To the Editor,
Chronic myelomonocytic leukemia (CMML) is a clonal hematopoietic stem cell disorder characterized by sustained peripheral blood (PB) monocytosis, bone marrow (BM) dysplasia, and an inherent risk for transformation to acute myeloid leukemia (AML); ~20–30% over 3–5 years1,2. Patients with CMML have ~10–15 gene mutations in coding DNA regions, with common mutations involving TET2 (~60%), ASXL1 (~40%), SRSF2 (~40%), and the oncogenic RAS pathway (~30%)3. Blastic plasmacytoid dendritic cell neoplasm (BPDCN) is a rare hematodermic malignancy of dendritic cell origin that is known to infiltrate the blood, BM, skin, and lymph nodes; a diagnosis of which is based on a blast immunophenotype characterized by the expression of CD4, CD43, CD56, CD123, BDCA-2/CD303, TCL1, and CTLA1. Cytogenetic studies, array-based comparative genomic hybridization, and gene expression profiling have demonstrated recurrent inactivation/losses/decreased expression of RB1 (retinoblastoma 1; 13q13-q21), LATS2, CDKN1B, CDKN2A, and TP53 genes,4 while next-generation sequencing (NGS) studies have demonstrated point mutations involving TET2 (~36%), ASXL1 (~32%), NRAS (~30%), ATM (21%), along with deletions involving RB1 and APC (~6%)5,6. In addition, recurrent MYB gene rearrangements have been documented in both children and adults, with fusion oncogenes including MYB-ZFAT, MYB-PLEKHO1, and MYB-DCPS7. Common clonal origins for CMML and BPDCN have been suggested based on similar genetic and epigenetic deregulations identified, along with sporadic case reports of CMML transformation to BPDCN8,9. We provide the first in-depth whole-exome sequencing (WES) results on a patient with CMML that transformed to BPDCN and provide information with regards to possible mechanisms of transformation.
A 61-year-old male was referred to the hematology clinic in 2014 for sustained PB monocytosis. A BM biopsy was suggestive of CMML-0 (World Health Organization 2016 criteria) with normal cytogenetics (Fig. 1). He did have plasmacytoid dendritic cell nodules comprising ~20% of the BM cellularity. NGS at diagnosis identified mutations involving TET2 (variant allele frequency (VAF) 40%) and SRSF2 (43%). He was conservatively managed with routine blood count checks. In 2017, he presented with worsening constitutional symptoms, a generalized skin rash, and progressive cytopenias. A BM biopsy and skin biopsy were suggestive of BPDCN with normal BM metaphase cytogenetics (Fig. 1). He was treated with AML-like induction chemotherapy using idarubicin and cytarabine and then received salvage chemotherapy with fludarabine, idarubicin, and cytarabine, but was found to have persistent disease and died secondary to infectious complications. BM DNA was available at the time of CMML diagnosis and at transformation to BPDCN. In addition, sorted T lymphocytes at CMML diagnosis were isolated and cultured to serve as a potential germline control for WES.
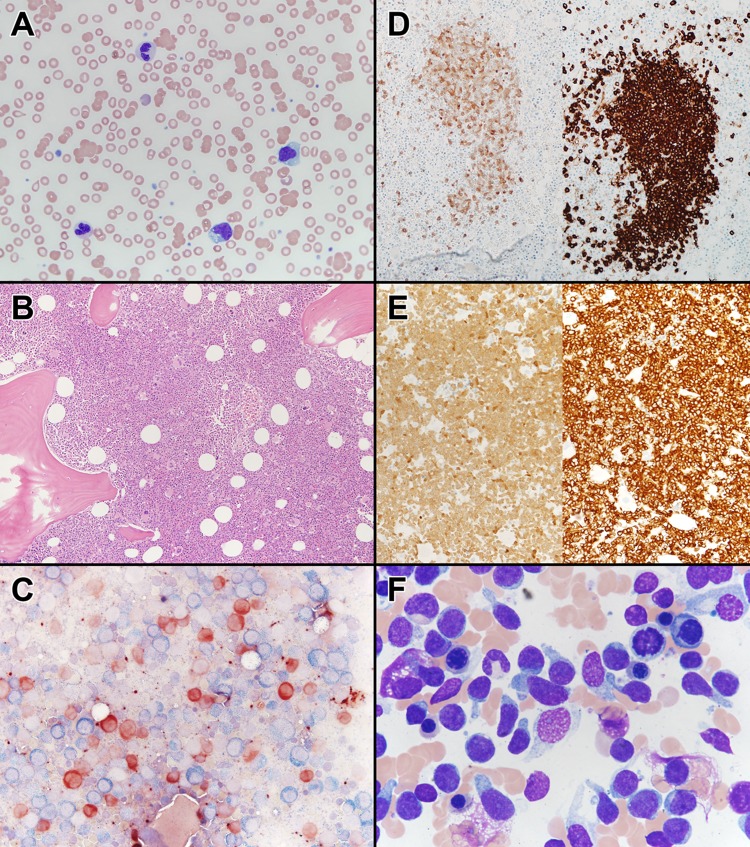
Fig. 1.
