Abstract
O-GlcNAcylation is a dynamic and functionally diverse post-translational modification shown to affect thousands of proteins, including the innate immune receptor nucleotide-binding oligomerization domain-containing protein 2 (Nod2). Mutations of Nod2 (R702W, G908R and 100fs) are associated with Crohn’s disease and have lower stabilities compared to wild type. Cycloheximide (CHX)-chase half-life assays have been used to show that O-GlcNAcylation increases the stability and response of both wild type and Crohn’s variant Nod2, R702W. A more rapid method to assess stability afforded by post-translational modifications is necessary to fully comprehend the correlation between NLR stability and O-GlcNAcylation. Here, a recently developed cellular thermal shift assay (CETSA) that is typically used to demonstrate protein-ligand binding was adapted to detect shifts in protein stabilization upon increasing O-GlcNAcylation levels in Nod2. This assay was used as a method to predict if other Crohn’s associated Nod2 variants were O-GlcNAcylated, and also identified the modification on another NLR, Nod1. Classical immunoprecipitations and NF-κB transcriptional assays were used to confirm the presence and effect of this modification on these proteins. The results presented here demonstrate that CETSA is a convenient method that can be used to detect the stability effect of O-GlcNAcylation on O-GlcNAc-transferase (OGT) client proteins and will be a powerful tool in studying post-translational modification.
Keywords: OGT, NLRs, Crohn’s Disease, CETSA, Peptidoglycan, O-GlcNAcylation
Introduction
O-GlcNAcylation is a dynamic post-translational modification (PTM) characterized by the addition of monosaccharides of O-linked N-acetylglucosamine (GlcNAc) to serine and/or threonine residues of a protein using the nucleotide sugar, uridine diphosphate (UDP)-GlcNAc (Groves et al. 2013). Two enzymes mediate this modification: O-GlcNAc Transferase (OGT), which adds GlcNAc to target proteins, and O-GlcNAcase (OGA), which removes GlcNAc (Torres and Hart 1984; Haltiwanger et al. 1990; Dong and Hart 1994). O-GlcNAcylation is a functionally diverse PTM, associated with transcriptional regulation, signaling in response to nutrients and stress, and prevention of nascent polypeptides from early degradation (Jackson and Tjian 1988; Hart et al. 2007; Yang and Qian 2017; Zhu et al. 2015; Worth et al. 2017). We, along with others, have shown that modification of a protein substrate by OGT often leads to increased cellular stability of the client protein (Hou et al. 2016; Qin et al. 2017; Yang and Qian 2017; Ruan et al. 2012; Chu et al. 2014).
In order to fully appreciate the correlation between stability and O-GlcNAcylation on OGT’s thousands of protein substrates, accessible, high throughput methods are needed. Here an easy to use cellular thermal shift assay (CETSA) is repurposed to probe the effect of the OGT-modification on protein stability. CETSA is a relatively new technique introduced by Par Nordlund and colleagues at the Karolinska Institutet to assess protein-ligand binding utilizing live cells (Martinez Molina et al. 2013; Jafari et al. 2014). This assay measures temperature-induced aggregation of proteins by incubating cells with a ligand at a temperature gradient and quantifying the amount of remaining soluble protein at each temperature. Non-covalent interactions between protein and ligand stabilize the protein, leading to an increase in melting temperature. We speculated that this assay could be used for more than just ligand binding, in particular the stabilizing effect of O-GlcNAcylation on proteins. We have recently shown that nucleotide-binding oligomerization domain-containing protein 2 (Nod2), an intracellular human innate immune receptor that is mutated in Crohn’s disease, is stabilized by O-GlcNAcylation (Hou et al. 2016). Here, NOD-like receptors (NLRs) are used as a model system for assessing if CETSA could be used to study protein-stabilizing effects via post-translational OGT modification.
Nod2 and Nod1 are members of the NLR family, which is comprised of cytosolic proteins that contain a common nucleotide-binding and oligomerization domain (NOD or NBD) (Franchi et al. 2009). These receptors specifically recognize microbe associated molecular patterns (MAMPs) that are derived from bacterial peptidoglycan (PG). PG is a rich source of MAMPs that are specifically recognized by pattern recognition receptors (PRRs) of the innate immune system, which allows the body to distinguish harmful, pathogenic bacteria from the trillions of commensal bacteria that inhabit it (Janeway and Medzhitov 2002; Tosi 2005; Mackey and McFall 2006). PG is a polymer composed of alternating GlcNAc and N-acetylmuramic acid (MurNAc) sugars that completely coats the bacterial cell (van Heijenoort 2001; Lovering et al. 2012). PG fragments such as γ-D-iso-glutamyl-meso-diaminopimelic acid (iE-DAP), muramyl dipeptide (MDP), and GlcNAc have been shown to elicit an innate immune response via NLR activation (Chamaillard et al. 2003; Inohara et al. 2003; Girardin, Boneca, Carneiro et al. 2003; Girardin, Boneca, Viala et al. 2003; Wolf et al. 2016) (Figure 1).
Fig. 1.
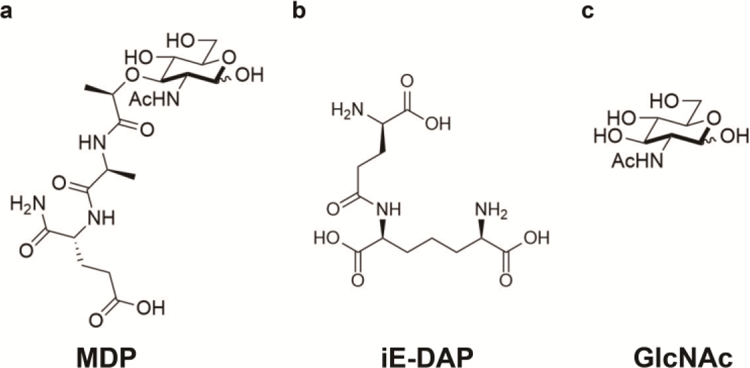
NLR Activators (A) MDP consists of N-acetylmuramic acid with an L-Ala D-isoGln dipeptide chain. It is a ligand derived from peptidoglycan detected by Nod2 (B) iE-DAP is a dipeptide derived from peptidoglycan detected by Nod1 (C) GlcNAc, a carbohydrate, leads to inflammasome formation for NLRP3 and is also O-linked to protein serines and threonines by OGT, with UDP-GlcNAc serving as the donor
Misregulation of NLR activation is associated with a variety of diseases (Martinon et al. 2009). Multiple Nod2 point mutations (R702W, G908R, and 1007fs) have been linked to an increased susceptibility for Crohn’s disease (Ogura, Bonen et al. 2001; Ogura, Inohara et al. 2001). These Crohn’s associated variants appear to be especially unstable compared to wild type Nod2 (Mohanan and Grimes 2014). Whereas, Nod1, another NLR family member, is expressed throughout the body and is associated with diverse diseases, including stomach cancer, lung cancer, and inflammatory bowel syndrome (Molnar et al. 2007; Kutikhin 2011; Bouskra et al. 2008).
