Abstract
The Mexican tetra, Astyanax mexicanus, comes in two forms: a classical river-dwelling fish and a blind and depigmented cave-dwelling fish. The two morphotypes are used as models for evolutionary biology, to decipher mechanisms of morphological and behavioural evolution in response to environmental change. Over the past 40 years, insights have been obtained from genetics, developmental biology, physiology and metabolism, neuroscience, genomics, population biology and ecology. Here, we promote the idea that A. mexicanus, as a model, has reached a stage where an integrated approach or a multi-disciplinary method of analysis, whereby a phenomenon is examined from several angles, is a powerful tool that can be applied to understand general evolutionary processes. Mexican cavefish have undergone considerable selective pressure and extreme morphological evolution, an obvious advantage to contribute to our understanding of evolution through comparative analyses and to pinpoint the specific traits that may have helped their ancestors to colonize caves.
Keywords: Astyanax mexicanus, evolutionary biology, multi-disciplinary
1. Introduction
Mexican tetras in rivers of Central America and in caves of central Mexico are the same species: Astyanax mexicanus. The former are ‘wild-type’ multi-coloured tropical freshwater fish. The latter are ‘natural mutants’ that live in total and permanent darkness, and they lack eyes and pigmentation (figure 1). They can be found in 30 karst caves in a restricted region of Mexico [2]. The two types of morphs can breed and give a fertile progeny, and cavefish from different caves are also inter-fertile [3]. Hence, after a drastic and sudden environmental change, their phenotypic evolution occurred rapidly within about 20 000 years [4] and is probably still ongoing. Of note, based on morphological distinctions, surface and cave morphs were originally proposed as different species and even genus, and the eyeless fish was given the name Anoptichthys jordani [5]. Speciation between the two morphs may be on its way [6], but has not happened yet, based on inter-fertility criteria.
Figure 1.
Astyanax mexicanus cavefish and surface fish. The morphology of the fish and the environment where they live are illustrated in a river and three different caves. Important differences, including between caves, are described below. Scale bars, 1 cm. (a) Surface fish. A specimen caught in the Nacimiento del Rio Choy. This environment is characterized by ambient light and its associated primary production. There, fish live in large schools, together with many other fish species (example shown: poecilids). (b) Pachón cavefish. The Pachón cave is isolated, and the water enters exclusively by percolation during the rainy season. The bottom of the pool is mud, on which the adult fish feed [1]. They cohabit with many troglomorphic arthropods, including large ones like the Speocirolana pelaezi (3 cm) shown at the bottom left, on which they can also feed. (c) Chica cavefish. The Chica cave regularly receives external water flows and hosts a large bat colony. Fish (arrow) swim in waters with high organic contents, including decomposing bat cadavers (bottom pictures, arrowheads) and detritus brought from the surface by incoming water. (d) Subterráneo cavefish. The Subterráneo cave offers a rocky substrate. Seasonal floods bring surface fish into the cave, and hybridization sometimes occurs. The bottom right picture shows an eyed, F2-type hybrid individual, cohabiting with blind cavefish in this cave. Here, predators are found, such as the non-troglomorphic crayfish shown at bottom left.
The A. mexicanus system presents many advantageous features and appears an outstanding model system to study evolutionary forces (distal mechanisms) and evolutionary mechanisms (proximal mechanisms) underlying morphological, behavioural and physiological adaptations in response to environmental change. When compared with other fish such as sticklebacks, cichlids or whitefish that have also undergone recent radiations and have significantly contributed to evolutionary biology [7–9], A. mexicanus cavefish underwent considerable selective pressure and extreme morphological evolution. Obvious advantages of tetras are the insights that can be gained into understanding how circadian rhythms influence physiology and how other senses can compensate for loss of sight. The behavioural and genetic bases of their apparently easy colonization of caves—when compared with other endemic fish species living in Mexican rivers—sometimes called ‘pre-adaptations’, can be studied. Moreover, the cavefish model, like other fish evolutionary models, fulfils the prerequisites to any research: it has entered the genome era [10,11], can be bred and manipulated relatively easily [12,13] and its population biology has been studied for decades [2,14].
Several reviews, including a book on the biology of Mexican cavefish by contributors from the whole Astyanax scientific community, have been published recently [15–17], dealing with all the many facets of cavefish evolutionary biology. Instead of specifically reviewing one aspect, here we will focus on combining approaches to better understand cavefish evolution and to illuminate evolutionary mechanisms in general.
2. Towards an integrated approach
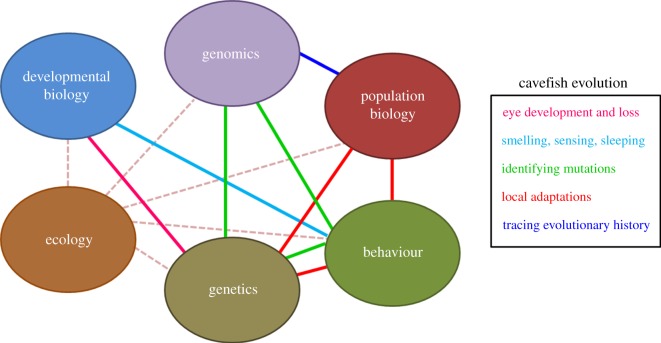
Researchers of Mexican cavefish have various disciplinary backgrounds: genetics, developmental biology, neuroscience, physiology, metabolism, genomics, population biology, ecology, cave biology or speleology. An integrated approach is by essence a multi-disciplinary method of analysis, whereby a phenomenon is examined from several angles (figure 2). In sticklebacks, for example, the combination of population biology, quantitative genetics and developmental biology has revealed strikingly convergent processes occurring in the non-coding regulatory sequences of a developmental transcription factor (Pitx1) and underlying the morphological evolution (loss of pelvic spine) of multiple freshwater populations from marine ancestors [18–21]. Here, we provide examples of questions that strongly benefit from an integrated approach in the cavefish system.
