Abstract
Background
The prognostic significance of plasma cell-free DNA (cfDNA) androgen receptor amplification (ARamp) in metastatic castration resistant prostate cancer (mCRPC) compared with circulating tumor cell (CTC) counts is not known.
Methods
As part of correlative aims of a prospective study in mCRPC, concurrent and serial collections of plasma and CTCs were performed. Specimen collections were performed at baseline after progression on androgen deprivation therapy and then 12 weeks later. QuantStudio3D digital PCR system (dPCR) was used to determine plasma cfDNA AR copy number variations and Cell search assay for enumerating CTC counts. Association of baseline cfDNA ARamp status/CTC counts with overall survival (OS) (primary goal) was evaluated using Kaplan–Meier method and log-rank test (p ≤ 0.05 for significance) and Receiver Operator Curves (ROC) for ARamp status and CTCs ≥ 5. A multivariate analysis was performed using Cox regression models that included ARamp, CTC counts and other clinical factors.
Results
ARamp was detected in 19/70 patients at baseline. At the time of analysis, 28/70 patients had died (median follow-up 806 days (IQR: 535–966)). ARamp was associated with poor OS (2 year OS of 35% in ARamp vs. 71% in non-ARamp; log-rank p-value= <0.0001). Baseline CTC count ≥ 5 (vs < 5) was also associated with poor survival (2 year OS of 44% vs 74%; log-rank p=0.001). ROC analysis demonstrated area under the curve (AUC) of 0.66 for ARamp- and 0.68 for CTC counts-based prognosis (p=0.84 for difference). The best two variables included for multivariable analysis were ARamp and CTC ≥ 5, however the two factor model was not significantly better than using ARamp alone for predicting survival (HR=5.25; p=0.0002).
Conclusions
CTCs and plasma cfDNA ARamp were observed to have equal prognostic value in mCRPC. Larger cohorts that incorporate molecular and clinical factors are needed to further refine prognosis in CRPC.
Keywords: Androgen receptor, cell-free DNA, prognostication, amplification
Introduction
Among men in the US prostate cancer is the second most common cause of cancer related deaths1. Progression from advanced castration sensitive disease treated with androgen deprivation therapy (ADT) to a castrate resistant state is inevitable2. Metastatic castration resistant prostate cancer (mCRPC) includes a heterogeneous group of patients for whom novel therapeutic options include drugs which target the AR axis3–5. Despite therapeutic advances challenges in predicting survival or matching treatments with drug response or resistance (primary or acquired) in mCRPC remain and disease progression occurs in majority of the patients. The underlying heterogeneity of cancer biology in castrate resistance6–8 can impact treatment response as well as survival. At present while several non-FDA cleared clinical prognostic factor based models have been proposed9, no genome-based prognostic markers are used to predict survival in castrate resistance state and the only FDA cleared prognostic marker of survival in mCRPC is the circulating tumor cell (CTC) count with CTCs ≥ 5 per 7.5ml whole blood as a predictor for inferior survival10. The widespread use of CTC count-based prognostication has its limitations that prevent it from being widely adopted in clinical practice. These include the lack of a single objective and agreeable definition for a CTC for detecting these rare cells, the inability to reliably probe molecular profiles in CTCs to evaluate underlying tumor biology during treatments, and the clinical observation that CTCs are not shed or captured universally in all advanced stage patients.
