IRs in at least one patient’s samples (Dataset S38). Interestingly, however, distinct introns of these splicing factors tend to be affected by IR in different patients (Dataset S38). To evaluate IR heterogeneity that perturbs splicing factors in cancer, we profiled a total set of 30 IR events (affecting 21 splicing factors) that are significantly different in tumor versus matched normal tissues in at least three patients and plotted the magnitude and direction of retained intron isoform-level changes (ΔΨ; IRtumor–normal) across all patients (Fig. 5C and Dataset S39). Again, we found that in general each patient’s tumor sample possesses its own specific spectrum of IR events for the identity of retained introns, direction of IR splicing changes, as well as the extent of IR changes affecting these splicing factors (Fig. 5C and Dataset S39). However, for four factors, FASTK, METTL3, SLC39A5, and SRSF2, the same intron and similar direction of changes in IR were observed in most patients, but still with variations in the extent of change in IR across patients (Fig. 5C). Thus, we conclude that aberrant IR splicing events in cancer are highly heterogeneous across individual patient tumors even within the same cancer type and that the functional effects of IR splicing events in cancer are probably more random and diverse than previously thought. There is indeed an association of IR changes with splicing regulators in colon tumors; however, the variance in how IR can affect the splicing program in different patients’ tumors is also high.
Fig. 5.
JUM revealed striking diversity and high variance of intron retention splicing in colon cancer patients’ tumor samples versus matched normal tissues. (A) Volcano plots showing the magnitude and direction of retained intron isoform-level changes (ΔΨ; IRtumor–normal) between tumor and matched normal tissues on the x axis and the statistical significance of change [−log10(P value)] on the y axis for every JUM-profiled IR event in each of the five male patients AA3511, AA3655, AA3712, AA3713, and F46704. Every dot is an IR event, and red dots mark IR events that are significantly changed between tumor and matched normal tissues (adjusted P value ≤ 0.05 and ΔΨ ≥ 0.1). Dots (IR events) plotted on the right side of the dashed line in each panel are IR events with more retained intron isoform in the tumor, while dots on the left side are IR events with more retained intron isoform in normal tissues. (B) Heatmap plot showing the magnitude and direction of retained intron isoform-level changes (ΔΨ; IRtumor–normal) between tumor and matched normal tissues for the 414 IR events that are significantly changed in at least one patient’s tumor versus matched normal tissues identified by JUM and are also annotated in the MISO-provided annotation library across all 11 patients. Each row is an IR event and each column is a patient as specified below the plot, with male patients’ ID shown in blue and female patients’ ID shown in red. Positive ΔΨ values are plotted in blue showing more retained intron in the tumor tissue for that IR event in the corresponding patient’s samples, and negative ΔΨ values are plotted in red showing more retained intron in normal tissue in the corresponding patient’s samples. The color key is shown (Right). The bigger the absolute value of ΔΨ, the deeper the color. (C) Heatmap plot showing the magnitude and direction of retained intron isoform-level changes (ΔΨ; IRtumor–normal) between tumor and matched normal tissues for the 30 IR events affecting 21 splicing factors that are significantly changed in at least three patients’ tumors versus matched normal tissues. Each row is an IR event affecting a splicing factor, with the name of the splicing factor listed in each row. If multiple introns in the splicing factor are involved, the IR event is labeled as “SplicingFactor_IR_#.” The color code is as specified in B and the color key is listed (Left).
We also compared the performance of JUM in analyzing IR events in these patient datasets with MISO, rMATS, as well as IRFinder, a tool tailored for IR analyses (46) (SI Appendix, Fig. S3 B–D and Datasets S1–S34). We found that JUM and IRFinder identified the most number of significantly changed IR events (∼200 to 600) while rMATS identified the lowest (11 to 99) (SI Appendix, Fig. S3B and Tables S5 and S6). Importantly, JUM analysis yields the lowest false positive rate in calling IR (SI Appendix, Figs. S3C and S4).