Figure 2.
Towards an integrated approach. The mechanisms underlying cavefish evolution—and the evolution of any species—are better understood by combining insights from different disciplines. Connections and combinations between different disciplines are represented by coloured lines. The corresponding questions that can be addressed and are reviewed in text are indicated in the same colour.
(a). Developmental biology and genetics: eye development and loss
A typical question at the frontiers of these two disciplines is that of eye development and loss, i.e. the mechanisms of morphological evolution. The problem was first examined through crosses between surface and cavefish or between cavefish originating from different caves [15]. F1 hybrids from cave × surface parents have slightly reduced but functional eyes, indicating that cave alleles are mostly recessive. Sometimes, F1 hybrids generated from parents originating from different caves develop small eyes, and such partial complementation suggests that different genes are affected in different caves, highlighting convergent evolution [22,23]. Quantitative trait loci (QTL) analyses show that the control of eye size in F2 families is a complex trait, involving 12–15 loci [24]. As cave alleles at every eye or lens QTL cause size reductions, the authors suggested that eye loss evolved by natural selection, rather than by drift. A recent study examining cavefish retinal degeneration in the context of QTL mapping identified four regions, each involved in the control of the thickness of one distinct retinal layer, and in which several candidate genes, mostly corresponding to developmental transcription factors and regulators, were identified [25].
From the developmental biology standpoint, cavefish eye degeneration is triggered by the defective embryonic lens, which rapidly becomes apoptotic and causes the collapse of the entire eye [26]. Lens apoptosis is indirectly due to an increase in Shh (Sonic Hedgehog) signalling from the embryonic midline [27]. Importantly, increased Shh signalling also affects pleiotropically other tissues of the future head and forebrain: jaw size and taste bud numbers are increased [28], the olfactory placode is enlarged at the expense of the lens placode [29], the preoptic region and hypothalamus in the forebrain are enlarged, partly at the expense of the ventral retina [30], and development is significantly changed [31,32]. These findings suggested that indirect selection might be at work during developmental evolution of eye loss in cavefish, i.e. that the eyes would be lost as secondary collateral damage resulting from the constructive evolution of other beneficial traits. Therefore, the evo-devo approach and the genetic approach converge in proposing selection acting as one of the distal mechanisms underlying eye loss. Of note, other evidence from studies of molecular evolution of the developmental transcriptome suggests genetic drift as a mechanism for eye loss: in cavefish, radical mutations (i.e. giving rise to amino acids with distinct physico-chemical properties in the two morphs) are over-represented in ‘eye genes’, which can be explained by relaxed selection for vision during evolution in the absence of light [10,33]. Therefore, there are probably two components underlying eye loss in cavefish: selection (direct or indirect) to improve survival in the dark and drift by random accumulation of mutations in ‘eye genes’ that are not counter-selected [34].
(b). Developmental biology and behaviour: smelling, sensing and sleeping
Embryogenesis proceeds through a hierarchical cascade of events starting before fecundation. Changes in brain shape, and consequently function, could arise from natural selection acting at different levels during embryogenesis: maternal gene products in the oocyte, expression of morphogens and patterning genes and timing of cell proliferation/differentiation. Emergence of novel behaviours through developmental variations is essential for evolutionary radiation [35,36]. Recent advances in sequencing, genome editing and transgenesis are helping to unravel the molecular mechanisms of this evolution. Studying variations in the regulatory genome and epigenome in the near future will shed light on the mechanisms linking gene function and evolution of development.
Differential behavioural performances may stem from sensory specializations that have a developmental basis. In parallel to eye regression, cavefish evolved larger olfactory epithelia, sensory organs specialized in the detection of molecules related to feeding and reproduction. The enlargement in the embryonic tissue fated as olfactory epithelia results from subtle changes in the expression of the signalling molecules Bmp4 and Shh in the embryonic prechordal mesoderm. In parallel, behavioural assays have demonstrated the cavefish's remarkable olfactory capabilities [29]. Increased olfactory sensitivity is probably not determined only by the size of the sensory organ and developmental plasticity and changes in central processing must also contribute to this adaptation [37].
Some cavefish populations have evolved a vibration attraction behaviour (VAB) towards oscillating objects in the water, mediated by neuromasts of the lateral line, particularly those located in the suborbital eye region [38,39]. Numeric differences in neuromasts appear at larval stages [40], but the embryological and genetic bases for such modifications remain unknown. Increased neuromast numbers in the optic region may result from eye degeneration, but this is independent of the pleiotropic Shh hypersignalling leading to eye regression [39].