Nod2 has been shown to be post-translationally O-GlcNAcylated, regulating stability and MDP mediated NF-κB activation in the wild type and one of the Crohn’s variants (Hou et al. 2016). Increases in Nod2 stability were determined using a half-life assay, where cells were treated with the translational inhibitor cycloheximide (CHX) and lysates were collected over time, measuring changes in protein concentration by quantitative Western blotting (Belle et al. 2006; Hou et. al 2016). Here, a novel use of a cellular thermal shift assay is described to assess OGT’s effects on Nod2 and Nod1’s cellular stabilities, demonstrating that this technique is a useful method for identifying post-translational modifications that stabilize proteins.
Materials and methods
Materials
All antibodies were purchased from Cell Signaling Technology, except CTD110.6, which was produced and gifted by Core C4 (Department of Biological Chemistry, Johns Hopkins University), and Flag, which was purchased from Sigma-Aldrich. Thiamet-G was purchased from Sigma Aldrich. MDP was purchased from Bachem. iE-DAP was purchased from Invivogen. cOmplete EDTA-free protease inhibitor cocktail was purchased from Sigma-Aldrich.
Cell Culture
HEK293T cells were obtained from American Type Culture Collection (ATCC, Manassas, VA). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM), 10% (v/v) fetal bovine serum (FBS) (Atlantic Biologicals), 2 mM L-glutamine, 1% (v/v) penicillin/streptomycin and grown in a humidified incubator at 37°C and 5% CO2.
HEK293T-Nod2-Myc/tet-op cells were used and have been previously described (Mohanan and Grimes 2014). Tetracycline-inducible cell lines, HEK293T-Nod2-G908RMyc/tet-op, HEK293T-Nod2–1007fs-Myc/tet-op, and HEK293T-Nod1-Flag/tet-op cell lines were created using a RetroTet ART expression system (Springer and Blau 1997; Rossi et al. 1998) and the appropriate restriction enzymes ((Nod2 and variants: BglII and AscI) (Nod1: EcoRI and AscI) (New England BioLabs))
HEK293T-Nod2-G908R-Myc, HEK293T-Nod2–1007fs-Myc, HEK293T-Nod2-S933A-Myc, and HEK293T-Nod1-Flag cell lines were created using K2605 lentiviral vector with the appropriate insert and the appropriate restriction enzymes (Nod2 and variants: BamHI and XhoI, Nod1: SpeI and XhoI) (New England BioLabs)) (Mohanan et al. 2013).
Cellular Thermal Shift Assay
Cells were seeded in 100 mm dishes at a density of 8 × 106. The next day, they were treated with 1 μM Thiamet-G or a DMSO control for 4–8 hours. The cells were then washed with phosphate-buffered saline (PBS) plus protease inhibitors, then resuspended in PBS plus protease inhibitors and centrifuged at 150 x g for 5 minutes. The cell pellet was then resuspended in 500 µL PBS plus protease inhibitors and 27 µL fractions were aliquoted into PCR-tubes. The cells were incubated in a thermocycler (Bio-Rad) at a temperature gradient of 40 – 68.5°C for three minutes, and then were allowed to rest at room temperature for three minutes before being flash frozen in liquid nitrogen. Cells were lysed by freezing in liquid nitrogen and thawing in a 25°C water bath three times, and were spun down at 20,000 x g for 20 minutes. The supernatant was carefully removed and mixed with 5x Laemmli loading dye (100 mM Tris-HCl, pH 6.8, 4% SDS, 12% glycerol, 0.008% bromophenol blue, 2% β-mercaptoethanol), and boiled for 8 minutes. Samples were then subjected to Western blot analysis.
CETSA data was obtained by measuring band intensities of Myc/Flag normalized to Sod1 loading control, using Image Lab 5.0. The intensities were also normalized to the lowest temperature (40°C) and a sigmoidal curve was fit using Graphpad Prism. Melting temperatures were obtained by using the fitted line equation when the ratio of protein remaining was 0.5.
Data was analyzed for statistical significance using an F-test to compare population variances. The F-test is considered significant at the 0.05 level if both the p-value and F-statistic are smaller than 0.05 and greater than F-critical, respectively. The F-test was done in a pairwise manner for the 4 different cell lines using the Prism software.
Co-immunoprecipitation
Co-immunoprecipitations were performed similarly to previous reports (Mohanan and Grimes 2014; Hou et al. 2016). Cells were washed twice with PBS and harvested with lysis buffer (1% Triton X-100, 2 mM EDTA, 4 mM Na3PO4, 100 mM MES, pH 5.8, 10 mM NaF, 1 protease inhibitor tablet), before being lysed with a 20-gauge needle and centrifuged at 10,000 rpm for 10 minutes. A Bradford assay (Bio-Rad) was used to quantify protein concentrations. 1.2 mg of lysates were mixed with 2 μL primary antibody (Anti-Myc or Anti-Flag) and incubated overnight at 4°C. The lysates were then incubated with 40 µl of protein A Dynabeads (Invitrogen) for 3 hours at 4°C. Next, the mixture was washed three times with 200 μl of lysis buffer and the proteins eluted by boiling at 100°C for 5 minutes with 2X Laemmli loading dye. Samples were then subjected to Western blot analysis.
Western blot analysis
Samples were run on SDS-page gels, either 4–20% Mini-PROTEAN Precast (Bio-Rad) gels for CETSA or 7.5% polyacrylamide for immunoprecipitations, before transferring to a nitrocellulose membrane. The membrane was blocked with 10% nonfat dry milk in Tris-buffered saline Tween-20 (TBST) for 1 hour at room temperature, washed three times with TBST for five minutes each, and incubated with primary antibody overnight at 4°C. The next day, the membranes were washed 3 times with TBST for five minutes each and incubated with the secondary antibody for 1 hour at room temperature. The membranes were washed 3 times more with TBST for 5 minutes each. The bands were detected using an ECL kit (Bio-Rad).