JUM Detected Significantly More Differentially Spliced AS Events in Human Cell Lines Bearing Cancer-Associated Mutation in the Splicing Factor SRSF2 with High Accuracy.
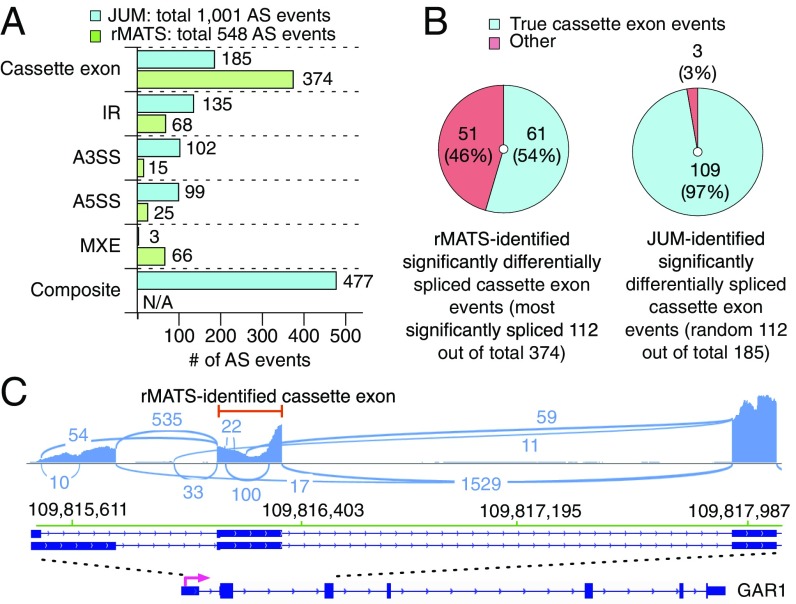
To further evaluate the sensitivity of JUM in detecting global AS changes in cell samples with complex AS patterns, we used JUM to analyze global AS changes caused by a cancer-associated point mutation (P95H) in the splicing factor SRSF2 in endogenously CRISPR-edited human K562 cell lines (47). Previously, Zhang et al. used rMATS to profile AS changes in the datasets and reported a total of 548 significantly changed AS events, including 374 SE events, 68 IR events, 15 A3SS events, 25 A5SS events, and 66 MXE events (47) (Fig. 6A). The numbers of differentially spliced AS events are distributed with high bias among AS pattern categories, with the majority reported in SE (∼68%) and only 5 and 3% in A5SS and A3SS patterns, respectively. By contrast, using JUM with the same statistical cutoff reported in the previous study (47) (adjusted P value ≤ 0.1, ΔΨ ≤ 0.1), we found a total of 1,001 AS events that are differentially spliced in cells carrying the point mutation in SRSF2, almost double the number of events found by rMATS (Dataset S40). Among them, JUM found 185 SE events, 135 IR events, 102 A3SS events, 99 A5SS events, 3 MXE events, and 477 composite events, with significantly less bias in the detection of AS events across different AS categories compared with rMATS. Moreover, to test if the distinctively high number of SE events reported by rMATS is real, we visually examined the top 112 most significantly differentially spliced SE events reported by rMATS using IGV (48) (Fig. 6B). Interestingly, we found about 46% (51 out of 112) of these events are not SE events but occur in combination with other AS patterns, similar to what JUM classifies as composite (Fig. 6 B and C). We also examined 112 randomly chosen JUM-reported differentially spliced SE events out of 185 and found 97% of these are indeed true SE events (Fig. 6B). In summary, these results demonstrate that JUM can detect significantly more differentially spliced AS events with high accuracy in cell samples with complex AS patterns in comparison with other annotation-based methods like rMATS.
Fig. 6.
Comparison of JUM and rMATS in analyzing global AS changes brought about by a cancer-associated point mutation in the splicing factor SRSF2 in human K562 cell lines. (A) Number of significantly differentially spliced AS events reported in every AS pattern category by the two methods. N/A, not available. (B) Number of cassette exons that are true SE events reported by each method. (C) An example of an incorrectly classified SE event reported by rMATS in the gene GAR1. The reported SE exon is specified by a red line.
