Abstract
Objective
To investigate the molecular underpinnings of thyroid cancer, preclinical cell line models are crucial; however, ∼40% of these have been proven to be either duplicates of existing thyroid lines or even nonthyroid-derived lines or are not derived from humans at all. Therefore, we set out to establish procedures and guidelines that should proactively avoid these problems, which facilitated the creation of criteria to make valid preclinical models for thyroid cancer research.
Design
Based on our recommendations, we systematically characterized all new cell lines that we generated by a standardized approach that included (1) determination of human origin, (2) exclusion of lymphoma, (3) DNA fingerprinting and histological comparisons to establish linkage to presumed tissue of origin, (4) examining thyroid differentiation by screening two to three thyroid markers, (5) examination of biological behavior (growth rate, tumorigenicity), and (6) presence of common thyroid cancer genetic changes (TP53, BRAF, PTEN, PIK3CA, RAS, TERT promoter, RET/PTC, PAX8/PPARγ, NF1, and EIF1AX).
Results
We established seven new thyroid cell lines (LAM136, EAM306, SDAR1, SDAR2, JEM493, THJ529, and THJ560) out of 294 primary culture attempts, and 10 patient-derived tumor xenografts (PDTXs; MC-Th-95, MC-Th-374, MC-Th-467, MC-Th-491, MC-Th-493, MC-Th-504, MC-Th-524, MC-Th-529, MC-Th-560, and MC-Th-562) out of 67 attempts. All were successfully validated by our protocols.
Conclusions
This standardized approach for cell line and PDTX characterization should prevent (or detect) future cross-contamination and ensure that only valid preclinical models are used for thyroid cancer research.
We generated seven new thyroid cell lines as well as 10 PDTX mouse models. We discuss methodology and describe optimal criteria for creating patient tumor tissue–derived preclinical thyroid cancer models.
Thyroid cancer is the most common endocrine malignancy with a female-to-male ratio of 3:1 (1) and median age of 51 years. The well-differentiated thyroid cancer (WDTC) subtypes [papillary (PTC), follicular (FTC), Hürthle cell cancer (HCC), insular], medullary, poorly differentiated (PDTC), and anaplastic (ATC), differ from each other in clinical course, underlying tumor genetic changes and therapeutic options. The most common driver mutations found in PTC are BRAF (61.7%), RAS (12.9%), TERT promoter (9.4%), RET/PTC rearrangements (6.3%), EIF1AX (1.5%), PPM1D (1.2%), and CHEK2 (1.2%) (2), whereas FTC includes RAS (45%), PAX8/PPARγ (PPFP) rearrangements (35%), and the TERT promoter (11.4%) (3, 4). PDTC involves the TERT promoter (40% to 52%), RAS (28%), BRAF (33%), EIF1AX (11%), TP53 (8% to 25%), and RET/PTC rearrangements (6% to 13%) (5–7); ATC includes TP53 (73%), TERT promoter (73%), BRAF (45%), RAS (24%), PIK3CA (18%), PTEN (15%), NF1 (9%), EIF1AX (9%), and STK11 (6%) (7, 8). ATC differs from PDTC by the frequency of alterations in four pathways: PI3K/AKT/mTOR (39% vs 11%), SWI/SNF chromatin remodeling (36% vs 6%), histone methyltransferases (24% vs 7%), and mismatch excision repair (12% vs 2%) (7, 9).
WDTCs, usually managed by surgery and radioactive iodine remnant ablation and thyroxine replacement, have an excellent prognosis (>90% 5-year survival). For radioiodine refractory tumors (excluding medullary thyroid cancer), chemotherapy and targeted therapy options include: doxorubicin, cisplatin, lenvatinib, and sorafenib (1, 10). In contrast, PDTC and ATC have a poorer prognosis, with little response to therapy with 5-year survivals of 62% and 5% (8, 11), respectively. This is indicative of a true lack of understanding of the biological underpinnings of each thyroid tumor subytype.
To investigate therapy-resistant WDTCs, PDTC and ATC, preclinical models are needed that cover the entire spectrum of thyroid tumor types, including primary normal thyrocytes. Despite some limitations, human cell lines are essential for testing biological mechanisms and new therapies in vitro prior to moving forward into animal models. Unfortunately, studies have shown that 18 of 36 cell lines (40% thyroid) are either misidentified or cross-contaminated, a situation that continues to bedevil the field (12). Even when cell lines have been characterized with unique short tandem repeat (STR) DNA profiles, there is no guarantee that they are descendent from a particular tumor, as those have often not been characterized in the first place. For example, the FTC origin of WRO (13) is questionable because one version of the cell line in distribution harbors a BRAFT1799A (V600E) mutation, which is specific to PTC (3, 14), whereas another version is BRAF wild-type. Both WRO versions have unique STR profiles (14), but the question remains: which version is the real WRO? Owing to problems such as this, the number of unique thyroid cell lines, especially FTC-derived lines, is quite small. Therefore, proper criteria need to be established to create new cell line models for research.
In vitro cell lines that are engrafted in immune-compromised mouse models often bear little resemblance to the original tumors, due to loss of tumor heterogeneity and the human microenvironment, or drug resistance mechanisms. Genetically engineered mouse models address this problem partially; however, they recapitulate neither the human microenvironment nor the genetic/epigenetic complexity of human tumors, and they tend to have amplified responses to novel drugs. These limitations contribute to the only 5% effectiveness rate of anticancer agents in phase III trials (15).
As a response, new preclinical patient-derived tumor xenograft (PDTX) models have emerged (also called PDX models or avatars) (16). PDTX models resemble the original patient tumor genetically, maintain cell-autonomous heterogeneity, maintain tumor architecture with stromal elements, and display drug responses that correlate strongly with clinical data. However, this model also has limitations: long engraftment time (1 to 4 months), loss of extrinsic non–cell-autonomous heterogeneity, inability to test immune-based therapies (the mice are immunocompromised), and changes in tumor characteristics over passaging (17, 18).
Thus, more models need to be created and they need to be well characterized and quality controlled during their creation process. In this study, we set out to demonstrate how one can apply common sets of experimental tests to primary tumor cell line models and PDTX models, which allow their unequivocal identification as specific human thyroid-derived lines (linked back to the original tissue/cell of origin), as well as permitting biological response assessment and tumor genetic characterization.
Materials and Methods
Patients, cell lines, and PDTXs
Thyroid patient tissue was collected in accordance with Mayo Clinic Institutional Review Board protocol. LAM136 originated from an 81-year-old female (4.3-cm conventional PTC; T3N1bM0). EAM306 originated from a 59-year-old female (7.2-cm angioinvasive FTC; T3N0M0). SDAR1 and SDAR2 both originated from a 66-year-old male (6.3-cm widely invasive FTC with areas of de-differentiation, as well as epithelioid and spindle cell features; T4aN0M1). SDAR1 was derived from the primary tumor site and SDAR2 from locally invasive regions in the neck. JEM493 (and MC-Th-493) originated from a 75-year-old male (6.0-cm ATC; T4bN1aM1; microscopic PTC foci and mild lymphocytic thyroiditis were also present). THJ529 (and MC-Th-529) originated from a 45-year-old male (8-cm PDTC involving 16 of 37 lymph nodes; T3N1bMx). THJ560 (and MC-Th-560) originated from a 49-year-old female (6.5-cm ATC arising in association with PTC; T4bN1bMx). Information on these cell lines was made available to the Cellosaurus (web.expasy.org/cellosaurus/). LAM136, EAM306, and SDAR1 were previously published in reference to SCD1 expression (19).
For PDTX only models, MC-Th-95 originated from a male patient who presented with rare squamous cell thyroid carcinoma (SCTC). MC-Th-374 originated from a female [4.5 cm follicular variant PTC (FVPTC); T3NxMx]. MC-Th-467 originated from a 68-year-old male (3-cm PDTC arising from insular carcinoma; T2N1aMx). MC-Th-491 originated from a 68-year-old female (3.5-cm ATC; T4bN1aMx). MC-Th-504 originated from a 59-year-old male (5-cm ATC in association with PTC; T4bN1aMx). MC-Th-524 originated from an 80-year-old male (8-cm ATC in association with PTC and poorly differentiated SCTC). MC-Th-562 originated from a 69-year-old male (6-cm ATC with sarcomatoid features).
Tissue processing and establishment of PDTXs
Tumor tissues were collected in RPMI 1640 (catalog no. 10-040-CV; Corning, Manassas, VA) and processed within 4 hours. Tissues were cut into 4 × 4 × 4-mm cubes and were allocated as follows, as applicable: (1) formalin fixation for paraffin embedding (FFPE); (2) cryogenically frozen in RPMI 1640 supplemented with 5% FBS (catalog no. 16000-044, lot no. 1891471; Life Technologies, Grand Island, NY) plus 5% dimethyl sulfoxide (catalog no. D2650; Sigma-Aldrich, St. Louis, MO) plus 200 mM trehalose dihydrate (catalog no. T0167; Sigma-Adrich) plus 1% penicillin–streptomycin–amphotericin B (PSA; catalog no. 30-004-CI; Corning); (3) flash frozen in liquid nitrogen; (4) finely minced for primary culture; and (5) surgically implanted under anesthesia with 100 μL of Matrigel® (catalog no. 354234; Corning) into 4- to 6-week-old athymic nude female mice (strain 069; Envigo, Indianapolis, IN) for PDTX models. The take rate of engraftment is usually between 1 and 4 months with attempts up to 6 months. Take heed that longer engraftments increase the risk of the tumors actually arising from lymphoma (20). Epithelial founder generation models (G0) were then passaged using 5 × 5 × 5-mm fragments into new mice (G1 and so forth) for expansion and for cryogenically frozen stocks. Only 67 preselected tissues were attempted for generating PDTX models (29.9% metastatic PTC, 23.9% PTC, 13.4% FTC, 11.9% ATC, 10.4% FVPTC, 10.5% other) and the resulting JEM493, THJ529, and THJ560 cell lines with patient-matched PDTXs included: MC-Th-493, MC-Th-529, and MC-Th-560, respectively. Seven other patient-matched thyroid PDTX models were developed and these included MC-Th-95 (KRASG12V mutation), MC-Th-374 (HRASQ61R mutation), MC-Th-467, MC-Th-491, MC-Th-504 (BRAFV600E mutation), MC-Th-524, and MC-Th-562. Six of the 10 models were deposited and made available with Charles River Laboratories (www.criver.com/products-services/basic-research).