NF-κB Assay
NF-κB activation was measured as described previously (Hou et al. 2014). Briefly, Hek293T cells were transfected with pGL4.32 [luc2P/NF-κB-RE/Hygro] vector (10 ng) and pRL Renilla luciferase reporter vector (1 ng), along with CMV control plasmid or Nod1, Nod2, or Nod2 Crohn’s variants (0.1 ng) in CMV vectors. Cells were pretreated with 100 nM Thiamet-G for 4 hours, then 20 μM iE-DAP (Nod1) or 20 μM MDP (Nod2 and Crohn’s variants) for 8 or 4 hours, respectively. Relative luciferase activity was detected using a Dual-Luciferase Reporter Assay (Promega) and measured on a luminometer (Perkin Elmer). All assays were performed using three biological replicates.
Cycloheximide-chase Half-life Assay
Cycloheximide (Calbiochem) was used at a concentration of 50 μg/ml, and the lysates were collected every 4 hours. Thiamet-G (1 μM) was incubated 4 hours prior to the addition of cycloheximide. Cells were treated, lysed, quantified for protein content, and analyzed by Western blot.
The protein bands for Nod1-Flag and Actin were quantified using Image Lab 5.0. Actin is used as the loading control and the ratio of the intensity of flag to actin bands (Ir) were used to analyze the half-life values as previously described (Hou et al. 2016). Briefly, relative flag band intensities were plotted against time assuming first-order decay (ln(Ir) vs. time). The rate constant was calculated using the negative slope of the line (k = - slope), and the corresponding half-life was calculated (T1/2 = ln(2)/k). Each condition was performed in triplicate and the Student’s t-test was used to determine statistical significance. P < 0.05 was considered as statistically significant
Results
Nod2 is stabilized by O-GlcNAc modification
Previously, HEK293T cells expressing Nod2 were treated with Thiamet-G (1 uM, 4 hours), an inhibitor of OGA that leads to an increase in O-GlcNAcylation levels, and showed an increased Nod2 half-life (Hou et al. 2016). The development of CETSA has provided a new assay that can monitor protein stability in live cells, but to date has only been used to observe the stabilizing effect caused by protein-ligand binding. In this study we sought to use this assay to assess the stabilizing effect of O-GlcNAcylation on proteins. In order to test the application of CETSA on post-translational modifications, data was collected on Nod2 in the presence of Thiamet-G and compared to CHX-chase half-life analysis.
Briefly, HEK293T-Nod2-Myc/tet-op cells were treated with or without Thiamet-G, collected, and incubated at a temperature gradient of 40 – 68.5°C. Cells treated with Thiamet-G saw an increase in melting temperature, from 49.10 ± 0.37 (R2 value = 0.94) to 51.01 ± 0.66 (R2 value = 0.90), and showing statistical significance (F-statistic [4.057] > F-critical [2.49] and p value < 0.05) (Figure 2, Figure 3D). These data support the previously published CHX-chase assay data, demonstrating that increasing O-GlcNAcylation levels of Nod2 increases its stability and confirmed that CETSA is a viable method to study the stabilization effects of post-translational modifications.
Fig. 2.
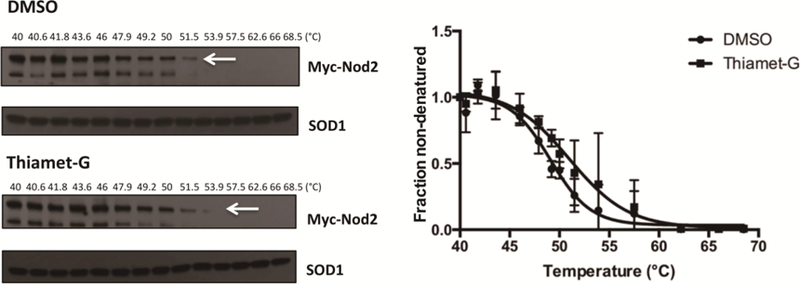
CETSA of wild type Nod2. HEK293T-Nod2-Myc/Tet-op cells were incubated with 1 µM Thiamet-G or DMSO for 8 h, and CETSA and immunoblots were performed (see materials and methods). Relative amounts of Nod2-Myc to SOD1 were plotted from three independent experiments as the means ± S.D. using GraphPad Prism (Boltzmann Sigmoidal model)
Fig. 3.
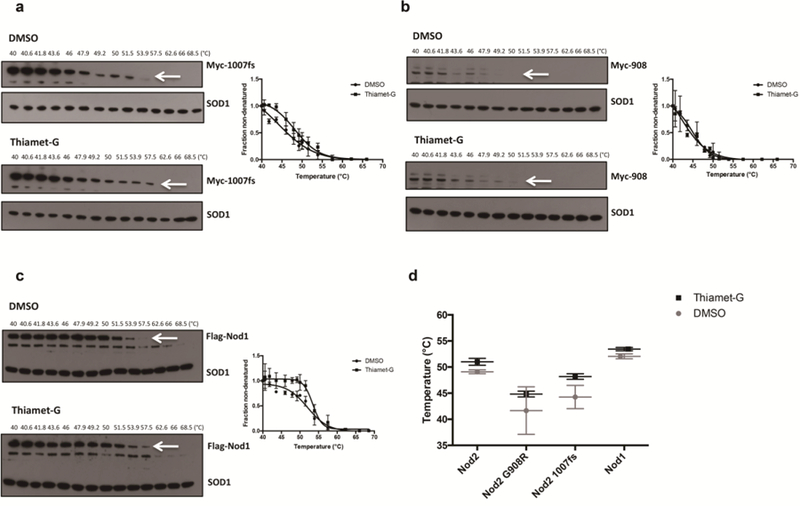
CETSA of Nod2 Crohn’s associated variants and Nod1 (A) HEK293T-Nod2–1007fs-Myc/Tet-op and (B) HEK293T-Nod2-G908R-Myc/Tet-op cells were incubated with 1 µM Thiamet-G or DMSO for 4 h, and CETSA and immunoblots were performed (see materials and methods). (C) HEK293T-Nod1-Flag/Tet-op cells were incubated with 1 µM Thiamet-G or DMSO for 8 h, and CETSA and immunoblots were performed. Relative amounts of Nod2 Myc/Nod1 Flag to SOD1 were plotted from three independent experiments as the means ± S.D. using GraphPad Prism (Boltzmann Sigmoidal model (D) Melting temperature comparison between DMSO and Thiamet-G in Nod1, Nod2 and Nod2 variant cells obtained by CETSA. * indicates significance according to F-test
O-GlcNAcylation regulates the stability of a Crohn’s variant of Nod2 and another NLR, Nod1
We speculated that since CETSA allowed the detection of protein stability via shifts in melting temperature for Nod2 upon Thiamet-G treatment, this assay could be used on other proteins to identify an effect of the O-GlcNAc modification. CETSA was performed similarly on the Nod2–1007fs and Nod2-G908R Nod2 Crohn’s variants, using HEK293T-Nod2–1007fs-Myc/tet-op and HEK293T-Nod2-G908R-Myc/tet-op cell lines, respectively. The Nod2–1007fs mutant showed an increase in melting temperature upon Thiamet-G treatment, increasing from 44.26 ± 2.22 (R2 value = 0.95) to 48.18 ± 0.53 (R2 value = 0.96), and showing statistical significance (F-statistic [12.73] > F-critical [2.49] and p value < 0.05). Together these data suggest an increase in stabilization upon increasing O-GlcNAcylation (Figure 3A, 3D). There was no significant shift in melting temperature for the Nod2-G908R variant; the melting temperatures were 41.66 ± 4.55 (R2 value = 0.90) and 44.83 ± 0.55 (R2 value = 0.98) for DMSO and Thiamet-G treatment (Figure 3B, 3D), respectively, and no statistical significance was observed (F-statistic [0.4157] < F-critical [2.49] and p value > 0.05). These data suggest that increasing O-GlcNAc levels does not increase the stability of this mutant, or that this mutant is not post-translationally modified.