JUM Identified Significantly More Real, Novel, and Functionally Important AS Events in the Head Sample of a Drosophila Strain Carrying a Mutation in the Splicing Factor PSI and Is Capable of Predicting the Regulatory Function of PSI Based on the Splicing Pattern Changes.
To assess JUM’s performance in profiling AS changes in tissues with complex AS patterns, we compared the performance of JUM, MISO, and rMATS in identifying global AS changes in the male head transcriptome of a Drosophila strain that carries a mutation in the splicing factor PSI, leading to the expression of truncated PSI protein (41, 49) (Datasets S41–S43). The resultant strain exhibits male courtship behavior defects (41). Importantly, the specific mutation in PSI disrupts its interaction with U1 small nuclear ribonucleoprotein particle (U1 snRNP), and thus is expected to affect splicing decisions on a set of target 5′ splice sites (50).
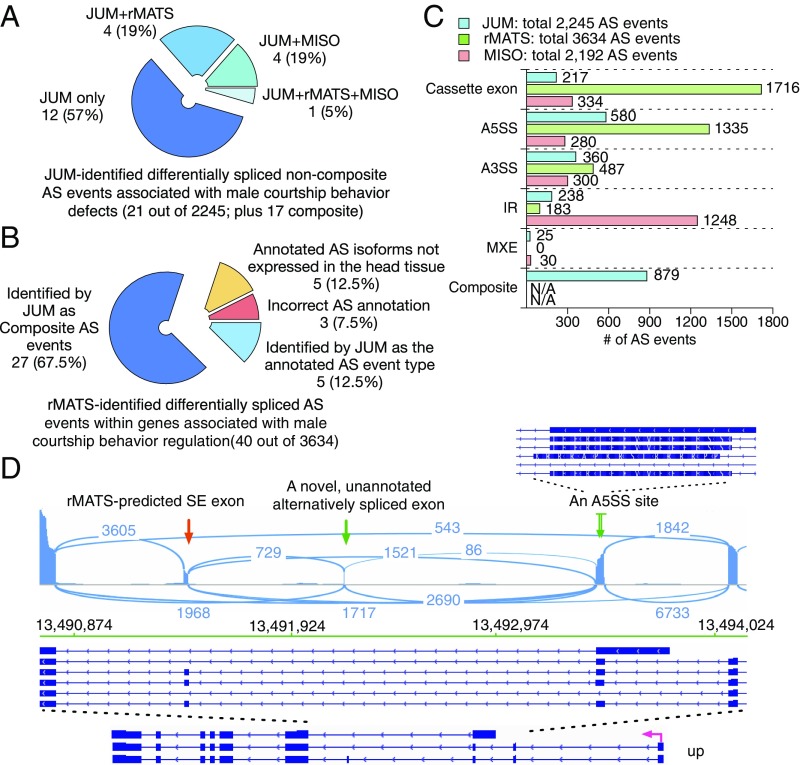
We performed two single-blind, counter tests that compared the performance of JUM with MISO and rMATS. For the first test, we took the set of 21 JUM-identified differentially spliced noncomposite AS events that are functionally linked to the male courtship behavior defects observed in the male Drosophila PSI mutant flies (49), and asked if rMATS and MISO can identify these phenotypically related AS events as well (Fig. 7A and SI Appendix, Table S8). A visual validation using IGV showed that all of these 21 AS events are correctly classified in the corresponding AS pattern category by JUM. Among them, we found that the majority of these AS events (12 out of 21; 57%) were identified exclusively by JUM, since neither the annotated AS library for MISO nor the novel splicing-aided mode of rMATS was able to identify these true, novel, and phenotypically associated AS events in the male PSI mutant fly head samples (Fig. 7A and SI Appendix, Table S8). Only 1 AS event (5%) was identified by all three methods. Interestingly, this is an SE event which is the most well-annotated AS pattern among all AS pattern categories (Fig. 7A and SI Appendix, Table S8). Four AS events are identified by rMATS and JUM but not MISO (19%) and four by MISO and JUM but not rMATS (19%) (Fig. 7A and SI Appendix, Table S8). To confirm that JUM is capable of detecting true AS events that are missed by other software, we performed experimental qRT-PCR validation of the 12 male courtship-associated AS events that were only identified by JUM but not rMATS or MISO (SI Appendix, Fig. S7). Interestingly, all 12 events were validated as true, significantly changed AS events in the PSI mutant male fly head tissue compared with wild type (SI Appendix, Fig. S7). These results suggest that JUM is clearly capable of identifying significantly more (in this case two times) functionally relevant, novel, and tissue-specific AS events that are not recognized by other annotation-based techniques, even when the annotation-based software is aided by a novel splice junction-detection mode.