Primary culture
Minced tissue from 294 patients (59.5% PTC, 18.7% metastatic PTC, 10.2% FVPTC, 6.1% FTC, 3.1% ATC, 1.7% PDTC, and 0.7% other) were placed into 6-cm Cellbind® plates (catalog no. 3295; Corning) and cultured with 3 mL of αBROV medium (patent filed, US 14/190,733) at 37°C in a humidified atmosphere with 5% CO2 until cells were attached and confluent. Once confluent, cells were trypsinized (catalog no. 25-053-CI; Corning) and placed into six-well ultra-low attachment plates (catalog no. 3471; Corning) in 2 mL of αBROV medium for 5 days. If spheroids were present, they were collected and plated into new Cellbind® plates. The cells were then propagated in αBROV medium until passage 25. Surviving primary cells were changed into RPMI 1640 supplemented with 5% FBS (catalog no. 16000-044, lot no. 1891471; Life Technologies), 1% nonessential amino acids (catalog no. 25-025-CI; Corning), 1 mM sodium pyruvate (catalog no. 25-000-CI; Corning), 10 mM HEPES (catalog no. 25-060-CI; Corning), and 1% PSA. Primary cells were considered “immortal or continuous” cell lines when they reached passage 40+ and cell stocks were regularly cryogenically frozen to avoid overpassaging (21). The remaining primary cultures senesced and were deemed mortal or noncontinuous cell lines. Live phase images were taken at magnification ×10 under bright field on EVOS FLc (Life Technologies). As controls, either benign or normal thyrocytes (would senesce between passage 5 and 8) were either propagated in αBROV medium or in DMEM/F12 (catalog no. 10-090-CV; Corning) supplemented with 5% FBS, 1 mM sodium pyruvate, 10 mM HEPES, 1% nonessential amino acids, 1 nM triiodo-l-thyronine (catalog no. T6397; Sigma-Aldrich), 8 ng/mL epidermal growth factor (catalog no. AF-100-15; PeproTech Rocky Hill, NJ), 5 μg/mL insulin–transferrin–sodium selenite (catalog no. 354350; Corning), 0.45 μg/mL iron(II) sulfate heptahydrate (catalog no. F8633; Sigma-Aldrich), and 1% PSA. For cost savings, the FBS and iron sulfate can be substituted with iron-fortified bovine calf serum (catalog no. SH3007203; HyClone Laboratories, Lohan, UT). Please note that switching between serums or even lot numbers also requires quality control to ensure reproducibility (21).
DNA/RNA isolation and analysis
For FFPE patient and PDTX tissues, five-10 μm sections were cut and deparaffinized, followed by DNA extraction (QIAamp DNA FFPE tissue kit, catalog no. 56404; Qiagen, Hilden, Germany). For cell lines, confluent 10-cm plates were scraped and pelleted. DNA was isolated using Purelink® genomic DNA kits (catalog no. K1820-01; Invitrogen, Carlsbad, CA). RNA was isolated for frozen tissues and cell lines using Purelink® RNA mini kits (catalog no. 12183018A; Invitrogen, Waltham, MA).
STR testing was performed by the Mayo Clinic Medical Genome Facility Genotyping Core (Rochester, MN) using a custom-designed STR profile based on the Combined DNA Index System of 13 loci set by the Federal Bureau of Investigation in 2016 (strbase.nist.gov). The primer sequences were verified with University of California, Santa Cruz (UCSC) and National Center for Biotechnology Information (NCBI) databases and validated using Centre d’Étude du Polymorphisme Humain (CEPH) cell line DNA GM12328, which matched known results conducted by commercial kits. Centre d’Étude du Polymorphisme Humain (CEPH) control sample NA10847 was included with every processing run and used as a control for all markers. Additionally, the primer mix was balanced in the multiplex to reduce spill over between markers and to provide comparable signal strengths. For analysis, the homozygous or heterozygous peaks at the loci were identified via bin alignment and exclusion of artifact peaks, which include spectral bleed and stutter peaks. Stutter peaks arise from polymerase slippage and were considered irrelevant unless there was overlap with nearby alleles. Thus, for split peaks, control DNA was used to determine which split peak (first or last) was the correct peak. For confirmation of allele loss at specific loci, DNA samples were reassayed using the PowerPlex 16HS (catalog no. DC2101; Promega, Madison, WI) and independently evaluated externally by the Genetics Resources Core Facility at Johns Hopkins University.
For BRAF, RAS (codons 12, 13, and 61) and RET/PTC analysis, real-time PCR methods were used as previously described (22, 23). PIK3CA (exons 9 and 20), TERT promotor mutations, and PAX8/PPARγ fusion (PPFP) were analyzed using primers listed in Supplemental Table 1. All amplicons were purified using Purelink® PCR purification kits (catalog no. K310001; Invitrogen) and bidirectionally sequenced using Sanger sequencing (Big Dye Terminator v1.1) by the Mayo Clinic Medical Genome Facility Genotyping Core. For the remaining mutations (TP53, PTEN, NF1, EIF1AX), MSK-IMPACT screening was performed at Dr. J.A. Fagin’s laboratory, as previously described (24). For RT-PCR, cDNA was generated from 3 μg of RNA using a high-capacity cDNA reverse transcription kit (catalog no. 4368814; Applied Biosystems, Foster City, CA) and 100 ng of cDNA was amplified with a ThermalAce DNA polymerase kit (catalog no. 45-0116; Invitrogen) using primers listed in Supplemental Table 1 at the indicated conditions. For quantitative PCR (qPCR), 20 ng of cDNA was amplified with TaqMan Fast universal PCR master mix (catalog no. 4352042; Applied Biosystems) at 60°C annealing temperature with a 20× human-specific Src TaqMan probe (Hs00178494_m1) and a 20× mouse-specific Src TaqMan probe (Mm00436783_m1) on the QuantStudio 7 Flex PCR system (Applied Biosystems). Absolute cycle threshold (CT) values were recorded and the difference (Δ) of CT values between species control and sample were calculated.
Immunohistochemistry
FFPE tissues were cut into 5-μm sections, deparaffinized, and hydrated and antigen was retrieved for 25 minutes at pH 6.0 and blocked for 5 minutes with Dako diluent (S3022) (Dako, Carpinteria, CA). Immunocytochemistry for cell lines was performed at 75% confluence in four-well Laboratory-Tek chamber slides coated with poly-d-lysine (Nunc, Roshester, NY). Fixation was done with 2% paraformaldehyde. Immunostaining was done for 1 hour with either lamin A/C (catalog no. NBP1-95336, RRID: AB_11034444, at 1:400; Novus Biologicals, Littleton, CO), cytokeratin (catalog no. M0821; Dako), predilute human CD45 (catalog no. GA75161-2, RRID: AB_2661839; Dako), mouse CD45 (catalog no. 05-1416, RRID: AB_10562966; Millipore) at 1:200, thyroid transcription factor-1 (TTF1; catalog no. M3575; Dako) at 1:50, thyroid-stimulating hormone receptor (TSHR; catalog no. ab13399) at 1:100 (Abcam, Cambridge, MA), and sodium iodide symporter (NIS; catalog no. MA5-12308, RRID: AB_10977040; Thermo Fisher Scientific) at 1:100. Normal thyroid tissue and cells were used as positive controls, and mouse pancreatic cancer tissue was used as a negative control. For detection, the EnVision dual-labeled polymer kit (Dako) was used for 30 minutes for anti-species secondary antibodies and stained with diaminobenzidine chromagen (DAB) for 5 minutes and then lightly counterstained with Gill I hematoxylin (Sigma-Aldrich) for 30 seconds before dehydration and mounting. Images were obtained at magnification ×20 using an Aperio AT2 Scanscope (Leica Biosystems, Buffalo Grove, IL). Immunohistochemistry (IHC) scoring of thyroid markers was done using an algorithm in the ImageScope software (Leica Biosystems) based on signal intensity of weak (1+), moderate (2+) or strong staining (3+) with criteria of at least 20% positivity.
Growth curves
For growth curves, 20,000 cells per well were plated in 12-well plates (catalog no. 25-106; Genesee Scientific, San Diego, CA) and counted using a Coulter particle counter (Beckman Coulter, Brea, CA) at the following time points: 24 hours, 72 hours, 120 hours, and 168 hours. For tumorigenicity, cells were suspended in 50% Matrigel® (catalog no. 354234; Corning) in thyroid media and subcutaneously injected on the right flank in either 4- to 6-week-old athymic nude female mice (strain 069; Envigo) or 4- to 6-week-old NOD scid gamma female mice (strain 005557; The Jackson Laboratory, Bar Harbor, ME). Tumors were measured once or twice per week using calipers, and volumes were calculated by 0.536 × (length × width × height). Data were graphed using GraphPad Prism (GraphPad Software, La Jolla, CA). Population doubling time was calculated using the Doubling Time online software Cell calculator++ (available at: www.doubling-time.com/compute.php).
Databases
In addition to searching published references for information on thyroid cell lines, the following databases were also examined: Cellosaurus, American Type Culture Collection, German Collection of Microorganisms and Cell Cultures, Japanese Collection of Research Bioresources, European Collection for Cell Cultures, and Rikagaku Kenkyūsho. The following databases were also examined for PDTX models: Charles River, CrownBio, Wuxi AppTec, Horizon Discovery, The Jackson Laboratory, Center for Developmental Therapeutics, Champions Oncology, and Children’s Oncology Group.