We hypothesized that the O-GlcNAc modification may not be unique to Nod2, and may also affect other NLRs. In order to test this, a cell line expressing Nod1 under the transcriptional control of tetracycline (HEK293T-Nod1-Flag/tet-op) was created and used in CETSA. Cells containing Nod1 that were treated with Thiamet-G saw an increase in melting temperature for this protein, rising from 52.06 + 0.47 °C (R2 value = 0.95) to 53.44 + 0.32 °C (R2 value = 0.94), and showing statistical significance (F-statistic [17.03] > F-critical [2.49] and p value < 0.05). (Figure 3C, 3D). To confirm the result, a CHX-chase half-life assay was performed using Nod1. Nod1-expressing cells were treated with either 1 μM Thiamet-G or DMSO for 4 hours, then added cycloheximide and collected cells every 4 hours. In cells that were treated with Thiamet-G, Nod1 had a higher half-lives, rising from 15.55 +/− 6.17 hours (DMSO treatment) to 27.45 +/− 5.37 hours (Figure 4). These data suggest that Nod1 could be similarly stabilized by O-GlcNAcylation as Nod2
Fig. 4.
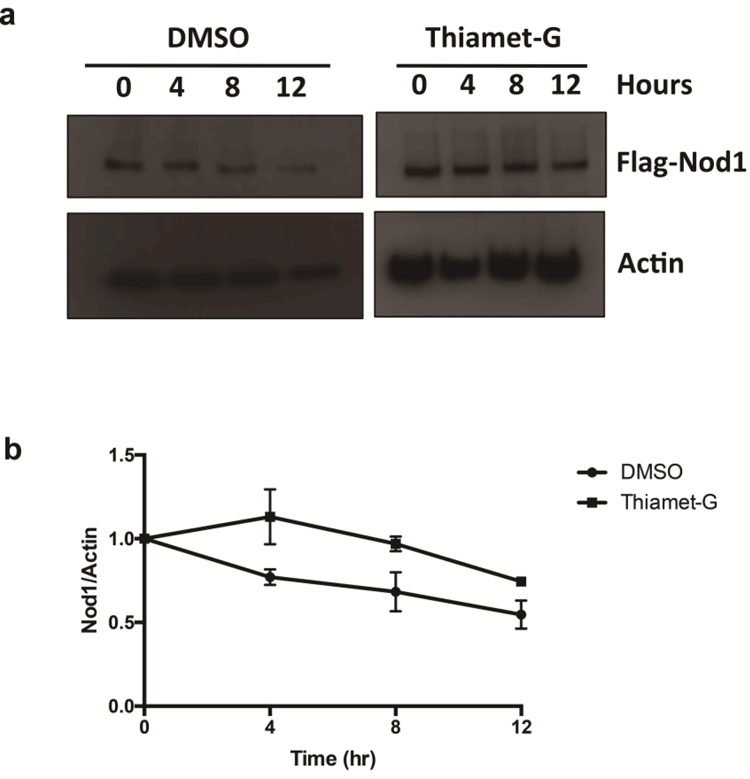
O-GlcNAcylation regulates the half-life of Nod1. (A) HEK293T-Nod1-expressing cells were incubated with 1 μM Thiamet-G or DMSO for 4 h, prior to cycloheximide treatment (50 μg/mL) and cells were collected at the indicated time intervals. Nod1 was detected by anti-Flag western blotting. (B) Half-lives were determined by plotting relative Flag intensities vs. time assuming first order decay, and calculating T1/2 = ln(2)/k
Nod1 and Crohn’s variants Nod2-G908R and Nod2–1007fs are directly modified by O-GlcNAc
In order to confirm that Nod1 and the Crohn’s variants are directly O-GlcNAcylated, cells expressing these proteins were treated with Thiamet-G and lysed. Nod1 was immunoprecipitated using a Flag-tag, while the mutants Nod2–1007fs and Nod2-G908R were immunoprecipitated using the Myc-tag. An immunoblot was performed with CTD110.6, an antibody that detects O-GlcNAc. These data suggest that the two Crohn’s associated variants, Nod2-G908R and Nod2–1007fs, and Nod1 are modified by OGT (Figure 5). The CETSA data predicted that Nod1 and Nod2–1007fs were modified, but suggested that Nod2-G908R was not or did not cause a shift in stability (Figure 3). These two data sets suggest that the stability afforded by the PTM on both Nod1 and Nod2–1007fs could affect their abilities to signal the presence of a PG ligand.
Fig. 5.