Fig. 7.
Comparison of JUM, rMATS, and MISO in analyzing global AS changes brought about by a truncation mutation in the splicing factor PSI in Drosophila male heads. (A) Test of whether rMATS and MISO can also identify JUM-predicted, experimentally validated, and functionally crucial differentially spliced noncomposite AS events that are associated with the male courtship defect phenotype in the male head sample of a PSI mutant Drosophila strain. (B) Test of whether JUM can also identify rMATS-predicted, significantly changed AS events in genes associated with male courtship regulation in the male head sample of a PSI mutant Drosophila strain. (C) Number of significantly differentially spliced AS events reported in every AS pattern category by the three methods. (D) An example of an rMATS-predicted “SE” event that actually represents a much more complicated AS pattern in the male fly head and was reclassified correctly by JUM as a composite AS event is shown. Exon coverage from RNA-seq data is shown in blue; arcs represent splice junctions identified from the RNA-seq data; Drosophila annotation (dm3) of the transcripts is shown (Bottom). The rMATS-predicted SE exon is specified by a red arrow. This SE exon is in fact alternatively spliced in combination with an upstream novel, unannotated alternatively spliced exon (left green arrow; whose existence was proven by the RNA-seq tracks and splice junction reads), as well as an A5SS site in the upstream exon (right green arrow; a zoom-in at that upstream exon is shown to provide a detailed view of the A5SS site).
For the counter test, we took the set of 40 rMATS-identified, significantly changed AS events that are within genes associated with male courtship behavior regulation and asked if JUM can identify these AS events as well (Fig. 7B and SI Appendix, Table S9). We found that among them, five events (12.5%) are identified by JUM also as significantly changed AS events in the category classified by rMATS (Fig. 7B and SI Appendix, Table S9); 27 events (67.5%) are identified by JUM also as differentially spliced AS events but reclassified as composite AS events, and a visual examination using the IGV browser confirmed the predictions of JUM (Fig. 7 B and D and SI Appendix, Fig. S12A and Table S9). Eight events (20%) are not identified by JUM. However, when we examined these events individually by the genome browser, we found that three events (7.5%) are incorrectly annotated AS events called by rMATS in the first place (Fig. 7B and SI Appendix, Fig. S12B and Table S9). As for the rest, five events (12.5%) that rMATS reported/annotated as AS isoforms are either not expressed or too poorly expressed to be detected by RNA-seq in the head tissue samples under study (Fig. 7B and SI Appendix, Fig. S12C and Table S9). These results suggest that JUM is capable of identifying true differentially spliced AS events and profiling the events into the correct AS pattern category compared with other annotation-based software.