Several attempts for improving the limitations of CTC-based assays are under development11 while alternative methods for prognostication or predicting treatment outcomes by molecular characterization of the metastatic prostate cancer genome are also being developed. One such novel method involves the evaluation of cell free DNA (cfDNA) in blood plasma. Such a concept of “liquid biopsies” has begun to be evaluated for use in clinical practice in many tumor types including advanced prostate cancer where prospective studies with limited patient populations have evaluated cfDNA based markers for prognosis and prediction in mCRPC12–16. Since AR copy number variations (CNVs), particularly AR amplification (ARamp), is a key genomic aberration signature observed at the time of developing castrate resistance, ARamp in plasma cfDNA has been investigated in mCRPC, but after the use of chemotherapy or AR axis directed therapeutic agents for predictive and prognostic value12,16,17. Since it is unclear if plasma AR CNVs in mCRPC immediately following progression on ADT prior to initiating any standard of care mCRPC treatments can also be used to prognosticate survival we determined the prognostic value of plasma AR CNVs and CTC count specimens collected concurrently as part of correlative studies in an on-going prospective trial (https://clinicaltrials.gov/ identifier NCT # 01953640) in pre-chemotherapy mCRPC patients progressing on ADT. These correlative aims were considered exploratory and hypothesis generating in nature.
Materials and Methods
Patient recruitment and blood collection methods
Metastatic CRPC patients with progressive disease on continuous ADT were enrolled in a prospective trial in which patients underwent uniform collection and processing of blood, CTCs and urine specimens as part of the correlative aims of the study at baseline, before initiating abiraterone acetate and prednisone therapy (AA/P). Prospective collections of specimens were repeated after 12 weeks of AA/P treatment. The goal of the correlative study presented here was to determine prognostic outcomes based on CNVs observed in plasma cfDNA AR gene and CTC counts. The study was approved by Institutional Review Boards at Mayo Clinic and Medical College of Wisconsin and all patients signed an informed consent at the time of enrollment.
Plasma preparation and cfDNA extraction
Plasma blood collection was performed in 4 ml K2-EDTA plasma separator tubes and centrifuged at 2000 rpm for 10 minutes within 2 hours of collection for generating platelet rich plasma, followed by a second round of centrifugation of the supernatant for generating platelet poor plasma. The supernatant was fractioned into multiple 500 uL aliquots for storage at −80°C. Aliquots did not undergo any freeze-thaw cycles. QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA) was used for extracting cfDNA from 500 uL plasma. The DNA concentrations were quantified by a Qubit 2.0 Fluorometer following the standard protocol (Life Technologies, Carlsbad, CA).
Quantification of AR copy number by digital PCR and CTC assays
To quantify AR copy numbers (CN) in plasma cfDNA, Taqman-based AR CN assays was utilized including FAM-AR assay ID: Hs04511283_cn (cat 4400291) (Life Technologies) and VIC Copy Number Reference Assay: RNase P (cat 4403326) (Life Technologies). Details of digital PCR methods used for quantitation of AR copy numbers are provided separately under “Supplementary Methods”.
To enumerate CTCs, 7.5 ml whole blood was collected in CELLSEARCH® Circulating Tumor Cell Kits as per manufacturer’s direction. CTC enumeration was performed using the FDA cleared CELLSEARCH® CTC Test18.
Statistical Methods
As part of the correlative aims in the study cohort enrolled, association of baseline plasma cfDNA ARamp and CTC counts with overall survival (OS) at 19 months was explored. OS was defined as the time between registration on study and the date of death from any cause or the date of the last follow-up visit. Kaplan-Meier (KM) method was used to show overall survival distributions by ARamp status and CTC counts (<5 vs ≥ 5 cells). Tests of significance for AR amplification status and CTC counts association with OS were performed using log-rank test. Receiver operator curves (ROC) for both variables evaluated area under the curve (AUC) for predicting survival at 19 months. For the ROC analyses AUCs for both markers were calculated and compared when used alone and in combination. A ROC sensitivity analysis was also performed where AUCs were determined at various time points (15, 18, 21, 24 months). In order to determine the effect of multiple factors on survival including volume of metastatic disease (high versus low), a multivariate Cox regression model was utilized to assess association of several covariates measured at study enrollment (baseline) with OS as detailed under “Supplementary Methods”.
A secondary analysis was performed to explore if baseline ARamp status was predictive for developing treatment resistance at 12 weeks (primary resistance) using Chi-square tests and with progression-free survival (PFS) using log-rank test. Further details for the secondary analysis are provided under “Supplementary Methods”. All statistical analyses were performed with SAS 9.3 software (SAS Institute, Cary, NC, USA) and all P-values were two-sided with a cutoff for significance at P≤0.05.