Results
Histology, morphology, and species verification
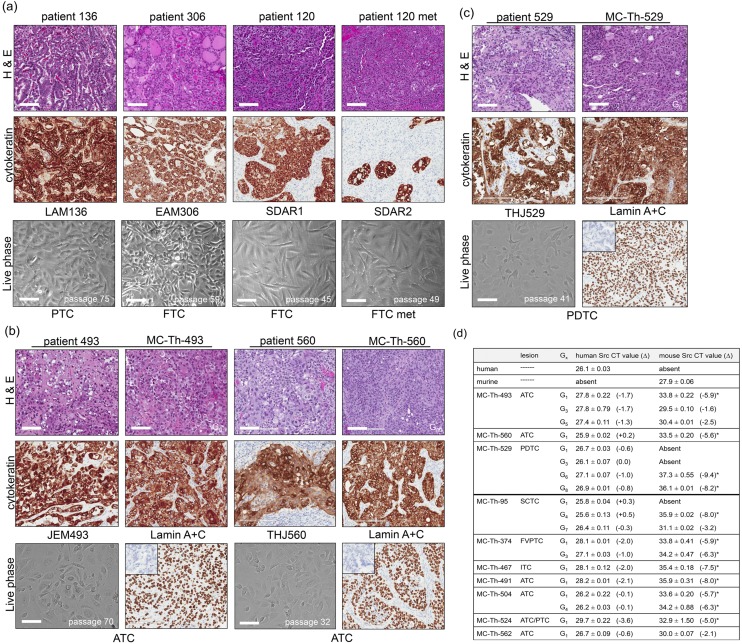
Tumor tissues from the patients and the derived PDTX tissues were either stained with hematoxylin and eosin (H&E) or pan-cytokeratin for visualization of tumor architecture and for verification of cytokeratin positivity in the thyroid. Phase images of corresponding immortal cell lines were captured to examine cellular morphology. WDTC-derived cell lines (LAM136 and EAM306) both expressed abundant cytokeratin, whereas the invasive FTC-derived lines SDAR1 and SDAR2 showed somewhat lesser cytokeratin expression [Fig. 1(a)]. The ATC-derived cell lines JEM493 and THJ560 [Fig. 1(b)] and PDTC THJ529 are depicted with their corresponding PDTX models in Fig. 1(b) and 1(c), respectively. The PDTX models (MC-Th-493, MC-Th-560, and MC-Th-529) showed similar histology and cytokeratin expression as their originating tumors. In addition to cytokeratin, the tumors were screened at G0 for both human-specific CD45 and mouse-specific CD45 to ensure that the tumors did not arise from lymphomas as reported by Wetterauer et al. (20) in NOD scid gamma mice. Human spleen and mouse spleen were used as controls (data not shown). Lamin A/C expression confirmed that the tumor cells are of human origin. The mouse stroma was negative and murine pancreatic tumors were used as a negative control (inlay) [Fig. 1(b) and 1(c)]. Every PDTX generation (one to two individuals) was checked by H&E and for human lamin A/C expression to ensure integrity of the models. Using MC-Th-529, we carried out the generations out farther up to G8 and showed that the PDTX model conserved its histological phenotype and continued to express human lamin A/C [Supplemental Fig. 1(a)].
Figure 1.
Histology, morphology, and species verification. (a) H&E and cytokeratin staining of well-differentiated patient tumors (patients 136, 306, and 120 and its metastasis) along with live images of corresponding patient-matched cell lines (LAM136, EAM306, SDAR1, and SDAR2). SDAR1 was derived from the primary tumor site and SDAR2 was derived from the locally invasive regions in the neck. The images were captured of LAM136 at passage 75, EAM306 at passage 59, SDAR1 at passage 45, and SDAR2 at passage 49. (b) H&E and cytokeratin staining of ATC patients 493 and 560 alongside their PDTX as well as live images of its matched cell lines (JEM493 and THJ560). The images were captured of JEM493 at passage 70 and THJ560 at passage 32. (c) H&E and cytokeratin staining of PDTC patient 529 alongside his PDTX and corresponding cell line (THJ529). The image captured of THJ529 was at passage 41. All PDTX images were at generation 0 (G0). Human lamin A/C was used as a marker for human species verification. Murine pancreatic cancer was used as a negative control (inlay). IHC images were obtained at original magnification ×20 using an Aperio AT2 Scanscope. Live phase images were taken at original magnification ×10 under bright field on EVOS FLc. Scale bars, 100 μm. (d) Table of PDTX species content assessed by qPCR of human- and murine-specific Src probes. The tumor generation is as indicated. The average absolute CT value is shown ± SD (n = 3). The Δ of CT values between species control and sample is shown. The source for the human control was human thyroid tumor tissue, whereas the murine control was mouse pancreatic cancer. The asterisk (*) indicates Δ >4.0 (eightfold).
Species content was further confirmed by qPCR on cDNA ranging from G1 to G8 tumors using either human- or murine-specific Src probes. The average absolute CT value was reported as well as the Δ between CT values for the species control and the PDTXs. For human Src, all of the PDTXs had similar low CT values as the control, indicative of abundant human Src mRNA levels and predominately human tissue with Δ <4.0 (under eightfold) [Fig. 1(d)]. For murine Src in G1 tumors, the CT values were significantly higher, indicative of very low levels of mRNA present than the control with Δ >4.0 (over eightfold) with the exception of MC-Th-562. MC-Th-529, MC-Th-374, and MC-Th-504 models continued to maintain low levels of murine Src even up to G8, G3, and G4, respectively. Alternatively, murine Src was initially completely absent in MC-Th-95, but during multiple generations the levels of murine Src increased by G7 (Δ <4.0). In MC-Th-493, murine Src levels had increased as early as G3 (Δ <4.0) [Fig. 1(d)].
DNA fingerprinting and thyroid markers
STR analysis of all seven cell lines and 10 PDTX model profiles matched their originating tumor profiles (Table 1). Also, all male-derived cell lines and PDTX models carried the amelogenin Y-allele. There were a few loss of heterogeneity (LOH) events observed at specific loci, which included both JEM493 and MC-Th-493 at locus TH01 with loss of variant allele 6, and MC-Th-467 at locus D21s11 with loss of variant allele 31.2 [Table 1; Supplemental Fig. 2(a)]. Both THJ560 and MC-Th-560 independently had LOH events at the following: loci D13S317 with loss of variant allele 8, loci D18S51 with loss of variant allele 16, and loci FGA with loss of variant allele 21 [Table 1; Supplemental Fig. 2(b)]. MC-Th-374 had an LOH event at locus D21s11 with a loss of variant allele 28 and at locus D16S539 with a loss of variant allele 13 [Table 1; Supplemental Fig. 2(c)]. Patient 562 at locus D16S539 had variant alleles 11, 12, and 14 whereas MC-Th-562 only had variant alleles 11 and 12. Additional LOH was also observed at multiple loci, which included the following loci: TH01 for variant allele 8, D18S51 for variant allele 13, D21s11 for variant allele 28, D3S1358 for variant allele 15, and FGA for variant allele 21 [Table 1; Supplemental Fig. 2(d)]. These LOH events were verified by two independent facilities for confirmation.
Table 1.
STR DNA Fingerprint Profiles of Seven Thyroid Cell Lines, 10 PDTX Models, and the Originating Patient Tumor
| AM | D5S818 | D13S317 | D7S820 | D16S539 | vWA | TH01 | TPOX | CSF1PO | D18S51 | D21s11 | D3S1358 | D8S1179 | FGA | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient 136 | X | 8, 11 | 11, 12 | 9, 11 | 11, 12 | 14, 16 | 6, 9.3 | 8 | 10, 15 | 10, 13 | 31.2, 32.2 | 14, 18 | 10, 12 | 22, 26 |
| LAM136 p75 | X | 8, 11 | 11, 12 | 9, 11 | 11, 12 | 14, 16 | 6, 9.3 | 8 | 10, 15 | 10, 13 | 31.2, 32.2 | 14, 18 | 10, 12 | 22, 26 |
| Patient 306 | X | 11 | 8, 12 | 11, 12 | 12 | 16, 17 | 6, 9.3 | 9, 11 | 10, 13 | 12, 15 | 28, 31.2 | 15, 18 | 14, 15 | 21 |
| EAM306 p52 | X | 11 | 8, 12 | 11, 12 | 12 | 16, 17 | 6, 9.3 | 9, 11 | 10, 13 | 12, 15 | 28, 31.2 | 15, 18 | 14, 15 | 21 |
| Patient 120 | XY | 13 | 8, 12 | 9, 10 | 11 | 17 | 9.3 | 8 | 11 | 16 | 30 | 17 | 14 | 27 |
| SDAR1 p62 | XY | 13 | 8, 12 | 9, 10 | 11 | 17 | 9.3 | 8 | 11 | 16 | 30 | 17 | 14 | 27 |
| SDAR2 p62 | XY | 13 | 8, 12 | 9, 10 | 11 | 17 | 9.3 | 8 | 11 | 16 | 30 | 17 | 14 | 27 |
| Patient 493 | XY | 12, 14 | 11, 12 | 12 | 12 | 14, 15 | 6, 7 | 8, 11 | 10, 13 | 12, 19 | 28, 29 | 16, 17 | 11, 14 | 19, 24 |
| JEM493 p68 | XY | 12, 14 | 11, 12 | 12 | 12 | 14, 15 | 7 | 8, 11 | 10, 13 | 12, 19 | 28, 29 | 16, 17 | 11, 14 | 19, 24 |
| MC-Th-493 | XY | 12, 14 | 11, 12 | 12 | 12 | 14, 15 | 7 | 8, 11 | 10, 13 | 12, 19 | 28, 29 | 16, 17 | 11, 14 | 19, 24 |
| Patient 529 | XY | 10, 12 | 12 | 10, 12 | 9, 12 | 16, 18 | 6, 9 | 8 | 10, 11 | 14, 18 | 31.2, 32.2 | 15, 19 | 13, 14 | 24, 24.2 |
| THJ529 p41 | XY | 10, 12 | 12 | 10, 12 | 9, 12 | 16, 18 | 6, 9 | 8 | 10, 11 | 14, 18 | 31.2, 32.2 | 15, 19 | 13, 14 | 24, 24.2 |
| MC-Th-529 | XY | 10, 12 | 12 | 10, 12 | 9, 12 | 16, 18 | 6, 9 | 8 | 10, 11 | 14, 18 | 31.2, 32.2 | 15, 19 | 13, 14 | 24, 24.2 |
| Patient 560 | X | 12 | 8, 12 | 10, 12 | 9, 14 | 14 | 6, 7 | 8 | 9, 10 | 15, 16 | 29, 32.2 | 16, 17 | 12, 14 | 21, 24 |
| THJ560 p34 | X | 12 | 12 | 10, 12 | 9, 14 | 14 | 6, 7 | 8 | 9, 10 | 15 | 29, 32.2 | 16, 17 | 12, 14 | 24 |
| MC-Th-560 | X | 12 | 12 | 10, 12 | 9, 14 | 14 | 6, 7 | 8 | 9, 10 | 15 | 29, 32.2 | 16, 17 | 12, 14 | 24 |
| Patient 95 | XY | 12, 13 | 9 | 10, 11 | 12, 13 | 16, 17 | 6, 9 | 8, 11 | 12 | 15 | 29, 30 | 14, 17 | 13, 14 | 22, 23 |
| MC-Th-95 | XY | 12, 13 | 9 | 10, 11 | —a | 16, 17 | 6, 9 | 8, 11 | 12 | 15 | —a | 14, 17 | 13, 14 | 22, 23 |
| Patient 374 | X | 11, 13 | 8, 12 | 10, 12 | 9, 13 | 14, 18 | 6 | 8, 11 | 10, 12 | 11, 14 | 28, 29 | 15, 17 | 12, 15 | 21, 24 |
| MC-Th-374 | X | 11, 13 | 8, 12 | 10, 12 | 9 | 14, 18 | 6 | 8, 11 | 10, 12 | 11, 14 | 29 | 15, 17 | 12, 15 | 21, 24 |
| Patient 467 | XY | 13 | 11 | 10 | 9, 11 | 15, 17 | 9, 9.3 | 8, 9 | 10, 11 | 12, 15 | 31, 32.2 | 14, 15 | 14, 15 | 19, 20 |
| MC-Th-467 | XY | 13 | 11 | 10 | 9, 11 | 15, 17 | 9, 9.3 | 8, 9 | 10, 11 | 12, 15 | 31 | 14, 15 | 14, 15 | 19, 20 |
| Patient 491 | X | 11, 13 | 9, 11 | 9, 11 | 8, 12 | 17, 18 | 9.3 | 8, 11 | 11 | 11, 16 | 29, 30.2 | 16 | 12, 13 | 21, 24 |
| MC-Th-491 | X | 11, 13 | 9, 11 | 9, 11 | 8, 12 | 17, 18 | 9.3 | 8, 11 | 11 | 11, 16 | 29, 30.2 | 16 | 12, 13 | 21, 24 |
| Patient 504 | XY | 11 | 11, 12 | 10, 11 | 12 | 15, 17 | 9.3 | 10, 12 | 10 | 12, 15 | 30, 30.2 | 15, 17 | 10, 13 | 19, 21 |
| MC-Th-504 | XY | 11 | 11, 12 | 10, 11 | 12 | 15, 17 | 9.3 | 10, 12 | 10 | 12, 15 | 30, 30.2 | 15, 17 | 10, 13 | 19, 21 |
| Patient 524 | XY | 11, 13 | 9, 10 | 9, 10 | 8, 10 | 14, 16 | 6, 7 | 11 | 10, 12 | 12, 15 | 30, 30.2 | 15, 18 | 13 | 20, 24.2 |
| MC-Th-524 | XY | 11, 13 | 9, 10 | 9, 10 | 8, 10 | 14, 16 | 6, 7 | 11 | 10, 12 | 12, 15 | 30, 30.2 | 15, 18 | 13 | 20, 24.2 |
| Patient 562 | XY | 11, 12 | 11, 12 | 9, 12 | 11, 12, 14 | 17 | 6, 8 | 8, 12 | 11, 12 | 12, 13 | 28, 29 | 14, 15 | 12, 13 | 21, 21.2 |
| MC-Th-562 | XY | 11, 12 | 11, 12 | 9, 12 | 11, 12 | 17 | 6 | 8, 12 | 11, 12 | 12 | 29 | 14 | 12, 13 | 21.2 |
All male-derived cell lines and PDTX models carried the amelogenin Y-allele.