We further examined the distribution of differentially spliced AS events predicted by each method across different AS pattern categories (Fig. 7C). rMATS again reported the highest number of differentially spliced AS events in SE (1,716 out of a total of 3,634; 47%), and MISO reported the highest number of AS changes in IR (1,248 out of a total of 2,192; 57%). JUM, on the other hand, reported the highest number of changes in A5SS (580 out of a total of 2,245; 26%) and again reported a much less skewed distribution in the other AS pattern categories (217 in SE, 360 in A3SS, 183 in IR, and 25 in MXE) than rMATS and MISO. Importantly, the changed AS event distribution reported by JUM correctly reflects the functional association of the specific PSI mutation with U1 snRNP and the regulation of 5′ splice site usage. Taken together, we conclude that JUM is not only able to accurately detect novel, tissue-specific AS events that are missed or misclassified by other annotation-based methods but JUM’s unique feature of accurately assembling AS patterns directly from RNA-seq data can be useful in predicting the functions of splicing regulators from the global AS changes caused by the regulator. Such unique features are not found in other currently available AS analysis methods.
Discussion
As a major mechanism for eukaryotic gene regulation, AS generates exceptionally diverse patterns of mRNA populations and their encoded proteins in metazoans. Different tissues, even subcellular populations within a given tissue or organ, possess their own distinct AS profiles that are dynamically altered over temporal stages of development and cellular activities (SI Appendix, Fig. S1). The diversity and dynamics of AS patterns impose a major challenge for computational tools to quantify and compare AS profiles from RNA-seq data. Currently available AS analysis software tools commonly employ a top-down strategy based on prebuilt annotated collections of known AS events to outline the general picture of splicing patterns for downstream analysis. This strategy greatly facilitates downstream quantitation, but at the same time fails to address the diversity and tissue specificity in AS patterns, even when aided with workarounds to include novel splicing events specific to the sample under study.
With JUM, we approach the problem of tissue-specific AS pattern analysis with a different philosophy, the bottom-up approach that profiles, quantitates, and analyzes AS patterns directly from the sample under study (Figs. 1 and 2). By utilizing the unique topological features of the splicing graph representing each AS pattern, JUM is able to accurately construct and quantitate the sample-specific AS atlas through assembling the basic graphical nodes of the atlas-called AS structures that are profiled directly from the sample (Fig. 1). It should be noted that two other methods, MAJIQ (18) and LeafCutter (19), share similar conceptual designs as JUM in which they utilize graphs of splicing junction clusters for splicing quantification. However, MAJIQ is not annotation-free and is still dependent on a preannotated transcriptome for the construction of splicing graphs; LeafCutter, although annotation-free, only emphasizes quantifying levels of intron excision, without regard to detection, quantification, or analysis of AS event patterns; LeafCutter also does not detect intron retention events, an important class of AS.
JUM is further equipped with three unique features. First, JUM applies a well-developed approach to analyzing IR events and significantly decreases the high false positive rate of IR classification in currently available AS analysis tools (Figs. 3 and 5 and SI Appendix, Fig. S4). Second, JUM profiles the composite AS events as a separate AS pattern category, which is widespread in various tissue types and can play important roles in shaping cellular physiology but is not usually covered in other currently available AS methods (Fig. 2). Last but not least, JUM accurately profiles changed AS events in terms of the standard AS pattern categories of SE, A5SS, A3SS, IR, and MXE directly from the RNA-seq datasets (Figs. 6 and 7). This feature is especially important when investigating regulatory mechanisms of splicing regulators, which can be reflected in the distribution of changed AS events among the different AS pattern categories (Fig. 7).
In conclusion, JUM presents a statistically rigorous approach to address, evaluate, quantitate, and classify the complex and diverse patterns of AS profiles in eukaryotic transcriptomes. We are confident that this approach will provide important insights into the dynamic regulation of AS and gene expression, particularly in poorly annotated genomes and complex cell or tissue types that are already known to generate extremely diverse mRNA isoform profiles, such as gonads (testes and ovaries), pluripotent stem cells, and a variety of neuronal cell types and nervous system tissues.
Materials and Methods
RNA-Seq Data.