a Peripheral blood smear of the patient with a diagnosis of chronic myelomonocytic leukemia, demonstrating atypical dysplastic granulocytes, atypical monocytes, and a blast. Wright Giemsa stain, ×400 magnification. b Bone marrow core biopsy of the patient with chronic myelomonocytic leukemia, demonstrating a hypercellular marrow (90%), with dysplastic megakaryocytes. Hematoxylin and eosin, ×100 magnification. c Butyrate esterase (brown) and chloroacetate esterase (blue) cytochemical dual stain, demonstrating increased butyrate esterase monocytes and dual esterase-positive bone marrow monocytes, suggestive of monocytic dysplasia (×400 magnification). d (Left) Bone marrow core biopsy, TCL1 immunohistochemistry, demonstrating TCL1-positive plasmacytoid dendritic cell nodules (×200 magnification). (Right) Bone marrow core biopsy, CD123 immunohistochemistry, demonstrating CD123-positive plasmacytoid dendritic cell nodules (×200 magnification). e (Left) Bone marrow biopsy, TCL1 immunohistochemistry, demonstrating diffusely TCL1-positive blasts, suggestive of a blastic plasmacytoid dendritic cell neoplasm (×100 magnification). (Right) Bone marrow core biopsy, CD123 immunohistochemistry, demonstrating diffusely positive CD123 blasts, suggestive of a blastic plasmacytoid dendritic cell neoplasm (×100 magnification). f Bone marrow aspirate obtained at the time of disease transformation demonstrating hand mirror-shaped blastic cells, characteristic for blastic plasmacytoid dendritic cell neoplasms. Wright Giemsa, ×1000 magnification
Two hundred nanogram of genomic DNA was sheared with the Covaris S2 system (LGC Genomics/KBioscience). DNA fragments were end repaired, extended with an “A” base on the 3′ end, ligated with paired-end adaptors with the Bravo Platform (Agilent), and amplified (six cycles). Exome-containing adaptor-ligated libraries were hybridized for 40 h with biotinylated oligo RNA baits, and enriched with streptavidin-conjugated magnetic beads using SureSelect Clinical Research (Agilent). The final libraries were indexed, pooled, and sequenced on Illumina HiSeq-2000 sequencer at the Institute Gustave Roussy (Paris, France). Raw reads in FASTQ format from each exome sequencing lane were aligned to the reference human genome (Hg19) using Burrows Wheeler-Alignment. Aligned reads were processed and sorted with SAMtools and PCR duplicates were removed. Nucleotide variants (single-nucleotide polymorphisms and indels) were called with VarScan and all variants with a Phred-based quality score <30.0 were called low quality and ignored. On an average, 369 million reads were sequenced per sample. A formal copy number variation (CNV) analysis was carried out on the WES data, both at CMML diagnosis and at BPDCN transformation, using the FACETS analysis software (https://sites.google.com/site/mskfacets).