Results
Clinical characteristics of the study cohort
Between 5/2013 and 9/2015, 92 patients were enrolled on the main study of which 70 had plasma samples for cfDNA isolation available at baseline and 12-weeks after initiating AA/P treatment. Demographic characteristics of the enrolled population are summarized in Table 1. These were used in the analysis of ARamp-based prognosis. Supplementary Table 1 provides the demographics of the 22 patients not included in this analysis for lack of specimens. We observed that 38/70 had high and 32/70 had low volume metastatic disease. The median study follow-up at the time of this analysis was 806 days (IQR: 535–966) during which time 28/70 patients had died. At twelve weeks of treatment 30/70 patients demonstrated progression using composite assessment criteria and during the follow up period at the time of this analysis a total of 55/70 patients had progressed with a median progression free period of 194 days.
Table 1.
Baseline Patient Characteristics and cfDNA, AR amplification and CTC results
| Characteristic | Total (N=70) |
|---|---|
| Race (N=69) – no. (%) | |
| White | 66 (96) |
| Black or African American | 1 (1) |
| Asian | 1 (1) |
| American Indian or Alaska Native | 1 (1) |
| Age - yr | |
| Median | 71.5 |
| Range | 39-91 |
| Gleason score at initial diagnosis – no. (%) | |
| 2 – 6 | 12 (17) |
| 7 | 19 (27) |
| 8 – 10 | 39 (56) |
| Primary radiation therapy at initial diagnosis – no. (%) | |
| Yes | 41 (59) |
| No | 29 (41) |
| Primary radical prostatectomy at initial diagnosis – no. (%) | |
| Yes | 33 (47) |
| No | 37 (53) |
| Volume of metastatic disease – no. (%) | |
| Low | 32 (46) |
| High | 38 (54) |
| Time from starting ADT to CPRC - yr | |
| Median | 2.5 |
| Interquartile range | 1.1–4.6 |
| Metastatic Biopsy site at study enrollment – no. (%) | |
| Bone | 49 (70) |
| Lymph nodes | 13 (19) |
| Liver/lung | 3 (4) |
| Others | 5 (7) |
| PSA at study enrollment – ng/ml | |
| Median | 16.2 |
| Interquartile range | 8.0-38.9 |
| Serum Chromogranin levels at study enrollment (N=68) – ng/ml | |
| Median | 91.0 |
| Interquartile range | 55.0–235.5 |
| Testosterone at study enrollment (N=68) – ng/dl | |
| Median | 7.0 |
| Interquartile range | 6.9–10.0 |
| LDH at study enrollment (N=66) – U/L | |
| Median | 187 |
| Interquartile range | 170–209 |
| FACT-P: Physical Well Being score at study enrollment | |
| Median | 23.5 |
| Interquartile range | 20–26 |
| FACT-P: Total Score at study enrollment | |
| Median | 118 |
| Interquartile range | 106–131 |
| Opiate/pain medication use (N=59) – no. (%) | |
| Yes | 43 (73%) |
| No | 16 (27%) |
| Study follow-up | |
| Median days of follow-up (IQR) | 806 (535-966) |
| Number of deaths (%) | 28 (40) |
| Median time to death | 805 |
|
Baseline and 12 week cfDNA, AR amplification and CTC Levels | |
| cfDNA (N=70)/AR amp (N=70)/CTC (N=66) | Value |
|
| |
| Baseline cfDNA yield (ng) | |
| Mean (SD) | 5.7 (6.3) |
| Median (IQR) | 3.7 (1.5–6.8) |
| Baseline AR amplification (%) | |
| Yes | 27 |
| No | 73 |
| Baseline CTC count | |
| Mean (SD) | 16.3 (53.4) |
| Median (IQR) | 2 (1–9) |
| Baseline CTC >= 5 cells (%) | |
| Yes | 36 |
| No | 64 |
| 12-week cfDNA purified amount | |
| Mean (SD) | 5.4 (5.6) |
| Median (IQR) | 3.5 (1.8-7.5) |
| 12-week AR amplification (%) | |
| Yes | 17 |
| No | 83 |
| 12-week CTC count (N=55) | |
| Mean (SD) | 5.4 (17.3) |
| Median (IQR) | 0 (0-2) |
| 12-week CTC >= 5 cells (N=55) (%) | |