Raw RNA-seq data (FASTQ format) for Drosophila male fly heads and K562 cell lines with SRSF2 mutations described in this paper were derived as previously described (47, 49). Human colon tumor and matched normal tissue poly-A–selected RNA-seq data (in BAM format) were acquired from TCGA database. A detailed description of the patient tumor and normal samples used in this study is given in SI Appendix, Table S4. The downloaded BAM files were transformed back to FASTQ format by using the SamToFastq function in PICARD tools (broadinstitute.github.io/picard/) before analysis. The FASTQ data were then mapped to human genome hg38 as described below. The sequencing read mapping results are summarized in SI Appendix, Table S7 for each patient. Approval from the NCBI Database of Genotypes and Phenotypes (dbGaP) was obtained for TCGA controlled data access.
RNA-Seq Data Mapping for JUM.
RNA-seq reads are mapped to the human (hg38) (or hg19 for MISO, as the MISO-provided annotation library is in hg19) and Drosophila (dm3) genomes using STAR (51) in the two-pass mode, as instructed in the STAR manual as well as the JUM manual on Github. Only unique mapped reads are kept in the output for downstream JUM analysis.
RNA-Seq Data Experiment Simulation.
We used the ASmethodBenchmarking software as described in ref. 42 to simulate RNA-seq datasets; triplicates of ∼80 million 100-bp reads are simulated for each condition, with three levels of AS changes. Parameters are listed in SI Appendix, Table S1.
Running JUM, MISO, MAJIQ, Cufflinks, Whippet, rMATS, and IRFinder in This Study for Differential AS Analysis.
The detailed commands for each software tool as well as the versions of each software tool used in this study are listed in SI Appendix, Tables S2 and S3. Detailed statistical cutoffs applied in each case for all software tools are listed in SI Appendix, Materials and Methods.
ROC Curve Plotting and AUC Calculation.
The ranking score for ROC curve plotting is chosen to be 1-adjusted_pvalue for JUM, Cufflinks, and rMATS, as the three methods provide adjusted P values from multiple testing correction. For the rest, the ranking score is set to be the maximum value of E(dPSI) among local splicing variations in an AS event for MAJIQ (18), the value of the Bayes factor for MISO (10), and the probability value for Whippet (21). The R package ROCR (52) is used to plot ROC curves and calculate the AUC metric.
Algorithm to Construct AS Patterns from Profiled AS Structures.
We first profile all AS structures from the RNA-seq data and calculate the SI value for each sub-AS-junction in these AS structures. Two AS structures are defined as “linked” if they share one specific sub-AS-junction, and a “path” is drawn between the two AS structures. Under this definition, a “loop” of AS structures is searched in the whole pool of AS structures, with every AS structure in the loop linked to one other by a path. Each profiled loop of AS structures corresponds to an AS event, and is allocated to each AS pattern category based on the features of the sub-AS-junction SI value distributions.
Visualization.
All RNA-seq track data and junction reads were visualized using IGV (48) and the Sashimi plots tool (53).
Gene Ontology Analysis.
Gene Ontology analyses were performed using GOrilla (cbl-gorilla.cs.technion.ac.il/) (54). For each patient, a list of transcripts expressed at greater than 10 reads was used as a background dataset.
Supplementary Material
Acknowledgments
We thank Jeffrey Paulsen, Kate Abruzzi, and Chao Di for helpful critiques and comments. We thank Yeon Lee for help testing the first user-friendly version of the JUM package. We thank Ashley Albright and the Michael Eisen lab for help with the qRT-PCR experiments. The results shown here are in part based upon data generated by TCGA Research Network (https://cancergenome.nih.gov/). This work was supported by NIH Grants R01GM097352 and NIH R35GM118121 (to D.C.R., Principal Investigator) and by the NIH Center for RNA Systems Biology at UC, Berkeley (P50GM102706; to D.C.R., Coprincipal Investigator). Q.W. is supported by an Arnold O. Beckman postdoctoral fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The simulated datasets used in this paper have been deposited on Github and are available at https://github.com/qqwang-berkeley/JUM.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1806018115/-/DCSupplemental.