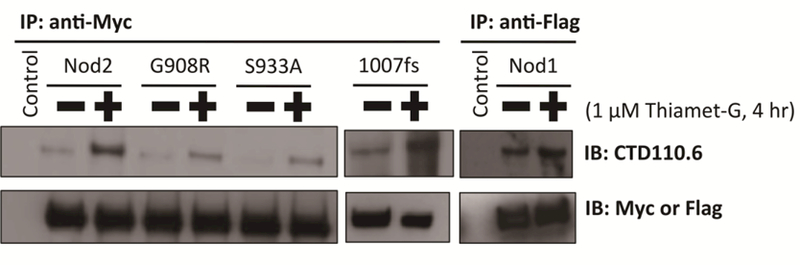
OGT modifies Nod2 mutants and Nod1. HEK293T-Nod2-Myc, HEK293T-Nod2-G908R-Myc, HEK293T-Nod2-S933A-Myc, and HEK293T-Nod2–1007fs-Myc cell lines were immunopurified with Myc antibody and blotted with CTD110.6 and Myc antibodies; control cells do not express Myc-Nod2. HEK293T-Nod1-Flag cell line was immunopurified with Flag antibody and blotted with CTD110.6 and Flag antibodies. Control cells do not express Flag-Nod1. Thiamet-G inhibits OGA, elevating O-GlcNAcylation
O-GlcNAcylation regulates NF-κB activation of Nod1 and Nod2–1007fs, but not Nod2-G908R
Upon MAMP stimulation, Nod1 and Nod2 are known to initiate the NF-κB inflammatory cascade, which ends with the production of proinflammatory cytokines and chemokines (Kobayashi et al. 2002; McCarthy et al. 1998; Nembrini et al. 2009). A standard luciferase assay has previously been used to show that the three Crohn’s associated variants have decreased NF-κB activity when stimulated with MDP but the response can be increased if Hsp70 is over-expressed (Inohara et al. 2003; Mohanan and Grimes 2014). We have previously shown that Nod2 and the Crohn’s associated variant, Nod2-R702W, have increased NF-κB activation when OGA is inhibited (Hou et al. 2016). To test if O-GlcNAcylation can restore NF-κB activation by the other mutants (Nod2-G908R and Nod2–1007fs), cells were treated with a low dose (100 nM) of Thiamet-G that still causes a shift in O-GlcNAc levels (Supporting Information, Figure S1A-B) but will not cause off target effects in the NF-κB assay (Hou et al. 2016). Cells were treated with 100 nM Thiamet-G for 4 hours, and then 20 μM MDP for 5 hours. In wild type, Nod2-R702W, and Nod2–1007fs, Thiamet-G treatment led to an increase in NF-κB activation, while the Nod2-G908R mutant saw no change (Figure 6A). Similarly, Nod1 was tested, and when stimulated with 20 μM iE-DAP for 8 hours after a 4 hour, 100 nM Thiamet-G pretreatment, an increase in iE-DAP induced NF-κB activation was observed (Figure 6B). These data suggest that variants that are stabilized by the O-GlcNAc modification (Figure 3D) yield more robust NF-κB activity. Thus an increase in melting temperature in CETSA (Figure 3) is correlated with an increase in NF-κB activation.
Fig. 6.
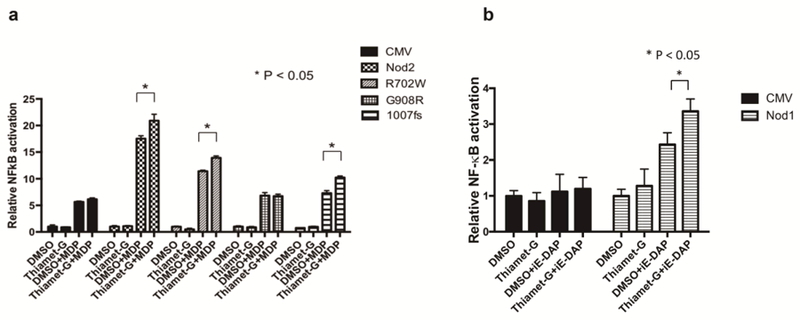
NF-κB assay of Nod2 variants and Nod1. (A) Dual-luciferase assay performed on HEK293T cells transfected with 0.1 ng Nod2, 702, 908 or 1007fs plasmid in the presence of 100 nM Thiamet-G or DMSO for 4 h. After 4 h, cells were incubated with 20 µM MDP for 5h, harvested, and tested for luciferase activity (B) Dual-luciferase assay performed on HEK293T transfected with 0.1 ng Nod1 in the presence of 100 nM Thiamet-G or DMSO for 4 h. After 4 h, cells were incubated with 20 µM iE-DAP for 8h, harvested, and tested for luciferase activity
The Nod2-G908R variant has a serine residue near the mutation site
The Nod2 Crohn’s variant, Nod2-G908R, appears to be O-GlcNAcylated (Figure 4), but no change in stability (Figure 3B, 3D) or activity (Figure 6A) was observed when O-GlcNAcylation levels were increased using the OGA inhibitor. Point mutants of residues near the proposed binding site for the MAMP, including R877, G905, W907, W931, and S933, have been shown to decrease NF-κB activation (Maekawa et al. 2016). Residues R877, W931, and S933 have also been predicted through computational modeling and binding studies to comprise the binding pocket for the MAMP (Lauro et al. 2017). The model suggests that when bound to MDP, residue S933 is very close to G908. S933 may be O-GlcNAcylated, and because a large structural change occurs when residue 908 is mutated from glycine to arginine, the mutation may affect the nearby GlcNAc modification by obstructing access for the enzyme OGT to modify S933. A HEK293T cell line that stably expresses a Myc-tagged Nod2-S933A point mutant was created. Myc antibody was used to immunoprecipitate the Nod2 mutant, and CTD110.6 antibody was used to Western blot for O-GlcNAcylation.
While the Nod2-S933A mutant still appears to be O-GlcNAcylated, the bands were decreased compared to wild type (Figure 5). The bands appear similar to those of Nod2-G908R, which also had decreased CTD110.6 levels relative to wild type. The S933A Nod2 mutant shows no NF-κB activity but retains the ability to bind to MDP (Maekawa et al. 2016; Lauro et al. 2016). These data suggest that S933 may be one of multiple O-GlcNAcylated residues in Nod2, and that replacing glycine with arginine at residue 908 may occlude OGT from accessing and modifying S933 (Figure 7).
Fig. 7.