| Yes | 13 |
| No | 87 |
Association of AR amplification with overall survival and other clinical factors
The median amount of cfDNA isolated from 500 μL plasma samples at baseline and at 12 weeks was 3.7 ng and 3.5 ng (Table 1), respectively. cfDNA amounts in pre-treatment plasma specimens did not differ by volume of disease (median amount=3.3 ng for low volume, and =3.8 ng for high volume, p-value for difference between groups = 0.86) or by CTC counts (median cfDNA amount of 3.7 for CTC < 5 and 3.0 for CTC >= 5; p-value for difference between group =0.76). The amount of cfDNA in plasma at baseline was not associated with OS after dividing the cohort into low cfDNA vs high cfDNA based on less than or greater than median (3.7 ng) amount (log-rank p-value=0.33). Using the criterion for defining ARamp we observed 19/70 patients at baseline and 12/70 at 12 weeks with ARamp, respectively (Table 1). Baseline ARamp status was not associated with age or Gleason score at diagnosis but associated with PSA levels at study entry (baseline) (p=0.0064) and 12- week plasma (p=0.0032). In baseline plasma samples, 16/38 (42%) patients with high volume disease showed ARamp while 3/32 (9%) patients with low volume disease demonstrated ARamp. Plasma ARamp status was associated with metastatic volume of disease at study enrolment (p=0.002). We did not detect baseline ARamp to be associated with duration of prior ADT (non-ARamp group =2.6 years vs 2.0 years in ARamp group; p=0.75). Prior exposure to anti-androgens was also not associated with baseline ARamp (28% in those with prior anti-androgen and 20% with no prior anti-androgen; p=0.69).
Predictive performance of prognosis using ARamp and CTC counts
Survival of the 19/70 (27%) patients with baseline ARamp at 10, 20 and 30 months follow up was 100%, 53% and 0%, respectively. Among the remaining 51 (73%) patients without ARamp the survival rate was 96%, 80% and 58%, respectively. Plasma ARamp was significantly associated with poor survival (2 year OS of 35% vs 71% in non-AR-amp; log-rank p=<0.0001) (Figure 1A). Baseline CTC count ≥ 5 (vs < 5) was also associated with poor OS (2 year OS of 44% vs 74%; log-rank p=0.001) (Figure 1B). ROC for plasma ARamp-based prognosis had an AUC of 0.66 (95% CI: 0.52-0.81) and for CTC had an AUC of 0.68 (95% CI: 0.54-0.83). There was no difference between AUCs for ARamp status (0.66) and CTC (0.68) (difference = 0.02, 95% CI: 0.21-0.18; p=0.84) (Figure 2A). By combining both ARamp and CTC the AUC was 0.75 (95% CI: 0.61-0.89) (Figure 2B). Compared to the AUC of CTC alone this increased AUC was not statistically significant (difference=0.07, 95% CI: 0.01-0.14). A sensitivity analysis investigating the AUCs of ARamp and CTC at different time points were consistent to those observed. AUCs ranged from 0.65 to 0.75 with no significant differences indicated at any time point. To determine the frequency of AR-amp based on the number of CTCs (<5 or ≥5) at baseline, 62.5% of patients (15/24) with CTC ≥5 were detected with ARamp while only 28.5% of patients (12/42) with CTC<5 were detected to have ARamp (P=0.014). Four patients had no CTC data at visit 1. For visit 2 while the number of patients with CTC data was lower than baseline collections, 57.1% of patients (4/7) with CTC ≥5 had AR amplification while only 16.6% of patients (8/48) with CTC<5 had AR-amp (p=0.05). Fifteen patients had no CTC data at visit 2.