Another behavioural adaptation of cavefish is sleep loss [41,42]. Sleep is a reversible state of elevated arousal threshold and quiescence. Its duration and intensity differ greatly across species, depending on ecological variables and evolutionary history [43]. It is regulated by the daily light–dark cycle (absent in caves), as well as by sensory processing and metabolic balance [44,45]. Neuromast hypersensitivity is implicated in sleep loss in cavefish [46]. The hypothalamic hypocretin/orexinergic system has a conserved role in the regulation of the sleep/wake cycle in vertebrates [47,48]. Higher hypocretin expression levels and numbers of neurons were reported in adult cavefish compared with surface fish, and functional assays confirmed that increased orexinergic neurotransmission is associated with the sleep-loss phenotype [49]. Neuroanatomical variations in the hypocretin neuronal cluster exist from early embryogenesis, resulting from morphogen modulations at the end of gastrulation [31]. Developmental manipulations reducing the number of hypocretin neurons in one-week-old larvae decrease activity in these larvae, mimicking surface fish. This demonstrates that developmental evolution of the hypocretin cluster impacts cavefish behaviour. Variations in the establishment of other hypothalamic neuronal systems (neuropeptide Y (NPY), proopiomelanocortin) were also noted, and their contribution to behavioural changes must be explored.
(c). Behaviour, genetics and functional genomics: identifying mutations in cavefish
Cavefish display multiple behavioural traits that are different from surface fish and these appear to be advantageous for life in subterranean habitat, and have probably been selected for. This implies that they have a genetic basis.
QTL approaches indicate that the loss of sleep [41], the loss of schooling [50], the special feeding posture with a 45° angle to the substrate [51] and the VAB [39,42] have a multi-genic determinism, as F2 fish resulting from crosses between two F1 hybrids present diverse levels of these behaviours. Several QTL intervals were identified, allowing mapping of genomic regions associated with particular behaviours. The regions related to the control of sleep/locomotor activity and VAB are different [42]. Moreover, QTL analysis suggests candidate genes, such as the narcolepsy-associated tp53-inducible protein gene, a potential regulator of sleep located in the QTL interval identified as regulating locomotor activity [42].
Sometimes, the study of behavioural phenotypes themselves can suggest candidate genes. For example, Tinaja cavefish hyperphagia suggested investigating the leptin pathway, which led to the identification of mutations in the Mc4r (melanocortin 4 receptor) gene [52]. Or, the effect of deprenyl (inhibitor of serotonin degradation) on aggressiveness, together with comparative neuroanatomical analyses, implicated the serotonergic system in aggressiveness differences between the two morphs, and pinpointed a mutation in the MAO (monoamine oxidase) enzyme [32,53]. Therefore, the study of behavioural phenotypes, but also neurochemical and neuroanatomical studies, allows better targeting of candidate genes responsible for behavioural changes between the two morphs.
Once a candidate gene is identified, functional genomics should test the role of the gene in behavioural control. When cavefish and surface fish possess different alleles, crosses or genome edition by CRISPR/Cas9 can be used to insert a surface allele in cavefish and vice versa. When the gene differs in terms of expression level between cavefish and surface fish, inactivation or overexpression can be achieved. For example, the hypocretin gene is overexpressed in cavefish. The knock-down of its expression by morpholinos has demonstrated its involvement in sleep loss in cavefish [49].
Behavioural studies, genetics and functional genomics identify the genetic bases of behavioural changes. Then, interesting contributions can arise from population and phylogeographical studies to draw hypotheses on the evolutionary history of the trait. For example, the distribution of mutated alleles of Mc4r among caves and rivers suggests that the derived allele entered the cave starting from the standing genetic variation existing in surface fish at the time of cave colonization [52] (see also §2d). Of note, genetic bases are one of several possible proximal causes for the behavioural difference between the two morphs. We also need to study the effects of the environment alone (e.g. absence of light, water physico-chemical parameters or trophic support), corresponding to the potential involvement of phenotypic plasticity or epigenetic mechanisms.
(d). Genomics and population biology: tracing back evolutionary history
The caves hosting cavefish have a broad distribution and have been grouped into three clusters: the El Abra, the Guatemala and the Micos groups. The evolutionary history and the origins of these cavefish populations are still poorly understood. While a rather ancient origin, millions of years ago, of the cavefish populations was generally quoted in the literature, recent analyses and modelling using two independent datasets (microsatellites and single nucleotide polymorphisms) suggest that cavefish invaded caves less than 25 000 years ago [4]. Such a recent origin should be verifiable and have interesting consequences in terms of population genomics approaches. The results should be comparable to those obtained in the threespine stickleback system (e.g. [54,55]). First, a recent origin and rapid evolution of cavefish implies that phenotypic evolution mainly relied on the fixation of genetic variants already existing in the ancestral surface population (as opposed to novel mutations arising, which takes more time), and many of them should still be found in the extant surface fish. Genome-wide scans should be performed at population level to estimate shared polymorphism [10], but we already have one example of such a case: the point mutation described in Mc4r, and responsible for increased appetite in some cavefish populations, has been found at low frequency (3%) in river-dwelling individuals [52]. Second, a recent origin also implies that gene loss/pseudogenization should not have had time to occur significantly on ‘eye genes’, which are not counter-selected in the dark. Genome-wide surveys on high-quality genomes are also needed to confirm this, but published data seem to support this prediction: cavefish crystallins [33] and opsins [56] do not show major loss of function mutations in their coding sequence, to be compared, for example, with two crystallins and two opsins that are inactivated/missing in the naked mole rat genome [57]. Third, if cavefish populations are young, it should be possible to find traces of selection genome-wide through selective sweep analyses. Selective sweep is the fixation of an allele through selection, and the fixation of neutral alleles at closely linked loci through genetic hitchhiking, resulting in reduced genetic diversity around the position under selection. Such analyses can be performed only if selection is recent; otherwise, novel polymorphisms arise with time and mask the swept region [58]. Again, population genomics approaches are promising, as an alternative/complement to QTL approaches, to identify loci under selection during cavefish phenotypic evolution.