Locus data were inconclusive.
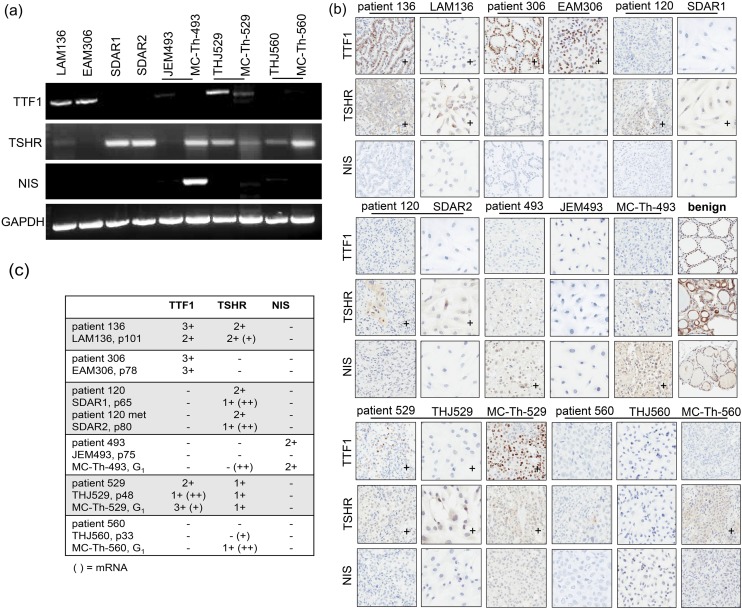
Once human origin and authenticity were confirmed, three thyroid-specific differentiation markers were examined at mRNA and protein levels in the cell lines and their corresponding PDTX models. By RT-PCR of cell lines and PDTX models, TTF1 and TSHR mRNA was observed at various amounts whereas NIS mRNA was only seen in MC-Th-493 [Fig. 2(a)]. For IHC of patient tumor tissue, cell lines, and PDTX models, protein expression varied and did not necessarily resemble the mRNA levels. PDTX models more closely resembled the patients’ expression pattern and intensity than did the cell lines [Fig. 2(b)]. Thus, both mRNA and protein levels were scored based on signal intensity and summarized for comparison [Fig. 2(c)]. LAM136, as expected, was the most differentiated as a PTC because it expressed both TTF1 and TSHR. The expression of TTF1 was to a lesser degree than the patient and despite having barely detectable TSHR mRNA; the protein expression was moderately expressed as in the patient. EAM306 only expressed TTF1, which was comparable to the patient. Both SDAR1 and SDAR2 only expressed TSHR, which was weaker than was seen in the patient despite having strong intensity levels of mRNA. JEM493 showed none of the three markers, whereas its PDTX counterpart expressed NIS comparable to the original patient. However, MC-Th-493 had moderate TSHR mRNA levels and had some small focal areas of positive staining (<20%) along with the patient. Despite having TSHR mRNA, THJ560 showed no TSHR at the protein level, whereas its corresponding PDTX expressed the receptor and the patient had some small focal areas of staining (<20%). THJ529 expressed comparable TSHR whereas TTF1 was reduced as compared with its PDTX counterpart and the original patient regardless of mRNA levels (Fig. 2). To examine whether thyroid marker differentiation changes over generations, we examined TTF1 in MC-Th-529 up to G8 and did not see any changes in overall TTF1 expression levels [Supplemental Fig. 1(b)].
Figure 2.
Thyroid marker analysis and comparison. (a) RT-PCR of TTF1, TSHR, NIS, and GAPDH loading control in thyroid cell lines and PDTX models. (b) IHC of TTF1, TSHR, and NIS in thyroid patients along with corresponding cell lines and PDTX models. Benign thyroid tissue was used as a positive control. IHC images were obtained at original magnification ×20 using an Aperio AT2 Scanscope. + indicated at least 20% positive signal intensity as described in “Materials and Methods.” (c) Summary of thyroid marker mRNA and protein expression (n = 3). IHC expression was scored as either weak (1+), moderate (2+), or strong (3+) with criteria of at least 20% positivity [mRNA when data deviated from protein expression and were categorized as either weak (+) or moderate–strong (++)]. Passage and generation numbers of the models are as indicated.
Common thyroid cancer mutations
Table 2 summarizes the common tumor genetic changes that we found in the cell lines and their corresponding PDTX models for comparison with the originating patient tumor. All models harbored TERT promoter mutations with C228T (EAM306, JEM493, MC-Th-493, THJ529, MC-Th-529, THJ560, and MC-Th-560) and C250T (LAM136, SDAR1, and SDAR2) being mutually exclusive (4, 6). Moreover, SDAR1 and SDAR2 had a homozygous C250T mutation as seen by a single peak detected by Sanger sequencing (Supplemental Fig. 3) and confirmed by MSK-IMPACT testing. An additional T349C mutation was detected in LAM136, EAM306, THJ560, and MC-Th-560 that was verified in the patient tumor source.
Table 2.
Thyroid Cell Lines and Their Corresponding PDTX Models Encompassing Common Mutations Found in Thyroid Tumors
| Lesion | Sex | TP53 | PTEN | PIK3CA | RAS | BRAF | TERT | |
|---|---|---|---|---|---|---|---|---|
| LAM136 | PTC | F | C577T [p.H193Y] | WT | WT | WT | T1799A [p.V600E] | T349C |
| A599Ta [p.N200I] | C250T | |||||||
| EAM306 | FTC | F | G976T [p.E326b] | WT | WT | NRASA182G [p.Q61R] | WT | T349C |
| C228T | ||||||||
| SDAR1 | FTC | M | G845C [p.R282P] | G160 [p.V54fs] | WT | WT | WT | C250Tc |
| G648a [p.V217fs] | ||||||||
| SDAR2 | FTC met | M | G648 [p.V217fs] | G160 [p.V54fs] | WT | WT | WT | C250Tc |
| JEM493 | ATC | M | WT | WT | WT | HRASA182G [p.Q61R] | WT | C228T |
| MC-Th-493 | ATC PDTX | M | WT | WT | WT | HRASA182G [p.Q61R] | WT | C228T |
| THJ529 | PDTC | M | C722G [p.S241C] | WT | WT | WT | T1799A [p.V600E] | C228T |
| MC-Th-529 | PDTC PDTX | M | WT | WT | WT | WT | T1799A [p.V600E] | C228T |
| THJ560 | ATC | F | G820T [p.V274F] | WT | WT | WT | T1799A [p.V600E] | T349C C228T |
| MC-Th-560 | ATC PDTX | F | G820T [p.V274F] | WT | WT | WT | T1799A [p.V600E] | T349C C228T |
Mutations for TP53, PTEN, NF1, and EIF1AX were determined by MSK-IMPACT testing. EIF1AX, RET/PTC, and PAX8/PPARγ (PPFP) were wild-type in all models.
Abbreviation: WT, wild-type.
Low allelic frequencies (<0.16), which may not be detectable via Sanger sequencing.
Nonsense mutation.
Homozygous mutation as seen by a single peak detected by Sanger sequencing and confirmed by MSK-IMPACT testing.