Figure 1.
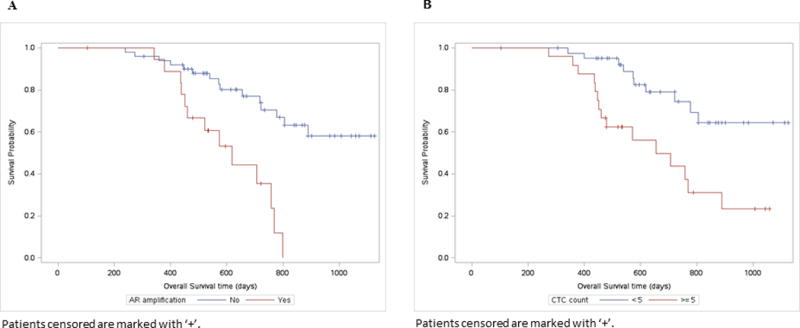
A: Kaplan Meier curves for baseline plasma cfDNA AR amplification with overall survival.
B: Overall Survival by CTC Count at baseline. Patients with CTC counts greater or equal to 5 cells are at higher risk of death than those with < 5 cells (log-rank p-value=0.001).
Figure 2.
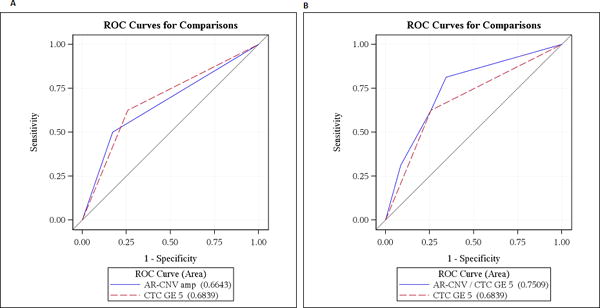
A: ROC curves comparing AR amplification and CTC based overall survival prognostication. No difference between AUCs for AR amp (0.66) and CTC (0.68), p=0.84
B: No difference between AUCs for AR amp + CTC (0.75) vs. CTC alone (0.68), p=0.08
Multivariate predictive model of prognosis in pre-chemotherapy mCRPC
At the univariate level using seven known and promising prognostic factors in mCRPC Cox regression analysis showed association of OS with ARamp (HR=5.25; p=0.0002), CTC ≥ 5 (HR=3.42; p=0.003), log PSA (HR=1.35; p=0.009) and high metastatic disease volume (HR=2.36; p=0.04) (Figure 3). All possible combinations of these 4 covariates were considered in the search for the most parsimonious model. No two factor or higher multivariate model was identified that was statistically better than univariate model with ARamp alone (HR=5.25; p=0.0002). The best two variable multivariable model included ARamp and CTC >= 5. However, this two factor model was not significantly better than ARamp alone (p-value for difference in models of 0.06).
Figure 3.