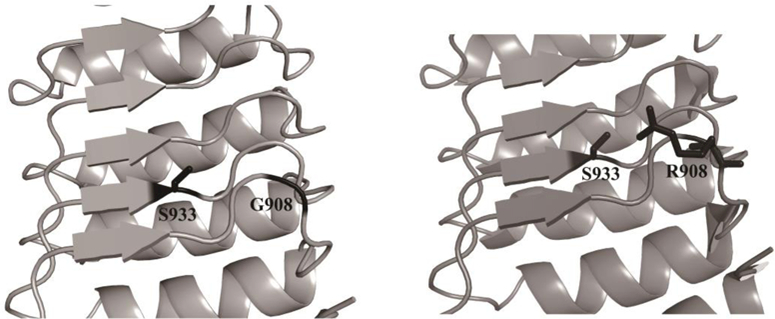
Residue 908 is in close proximity to S933. In the wild type G908 Nod2, S933 is free and solvent exposed. In the Crohn’s associated variant, Nod2-G908R, the bulky arginine side chain can be situated directly nearby S933. Images were created using MacPyMol and PDB 5IRN of crystallized rabbit Nod2
Discussion
The initial curiosity in a link between NLRs and OGT was prompted by the fact that OGT uses UDP-GlcNAc as a substrate, and that UDP-GlcNAc serves as a precursor for PG biosynthesis, which contains the ligands detected by NLRs when initiating an inflammatory response. UDP-GlcNAc is also released from cells during bacterial growth and host invasion (Park and Uehara 2008). We have previously demonstrated that wild type and Nod2-R702W Crohn’s associated variant are O-GlcNAcylated, which leads to an increase in stability according to cycloheximide-chase half-life assays. Now, we have expanded this study using CETSA as a feasible tool to study protein stability and confirmed that all Crohn’s associated variants, including G908R and 1007fs, are O-GlcNAcylated. Additionally, this technology has led us to explore other NLR proteins such as Nod1 and determined that it too is O-GlcNAcylated, suggesting that this modification may be common among NLRs or PG-detecting proteins.
CETSA proved to be a useful tool for assaying if a particular NLR is stabilized when global GlcNAcylation levels increase. This method is very sensitive, with capabilities of detecting changes in melting temperatures that are less than 2̜°C. CETSA serves as a good complement to CHX-chase half-life assays. While both assays give insight on a protein’s stability, they do so in different regards. CETSA measures a protein’s thermal stability, or the temperature in which it aggregates and becomes insoluble. CHX-chase assays halt protein translation and measure the rate at which the cell degrades the target protein. Both assays have inherent drawbacks in that they cause stress to cells, as CETSA exposes a fraction of cells to temperatures significantly higher than physiological and CHX inhibits the ribosome, affecting every cellular process. CETSA has been coupled with quantitative mass spectrometry to study the effects of a complete cellular proteome when treated with a drug, demonstrating this assay’s highthroughput potential (Savitski et al. 2014).
The measured stability improvements observed by raising O-GlcNAcylation levels could be caused by different mechanisms. The O-GlcNAc modification can directly stabilize the protein, allowing it to fold into a more stable conformation. Additionally, it is possible that the PTM allows for the recruitment of other proteins and the formation of a larger protein complex. Increasing the number of protein-protein interactions would also lead to an increase in melting temperature detectable by CETSA. In order to increase O-GlcNAcylation levels on NLRs, we used Thiamet-G, which inhibits the enzyme OGA and thus prevents GlcNAc from being removed from target proteins (Yuzwa et al. 2008). Thiamet-G is one of the most frequently used compounds for increasing O-GlcNAc levels in cells because it is commercially available and more specific compared to other OGA inhibitors (Zachara et al. 2012). However, since it increases O-GlcNAc levels across all proteins, it is possible that Thiamet-G is causing an off-target effect that plays a role in NLR stability. Therefore, more work is necessary to determine the physiological effect O-GlcNAc has on NLRs.
Wild type Nod2 and Nod1 have smaller shifts in melting temperature compared to Nod2–1007fs. Since these proteins already had higher melting temperatures, this suggests that they are naturally more stable compared to the mutated proteins. A more dramatic rescue in stability upon OGA inhibition is observed with the unstable Crohn’s associated variant, Nod2–1007fs. Interestingly, Nod1 had a slightly higher melting temperature than Nod2, suggesting that it may be more stable. The CHX-chase experiment performed on Nod1 also suggests it has a longer half-life relative to Nod2, as previously measured (Hou et al. 2016). It is important to note that the cell lines tested were not identical, as Nod1 had a Flag-tag and Nod2 a Myc-tag. Therefore, it is possible that other factors contributed to that difference.
Nod2-G908R, despite showing O-GlcNAcylation in immunoblot assays (Figure 5), did not see the same increase in stability (Figure 3B) and NF-κB activation (Figure 6A) as did all of the other NLRs that were tested. Recently, a crystal structure of the ligand-binding domain of Nod2 was solved (Maekawa et al. 2016). Although no ligand density was observed in the crystal structure, a region of unknown electron density was detected. Mutational analysis suggested that this could be a potential binding pocket for the MAMP, MDP. Using this model (Figure 7), it is predicted that a serine residue, S933, is located near the binding pocket and the G908R mutation. Mutation from glycine to arginine at position 908 could possibly create a steric block that obstructs OGT from modifying S933, as multiple rotomers of R908 are predicted by PyMol to come in close contact with the serine residue (Figure 7). The S933A mutant fails to activate an NF-κB response (Maekawa et al. 2016. If S933 is the site of PTM, then when cells are treated with S933A Nod2 and Thiamet-G, which prevents the removal of O-GlcNAc from client proteins, an increase of O-GlcNAcylation on mutant Nod2 would not be observed. However, O-GlcNAc was still detected in immunoblotting of Nod2-G908R cells (Figure 5), albeit at diminished levels, suggesting that this cannot be the lone modified residue or that there is enough flexibility such that the G908R mutant does not entirely obstruct OGT from accessing S933.
CETSA is a powerful method for assaying ligand binding in cells. Here we expanded this user friendly, highly reproducible assay and demonstrated its capability of detecting changes in NLR stability upon post-translational modification by OGT. This cell-based assay accurately predicted the NLR constructs that give an increase in ligand induced NF-κB signaling upon increasing the post-translational modification. We will use this assay in the future to identify additional NLRs or other proteins that could be stabilized by O-GlcNAcylation, ultimately providing a deeper understanding of innate immune regulation.