(e). From population biology to behaviour and genetics: highlighting local adaptations
There are 30 described cave populations of A. mexicanus, named after the cave where they live [2]. All populations are blind and depigmented, but these striking phenotypes are achieved, at least partially, through convergences. The set of genes involved in eye degeneration is not identical in different cave populations (§2a), and the mutations/deletions in Oca2 (ocular and cutaneous albinism 2) responsible for the albino phenotype are not the same in the Molino (Guatemala group), Pachón or Japonés (both from the El Abra group) caves [59]. Other, less obvious phenotypes also show significant variations among caves. VAB [38] is present in Pachón and Sabinos cavefish (both from El Abra group), but does not have the same parental genetic determinism, paternal or maternal inheritance, in these two populations [60]. Additionally, some cave populations show VAB (Pachón, Sabinos, Piedras) but some do not (Molino) [39,42]. Finally, a small proportion of surface fish show a weak form of VAB, suggesting that the surface ancestors became adapted to caves by positive selection for VAB, followed by continued selection for amplification of this behaviour and its underlying sensory receptors, the neuromasts [38]. Similar phenotypic variations exist for sleep, aggressiveness or feeding behaviour among cave populations. Varying degrees of evolutionary-derived sleep reduction are described in Pachón, Tinaja, Molino, Sabinos and Chica populations [41], but this feature is dependent on sensory inputs from the lateral line only in the Pachón population [46], indicating that the phenotype may have emerged by different mechanisms. The loss of territorial and hierarchical aggressiveness—a surface fish hallmark—is described in Pachón cavefish, while Molino cavefish have conserved some degree of aggressiveness [32,61,62], and this phenotypic difference might be related to the presence or absence of an MAO mutation in the genomes of these two populations [53]. The increased appetite of Tinaja cavefish is not seen in Pachón cavefish [31,52], possibly related to the presence or absence in their genomes of mutated Mc4r alleles [52]. These examples highlight the complex genotype/phenotype relationships and the complex origins of Astyanax cavefish populations. Each cave seems to be a special case in terms of phenotype and combinations of alleles, and the possible scenario to explain allelic distributions in terms of origins, migrations and gene flows will necessitate further efforts in population biology and population genomics. Also, the impact of the specific environments found in each cave will have to be analysed, as the underestimated diversity in different caves (e.g. presence/absence of bats, predators or surface fish, trophic support, water entering by flooding or percolation, figure 1) may well contribute to the selection and fixation of different alleles, depending on local ecological conditions.
(f). Back to fieldwork and ecology of the species
Despite the fact that 30 cavefish caves are described and relatively accessible [2,63], few laboratory findings have been corroborated by experiments in the wild. Validating interpretations by assessing wild life cycle and cave ecology is important to avoid mis-/over-interpretations and to understand ecological diversity of caves, which may be related to genetic and behavioural differences (figure 1).
Subterranean habitats receive energy from external sources (bat colonies, insects, stream and percolating water charged with microfauna, particles and organic carbon) [2,64]. Food supplies vary depending on season and locality, hence the variable mud carbon contents in different caves [65]. Nevertheless, examination of wild cavefish gut contents containing bat guano, mud, plankton, insects and congener fragments in different seasons suggested well-nourished adults with non-specialized dietary regime in the Chica, Sabinos, Subterráneo and Pachón caves [1,66–68]. Pachón cavefish growth rate is comparable to their surface conspecifics [65] and healthy juveniles with digestive systems containing large numbers and varieties of micro-arthropods are found [1], suggesting excellent hunting skills in blind fry, like in laboratory conditions [69]. Subterráneo wild cavefish are more responsive to olfactory stimuli than eyed hybrids cohabiting in the cave [70], suggesting that olfactory skills described in the laboratory [29] are also at work in the natural environment. VAB or acoustic capabilities should be investigated as well in several caves, as food-finding skills may depend on cave ecosystems.
Laboratory and wild cavefish have higher fat content than surface fish (S Rétaux 2004--2018, personal observations). Cavefish metabolism is also modified at the circadian level. In wild Chica cavefish, but not in laboratory animals of the same origin, low expression levels of the oscillatory gene period1 are associated with abolished rhythmic functions, suggesting that ‘cavefish experience permanent light instead of perpetual darkness' [71]. Alterations of circadian rhythm are a common trait in cave animals [72] and might contribute to saving energy by lowering metabolic demand [73]. Finally, the hypothesis of a genetic limitation in body size (wild Pachón cavefish seem to have smaller maximal sizes than Tinaja and Subterráneo) to lower energy expenditure and food needs has yet to be investigated [65]. These studies on wild specimens enlighten the many modifications in complex functions that might have evolved in different cave populations.
Longitudinal investigations are also needed to better understand cavefish lifestyle. Cavefish breed easily in laboratories, yet little is known of their reproductive cycle and mating behaviour in the wild. In the Chica cave, Bridges described large females with eggs and small specimens that he estimated to be less than a month old, as reported in [66]. The dry season was suggested to correspond to the cavefish breeding period, which is consistent with the report of juveniles in Pachón in March 2016 [1]. If confirmed, this would suggest a ‘synchronized’ breeding season from South to North of the Sierra—different from the North America Amblyopsid cavefish, which breed after spring floods that introduce food into caves [74]. As laboratory spawning in Astyanax cavefish and surface fish is induced by variations in water temperature [12], following circadian and year-round climatic variations in caves will be crucial.