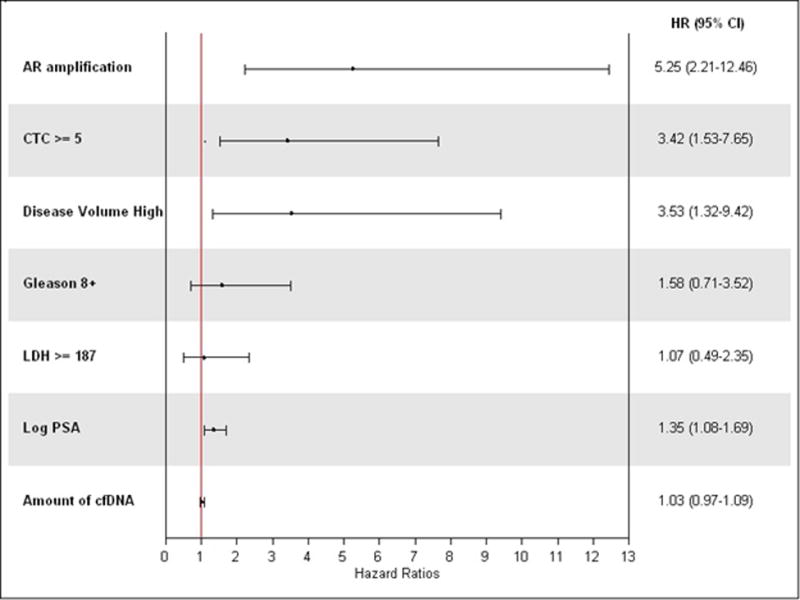
Forrest plot for univariate analysis of baseline covariates with survival
Association of ARamp as a predictive biomarker with treatment response to AA/P
An exploratory analysis was performed to determine the effect of ARamp status before initiating AA/P treatment and response at 12-weeks and with disease free progression. ARamp status at baseline was not associated with the composite progression at 12 weeks (p=0.49), PSA progression at 12 weeks (p=0.31), or progression free survival (p=0.27) (Supplementary Figure 3). In 30/70 patients with progression at 12-weeks, 23 patients did not have plasma ARamp at baseline (AR copy number neutral). Of these five converted to ARamp at 12 weeks while the remaining 18 did not. Of the remaining 7/30 with ARamp in the baseline specimen four patients retained AR amplification status at time of 12 week progression (Supplementary Figure 2).
Discussion
In a prospective cohort of pre-chemotherapy mCRPC patients we observed plasma cfDNA-based ARamp status and CTC counts (≥5 per 7.5 ml blood sample) to be independently prognostic for survival. We explored ARamp status using a limited set of seven clinical factors and CTC counts (Figure 3) for incorporating in a multivariate regression model and detected ARamp status detected at the time of ADT failure to independently predict survival. This allows for additional refinement of prognostic groups in mCRPC using emerging genomic biomarkers and for pursuing aggressive treatment interventions in poor prognosis patients more accurately. Assessing prognosis by developing validated molecular prognostic biomarkers in CRPC is an active and highly recommended area of research19. Since the cohort size is small, it limits the value of our observation and a more formal determination is needed in larger cohorts9.
We did not observe ARamp status to predict efficacy of pre-chemotherapy AA/P response. This result differs from a previous report in which ARamp was predictive of post chemotherapy efficacy for AA/P12. The difference could be because our study was performed in mCRPC patients immediately following progression on ADT from a hormone sensitive stage, at which time the full repertoire of AR axis-based genomic aberrations including focal amplifications in AR gene has not fully emerged. In fact the detected rate of AR amplification status in our study was slightly lower (27%), than the 40% incidence reported from mCRPC patients who have undergone several chemotherapy treatments12.
Serially obtained specimens allowed us to determine changes in plasma cfDNA AR copy numbers. Interestingly we did not detect ARamp status to change in the paired samples after 12 weeks of treatment using our pre-defined cut off ratio (of ≥ 2.0) to classify AR amplification status. At the same time while AR amplification did not change after 12-weeks of treatment AR copy number changes were observed based on absolute numerical values of AR copy numbers in the two serial samples. The numerical increase in AR copy numbers was found in 20 (67%) of the 30 patients who were observed to have a 3-month progression while in only 13 (33%) of 40 patients without 3-month progression. Shifts in AR copy numbers post treatment in advanced prostate cancer have been previously reported in plasma and may represent rapid adaptations to selection pressures under treatment17. Not all studies in advanced prostate cancer evaluating plasma cfDNA AR have reported consistently similar shifts in copy numbers in mCRPC patients for prediction of treatments outcomes. Results from only one large study suggest that plasma ARamp serially captured may have predictive value for determining AA/P efficacy12. At present it is not clear if the shift in AR copy numbers from a time-dependent clonal evolution process represents a steady accumulation of AR-based aberrations/amplifications over time as mCRPC progresses or if this represents a treatment effect.
