Abstract
Chemotherapeutic drugs have a common intent to activate apoptosis in tumor cells. However, master regulators of apoptosis (e.g., p53, p16/CDKN2A) are frequently genetically inactivated in cancers, resulting in multidrug resistance. An alternative, p53-independent method for terminating malignant proliferation is to engage terminal-differentiation. Normally, the exponential proliferation of lineage-committed progenitors, coordinated by the master transcription factor (TF) MYC, is self-limited by forward-differentiation to terminal lineage-fates. In cancers, however, this exponential proliferation is disengaged from terminal-differentiation. The mechanisms underlying this decoupling are mostly unknown. We performed a systematic review of published literature (January 2007–June 2018) to identify gene pathways linked to differentiation-failure in three treatment-recalcitrant cancers: hepatocellular carcinoma (HCC), ovarian cancer (OVC), and pancreatic ductal adenocarcinoma (PDAC). We analyzed key gene alterations in various apoptosis, proliferation and differentiation pathways to determine whether it is possible to predict treatment outcomes and suggest novel therapies. Poorly differentiated tumors were linked to poorer survival across histologies. Our analyses suggested loss-of-function events to master TF drivers of lineage-fates and their cofactors as being linked to differentiation-failure: genomic data in TCGA and ICGC databases demonstrated frequent haploinsufficiency of lineage master TFs (e.g., GATA4/6) in poorly differentiated tumors; the coactivators that these TFs use to activate genes (e.g. ARID1A, PBRM1) were also frequently inactivated by genetic mutation and/or deletion. By contrast, corepressor components (e.g., DNMT1, EED, UHRF1, and BAZ1A/B), that oppose coactivators to repress or turn off genes, were frequently amplified instead, and the level of amplification was highest in poorly differentiated lesions. This selection by neoplastic evolution towards unbalanced activity of transcriptional corepressors suggests these enzymes as candidate targets for inhibition aiming to re-engage forward-differentiation. This notion is supported by both pre-clinical and clinical trial literature.
Facts
Treatment outcomes for most disseminated p53 mutant solid tumors are poor.
The most lethal of these tumors are morphologically poorly differentiated.
Differentiation-restoring treatments are an emerging non-toxic, p53-independent treatment alternative.
Advances in omics big data can be used to define molecular targets for differentiation-restoring therapy.
Pharmacological inhibition of corepressor enzymes re-engages suppressed differentiation pathways.
Open questions
Which among the multiple corepressors identified in cells are rational molecular targets for pharmacologic manipulation?
What is the in vivo evidence of how corepressor inhibiting therapy triggers terminal-differentiation?
What are the key MYC-antagonists in any given lineage that mediate cell cycle exits by terminal-differentiation?
Introduction
Conventional chemotherapy aims to activate apoptosis even in tumors where master regulators of apoptosis are physically unavailable through inactivating gene mutations, leading to multi-drug resistance1. Therefore, alternative targets and pathways of therapy are needed. The complex process of cellular proliferation is coordinated by the master transcription factor (TF) MYC. Master TF that drive commitment into a lineage cooperate with MYC to drive exponential proliferation, but simultaneously drive forward-differentiation that culminates in cell cycle exits2–4. Malignant exponential self-replication involves decoupling of proliferation from this forward-differentiation5–7. One way of approaching the mechanisms underlying such decoupling is to consider three major modules in multi-cellular cell physiology: (i) proliferation or replication; (ii) apoptosis; (iii) lineage-differentiation7. Cell proliferation, the essence of all cancers, is coordinated by the master TF MYC2,8,9—amplification of the MYC gene, and activation of MYC or its paralogues is seen across all human malignancies10. The master regulator of cellular apoptosis p53 (TP53) or its key cofactors are almost universally inactivated in human malignancies11—while multiple copies of TP53 protect elephants from a high rate of cancer proportionate to their high cellular mass12, TP53 mutations can be viewed as “the elephant in the room” of all cancer therapy, since most treatments intend to activate this master regulator which ironically is absent from most cancers. The mechanisms underlying impeded lineage-differentiation in cancer are still mostly opaque and require wider mechanistic characterization to allow development of therapeutic interventions aiming to restore lineage-fates.
Differentiation-failure is used to distinguish malignant from benign tumors13, and the degree of differentiation-failure separates high from low aggressive transformations, e.g., pancreatic intraepithelial neoplasia (PanIN) lesions from pancreatic ductal adenocarcinoma (PDAC). Loss-of-differentiation may not be obvious by light microscopy, but can be detected using gene expression analysis of differentiation factors6. Tissue differentiation is usefully considered in three compartments: (1) tissue stem cells—cells with an intrinsically low proliferation rate but capable of self-renewal and of giving rise to daughter cells committed into various tissue-lineages14–16; (2) lineage-committed progenitors—cells with high levels of MYC activity17–20 and exponential proliferation that is coupled with forward differentiation towards lineage-fates; and (3) terminally differentiated cells that have transitioned from exponential proliferation to a focus on performing specialized tissue functions21–23. Each stage of differentiation is regulated by key master TFs—stem cell TFs, lineage-progenitor TFs and terminal-differentiation TFs. The purpose of this review and study is to define differentiation related molecular targets that can be used in the development of p53 independent therapies that are not toxic toward normal stem cells and that do not alter normal stem cell replication.