Despite the obvious difficulties of performing field experiments, moving towards a fully integrative and multi-disciplinary approach to better apprehend cavefish evolution requires further fieldwork. In particular, taking into account the specific ecological features of the different caves, or of the different ponds in a single cave, is essential. This would allow the establishment of mesocosm to study cavefish behaviour and lifestyle in more natural conditions, and also to perform more long-term artificial evolution studies on surface fish.
3. Conclusion
This review highlights the importance of considering cavefish evolution from multiple angles. Such an enterprise may sometimes appear long and difficult, but we believe it will turn out to be rewarding and insightful to understand the secrets of cavefish adaptations to their environment. The huge difference in environmental conditions experienced by surface and cave morphs, the existence of multiple, rapidly evolved cave populations, the variety of phenotypes exhibited by cavefish at morphological, physiological and behavioural levels, the experimental amenability exhibited by the species make Mexican tetras primary fish models to answer broader evolutionary questions. Major research directions will include deciphering the genetic and developmental evolutionary processes, including the type and number of genes involved, or whether coding or regulatory sequences are at play for morphological evolution. Fascinating questions also include the mechanisms of convergence, or how similar outcomes can result from different mechanisms, highlighting the power of ‘genetic tinkering’ in evolution [75].
Acknowledgements
We thank all past and present group members who contributed to an excellent scientific atmosphere and to the advancement of our research projects. We also thank Didier Casane and Luis Espinasa for fruitful long-term collaborations and scientific stimulation.
Data accessibility
This article has no additional data.
Authors' contributions
All four authors wrote the review.
Competing interests
We declare we have no competing interests.
Funding
Work in the group is supported by CNRS, FRM, UNADEV-AVIESAN, IDEEV and an Ecos-Nord Franco-Mexican exchange program.
References
- 1.Espinasa L, Bonaroti N, Wong J, Pottin K, Queinnec E, Rétaux S. 2017. Contrasting feeding habits of post-larval and adult Astyanax cavefish. Subterr. Biol. 21, 1–17. ( 10.3897/subtbiol.21.11046) [DOI] [Google Scholar]
- 2.Mitchell RW, Russell WH, Elliott WR. 1977. Mexican eyeless characin fishes, genus Astyanax: environment, distribution, and evolution. Spec. Publ. Mus. Texas Tech. Univ. 12, 1–89. [Google Scholar]
- 3.Wilkens H. 1988. Evolution and genetics of epigean and cave Astyanax fasciatus (Characidae, Pisces). Support for the neutral mutation theory. In Evolutionary biology, vol. 23 (eds Hecht MK, Wallace B), pp. 271–367. New York, NY: Plenum. [Google Scholar]
- 4.Fumey J, Hinaux H, Noirot C, Thermes C, Rétaux S, Casane D. 2018. Evidence for Late Pleistocene origin of Astyanax mexicanus cavefish. BMC Evol. Biol. 18, 43–62. ( 10.1186/s12862-018-1156-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hubbs C, Innes WT. 1936. The first known blind fish of the family Characidae: a new genus from Mexico . Occasional Papers of the Museum of Zoolology, University of Michigan 342, 1–7. [Google Scholar]
- 6.Borowsky R, Cohen D. 2013. Genomic consequences of ecological speciation in Astyanax cavefish. PLoS ONE 8, e79903 ( 10.1371/journal.pone.0079903) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernatchez L, et al. 2010. On the origin of species: insights from the ecological genomics of lake whitefish. Phil. Trans. R Soc. B 365, 1783–1800. ( 10.1098/rstb.2009.0274) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henning F, Meyer A. 2014. The evolutionary genomics of cichlid fishes: explosive speciation and adaptation in the postgenomic era. Annu. Rev. Genomics Hum. Genet. 15, 417–441. ( 10.1146/annurev-genom-090413-025412) [DOI] [PubMed] [Google Scholar]
- 9.Peichel CL, Marques DA. 2017. The genetic and molecular architecture of phenotypic diversity in sticklebacks. Phil. Trans. R. Soc. B 372, 20150486 ( 10.1098/rstb.2015.0486) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hinaux H, Poulain J, Da Silva C, Noirot C, Jeffery WR, Casane D, Rétaux S. 2013. De novo sequencing of Astyanax mexicanus surface fish and Pachon cavefish transcriptomes reveals enrichment of mutations in cavefish putative eye genes. PLoS ONE 8, e53553 ( 10.1371/journal.pone.0053553) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McGaugh SE, et al. 2014. The cavefish genome reveals candidate genes for eye loss. Nat. Commun. 5, 5307 ( 10.1038/ncomms6307) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elipot Y, Legendre L, Père S, Sohm F, Rétaux S. 2014. Astyanax transgenesis and husbandry: how cavefish enters the lab. Zebrafish 11, 291–299. ( 10.1089/zeb.2014.1005) [DOI] [PubMed] [Google Scholar]