In addition to the TERT mutations, LAM136 was also a BRAFT1799A mutant with TP53C577T and TP53A599T mutations with only a 0.21 and 0.16 frequency, respectively. EAM306 was an NRASA182G mutant and a TP53G976T mutant. The partnered FTC cell line models SDAR1 and SDAR2 were both PTENG160 and TP53 mutant. SDAR2 was a TP53G648 mutant whereas SDAR1 was a TP53G845C mutant that was not detected in the primary tumor site of the patient. However, by MSK-IMPACT testing, SDAR1 also had a low TP53G648 allele frequency of 0.11, below the 15% to 16% required for Sanger sequencing detection. THJ529 and its PDTX both harbor a BRAFT1799A mutation, but only the cell line had an additional TP53C722G mutation. JEM493 and its PDTX both only have an HRASA182G mutation whereas THJ560 and its PDTX have BRAFT1799A mutations and TP53G820T mutations. EIF1AX, RET/PTC, and PPFP were also examined and were wild-type in both the cell lines and three PDTX models.
Growth rates and tumorigenicity
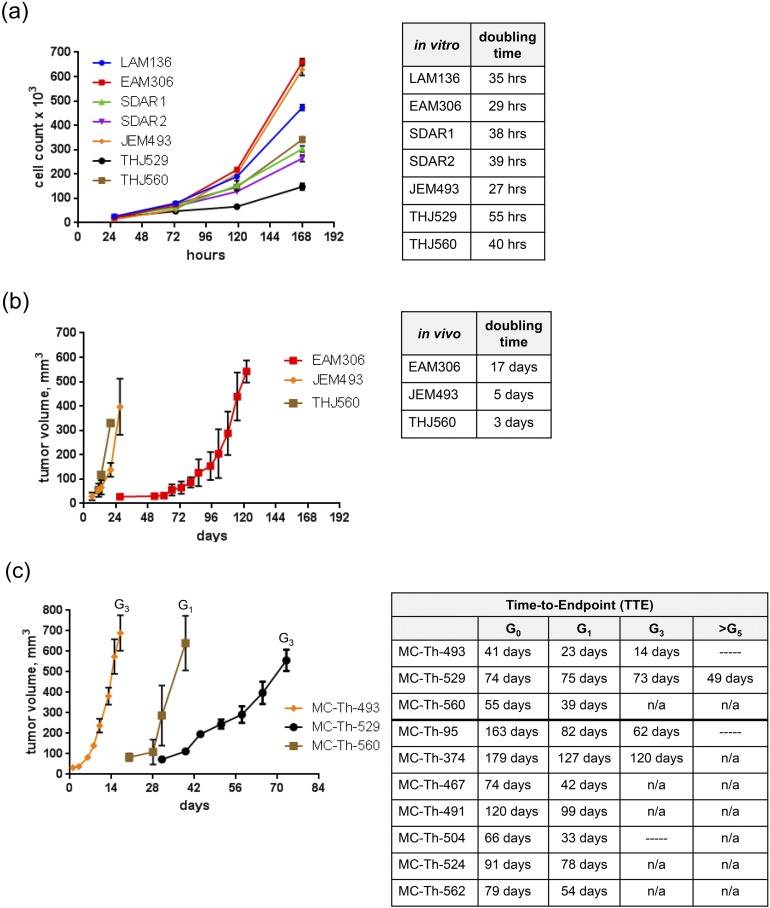
All seven cell lines were examined for population doubling time and tumorigenicity. In vitro, EAM306 and JEM493 were the most rapidly proliferating cell lines with doubling times <30 hours, whereas THJ529 was the slowest with a doubling time of 55 hours [Fig. 3(a)]. For tumorigenicity, 2 million cells in 50% Matrigel® were injected into the flanks of female athymic nude mice. The ATC lines (JEM493 and THJ560) grew rapidly in vivo with doubling times <5 days whereas EAM306 (FTC) had a doubling time of 17 days. The remaining cell lines did not grow in vivo even when 10 million cells were injected into the flanks of female NOD scid gamma mice [Fig. 3(b)]. Only the undifferentiated patient tumors gave rise to both cell lines and PDTX models (MC-Th-493, MC-Th-529, and MC-Th-560) within the 1- to 4-month time frame. Tumor volumes were measured once or twice weekly, with volumes between 600 and 800 mm3 for each generation after tumor implantation as time to endpoint (TTE). The most rapidly growing tumor, MC-Th-493, had an initial G0 TTE of 41 days, whereas succeeding propagation generations ranged from 14 to 23 days [Fig. 3(c)]. MC-Th-529 for multiple generations sustained a TTE ∼73 to 75 days until G5, which had a change in growth (>10 days) of 49 days. Later generations in MC-Th-560 were not accessed, but the G0 TTE was 55 days followed by a G1 TTE of 39 days. An earlier model, MC-Th-95, was initially incubated past the 4-month time frame and had increased growth with the subsequent generations that ranged from 62 to 82 days. MC-Th-374 had an initial G0 of 179 days whereas the subsequent generations ranged from 120 to 127 days [Fig. 3(c)]. As seen in the other PDTX models (MC-Th-467, MC-Th-491, MC-Th-504, MC-Th-524, and MC-Th-562) the initial G0 incubation was >G1 because the human tumor needed to establish itself within a foreign mouse microenvironment. Thus, G1 should be used as the reference point for growth comparison for subsequent generations. Future generations were expanded even further for MC-Th-493, MC-Th-95, and MC-Th-504, but the growth rate was not accessed. No expansions for future generations (>G1) were done in MC-Th-467, MC-Th-491, MC-Th-504, MC-Th-524, and MC-Th-562 [Fig. 1(d) and 3(c)].
Figure 3.
In vitro growth rates and tumorigenicity in vivo. (a) In vitro growth curves of thyroid cell lines with the indicated population doubling times in hours. Cells were counted at 48-h intervals after the initial 24-h time point. Data are graphed as mean cell count ± SD (n = 3). Media were changed every 72 h. (b) In vivo growth curves of thyroid cell lines that established tumors in vivo with the indicated doubling times in days. For EAM306, JEM493, and THJ560, 2 million cells in 50% Matrigel® were injected into the flanks of female athymic nude mice. LAM136, SDAR1, SDAR2, and THJ529 did not grow even when 10 million cells were injected into the flanks of female NOD scid gamma mice. (c) In vivo growth curves of PDTX models surgically implanted with 5 × 5 × 5-mm fragments and 100 μL of Matrigel® into the flanks of athymic nude female mice are shown with the indicated generation TTE in days after tumor implantation. Endpoint was defined as a tumor volume between 600 and 800 mm3. Tumors were measured once or twice weekly with calipers. Data are graphed as mean volume ± SE (n = 5). Most of the PDTX models were established within the 1- to 4-mo range with attempts up to 6 mo in two earlier models. n/a, not tested; —, tumor growth was not determined.
Discussion
We have contributed 7 new thyroid cell lines and 10 PDTX models of various thyroid cancer subtypes for cancer research. More importantly, we have also established a comprehensive protocol that allows inclusive characterization and subsequent tracking of the generated cell lines and PDTX models, facilitating correlation of the performance of these cell lines in preclinical studies with the original tumors. The overall cell line success rate was 2.3%, which was largely attributed to failure by PTC-derived cell lines. The most successful histological subtypes included ATC (22.2%), PDTC (20%), and FTC (16.7%), with TERT mutations appearing to be the common parameter for thyroid cell line establishment success. The verification criteria we recommend should include: (1) determination of human content within PDTX models at every generation, (2) exclusion of lymphoma, (3) DNA fingerprinting and histological comparisons to establish linkage to originating tumor, (4) examining thyroid differentiation (particularly in WDTC) by screening two to three thyroid markers such as TSHR, TTF1, TG, NIS, PAX8, and TPO (25), (5) examination of biological behavior (growth rate, tumorigenicity), and (6) screening for two or more common thyroid cancer genetic changes, for example, TP53, PTEN, PIK3CA, RAS, BRAF, TERT promoter, RET/PTC, PPFP, NF1, and EIF1AX (Figs. 1–3; Supplemental Figs. 1–3; Supplemental Table 1; Tables 1 and 2).
To further validate our approach, we have also performed a comprehensive online search for all thyroid cell line models that have been at the minimum characterized and STR profiled or karyotyped as valid models (14, 23, 26, 27). Based on this, there are only the following valid cell lines:
PTC (10 patients) included: LAM1 (28), K1 (29) [derivative of GLAG-66 (30, 31)], MDA-T22 (26), MDA-T32 (26), MDA-T41 (26), MDA-T85 (26), MDA-T68 (26), MDA-T120 (26), TPC1 (32), and the newly characterized LAM136.
Conventional FTC (five patients): ML-1 (33), FTC-133 (34) partnered with FTC-236 (35) and FTC-238 (35), TT2609-C02 (36) partnered with TT2609-B02 (36), and the newly characterized EAM306 and SDAR1 partnered with SDAR2. WRO (13) as an FTC was excluded because two versions of BRAF status and STR profiles are in distribution (3, 14)
HCC: a single line, XTC.UC1 (37).
PDTC (five patients): KTC1 (38), T243 (39), T351 (39), BCPAP (40) (originally accepted as PTC) (3), and the newly characterized THJ529.
ATC (>25 lines); but only 7 cell lines have been matched to the original patients: THJ-11T (23), THJ-16T (23), THJ-21T (23), THJ-29T (23), OGK-P (27) partnered with OGK-M (27), and the newly characterized JEM493 and THJ560.
These cell lines are summarized in Table 3 (28–61).
Table 3.