References
- 1.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wahl MC, Will CL, Lührmann R. The spliceosome: Design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 4.Fu XD, Ares M., Jr Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet. 2014;15:689–701. doi: 10.1038/nrg3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 6.Shkreta L, et al. Cancer-associated perturbations in alternative pre-messenger RNA splicing. Cancer Treat Res. 2013;158:41–94. doi: 10.1007/978-3-642-31659-3_3. [DOI] [PubMed] [Google Scholar]
- 7.Li YI, et al. RNA splicing is a primary link between genetic variation and disease. Science. 2016;352:600–604. doi: 10.1126/science.aad9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dvinge H, Kim E, Abdel-Wahab O, Bradley RK. RNA splicing factors as oncoproteins and tumour suppressors. Nat Rev Cancer. 2016;16:413–430. doi: 10.1038/nrc.2016.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor JP, Brown RH, Jr, Cleveland DW. Decoding ALS: From genes to mechanism. Nature. 2016;539:197–206. doi: 10.1038/nature20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anders S, Reyes A, Huber W. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012;22:2008–2017. doi: 10.1101/gr.133744.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trapnell C, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu Y, et al. DiffSplice: The genome-wide detection of differential splicing events with RNA-seq. Nucleic Acids Res. 2013;41:e39. doi: 10.1093/nar/gks1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brooks AN, et al. Conservation of an RNA regulatory map between Drosophila and mammals. Genome Res. 2011;21:193–202. doi: 10.1101/gr.108662.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen S, et al. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-seq data. Proc Natl Acad Sci USA. 2014;111:E5593–E5601. doi: 10.1073/pnas.1419161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vitting-Seerup K, Porse BT, Sandelin A, Waage J. spliceR: An R package for classification of alternative splicing and prediction of coding potential from RNA-seq data. BMC Bioinformatics. 2014;15:81. doi: 10.1186/1471-2105-15-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aschoff M, et al. SplicingCompass: Differential splicing detection using RNA-seq data. Bioinformatics. 2013;29:1141–1148. doi: 10.1093/bioinformatics/btt101. [DOI] [PubMed] [Google Scholar]
- 18.Vaquero-Garcia J, et al. A new view of transcriptome complexity and regulation through the lens of local splicing variations. eLife. 2016;5:e11752. doi: 10.7554/eLife.11752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li YI, et al. Annotation-free quantification of RNA splicing using LeafCutter. Nat Genet. 2018;50:151–158. doi: 10.1038/s41588-017-0004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tapial J, et al. An atlas of alternative splicing profiles and functional associations reveals new regulatory programs and genes that simultaneously express multiple major isoforms. Genome Res. 2017;27:1759–1768. doi: 10.1101/gr.220962.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sterne-Weiler T, Weatheritt RJ, Best A, Ha KC, Blencowe BJ. 2017. Whippet: An efficient method for the detection and quantification of alternative splicing reveals extensive transcriptomic complexity. bioRxiv:10.1101/158519.