- 13.Ma L, Jeffery WR, Essner JJ, Kowalko JE. 2015. Genome editing using TALENs in blind Mexican cavefish, Astyanax mexicanus. PLoS ONE 10, e0119370 ( 10.1371/journal.pone.0119370) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Avise JC, Selander RK. 1972. Evolutionary genetics of cave-dwelling fishes of the genus Astyanax. Evolution 26, 1–19. ( 10.1111/j.1558-5646.1972.tb00170.x) [DOI] [PubMed] [Google Scholar]
- 15.Casane D, Rétaux S. 2016. Evolutionary genetics of the cavefish Astyanax mexicanus. Adv. Genet. 95, 117–159. ( 10.1016/bs.adgen.2016.03.001) [DOI] [PubMed] [Google Scholar]
- 16.Keene AC, Yoshizawa M, McGaugh SE (eds). 2016. Biology and evolution of the Mexican cavefish. San Diego, CA: Academic Press. [Google Scholar]
- 17.Wilkens H, Strecker U. 2017. Evolution in the dark. Darwin's loss without selection. Berlin, Germany: Springer. [Google Scholar]
- 18.Chan YF, et al. 2010. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science 327, 302–305. ( 10.1126/science.1182213) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peichel CL. 2005. Fishing for the secrets of vertebrate evolution in threespine sticklebacks. Dev. Dyn. 234, 815–823. ( 10.1002/dvdy.20564) [DOI] [PubMed] [Google Scholar]
- 20.Shapiro MD, Bell MA, Kingsley DM. 2006. Parallel genetic origins of pelvic reduction in vertebrates. Proc. Natl Acad. Sci. USA 103, 13 753–13 758. ( 10.1073/pnas.0604706103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shapiro MD, Marks ME, Peichel CL, Blackman BK, Nereng KS, Jonsson B, Schluter D, Kingsley DM. 2004. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature 428, 717–723. ( 10.1038/nature02415) [DOI] [PubMed] [Google Scholar]
- 22.Borowsky R. 2008. Restoring sight in blind cavefish. Curr. Biol. 18, R23–R24. ( 10.1016/j.cub.2007.11.023) [DOI] [PubMed] [Google Scholar]
- 23.Wilkens H, Strecker U. 2003. Convergent evolution of the cavefish Astyanax (Characidae. Teleostei): genetic evidence from reduced eye-size and pigmentation. Biol. J. Linn. Soc. 80, 545–554. ( 10.1111/j.1095-8312.2003.00230.x) [DOI] [Google Scholar]
- 24.Protas M, Conrad M, Gross JB, Tabin C, Borowsky R. 2007. Regressive evolution in the Mexican cave tetra, Astyanax mexicanus. Curr. Biol. 17, 452–454. ( 10.1016/j.cub.2007.01.051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Quin KE, Yoshizawa M, Doshi P, Jeffery WR. 2013. Quantitative genetic analysis of retinal degeneration in the blind cavefish Astyanax mexicanus. PLoS ONE 8, e57281 ( 10.1371/journal.pone.0057281) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamamoto Y, Jeffery WR. 2000. Central role for the lens in cave fish eye degeneration. Science 289, 631–633. ( 10.1126/science.289.5479.631) [DOI] [PubMed] [Google Scholar]
- 27.Yamamoto Y, Stock DW, Jeffery WR. 2004. Hedgehog signalling controls eye degeneration in blind cavefish. Nature 431, 844–847. ( 10.1038/nature02864) [DOI] [PubMed] [Google Scholar]
- 28.Yamamoto Y, Byerly MS, Jackman WR, Jeffery WR. 2009. Pleiotropic functions of embryonic sonic hedgehog expression link jaw and taste bud amplification with eye loss during cavefish evolution. Dev. Biol. 330, 200–211. ( 10.1016/j.ydbio.2009.03.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hinaux H, Devos L, Bibliowicz J, Elipot Y, Alié A, Blin M, Rétaux S. 2016. Sensory evolution in blind cavefish is driven by early events during gastrulation and neurulation. Development 143, 4521–4532. ( 10.1242/dev.141291) [DOI] [PubMed] [Google Scholar]
- 30.Pottin K, Hinaux H, Rétaux S. 2011. Restoring eye size in Astyanax mexicanus blind cavefish embryos through modulation of the Shh and Fgf8 forebrain organising centres. Development 138, 2467–2476. ( 10.1242/dev.054106) [DOI] [PubMed] [Google Scholar]
- 31.Alie A, Devos L, Torres-Paz J, Prunier L, Boulet F, Blin M, Elipot Y, Retaux S. 2018. Developmental evolution of the forebrain in cavefish, from natural variations in neuropeptides to behavior. eLife 7, e32808 ( 10.7554/eLife.32808) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elipot Y, Hinaux H, Callebert J, Rétaux S. 2013. Evolutionary shift from fighting to foraging in blind cavefish through changes in the serotonin network. Curr. Biol. 23, 1–10. ( 10.1016/j.cub.2012.10.044) [DOI] [PubMed] [Google Scholar]
- 33.Hinaux H, Blin M, Fumey J, Legendre L, Heuze A, Casane D, Rétaux S. 2015. Lens defects in Astyanax mexicanus cavefish: evolution of crystallins and a role for alphaA-crystallin. Dev. Neurobiol. 75, 505–521. ( 10.1002/dneu.22239) [DOI] [PubMed] [Google Scholar]