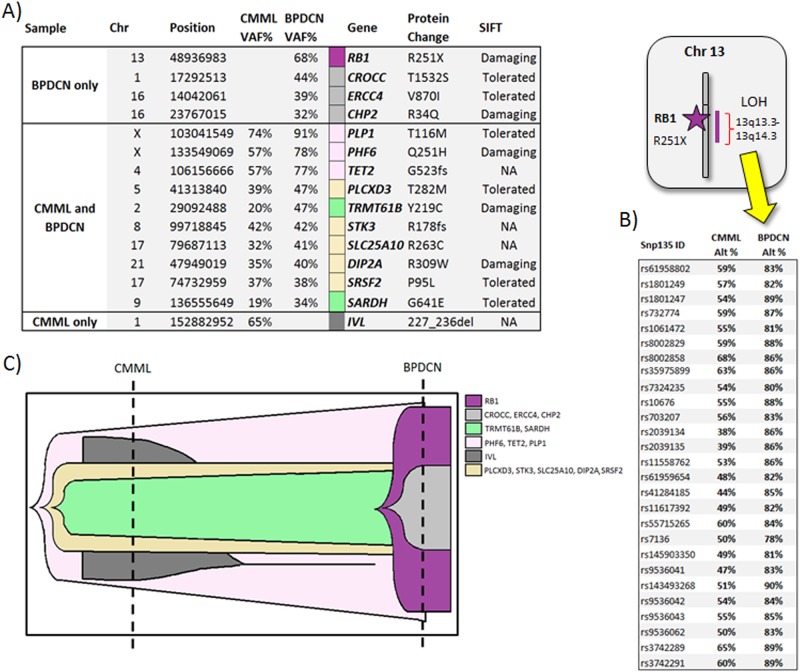
WES results on the DNA specimen obtained at CMML diagnosis identified the following pathogenic variants TET2 (c.1567_1568insA, p.G523Efs*44; VAF 57%), SRSF2 (c.284C>T, p.P95L; VAF 37%), PHF6 (c.753G>T, p.Q251H; VAF 57%), PLCXD3 (c.845C>T, p.T282M, VAF 39%), TRMT61B (c.656A>G, p.T219C, VAF 20%), STK3 (c.618del, p.N207Ifs*3, VAF 42%), SLC25A10 (c.787C>T, R263C, VAF 32%), DIP2A (c.1117C>T, R373W, VAF 35%), SARDH (c.1922G>A, p.G641E, VAF 19%), PLP1 (c.347C>T, T116M, VAF 74%), and IVL (c.706_735dup, P236_L245dup) (Fig. 2). At the time of BPDCN transformation, WES demonstrated acquisition of the following mutations: RB1 (c.751C>T, p.R251*; VAF 68% secondary to loss of heterozygosity of 13.q13-24), CROCC (c.4595C>G, T1532S, VAF 44%), ERCC4 (c.2608G>A, p.V870I, VAF 38%), and CHP2 (c.101G>A, R34Q, VAF 32%), while mutations involving the following genes were no longer detected, IVL and RFPL1. An increase in the VAF burdens in the following genes was also encountered at the time of BPDCN transformation, PHF6 (57–78%), TET2 (57–77%), PLCXD3 (39–43%), TRMT61B (20–40%), SLC25A10 (32–41%), DIP2A (35–40%), SARDH (19–34%), and PLP1 (74–91%). A formal CNV analysis also confirmed loss of heterozygosity of chromosome 13, involving the RB1 gene locus (supplemental figure).
Fig. 2.
Paired and germline whole-exome sequencing data obtained at the time of chronic myelomonocytic leukemia (CMML) diagnosis and at CMML transformation to a blastic plasmacytoid dendritic cell neoplasm (BPDCN). a Paired and germline whole-exome sequencing data obtained at the time of CMML diagnosis and at CMML transformation to BPDCN, demonstrating the biallelic inactivation of the RB1 gene. b Whole-exome sequencing data from chromosome 13, demonstrating biallelic inactivation of the RB1 gene secondary to loss of heterozygosity and the acquisition of a nonsense RB1 gene mutation (c.751C>T, p.R251*). c SciClone analysis demonstrating the clonal architecture and mutational evolution spectrum in a patient, at CMML diagnosis and at CMML transformation to BPDCN
CMML blast transformation morphologically almost always results in an AML phenotype and occurs due to the clonal acquisition of cytogenetic and molecular abnormalities2,3,10. In a large, two-center study of 171 patients with blast phase CMML, all patients met the morphological criteria for a diagnosis of AML (secondary AML) and cytogenetic clonal evolution was seen in 24% of patients, while molecular clonal evolution was seen in 50% of patients2. Blast transformation from CMML to BPDCN is an extremely rare event and is usually associated with CNVs and unique molecular changes9. In our patient, the main genetic event identified at BPDCN transformation was a truncating mutation in RB1, along with loss of heterozygosity of the RB1 gene, resulting in biallelic inactivation of RB1 (Fig. 2). The RB1 gene encodes a protein with tumor suppressor function that serves as a G1 checkpoint inhibitor (by inhibiting E2F transcription factors), a regulator of apoptosis and helps maintain permanent cell cycle arrest and chromosomal stability11. In addition, RB1 also acts as a transcription cofactor and an adaptor protein promoting the function of critical transcription factors11. Loss of RB1 function is associated with progression of human cancers via loss of cellular differentiation and chromosomal instability11. While RB1 mutations and deletions are common in BPDCN, they are infrequent in CMML or secondary AML arising from CMML, supporting our hypothesis that in our patient, biallelic loss of RB1 maybe a major contributor of a BPDCN phenotypic transformation3. Additional genetic events acquired at the time of BPDCN transformation included mutations involving ERCC4 (Excision Repair 1, Endonuclease Non-Catalytic Subunit) and CHP2 (Calcineurin B Homologous Protein 2). While these genes have important physiological functions and have been implicated in oncogenesis, their contribution to BPDCN transformation needs further elucidation. Increase in mutational frequencies of genes identified at CMML diagnosis, including TET2 and PHF6 are consistent with the process of clonal evolution and disease transformation. Interestingly, the VAF burden of SRSF2, a gene regulating pre-mRNA splicing, remained the same at CMML diagnosis and at BPDCN transformation12.