Published Thyroid Cell Lines Recommended for Biomedical Research as of 2017
| Cell Line | RRID | Sex | Gender | Lesion | Origin and Genotype References |
|---|---|---|---|---|---|
| K1 (GLAG-66) | CVCL_2537 | XY | M | PTC | Challeton et al. 1997 (14, 29) |
| CVCL_9918 | Antonini et al. 1993 (30) | ||||
| LAM1 | CVCL_W912 | XY | M | PTC met to lung/LN | Copland et al. 2006 (28); Supplemental Table 2 |
| LAM136 | CVCL_RP38 | X | F | PTC | Marlow et al. (this publication) (19) |
| MDA-T120 | CVCL_QW85 | X | F | PTC met to LN + ATC component | Henderson et al. 2015 (26) |
| MDA-T22 | CVCL_QW80 | X | F | PTC | Henderson et al. 2015 (26) |
| MDA-T32 | CVCL_W913 | XY | M | PTC | Henderson et al. 2015 (26) |
| MDA-T41 | CVCL_W914 | XY | M | PTC met to LN | Henderson et al. 2015 (26) |
| MDA-T68 | CVCL_QW83 | XY | M | FV-PTC | Henderson et al. 2015 (26) |
| MDA-T85 | CVCL_QW84 | XY | M | PTC met to LN | Henderson et al. 2015 (26) |
| TPC-1 | CVCL_6298 | X | F | PTC | Tanaka et al. 1987 (14, 32) |
| EAM306 | CVCL_AV62 | X | F | FTC | Marlow et al. (this publication) (19) |
| FTC-133 | CVCL_1219 | Xa | M | FTC met to LN | Goretzki et al. 1990 (14, 34) |
| FTC-236 | CVCL_2446 | Xa | M | FTC met to LN | Demeure et al. 1992 (35); European Collection of Authenticated Cell Cultures |
| FTC-238 | CVCL_2447 | Xa | M | FTC met to lung | Demeure et al. 1992 (35, 41) |
| ML-1 | CVCL_H525 | X | F | FTC | Schönberger et al. 2000 (14, 33) |
| SDAR1 | CVCL_AV63 | XY | M | FTC | Marlow et al. (this publication) (19) |
| SDAR2 | CVCL_AV64 | XY | M | FTC met to throat | Marlow et al. (this publication) |
| TT2609-B02b | CVCL_A595 | XY | M | FTC | Geldof et al. 2001 (36) |
| TT2609-CO2 | CVCL_2218 | XY | M | FTC | Geldof et al. 2001 (14, 36) |
| XTC.UC1 | CVCL_9916 | X | F | HCC variant of FTC | Zielke et al. 1998 (37); Supplemental Table 2 |
| BCPAP | CVCL_0153 | X | F | PDTC | Fabien et al. 1994 (14, 40) |
| KTC-1 | CVCL_6300 | XY | M | PDTC | Kurebayashi et al. 2000 (14, 38) |
| T243b | CVCL_M976 | XY | M | PDTC | Rodrigues et al. 2007 (39) |
| T351 | CVCL_M977 | XY | M | PDTC | Rodrigues et al. 2007 (27, 39) |
| THJ529 | CVCL_RP39 | XY | M | PDTC | Marlow et al. (this publication) |
| 8505C | CVCL_1054 | X | F | ATC | Ito et al. 1993 (14, 42) |
| ACT-1 | CVCL_6291 | X | F | ATC | Chung et al. 2002 (14, 43) |
| BHT101 | CVCL_1085 | X | F | ATC | Palyi et al. 1993 (14, 44) |
| C643 | CVCL_5969 | XY | M | ATC | Gustavsson et al. 1996 (14, 45) |
| Cal-62 | CVCL_1112 | X | F | ATC | Gioanni et al. 1991 (14, 46) |
| FF-1 | CVCL_RP34 | X | F | ATC | Frasca et al. 2001 (47); Supplemental Table 2 |
| FRO | CVCL_6287 | XY | M | ATC | Nishihara et al. 1997 (14, 48) |
| HTh7 | CVCL_6289 | X | F | ATC | Carlsson et al. 1983 (14, 49) |
| HTh74 | CVCL_6288 | X | F | ATC | Heldin et al. 1991 (14, 50) |
| HTh83 | CVCL_0046 | XY | M | ATC | Dahlman et al. 2000 (51, 52) |
| HTh104 | CVCL_A427 | X | F | ATC | Lee et al. 2007 (14, 53) |
| HTh112 | CVCL_A428 | XY | M | ATC | Lee et al. 2007 (51, 53) |
| JEM493 | CVCL_RP37 | XY | M | ATC | Marlow et al. (this publication) |
| KAT18 | CVCL_6303 | X | F | ATC | Ain et al. 1997 (14, 54) |
| KTC-2 | CVCL_6476 | X | F | ATC | Kurebayashi et al. 2003 (14, 55) |
| KTC-3 | CVCL_W911 | X | F | ATC | Pushkarev et al. 2004 (56); Supplemental Table 2 |
| LUTC-1 | CVCL_RP41 | X | F | ATC | Wennerberg et al. 2014 (57) |
| LUTC-2 | CVCL_RP42 | X | F | ATC | Wennerberg et al. 2014 (57) |
| LUTC-8 | CVCL_RP45 | XY | M | ATC | Wennerberg et al. 2014 (57) |
| LUTC-10 | CVCL_RP46 | XY | M | ATC | Wennerberg et al. 2014 (57) |
| LUTC-12 | CVCL_RP47 | X | F | ATC | Wennerberg et al. 2014 (57) |
| LUTC-14 | CVCL_RP49 | X | F | ATC | Wennerberg et al. 2014 (57) |
| MB-1 | CVCL_2109 | Xa | M | ATC | Stenner et al. 2008 (41, 58) |
| OCUT-1 | CVCL_6017 | X | F | ATC | Ogisawa et al. 2002 (59); Supplemental Table 2 |
| OGK-M | CVCL_RP35 | XY | M | ATC | Garg et al. 2015 (27) |
| OGK-P | CVCL_RP36 | XY | M | ATC | Garg et al. 2015 (27) |
| SW1736 | CVCL_3883 | X | F | ATC | Xu et al. 2003 (14, 60) |
| T235 | CVCL_6478 | X | F | ATC | Rodrigues et al. 2007 (14, 39) |
| T238 | CVCL_6299 | X | F | ATC | Rodrigues et al. 2007 (14, 39) |
| T241 | CVCL_M975 | X | F | ATC | Rodrigues et al. 2007 (27, 39) |
| THJ-11T | CVCL_W919 | XY | M | ATC | Marlow et al. 2010 (23) |
| THJ-16T | CVCL_W920 | X | F | ATC | Marlow et al. 2010 (23) |
| THJ-21T | CVCL_W921 | XY | M | ATC | Marlow et al. 2010 (23) |
| THJ-29T | CVCL_W922 | X | F | ATC | Marlow et al. 2010 (23) |
| THJ560 | CVCL_RP40 | X | F | ATC | Marlow et al. (this publication) |
| TTA1 | CVCL_6297 | X | F | ATC | Yano et al. 2007 (14, 61) |
Cellosaurus/resource identifications (RRIDs) are listed for each line. Sex was determined by genotype and gender was the phenotype of the patient.
Abbreviation: LN, lymph node.
Loss of the Y-linked allele.
No STR profiles, but has unique karyotypes. For cell lines without published STR genotypes, the STR profiles are listed in Supplemental Table 2 as indicated and cross-referenced to STR databases for authenticity.
Our study also confirmed that PDTX models better represented the patient with regard to differential expression of thyroid markers (Fig. 2) and mutations (Table 2). For example, MC-Th-529 was TP53 wild-type similar to the patient, but the cell line harbored a TP53C722G mutation that could have been either a subclone of the tumor tissue that would be difficult to detect by Sanger sequencing, or it might have been selected for two-dimensional cell culture. The PDTX models were characterized in similar fashion to the cell line models with the addition of quality control checkpoints. This was particularly important because PDTX models can be used as “avatars” for personalized medicine and as a screening platform for phase I clinical trials because they mimic clinical response to therapy (62). Thus, one to two individual mice for every generation were randomly checked for species content and heterogeneity for quality control (Fig. 1; Supplemental Fig. 1). This was particularly important because during subsequent generations, growth rate can change and the murine Src content in the PDTX models can increase as seen particularly in MC-Th-95 and MC-Th-529 [Figs. 1(d) and 3(c)]. With that said, the human Src content and human lamin A/C expression did not deviate (Fig. 1; Supplemental Fig. 1), so these PDTX models were still valid. In our experience with multiple tissue types, species content can change over generations and even completely lose human content when the initial engraftment reached 6 months (data not shown). More studies need to be conducted to explore this observation. Also, slower engraftments increase the risk of the tumors arising from lymphomas (20). Thus, to minimize the risk, engraftment time of PDTX models needs to be limited to the recommended 4 months and maintained at low passage or generation numbers to preserve genetic integrity of the original tumor (<G10) (17).
The 10 PDTX model success rate in athymic nude mice was 14.9%. Most of the models consisted of the anaplastic subtype (six models, 75% success) and the others included one FVPTC (14.2% success), one PDTC, one SCTC, and one insular TC with no success in FTC and conventional PTC. To our knowledge, there is only one other published thyroid PDTX model, which was of the anaplastic subtype, and it was used for therapy screening (63). Our group deposited 6 of the 10 newly developed models at Charles River Laboratories (www2.criver.com/l/60962/2016-07-13/9rjvzc). Another repository, CrownBio, also has seven thyroid PDTX models available. Champions Oncology did not have a publicly accessible database so we could not confirm whether thyroid PDTX models were deposited. Regardless, to improve the oncology problem of only a 5% efficacy in clinical trials (15), more translationally relevant PDTX models for thyroid cancer need to be developed, especially for the refractory papillary, follicular, Hürthle cell, poorly differentiated, and completely dedifferentiated anaplastic subtypes.
In conclusion, having a large resource of validated cell lines and PDTX models that cover the many unique mutation signatures found in patient thyroid tumors would enhance development of therapeutic options for radioiodine refractory and chemotherapy-resistant thyroid cancers. Furthermore, validating STR profiles back to the originating tumor source should be regularly practiced for verifying identification.
Supplementary Material
Acknowledgments
We thank Brandy Edenfield for embedding and processing tissues for immunohistochemistry as well as Dr. Jason Hall for providing mouse pancreatic tissue for mouse controls. We also thank the Mayo Clinic Medical Genome Facility Genotyping Core for expertise and help on acquiring the STR profiles.
Financial Support: This work was supported by National Institutes of Health and Medical Research Grants R01 CA136665 (to J.A.C. and R.C.S.) and P50 CA172012-01 (to J.A.F.), Florida Department of Health Bankhead-Coley Cancer Research Program Grant FL09B202 (to J.A.C. and R.C.S.), and Mayo Comprehensive Cancer Center Grant P30CA01508343 (to R.C.S), as well as support from Alfred D. and Audrey M. Petersen (to R.C.S.), the Francis and Miranda Childress Foundation Fund for Cancer Research (to J.A.C.), the John A. and Bette B. Klacsmann Fund for Cancer Research at the Mayo Clinic in Florida (to J.A.C.), the Bruno V. and Bruce E. Zanoni Endowed Research Fund (to J.A.C.), the Betty G. Castigliano Fund in Cancer Research Honoring S. Gordon Castigliano, MD, cancer research at the Mayo Clinic Florida (to J.A.C.), and by the Dewitt C. (Dash) Goff Fund for thyroid cancer research (to J.A.C.).
Disclosure Summary: L.A.M., R.C.S., and J.A.C. have received royalties for the thyroid PDTX models from Charles River Laboratories Inc. for research purposes. The remaining authors have nothing to disclose.