- 34.Rétaux S, Casane D. 2013. Evolution of eye development in the darkness of caves: adaptation, drift, or both? Evodevo 4, 26 ( 10.1186/2041-9139-4-26) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shumway CA. 2008. Habitat complexity, brain, and behavior. Brain Behav. Evol. 72, 123–134. ( 10.1159/000151472) [DOI] [PubMed] [Google Scholar]
- 36.Striedter GF. 2004. Principles of brain evolution. Oxford, UK: Oxford University Press. [Google Scholar]
- 37.Blin M, Tine E, Meister L, Elipot Y, Bibliowicz J, Espinasa L, Retaux S. 2018. Developmental evolution and developmental plasticity of the olfactory epithelium and olfactory skills in Mexican cavefish. Dev. Biol. S0012-1606(18)30089-7. ( 10.1016/j.ydbio.2018.04.019) [DOI] [PubMed] [Google Scholar]
- 38.Yoshizawa M, Goricki S, Soares D, Jeffery WR. 2010. Evolution of a behavioral shift mediated by superficial neuromasts helps cavefish find food in darkness. Curr. Biol. 20, 1631–1636. ( 10.1016/j.cub.2010.07.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshizawa M, Yamamoto Y, O'Quin KE, Jeffery WR. 2012. Evolution of an adaptive behavior and its sensory receptors promotes eye regression in blind cavefish. BMC Biol. 10, 108 ( 10.1186/1741-7007-10-108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeffery W, Strickler A, Guiney S, Heyser D, Tomarev S. 2000. Prox1 in eye degeneration and sensory organ compensation during development and evolution of the cavefish Astyanax. Dev. Genes Evol. 210, 223–230. ( 10.1007/s004270050308) [DOI] [PubMed] [Google Scholar]
- 41.Duboue ER, Keene AC, Borowsky RL. 2011. Evolutionary convergence on sleep loss in cavefish populations. Curr. Biol. 21, 671–676. ( 10.1016/j.cub.2011.03.020) [DOI] [PubMed] [Google Scholar]
- 42.Yoshizawa M, Robinson BG, Duboue ER, Masek P, Jaggard JB, O'Quin KE, Borowsky RL, Jeffery WR, Keene AC. 2015. Distinct genetic architecture underlies the emergence of sleep loss and prey-seeking behavior in the Mexican cavefish. BMC Biol. 13, 15 ( 10.1186/s12915-015-0119-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siegel JM. 2009. Sleep viewed as a state of adaptive inactivity. Nat. Rev. Neurosci. 10, 747–753. ( 10.1038/nrn2697) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dewasmes G, Duchamp C, Minaire Y. 1989. Sleep changes in fasting rats. Physiol. Behav. 46, 179–184. ( 10.1016/0031-9384(89)90252-7) [DOI] [PubMed] [Google Scholar]
- 45.Velluti RA. 1997. Interactions between sleep and sensory physiology. J. Sleep Res. 6, 61–77. ( 10.1046/j.1365-2869.1997.00031.x) [DOI] [PubMed] [Google Scholar]
- 46.Jaggard J, Robinson BG, Stahl BA, Oh I, Masek P, Yoshizawa M, Keene AC. 2017. The lateral line confers evolutionarily derived sleep loss in the Mexican cavefish. J. Exp. Biol. 220, 284–293. ( 10.1242/jeb.145128) [DOI] [PubMed] [Google Scholar]
- 47.Ohno K, Sakurai T. 2008. Orexin neuronal circuitry: role in the regulation of sleep and wakefulness. Front. Neuroendocrinol. 29, 70–87. ( 10.1016/j.yfrne.2007.08.001) [DOI] [PubMed] [Google Scholar]
- 48.Prober DA, Rihel J, Onah AA, Sung RJ, Schier AF. 2006. Hypocretin/orexin overexpression induces an insomnia-like phenotype in zebrafish. J. Neurosci. 26, 13 400–13 410. ( 10.1523/JNEUROSCI.4332-06.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jaggard JB, Stahl BA, Lloyd E, Prober DA, Duboue ER, Keene AC. 2018. Hypocretin underlies the evolution of sleep loss in the Mexican cavefish. eLife 7, e32637 ( 10.7554/eLife.32637) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kowalko JE, et al. 2013. Loss of schooling behavior in cavefish through sight-dependent and sight-independent mechanisms. Curr. Biol. 23, 1874–1883. ( 10.1016/j.cub.2013.07.056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kowalko JE, Rohner N, Linden TA, Rompani SB, Warren WC, Borowsky R, Tabin CJ, Jeffery WR, Yoshizawa M. 2013. Convergence in feeding posture occurs through different genetic loci in independently evolved cave populations of Astyanax mexicanus. Proc. Natl Acad. Sci. USA 110, 16 933–16 938. ( 10.1073/pnas.1317192110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aspiras AC, Rohner N, Martineau B, Borowsky RL, Tabin CJ. 2015. Melanocortin 4 receptor mutations contribute to the adaptation of cavefish to nutrient-poor conditions. Proc. Natl Acad. Sci. USA 112, 9668–9673. ( 10.1073/pnas.1510802112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Elipot Y, Hinaux H, Callebert J, Launay J, Blin M, Rétaux S. 2014. A mutation in the enzyme monoamine oxidase explains part of the Astyanax cavefish behavioral syndrome. Nat. Commun. 5, 3647 ( 10.1038/ncomms4647) [DOI] [PubMed] [Google Scholar]