Supplementary Material
Acknowledgments
MITZUTANI GLYCOSCIENCE FOUNDATION
NATIONAL SCIENCE FOUNDATION (CAREER CHE 1554967)
NIH NATIONAL HEART, LUNG, AND BLOOD INSTITUTE (P01HL107153 to NEZ)
NIH NIGMS COBRE P20GM104316–01A1
NIH NIGMS T32GM008550
This work was supported by the Mizutani Glycoscience Foundation, the Delaware COBRE program, with a grant from the National Institute of General Medical Sciences (NIGMS P20GM104316–01A1) the National Science Foundation (CAREER CHE 1554967), and the NIH Heart, Lung, and Blood Institute (P01HL107153 to N.E.Z.). W.R.D. thanks the NIH for support through a CBI training grant: 5T32GM008550. C.L.G is a Pew Biomedical Scholar. W.D. thanks the Chemistry-Biology Interface predoctoral training program (NIGMS 5T32 GM 08550–15).
We thank Professor John Koh for use of the tissue culture facility. CTD110.6 was gifted from Core C4 (Johns Hopkins University School of Medicine, P01HL107153). We would like to thank Professor April Kloxin for providing crucial insight and Jason Burch, Ophelia Ukaegbu, and Kristen DeMeester for detailed reading of the manuscript.
References
- Belle A, Tanay A, Bitincka L, Shamir R, O’Shea EK (2006) Quantification of protein half-lives in the budding yeast proteome. PNAS, 103(35):13004–13009. doi: 10.1073/pnas.0605420103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouskra D, Brezillon C, Berard M, Werts C, Varona R, Boneca IG, Eberl G (2008) Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature, 456(7221):507–510. doi: 10.1038/nature07450 [DOI] [PubMed] [Google Scholar]
- Chamaillard M, Hashimoto M, Horie Y, Masumoto J, Qiu S, Saab L, et al. (2003) An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol, 4(7):702–707, doi: 10.1038/ni945 [DOI] [PubMed] [Google Scholar]
- Chu CS, Lo PW, Yeh YH, Hsu PH, Peng SH, Teng YC, Kang ML, Wong CH, Juan LJ (2014) O-GlcNAcylation regulates EZH2 protein stability and function. Proc Natl Acad Sci USA, 111(4):1355–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai CL, Shi J, Chen Y, Iqbal K, Liu F, Gong CX (2013) Inhibition of protein synthesis alters protein degradation through activation of protein kinase B (AKT). J Biol Chem, 288(33):23875–23883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong DL, Hart GW (1994) Purification and characterization of an O-GlcNAc selective N-acetyl-beta-D-glucosaminidase from rat spleen cytosol. J Biol Chem, 269(30):19321–19330 [PubMed] [Google Scholar]
- Franchi L, Warner N, Viani K, Nunez G (2009) Function of Nod-like receptors in microbial recognition and host defense. Immunol Rev, 227(1):106–128. doi: 10.1111/j.1600-065X.2008.00734.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardin SE, Boneca IG, Carneiro LA, Antignac A, Jehanno M, Viala J, et al. (2003) Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science, 300(5625):1584–1587. doi: 10.1126/science.1084677 [DOI] [PubMed] [Google Scholar]
- Girardin SE, Boneca IG, Viala J, Chamaillard M, Labigne A, Thomas G, et al. (2003) Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem, 278(11):8869–8872. doi: 10.1074/jbc.C200651200 [DOI] [PubMed] [Google Scholar]
- Groves JA, Lee A, Yildirir G, Zachara NE (2013) Dynamic O-GlcNAcylation and its roles in the cellular stress response and homeostasis. Cell Stress Chaperones, 18(5):535–558. doi: 10.1007/s12192-013-0426-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haltiwanger RS, Holt GD, Hart GW (1990) Enzymatic addition of O-GlcNAc to nuclear and cytosolic proteins. Identification of a uridine-diphospho-N-acetylglucosamine: peptide beta-N-acetylglucosaminyltransferase. J Biol Chem, 265(5):2563–2568 [PubMed] [Google Scholar]
- Hart GW, Housley MP, Slawson C (2007) Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature, 446(7139):1017–1022. doi: 10.1038/nature05815 [DOI] [PubMed] [Google Scholar]
- Hou CW, Mohanan V, Zachara NE, Grimes CL (2016) Identification and biological consequences of the O-GlcNAc modification of the human innate immune receptor, Nod2. Glycobiology, 26(1):13–18. doi: 10.1093/glycob/cwv076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inohara N, Ogura R, Fontalba A, Gutierrez O, Pons F, et al. (2003) Host recognition of bacterial muramyl dipeptide mediated through NOD2. J Biol Chem, 278(8):5509–5512. doi: 10.1074/jbc.C200673200 [DOI] [PubMed] [Google Scholar]
- Jackson SP, Tijan R (1988) O-glycosylation of eukaryotic transcription factors: implications for mechanisms of transcriptional regulation. Cell, 55(1):125–133 [DOI] [PubMed] [Google Scholar]
- Jafari R, Almqvist H, Axelsson H, Ignatushchenko M, Lundback T, Nordlund P, Martinez Molina D (2014) The cellular thermal shift assay for evaluating drug target interactions in cells. Nat Protoc, 9(9):2100–2122. doi: 10.1038/nprot.2014.138 [DOI] [PubMed] [Google Scholar]
- Janeway CA, Medzhitov R (2002) Innate immune recognition. Annu Rev Immunol, 20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Inohara N, Hernandez LD, Galan JE, Nunez G, Janeway CA, et al. (2002) RICK/Rip2/CARDIAK mediates signaling for receptors of the innate and adaptive immune systems. Nature, 416(6877):194–199. doi: 10.1038/416194a [DOI] [PubMed] [Google Scholar]
- Kutikhin AG (2011) Role of NOD1/CARD4 and NOD2/CARD15 gene polymorphisms in cancer etiology. Hum Immunol, 72(10):955–968. doi: 10.1016/j.humimm.2011.06.003 [DOI] [PubMed] [Google Scholar]
- Lauro ML, D’Ambrosio EA, Bahnson BJ, Grimes CL (2017) Molecular Recognition of Muramyl Dipeptide Occurs in the Leucine-rich Repeat Domain of Nod2. ACS Infect Dis, 3(4):264–270. doi: 10.1021/acsinfecdis.6b00154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovering AL, Safadi SS, Strynadka NC (2012) Structural perspective of peptidoglycan biosynthesis and assembly. Annu Rev Biochem, 81:451–478. doi: 10.1146/annurev-biochem-061809-112742 [DOI] [PubMed] [Google Scholar]