The PCR-based cfDNA “liquid biopsy” assay for directly monitoring specific tumor-associated molecular changes has shown several advantages.15,20 In prostate cancer specifically it offers a chance to monitor putative tumor-specific genomic aberrations like chromosomal rearrangements and CN gains and losses such as the ETS gene family fusions, PTEN loss and ARamp. Studies show that cfDNA “liquid biopsy” clinical applications are promising and increasingly used in the clinic such as the EGFR gene-based T790M in non-small cell lung cancer.12,17,21
A limitation of this study is the relatively small sample size and the lack of comparison with known RNA-based biomarkers such as AR-V7 in CTCs which have been shown to be predictive biomarker for treatment response22,23. The value of AR-V7 CTC assay in mCRPC patients continues to evolve as a predictive and prognostic biomarker as was recently evaluated in a prospective cohort of 202 men of which at least forty percent men had previously received some form of mCRPC therapy23. It is unclear if the incorporation of this assay with cfDNA candidates and clinical prognostic factors including quality of life and pain scores will yield superior prognostic models than any one factor alone and definitive studies remain on-going. Another limitation is the use of one single reference gene. Since mCRPC is a genetically unstable disease, it will be more robust if using several reference genes to correct for possible genetic alterations in the targeted regions. Therefore, additional validation is needed in prospective and larger patient cohorts to confirm the prognostic value of circulating AR CNVs as a prognostic and predictive biomarker, preferably by comparing it to CTCs, existing clinical models and AR splice variants.
Supplementary Material
Acknowledgments
We are grateful for the administrative assistance provided by Bobbi-Ann Jebens for this submission. We also would like to thank all patients who participated in this study for their selfless contribution in bringing precision medicine to future advanced prostate cancer patients. We highly appreciate the support of family members as well.
Funding:
This study is funded in part by the Mayo Clinic Center for Individualized Medicine; National Institutes of Health - National Cancer Institute [R01CA21209] to MK and Liang W; Department of Defense [W81XWH-15-1-0634] to MK and SMD; and Mayo Clinic Schulze Center for Novel Therapeutics in Cancer Research to MK and Liewei W; Joseph and Gail Gassner to MK; and Advancing a Healthier Wisconsin Fund [#5520227] and National Institutes of Health [R01CA157881] to Liang W; National Institutes of Health - National Cancer Institute, [R01CA174777] to SMD.
Footnotes
Disclosure Statement: The authors have declared no conflicts of interest.
Supplementary information is available at PCAN’s website.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Prostate Cancer Trialists Collaborative Group. Maximum androgen blockade in advanced prostate cancer: an overview of the randomised trials. Prostate Cancer Trialists’ Collaborative Group. Lancet. 2000;355(9214):1491–1498. [PubMed] [Google Scholar]
- 3.de Bono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L, et al. Abiraterone and increased survival in metastatic prostate cancer. The New England journal of medicine. 2011;364(21):1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ryan CJ, Smith MR, de Bono JS, Molina A, Logothetis CJ, de Souza P, et al. Abiraterone in metastatic prostate cancer without previous chemotherapy. N Engl J Med. 2013;368(2):138–148. doi: 10.1056/NEJMoa1209096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beer TM, Tombal B. Enzalutamide in metastatic prostate cancer before chemotherapy. N Engl J Med. 2014;371(18):1755–1756. doi: 10.1056/NEJMc1410239. [DOI] [PubMed] [Google Scholar]