- 22.Yan Q, et al. Systematic discovery of regulated and conserved alternative exons in the mammalian brain reveals NMD modulating chromatin regulators. Proc Natl Acad Sci USA. 2015;112:3445–3450. doi: 10.1073/pnas.1502849112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schulz MH, Zerbino DR, Vingron M, Birney E. Oases: Robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics. 2012;28:1086–1092. doi: 10.1093/bioinformatics/bts094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grabherr MG, et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haas BJ, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie Y, et al. SOAPdenovo-Trans: De novo transcriptome assembly with short RNA-seq reads. Bioinformatics. 2014;30:1660–1666. doi: 10.1093/bioinformatics/btu077. [DOI] [PubMed] [Google Scholar]
- 27.Lu J, Tomfohr JK, Kepler TB. Identifying differential expression in multiple SAGE libraries: An overdispersed log-linear model approach. BMC Bioinformatics. 2005;6:165. doi: 10.1186/1471-2105-6-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson MD, Smyth GK. Moderated statistical tests for assessing differences in tag abundance. Bioinformatics. 2007;23:2881–2887. doi: 10.1093/bioinformatics/btm453. [DOI] [PubMed] [Google Scholar]
- 29.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson MD, Smyth GK. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics. 2008;9:321–332. doi: 10.1093/biostatistics/kxm030. [DOI] [PubMed] [Google Scholar]
- 32.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park E, Pan Z, Zhang Z, Lin L, Xing Y. The expanding landscape of alternative splicing variation in human populations. Am J Hum Genet. 2018;102:11–26. doi: 10.1016/j.ajhg.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jangi M, et al. SMN deficiency in severe models of spinal muscular atrophy causes widespread intron retention and DNA damage. Proc Natl Acad Sci USA. 2017;114:E2347–E2356. doi: 10.1073/pnas.1613181114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Braunschweig U, et al. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res. 2014;24:1774–1786. doi: 10.1101/gr.177790.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee Y, Rio DC. Mechanisms and regulation of alternative pre-mRNA splicing. Annu Rev Biochem. 2015;84:291–323. doi: 10.1146/annurev-biochem-060614-034316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Majumdar S, Rio DC. P transposable elements in Drosophila and other eukaryotic organisms. Microbiol Spectr. 2015;3:MDNA3-0004-2014. doi: 10.1128/microbiolspec.MDNA3-0004-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dvinge H, Bradley RK. Widespread intron retention diversifies most cancer transcriptomes. Genome Med. 2015;7:45. doi: 10.1186/s13073-015-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Solana J, et al. Conserved functional antagonism of CELF and MBNL proteins controls stem cell-specific alternative splicing in planarians. eLife. 2016;5:e16797. doi: 10.7554/eLife.16797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trapnell C, et al. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Labourier E, Blanchette M, Feiger JW, Adams MD, Rio DC. The KH-type RNA-binding protein PSI is required for Drosophila viability, male fertility, and cellular mRNA processing. Genes Dev. 2002;16:72–84. doi: 10.1101/gad.948602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu R, Loraine AE, Dickerson JA. Comparisons of computational methods for differential alternative splicing detection using RNA-seq in plant systems. BMC Bioinformatics. 2014;15:364. doi: 10.1186/s12859-014-0364-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chabot B, Shkreta L. Defective control of pre-messenger RNA splicing in human disease. J Cell Biol. 2016;212:13–27. doi: 10.1083/jcb.201510032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: Pathways and programs unhinged. Genes Dev. 2010;24:2343–2364. doi: 10.1101/gad.1973010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang J, Manley JL. Misregulation of pre-mRNA alternative splicing in cancer. Cancer Discov. 2013;3:1228–1237. doi: 10.1158/2159-8290.CD-13-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Middleton R, et al. IRFinder: Assessing the impact of intron retention on mammalian gene expression. Genome Biol. 2017;18:51. doi: 10.1186/s13059-017-1184-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang J, et al. Disease-associated mutation in SRSF2 misregulates splicing by altering RNA-binding affinities. Proc Natl Acad Sci USA. 2015;112:E4726–E4734. doi: 10.1073/pnas.1514105112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Q, et al. The PSI-U1 snRNP interaction regulates male mating behavior in Drosophila. Proc Natl Acad Sci USA. 2016;113:5269–5274. doi: 10.1073/pnas.1600936113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Labourier E, Adams MD, Rio DC. Modulation of P-element pre-mRNA splicing by a direct interaction between PSI and U1 snRNP 70K protein. Mol Cell. 2001;8:363–373. doi: 10.1016/s1097-2765(01)00311-2. [DOI] [PubMed] [Google Scholar]
- 51.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: Visualizing classifier performance in R. Bioinformatics. 2005;21:3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- 53.Katz Y, et al. Quantitative visualization of alternative exon expression from RNA-seq data. Bioinformatics. 2015;31:2400–2402. doi: 10.1093/bioinformatics/btv034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.