- Mackey D, McFall AJ (2006) MAMPs and MIMPs: proposed classifications for inducers of innate immunity. Mol Microbiol, 61(6):1365–1371. doi: 10.1111/j.1365-2958.2006.05311.x [DOI] [PubMed] [Google Scholar]
- Maekawa S, Ohto U, Shibata T, Miyake K, Shimizu T (2016) Crystal structure of NOD2 and its implications in human disease. Nat Commun, 7:11813. doi: 10.1038/ncomms11813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez Molina D, Jafari R, Ignatushchenko M, Seki T, Larsson EA, Dan C, Sreekumar L, Cao Y, Nordlund P (2013) Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science, 341(6141):84–87. doi: 10.1126/science.1233606 [DOI] [PubMed] [Google Scholar]
- Martinon F, Mayor A, Tschopp J (2009) The inflammasomes: guardians of the body. Annu Rev Immonol, 27:229–265 [DOI] [PubMed] [Google Scholar]
- McCarthy JV, Ni J, Dixit VM (1998) RIP2 is a novel NF-kappaB-activating and cell death-inducing kinase. J Biol Chem, 273(27):16968–16975 [DOI] [PubMed] [Google Scholar]
- Mohanan V, Grimes CL (2014) The molecular chaperone HSP70 binds to and stabilizes NOD2, an important protein involved in Crohn disease. J Biol Chem, 289(27):18987–18998. doi: 10.1074/jbc.M114.557686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanan V, Temburni MK, Kappes JC, Galileo DS (2013) L1CAM stimulates glioma cell motility and proliferation through the fibroblast growth factor receptor. Clin Exp Metastasis, 30(4):507–520. doi: 10.1007/s10585-012-9555-4 [DOI] [PubMed] [Google Scholar]
- Molnar T, Hofner P, Nagy F, Lakatos PL, Fischer S, Lakatos L, et al. (2007) NOD1 gene E266K polymorphism is associated with disease susceptibility but not with disease phenotype or NOD2/CARD15 in Hungarian patients with Crohn’s disease. Dig Liver Dis, 39(12):1064–1070. doi: 10.1016/j.dld.2007.09.003 [DOI] [PubMed] [Google Scholar]
- Nembrini C, Kisielow J, Shamshiev AT, Tortola L, Coyle AJ, Kopf M, et al. (2009) The kinase activity of Rip2 determines its stability and consequently Nod1- and Nod2-mediated immune responses. J Biol Chem, 284(29):19183–19188.doi: 10.1074/jbc.M109.006353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, et al. (2001) A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature, 411(6837):603–606. doi: 10.1038/35079114 [DOI] [PubMed] [Google Scholar]
- Ogura Y, Inohara N, Benito A, Chen FF, Yamaoka S, Nunez G (2001) Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-kappaB. J Biol Chem, 276(7):4812–4818. doi: 10.1074/jbc.M008072200 [DOI] [PubMed] [Google Scholar]
- Park JT, Uehara T (2008) How bacteria consume their own exoskeletons (turnover and recycling of cell wall peptidoglycan). Microbiol Mol Biol Rev, 72(2):211–227. doi: 10.1128/MMBR.00027-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W, Pinou L, Fan X, Quan B, Zhu Y, Qin K, Chen Y, Wang C, Chen X (2017) Quantitative time-resolved chemoproteomics reveals that stable O-GlcNAc regulates box C/D snoRNP biogenesis. PNAS, 114(33) E6749–E6758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi FMV, Guicherit OM, Spicher A, Kringstein AM, Fatyol K, Blakely BT, Blau HM (1998) Tetracycline-regulatable factors with distinct dimerization domains allow reversible growth inhibition by p16. Nature Genetics, 20(4):389–393 [DOI] [PubMed] [Google Scholar]
- Ruan HB, Han X, Li MD, Singh JP, Qian K, Azarhoush S, Zhao L, Bennett AM, Samuel VT, Wu J, Yates JR, Yang X (2012) O-GlcNAc transferase/host cell factor C1 complex regulates gluconeogenesis by modulating PGC-1a stability. Cell Metab, 16(2):226–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitski MM, Reinhard FBM, Franken H, Werner T, Savitski MF, Eberhard D, Martinez Molina D, Jafari R, Dovega RB, Klaeger S, Kuster B, Nordlund B, Bantscheff M, Drewes G (2014) Tracking cancer drugs in living cells by thermal profiling of the proteome. Science, 346(6205):1255784. [DOI] [PubMed] [Google Scholar]
- Springer ML, Blau HM (1997) High-efficiency retroviral infection of primary myoblasts. Somat Cell Mol Genet, 23(3):203–209 [DOI] [PubMed] [Google Scholar]
- Torres CR, Hart GW (1984) Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J Biol Chem, 259(5):3308–3317. [PubMed] [Google Scholar]
- Tosi MF (2005) Innate immune responses to infection. The Journal of allergy and clinical. Immunology, 116(2):241–249. [DOI] [PubMed] [Google Scholar]
- van Heijenoort J (2011) Formation of the glycan chains in the synthesis of bacterial peptidoglycan. Glycobiology, 11(3):25R–36R. [DOI] [PubMed] [Google Scholar]
- Wolf AJ, Reyes CN, Liang W, Becker C, Kenichi S, Wheeler ML, Cho HC, Popescu NI, Coggeshall KM, Arditi M, Underhill DM (2016) Hexokinase is an innate immune receptor for the detection of bacterial peptidoglycan. Cell, 166(3):624–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worth M, Li H, Jiang J (2017) Deciphering the functions of protein O-GlcNAcylation with chemistry. ACS Chem Biol, 12(2):326–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Qian K (2017) Protein O-GlcNAcylation: emerging mechanisms and functions. Nat Rev Mol Cell Biol, 18(7):452–465. doi: 10.1038/nrm.2017.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzwa SA, Macauley MS, Heinonen JE, Shan X, Dennis RJ, He Y, et al. (2008) A potent mechanism-inspired O-GlcNAcase inhibitor that blocks phosphorylation of tau in vivo. Nat Chem Biol, 4(8):483–490. doi: 10.1038/nchembio.96 [DOI] [PubMed] [Google Scholar]
- Zachara NE, Vosseller K, Hart GW. (2012) Detection and Analysis of Proteins Modified by O-Linked N-Acetyl Glucosamine. Curr Protoc Mol Biol, CHAPTER: Unit-176. doi: 10.1002/0471142727.mb1706s95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Liu TW, Cecioni S, Eskandari R, Zandberg WF, Vocadlo DJ (2015) O-GlcNAc occurs cotranslationally to stabilize nascent polypeptide chains. Nat Chem Biol, 11(5):319–25. doi: 10.1038/nchembio.1774 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