- 54.Marques DA, Lucek K, Meier JI, Mwaiko S, Wagner CE, Excoffier L, Seehausen O. 2016. Genomics of rapid incipient speciation in sympatric threespine stickleback. PLoS Genet. 12, e1005887 ( 10.1371/journal.pgen.1005887) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Terekhanova NV, Logacheva MD, Penin AA, Neretina TV, Barmintseva AE, Bazykin GA, Kondrashov AS, Mugue NS. 2014. Fast evolution from precast bricks: genomics of young freshwater populations of threespine stickleback Gasterosteus aculeatus. PLoS Genet. 10, e1004696 ( 10.1371/journal.pgen.1004696) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yokoyama S, Meany A, Wilkens H, Yokoyama R. 1995. Initial mutational steps toward loss of opsin gene function in cavefish. Mol. Biol. Evol. 12, 527–532. ( 10.1093/oxfordjournals.molbev.a040190) [DOI] [PubMed] [Google Scholar]
- 57.Kim EB, et al. 2011. Genome sequencing reveals insights into physiology and longevity of the naked mole rat. Nature 479, 223–227. ( 10.1038/nature10533) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rubin CJ, et al. 2012. Strong signatures of selection in the domestic pig genome. Proc. Natl Acad. Sci. USA 109, 19 529–19 536. ( 10.1073/pnas.1217149109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Protas ME, Hersey C, Kochanek D, Zhou Y, Wilkens H, Jeffery WR, Zon LI, Borowsky R, Tabin CJ. 2006. Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism. Nat. Genet. 38, 107–111. ( 10.1038/ng1700) [DOI] [PubMed] [Google Scholar]
- 60.Yoshizawa M, Ashida G, Jeffery WR. 2012. Parental genetic effects in a cavefish adaptive behavior explain disparity between nuclear and mitochondrial DNA. Evolution 66, 2975–2982. ( 10.1111/j.1558-5646.2012.01651.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Burchards H, Dölle A, Parzefall J. 1985. Aggressive behavior of an epigean population of Astyanax mexicanus and some observations on three subterranean populations. Behav. Processes 11, 225–235. ( 10.1016/0376-6357(85)90017-8) [DOI] [PubMed] [Google Scholar]
- 62.Espinasa L, Yamamoto Y, Jeffery WR. 2005. Non-optical releasers for aggressive behavior in blind and blinded Astyanax (Teleostei. Characidae). Behav. Processes 70, 144–148. ( 10.1016/j.beproc.2005.06.003) [DOI] [PubMed] [Google Scholar]
- 63.Elliott WR. 2016. Cave exploration and mapping in the Sierra de El Abra region. In Biology and evolution of the Mexican cavefish. (eds Keen AC, Yoshizawa M, McGaugh SE), pp. 9–40, Ch. 1. Academic Press. [Google Scholar]
- 64.Culver DC, Pipan T. 2009. The biology of caves and other subterranean habitats. Oxford, UK: Oxford University Press. [Google Scholar]
- 65.Simon V, Elleboode R, Mahe K, Legendre L, Ornelas-Garcia P, Espinasa L, Retaux S. 2017. Comparing growth in surface and cave morphs of the species Astyanax mexicanus: insights from scales. Evodevo 8, 23 ( 10.1186/s13227-017-0086-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Breder CM. 1942. Descriptive ecology of La Cueva Chica, with especial reference to the blind fish, Anoptichthys. Zoologica 27, 7–15. [Google Scholar]
- 67.Tafall BFO. 1943. Observaciones sobre la fauna acuatica de las cuevas de la región de Valles, San Luís Potosí (Mexico). Rev. Soc. Mex. Hist. Nat. 4, 43–71. [Google Scholar]
- 68.Wilkens H, Burns RJ. 1972. A new Anoptichthys cave population (Characidae. Pisces). Ann. Speleol. 27, 263–270. [Google Scholar]
- 69.Espinasa L, Bibliowicz J, Jeffery WR, Rétaux S. 2014. Enhanced prey capture skills in Astyanax cavefish larvae are independent from eye loss. Evodevo 5, 35 ( 10.1186/2041-9139-5-35) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bibliowicz J, Alie A, Espinasa L, Yoshizawa M, Blin M, Hinaux H, Legendre L, Pere S, Rétaux S. 2013. Differences in chemosensory response between eyed and eyeless Astyanax mexicanus of the Rio Subterraneo cave. Evodevo 4, 25 ( 10.1186/2041-9139-4-25) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Beale A, Guibal C, Tamai TK, Klotz L, Cowen S, Peyric E, Reynoso VH, Yamamoto Y, Whitmore D. 2013. Circadian rhythms in Mexican blind cavefish Astyanax mexicanus in the lab and in the field. Nat. Commun. 4, 2769 ( 10.1038/ncomms3769) [DOI] [PubMed] [Google Scholar]
- 72.Cavallari N, et al. 2011. A blind circadian clock in cavefish reveals that opsins mediate peripheral clock photoreception. PLoS Biol. 9, e1001142 ( 10.1371/journal.pbio.1001142) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moran D, Softley R, Warrant EJ. 2014. Eyeless Mexican cavefish save energy by eliminating the circadian rhythm in metabolism. PLoS ONE 9, e107877 ( 10.1371/journal.pone.0107877) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Niemiller ML, Poulson TL. 2010. Chapter 7: Subterranean fishes of North America: Amblyopsidae. In The biology of subterranean fishes (eds Trajano E, Bichuette ME, Kapoor BG), pp. 169–280. Boca Raton, FL: CRC Press. [Google Scholar]
- 75.Jacob F. 1977. Evolution and tinkering. Science 196, 1161–1166. ( 10.1126/science.860134) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.