By performing WES in paired samples and in the germline of a single patient with BPDCN transformation from an underlying CMML, we demonstrate (i) the common clonal origins of CMML and BPDCN, justifying the current inclusion of BPDCN as a myeloid neoplasm, (ii) the fact that the blast phenotype at CMML transformation is heavily dependent on the nature of genetic changes that occur at blast transformation, iii) the importance of the RB1 gene in BPDCN morphology and oncogenesis, and (iv) the role of clonal acquisitions and losses and increases in existing mutational allele burdens in disease progression/blast transformation.
Electronic supplementary material
Acknowledgements
Current publication is supported in part by grants from the “The Gerstner Family Career Development Award” and the Mayo Clinic Center for Individualized Medicine, Mayo Clinic, Rochester, MN, USA.” This publication was supported by CTSA Grant Number KL2 TR000136 from the National Center for Advancing Translational Science (NCATS). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41408-018-0120-5).
References
- 1.Arber DA, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–2405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 2.Patnaik M. M. et al. Blast phase chronic myelomonocytic leukemia: Mayo-MDACC Collaborative Study of 171 cases. Leukemia 2018 10.1038/s41375-018-0143-8. [DOI] [PMC free article] [PubMed]
- 3.Merlevede J, et al. Mutation allele burden remains unchanged in chronic myelomonocytic leukaemia responding to hypomethylating agents. Nat. Commun. 2016;7:10767. doi: 10.1038/ncomms10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leroux D, et al. CD4(+), CD56(+) DC2 acute leukemia is characterized by recurrent clonal chromosomal changes affecting 6 major targets: a study of 21 cases by the Groupe Francais de Cytogenetique Hematologique. Blood. 2002;99:4154–4159. doi: 10.1182/blood.V99.11.4154. [DOI] [PubMed] [Google Scholar]
- 5.Menezes J, et al. Exome sequencing reveals novel and recurrent mutations with clinical impact in blastic plasmacytoid dendritic cell neoplasm. Leukemia. 2014;28:823–829. doi: 10.1038/leu.2013.283. [DOI] [PubMed] [Google Scholar]
- 6.Stenzinger A, et al. Targeted ultra-deep sequencing reveals recurrent and mutually exclusive mutations of cancer genes in blastic plasmacytoid dendritic cell neoplasm. Oncotarget. 2014;5:6404–6413. doi: 10.18632/oncotarget.2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki K, et al. Recurrent MYB rearrangement in blastic plasmacytoid dendritic cell neoplasm. Leukemia. 2017;31:1629–1633. doi: 10.1038/leu.2017.101. [DOI] [PubMed] [Google Scholar]
- 8.Brunetti L, et al. Blastic plasmacytoid dendritic cell neoplasm and chronic myelomonocytic leukemia: a shared clonal origin. Leukemia. 2017;31:1238–1240. doi: 10.1038/leu.2017.38. [DOI] [PubMed] [Google Scholar]
- 9.Facchetti F, et al. Neoplasms derived from plasmacytoid dendritic cells. Mod. Pathol. 2016;29:98–111. doi: 10.1038/modpathol.2015.145. [DOI] [PubMed] [Google Scholar]
- 10.Patnaik MM, et al. Blast transformation in chronic myelomonocytic leukemia: risk factors, genetic features, survival, and treatment outcome. Am. J. Hematol. 2015;90:411–416. doi: 10.1002/ajh.23962. [DOI] [PubMed] [Google Scholar]
- 11.Burkhart DL, Sage J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat. Rev. Cancer. 2008;8:671–682. doi: 10.1038/nrc2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patnaik MM, et al. Spliceosome mutations involving SRSF2, SF3B1, and U2AF35 in chronic myelomonocytic leukemia: prevalence, clinical correlates, and prognostic relevance. Am. J. Hematol. 2013;88:201–206. doi: 10.1002/ajh.23373. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.