Glossary
Abbreviations:
- ATC
anaplastic thyroid cancer
- CT
cycle threshold
- FFPE
formalin fixation for paraffin embedding
- FTC
follicular thyroid cancer
- FVPTC
follicular variant papillary thyroid cancer
- HCC
Hürthle cell cancer
- H&E
hematoxylin and eosin
- IHC
immunohistochemistry
- LOH
loss of heterogeneity
- NIS
sodium iodide symporter
- PDTC
poorly differentiated thyroid cancer
- PDTX
patient-derived tumor xenograft
- PSA
penicillin–streptomycin–amphotericin B
- PTC
papillary thyroid cancer
- qPCR
quantitative PCR
- SCTC
squamous cell thyroid carcinoma
- STR
short tandem repeat
- TSHR
TSH receptor
- TTE
time to endpoint
- TTF1
thyroid transcription factor-1
- WDTC
well-differentiated thyroid cancer
- Δ
difference
References
- 1. National Cancer Institute. Cancer stat facts: thyroid cancer. Available at: seer.cancer.gov/statfacts/html/thyro.html. Accessed 1 August 2017.
- 2. Cancer Genome Atlas Research Network Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159(3):676–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Saiselet M, Floor S, Tarabichi M, Dom G, Hébrant A, van Staveren WC, Maenhaut C. Thyroid cancer cell lines: an overview. Front Endocrinol (Lausanne). 2012;3:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu X, Bishop J, Shan Y, Pai S, Liu D, Murugan AK, Sun H, El-Naggar AK, Xing M. Highly prevalent TERT promoter mutations in aggressive thyroid cancers. Endocr Relat Cancer. 2013;20(4):603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burman KD. Is poorly differentiated thyroid cancer poorly characterized? J Clin Endocrinol Metab. 2014;99(4):1167–1169. [DOI] [PubMed] [Google Scholar]
- 6. Landa I, Ganly I, Chan TA, Mitsutake N, Matsuse M, Ibrahimpasic T, Ghossein RA, Fagin JA. Frequent somatic TERT promoter mutations in thyroid cancer: higher prevalence in advanced forms of the disease. J Clin Endocrinol Metab. 2013;98(9):E1562–E1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Landa I, Ibrahimpasic T, Boucai L, Sinha R, Knauf JA, Shah RH, Dogan S, Ricarte-Filho JC, Krishnamoorthy GP, Xu B, Schultz N, Berger MF, Sander C, Taylor BS, Ghossein R, Ganly I, Fagin JA. Genomic and transcriptomic hallmarks of poorly differentiated and anaplastic thyroid cancers. J Clin Invest. 2016;126(3):1052–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Smallridge RC, Copland JA. Anaplastic thyroid carcinoma: pathogenesis and emerging therapies. Clin Oncol (R Coll Radiol). 2010;22(6):486–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kunstman JW, Juhlin CC, Goh G, Brown TC, Stenman A, Healy JM, Rubinstein JC, Choi M, Kiss N, Nelson-Williams C, Mane S, Rimm DL, Prasad ML, Höög A, Zedenius J, Larsson C, Korah R, Lifton RP, Carling T. Characterization of the mutational landscape of anaplastic thyroid cancer via whole-exome sequencing. Hum Mol Genet. 2015;24(8):2318–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tuttle RM. Overview of follicular thyroid cancer. Available at: www.uptodate.com/contents/follicular-thyroid-cancer-including-hurthle-cell-cancer. Accessed 1 August 2017.
- 11. Smallridge RC, Ain KB, Asa SL, Bible KC, Brierley JD, Burman KD, Kebebew E, Lee NY, Nikiforov YE, Rosenthal MS, Shah MH, Shaha AR, Tuttle RM; American Thyroid Association Anaplastic Thyroid Cancer Guidelines Taskforce . American Thyroid Association guidelines for management of patients with anaplastic thyroid cancer. Thyroid. 2012;22(11):1104–1139. [DOI] [PubMed] [Google Scholar]
- 12. Schweppe RE. Thyroid cancer cell lines: critical models to study thyroid cancer biology and new therapeutic targets. Front Endocrinol (Lausanne). 2012;3:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Estour B, Van Herle AJ, Juillard GJ, Totanes TL, Sparkes RS, Giuliano AE, Klandorf H. Characterization of a human follicular thyroid carcinoma cell line (UCLA RO 82 W-1). Virchows Arch B Cell Pathol Incl Mol Pathol. 1989;57(3):167–174. [DOI] [PubMed] [Google Scholar]
- 14. Schweppe RE, Klopper JP, Korch C, Pugazhenthi U, Benezra M, Knauf JA, Fagin JA, Marlow LA, Copland JA, Smallridge RC, Haugen BR. Deoxyribonucleic acid profiling analysis of 40 human thyroid cancer cell lines reveals cross-contamination resulting in cell line redundancy and misidentification. J Clin Endocrinol Metab. 2008;93(11):4331–4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hutchinson L, Kirk R. High drug attrition rates—where are we going wrong? Nat Rev Clin Oncol. 2011;8(4):189–190. [DOI] [PubMed] [Google Scholar]
- 16. Tentler JJ, Tan AC, Weekes CD, Jimeno A, Leong S, Pitts TM, Arcaroli JJ, Messersmith WA, Eckhardt SG. Patient-derived tumour xenografts as models for oncology drug development. Nat Rev Clin Oncol. 2012;9(6):338–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pompili L, Porru M, Caruso C, Biroccio A, Leonetti C. Patient-derived xenografts: a relevant preclinical model for drug development. J Exp Clin Cancer Res. 2016;35(1):189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cassidy JW, Caldas C, Bruna A. Maintaining tumor heterogeneity in patient-derived tumor xenografts. Cancer Res. 2015;75(15):2963–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. von Roemeling CA, Marlow LA, Pinkerton AB, Crist A, Miller J, Tun HW, Smallridge RC, Copland JA. Aberrant lipid metabolism in anaplastic thyroid carcinoma reveals stearoyl CoA desaturase 1 as a novel therapeutic target. J Clin Endocrinol Metab. 2015;100(5):E697–E709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wetterauer C, Vlajnic T, Schüler J, Gsponer JR, Thalmann GN, Cecchini M, Schneider J, Zellweger T, Pueschel H, Bachmann A, Ruiz C, Dirnhofer S, Bubendorf L, Rentsch CA. Early development of human lymphomas in a prostate cancer xenograft program using triple knock-out immunocompromised mice. Prostate. 2015;75(6):585–592. [DOI] [PubMed] [Google Scholar]
- 21. Baker M. Reproducibility: respect your cells! Nature. 2016;537(7620):433–435. [DOI] [PubMed] [Google Scholar]
- 22. Cradic KW, Milosevic D, Rosenberg AM, Erickson LA, McIver B, Grebe SK. Mutant BRAF(T1799A) can be detected in the blood of papillary thyroid carcinoma patients and correlates with disease status. J Clin Endocrinol Metab. 2009;94(12):5001–5009. [DOI] [PubMed] [Google Scholar]
- 23. Marlow LA, D’Innocenzi J, Zhang Y, Rohl SD, Cooper SJ, Sebo T, Grant C, McIver B, Kasperbauer JL, Wadsworth JT, Casler JD, Kennedy PW, Highsmith WE, Clark O, Milosevic D, Netzel B, Cradic K, Arora S, Beaudry C, Grebe SK, Silverberg ML, Azorsa DO, Smallridge RC, Copland JA. Detailed molecular fingerprinting of four new anaplastic thyroid carcinoma cell lines and their use for verification of RhoB as a molecular therapeutic target. J Clin Endocrinol Metab. 2010;95(12):5338–5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, Chandramohan R, Liu ZY, Won HH, Scott SN, Brannon AR, O’Reilly C, Sadowska J, Casanova J, Yannes A, Hechtman JF, Yao J, Song W, Ross DS, Oultache A, Dogan S, Borsu L, Hameed M, Nafa K, Arcila ME, Ladanyi M, Berger MF. Memorial Sloan Kettering-integrated mutation profiling of actionable cancer targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17(3):251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ma R, Latif R, Davies TF. Thyrotropin-independent induction of thyroid endoderm from embryonic stem cells by activin A. Endocrinology. 2009;150(4):1970–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Henderson YC, Ahn SH, Ryu J, Chen Y, Williams MD, El-Naggar AK, Gagea M, Schweppe RE, Haugen BR, Lai SY, Clayman GL. Development and characterization of six new human papillary thyroid carcinoma cell lines. J Clin Endocrinol Metab. 2015;100(2):E243–E252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Garg M, Okamoto R, Nagata Y, Kanojia D, Venkatesan S, MT A, Braunstein GD, Said JW, Doan NB, Ho Q, Akagi T, Gery S, Liu LZ, Tan KT, Chng WJ, Yang H, Ogawa S, Koeffler HP. Establishment and characterization of novel human primary and metastatic anaplastic thyroid cancer cell lines and their genomic evolution over a year as a primagraft. J Clin Endocrinol Metab. 2015;100(2):725–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Copland JA, Marlow LA, Williams SF, Grebe SK, Gumz ML, Maples WJ, Silverman VE, Smallridge RC. Molecular diagnosis of a BRAF papillary thyroid carcinoma with multiple chromosome abnormalities and rare adrenal and hypothalamic metastases. Thyroid. 2006;16(12):1293–1302. [DOI] [PubMed] [Google Scholar]
- 29. Challeton C, Branea F, Schlumberger M, Gaillard N, de Vathaire F, Badie C, Antonini P, Parmentier C. Characterization and radiosensitivity at high or low dose rate of four cell lines derived from human thyroid tumors. Int J Radiat Oncol Biol Phys. 1997;37(1):163–169. [DOI] [PubMed] [Google Scholar]
- 30. Antonini P, Linares G, Gaillard N, Venuat AM, Schlumberger M, Travagli JP, Caillou B, Berger R, Parmentier C. Cytogenetic characterization of a new human papillary thyroid carcinoma permanent cell line (GLAG-66). Cancer Genet Cytogenet. 1993;67(2):117–122. [DOI] [PubMed] [Google Scholar]