- 6.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161(5):1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487(7406):239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beltran H, Eng K, Mosquera JM, Sigaras A, Romanel A, Rennert H, et al. Whole-Exome Sequencing of Metastatic Cancer and Biomarkers of Treatment Response. JAMA Oncol. 2015;1(4):466–474. doi: 10.1001/jamaoncol.2015.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halabi S, Lin CY, Kelly WK, Fizazi KS, Moul JW, Kaplan EB, et al. Updated prognostic model for predicting overall survival in first-line chemotherapy for patients with metastatic castration-resistant prostate cancer. J Clin Oncol. 2014;32(7):671–677. doi: 10.1200/JCO.2013.52.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Danila DC, Heller G, Gignac GA, Gonzalez-Espinoza R, Anand A, Tanaka E, et al. Circulating tumor cell number and prognosis in progressive castration-resistant prostate cancer. Clin Cancer Res. 2007;13(23):7053–7058. doi: 10.1158/1078-0432.CCR-07-1506. [DOI] [PubMed] [Google Scholar]
- 11.van der Toom EE, Verdone JE, Gorin MA, Pienta KJ. Technical challenges in the isolation and analysis of circulating tumor cells. Oncotarget. 2016 doi: 10.18632/oncotarget.11191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Romanel A, Gasi Tandefelt D, Conteduca V, Jayaram A, Casiraghi N, Wetterskog D, et al. Plasma AR and abiraterone-resistant prostate cancer. Sci Transl Med. 2015;7(312):312re310. doi: 10.1126/scitranslmed.aac9511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heitzer E, Ulz P, Geigl JB. Circulating tumor DNA as a liquid biopsy for cancer. Clinical chemistry. 2015;61(1):112–123. doi: 10.1373/clinchem.2014.222679. [DOI] [PubMed] [Google Scholar]
- 14.Xia S, Kohli M, Du M, Dittmar RL, Lee A, Nandy D, et al. Plasma genetic and genomic abnormalities predict treatment response and clinical outcome in advanced prostate cancer. Oncotarget. 2015;6(18):16411–16421. doi: 10.18632/oncotarget.3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carreira S, Romanel A, Goodall J, Grist E, Ferraldeschi R, Miranda S, et al. Tumor clone dynamics in lethal prostate cancer. Sci Transl Med. 2014;6(254):254ra125. doi: 10.1126/scitranslmed.3009448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wyatt AW, Azad AA, Volik SV, Annala M, Beja K, McConeghy B, et al. Genomic Alterations in Cell-Free DNA and Enzalutamide Resistance in Castration-Resistant Prostate Cancer. JAMA Oncol. 2016;2(12):1598–1606. doi: 10.1001/jamaoncol.2016.0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ulz P, Belic J, Graf R, Auer M, Lafer I, Fischereder K, et al. Whole-genome plasma sequencing reveals focal amplifications as a driving force in metastatic prostate cancer. Nat Commun. 2016;7:12008. doi: 10.1038/ncomms12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10(20):6897–6904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- 19.Scher HI, Morris MJ, Stadler WM, Higano C, Basch E, Fizazi K, et al. Trial Design and Objectives for Castration-Resistant Prostate Cancer: Updated Recommendations From the Prostate Cancer Clinical Trials Working Group 3. J Clin Oncol. 2016;34(12):1402–1418. doi: 10.1200/JCO.2015.64.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu W, Laitinen S, Khan S, Vihinen M, Kowalski J, Yu G, et al. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nat Med. 2009;15(5):559–565. doi: 10.1038/nm.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heitzer E, Ulz P, Geigl JB, Speicher MR. Non-invasive detection of genome-wide somatic copy number alterations by liquid biopsies. Mol Oncol. 2016;10(3):494–502. doi: 10.1016/j.molonc.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371(11):1028–1038. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Antonarakis ES, Lu C, Luber B, Wang H, Chen Y, Zhu Y, et al. Clinical Significance of Androgen Receptor Splice Variant-7 mRNA Detection in Circulating Tumor Cells of Men With Metastatic Castration-Resistant Prostate Cancer Treated With First- and Second-Line Abiraterone and Enzalutamide. J Clin Oncol. 2017;35(19):2149–2156. doi: 10.1200/JCO.2016.70.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