- 31. Ribeiro FR, Meireles AM, Rocha AS, Teixeira MR. Conventional and molecular cytogenetics of human non-medullary thyroid carcinoma: characterization of eight cell line models and review of the literature on clinical samples. BMC Cancer. 2008;8(1):371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tanaka J, Ogura T, Sato H, Hatano M. Establishment and biological characterization of an in vitro human cytomegalovirus latency model. Virology. 1987;161(1):62–72. [DOI] [PubMed] [Google Scholar]
- 33. Schönberger J, Bauer J, Spruss T, Weber G, Chahoud I, Eilles C, Grimm D. Establishment and characterization of the follicular thyroid carcinoma cell line ML-1. J Mol Med (Berl). 2000;78(2):102–110. [DOI] [PubMed] [Google Scholar]
- 34. Goretzki PE, Frilling A, Simon D, Roeher HD. Growth regulation of normal thyroids and thyroid tumors in man. Recent Results Cancer Res. 1990;118:48–63. [DOI] [PubMed] [Google Scholar]
- 35. Demeure MJ, Damsky CH, Elfman F, Goretzki PE, Wong MG, Clark OH. Invasion by cultured human follicular thyroid cancer correlates with increased β1 integrins and production of proteases. World J Surg. 1992;16(4):770–776. [DOI] [PubMed] [Google Scholar]
- 36. Geldof AA, van Mourik JC, Rooimans MA, Arwert F, Hermsen MA, Schadee-Eestermans IL, van Dongen GA, van der Valk P, Lips P, Teule GJ Versteegh LRTvan der Clement EHP . Clonally related but phenotypically divergent human cancer cell lines derived from a single follicular thyroid cancer recurrence (TT2609). Thyroid. 2001;11(10):909–917. [DOI] [PubMed] [Google Scholar]
- 37. Zielke A, Tezelman S, Jossart GH, Wong M, Siperstein AE, Duh QY, Clark OH. Establishment of a highly differentiated thyroid cancer cell line of Hürthle cell origin. Thyroid. 1998;8(6):475–483. [DOI] [PubMed] [Google Scholar]
- 38. Kurebayashi J, Tanaka K, Otsuki T, Moriya T, Kunisue H, Uno M, Sonoo H. All-trans-retinoic acid modulates expression levels of thyroglobulin and cytokines in a new human poorly differentiated papillary thyroid carcinoma cell line, KTC-1. J Clin Endocrinol Metab. 2000;85(8):2889–2896. [DOI] [PubMed] [Google Scholar]
- 39. Rodrigues RF, Roque L, Krug T, Leite V. Poorly differentiated and anaplastic thyroid carcinomas: chromosomal and oligo-array profile of five new cell lines. Br J Cancer. 2007;96(8):1237–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fabien N, Fusco A, Santoro M, Barbier Y, Dubois PM, Paulin C. Description of a human papillary thyroid carcinoma cell line. Morphologic study and expression of tumoral markers. Cancer. 1994;73(8):2206–2212. [DOI] [PubMed] [Google Scholar]
- 41. Yu M, Selvaraj SK, Liang-Chu MM, Aghajani S, Busse M, Yuan J, Lee G, Peale F, Klijn C, Bourgon R, Kaminker JS, Neve RM. A resource for cell line authentication, annotation and quality control. Nature. 2015;520(7547):307–311. [DOI] [PubMed] [Google Scholar]
- 42. Ito T, Seyama T, Iwamoto KS, Hayashi T, Mizuno T, Tsuyama N, Dohi K, Nakamura N, Akiyama M. In vitro irradiation is able to cause RET oncogene rearrangement. Cancer Res. 1993;53(13):2940–2943. [PubMed] [Google Scholar]
- 43. Chung SH, Onoda N, Ishikawa T, Ogisawa K, Takenaka C, Yano Y, Hato F, Hirakawa K. Peroxisome proliferator-activated receptor gamma activation induces cell cycle arrest via the p53-independent pathway in human anaplastic thyroid cancer cells. Jpn J Cancer Res. 2002;93(12):1358–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pályi I, Péter I, Daubner D, Vincze B, Lõrincz I. Establishment, characterization and drug sensitivity of a new anaplastic thyroid carcinoma cell line (BHT-101). Virchows Arch B Cell Pathol Incl Mol Pathol. 1993;63(4):263–269. [DOI] [PubMed] [Google Scholar]
- 45. Gustavsson B, Hermansson A, Andersson AC, Grimelius L, Bergh J, Westermark B, Heldin NE. Decreased growth rate and tumour formation of human anaplastic thyroid carcinoma cells transfected with a human thyrotropin receptor cDNA in NMRI nude mice treated with propylthiouracil. Mol Cell Endocrinol. 1996;121(2):143–151. [DOI] [PubMed] [Google Scholar]
- 46. Gioanni J, Zanghellini E, Mazeau C, Zhang D, Courdi A, Farges M, Lambert JC, Duplay H, Schneider M. Characterization of a human cell line from an anaplastic carcinoma of the thyroid gland [in French]. Bull Cancer. 1991;78(11):1053–1062. [PubMed] [Google Scholar]
- 47. Frasca F, Vigneri P, Vella V, Vigneri R, Wang JY. Tyrosine kinase inhibitor STI571 enhances thyroid cancer cell motile response to hepatocyte growth factor. Oncogene. 2001;20(29):3845–3856. [DOI] [PubMed] [Google Scholar]
- 48. Nishihara E, Nagayama Y, Mawatari F, Tanaka K, Namba H, Niwa M, Yamashita S. Retrovirus-mediated herpes simplex virus thymidine kinase gene transduction renders human thyroid carcinoma cell lines sensitive to ganciclovir and radiation in vitro and in vivo. Endocrinology. 1997;138(11):4577–4583. [DOI] [PubMed] [Google Scholar]
- 49. Carlsson J, Nilsson K, Westermark B, Pontén J, Sundström C, Larsson E, Bergh J, Påhlman S, Busch C, Collins VP. Formation and growth of multicellular spheroids of human origin. Int J Cancer. 1983;31(5):523–533. [DOI] [PubMed] [Google Scholar]
- 50. Heldin NE, Cvejić D, Smeds S, Westermark B. Coexpression of functionally active receptors for thyrotropin and platelet-derived growth factor in human thyroid carcinoma cells. Endocrinology. 1991;129(4):2187–2193. [DOI] [PubMed] [Google Scholar]
- 51. Dahlman T, Lammerts E, Wik M, Bergström D, Grimelius L, Westermark K, Rubin K, Heldin NE. Fibrosis in undifferentiated (anaplastic) thyroid carcinomas: evidence for a dual action of tumour cells in collagen type I synthesis. J Pathol. 2000;191(4):376–386. [DOI] [PubMed] [Google Scholar]
- 52. Zhao M, Sano D, Pickering CR, Jasser SA, Henderson YC, Clayman GL, Sturgis EM, Ow TJ, Lotan R, Carey TE, Sacks PG, Grandis JR, Sidransky D, Heldin NE, Myers JN. Assembly and initial characterization of a panel of 85 genomically validated cell lines from diverse head and neck tumor sites. Clin Cancer Res. 2011;17(23):7248–7264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lee JJ, Foukakis T, Hashemi J, Grimelius L, Heldin NE, Wallin G, Rudduck C, Lui WO, Höög A, Larsson C. Molecular cytogenetic profiles of novel and established human anaplastic thyroid carcinoma models. Thyroid. 2007;17(4):289–301. [DOI] [PubMed] [Google Scholar]
- 54. Ain KB, Taylor KD, Tofiq S, Venkataraman G. Somatostatin receptor subtype expression in human thyroid and thyroid carcinoma cell lines. J Clin Endocrinol Metab. 1997;82(6):1857–1862. [DOI] [PubMed] [Google Scholar]
- 55. Kurebayashi J, Otsuki T, Tanaka K, Yamamoto Y, Moriya T, Sonoo H. Medroxyprogesterone acetate decreases secretion of interleukin-6 and parathyroid hormone-related protein in a new anaplastic thyroid cancer cell line, KTC-2. Thyroid. 2003;13(3):249–258. [DOI] [PubMed] [Google Scholar]
- 56. Pushkarev VM, Starenki DV, Saenko VA, Namba H, Kurebayashi J, Tronko MD, Yamashita S. Molecular mechanisms of the effects of low concentrations of taxol in anaplastic thyroid cancer cells. Endocrinology. 2004;145(7):3143–3152. [DOI] [PubMed] [Google Scholar]
- 57. Wennerberg E, Pfefferle A, Ekblad L, Yoshimoto Y, Kremer V, Kaminskyy VO, Juhlin CC, Höög A, Bodin I, Svjatoha V, Larsson C, Zedenius J, Wennerberg J, Lundqvist A. Human anaplastic thyroid carcinoma cells are sensitive to NK cell–mediated lysis via ULBP2/5/6 and chemoattract NK cells. Clin Cancer Res. 2014;20(22):5733–5744. [DOI] [PubMed] [Google Scholar]
- 58. Stenner F, Liewen H, Zweifel M, Weber A, Tchinda J, Bode B, Samaras P, Bauer S, Knuth A, Renner C. Targeted therapeutic approach for an anaplastic thyroid cancer in vitro and in vivo. Cancer Sci. 2008;99(9):1847–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ogisawa K, Onoda N, Ishikawa T, Takenaka C, Inaba M, Ogawa Y, Chung KH. Establishment and characterization of OCUT-1, an undifferentiated thyroid cancer cell line expressing high level of telomerase. J Surg Oncol. 2002;80(4):197–203. [DOI] [PubMed] [Google Scholar]
- 60. Xu X, Quiros RM, Gattuso P, Ain KB, Prinz RA. High prevalence of BRAF gene mutation in papillary thyroid carcinomas and thyroid tumor cell lines. Cancer Res. 2003;63(15):4561–4567. [PubMed] [Google Scholar]
- 61. Yano Y, Kamma H, Matsumoto H, Fujiwara M, Bando H, Hara H, Yashiro T, Ueno E, Ito K, Uchida K. Growth suppression of thyroid cancer cells by adenylcyclase activator. Oncol Rep. 2007;18(2):441–445. [PubMed] [Google Scholar]
- 62. Hidalgo M, Amant F, Biankin AV, Budinská E, Byrne AT, Caldas C, Clarke RB, de Jong S, Jonkers J, Mælandsmo GM, Roman-Roman S, Seoane J, Trusolino L, Villanueva A. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer Discov. 2014;4(9):998–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wunderlich A, Khoruzhyk M, Roth S, Ramaswamy A, Greene BH, Doll D, Bartsch DK, Hoffmann S. Pretherapeutic drug evaluation by tumor xenografting in anaplastic thyroid cancer. J Surg Res. 2013;185(2):676–683. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.