Methods
We conducted a systematic review in accordance to PRISMA guidelines (Fig. S1)24. We searched the national library of medicine through PubMed for literature containing cell proliferation, apoptosis, and differentiation in cancer (Fig. S1). Search terms included chemotherapy, cancer apoptosis, proliferation, and cell differentiation. We also searched the work cited in the identified articles for additional relevant literature. We then focused on three therapy-resistant cancers: Hepatocellular carcinoma (HCC), Ovarian cancer (OVC), and pancreatic ductal adenocarcinoma (PDAC). Additional terms of phase 1, 2, and 3 randomized clinical trials were searched in Web Science, Pubmed/MEDLINE, Embase, ClinicalTrials.gov and Google Scholar. The search strategy included studies published in English language from January 2007 to June 2018.
We then analyzed gene datasets from The Cancer Genome Atlas (TCGA) (https://cancergenome.nih.gov/) and International Cancer Genome Consortium (ICGC) (http://icgc.org/) to eliminate risk of bias such as selective reporting and publication bias of altered pathways. Search terms for altered genes in both databases were mut (missense, frameshift, inframe, truncating mutations), Hetloss (heterozygous deletion), Homdel (homozygous deletion), gain, and amplification25,26. We further searched protein–protein interactions in literature and in data deposited in UniProt (http://www.uniprot.org/) to identify master TFs and their interacting partners necessary for gene activation (coactivators) or repression (corepressors) (Table 1).
Table 1.
Lineage specific master transcription factors, coactivators and corepressors of various tissues and identified genetic alterations in human malignancies
| Tissue (n - TCGA) | Master transcription factor | Alteration frequency in TCGA database | Uniprot predicted coactivators | Alteration frequency in TCGA database | Uniprot predicted corepressors (http://www.uniprot.org/) | Alteration frequency in TCGA database |
|---|---|---|---|---|---|---|
| Liver (n = 442) | GATA4 6, 119 | 67% Hetloss | ARID1A | 44% Hetloss, fs* | KDM1B | 42% Amp, Gain |
| ARID2 | 17% Hetloss, fs* | BAZ1B | ||||
| FOXA1 120 | 3% Amp Gain | KMT2A | 28% Hetloss, fs* | SUZ12 | 32% Amp, Gain | |
| SMARCA4 | 24% Hetloss, fs* | DNMT1 | ||||
| FOXA2 120 | 30% Amp Gain | SMARCAD1 | 46% Hetloss | BAZ2A | 25% Amp, Gain | |
| 15% Amp, Gain | ||||||
| 14% Amp, Gain | ||||||
| Pancreas (n = 109) | GATA4 121 | 49% Hetloss | ARID1A | 49% Hetloss, fs* | BAZ1B | 57% Amp, Gain |
| ARID1B | 61% Hetloss | DNMT1 | ||||
| GATA6 121 | 23% Hetloss | ARID3C | 43% Hetloss | UHRF1 | 50% Amp, Gain | |
| SMARCD1 | 50% Hetloss | SUZ12 | ||||
| PTF1A 122 | 20% Hetloss | SMARCB1 | 34% Hetloss | BAZ2A | 45% Amp, Gain | |
| FOXA2 123 | 43% Amp Gain | 40% Amp, Gain | ||||
| PDX1 | 58% Hetloss | 39% Amp, Gain | ||||
| Ovary (n = 302) | GATA4 83 | 69% Hetloss | ARID3A | 91% Hetloss | EZH2 | 38% Gain amp |
| ARID3B | 50% Hetloss | DNMT1 | 39% Gain amp | |||
| FOXL1/2 82, 87 | 67% Amp, Gain | ARID3C | 37% Hetloss | BAZ1A | 18% Gain amp | |
| SMARCAD1 | 68% Hetloss | EED | 39% Gain amp | |||
| FOXO1 124 | 63% Hetloss | ARID1B | 67% Hetloss | |||
| ARID1A | 44% Hetloss | |||||
| SMARCA1 | 47% Hetloss |
We analyzed TCGA data in cBioPortal to determine genetic alterations in genes mediating differentiation pathways. Lineage specific transcription factors were identified using lineage tracing studies. Cofactors interacting with lineage specific transcription factors were determined using data deposited in UniProt database (http://www.uniprot.org/). Master transcription factors are lineage specific and they recruit various coactivators to cooperate and turn on differentiation genes. While heterozygous loss of GATA4 and inactivation by frameshift mutations of GATA4 coactivators are frequent in hepatocellular carcinoma, other master transcription factors such as FOXA1 are available to mediate differentiation pathways. However, corepressors such as KDM1B, which are also recruited by these TFs, are aberrant in HCC by copy number gains and amplification. Such alterations impair ability for differentiation to ensue in HCC through epigenetic suppression of target genes5, 88, 89. These forms of alterations are commonly observed also in PDAC, and OVC. Since corepressors are either gained or amplified in cancer but not inactivated by frameshift mutations, inhibition of these enzymes may serve as logical molecular targets of therapy (Fig. 5c, d)
*Frameshift mutation; hetloss, heterozygous deletion
Results
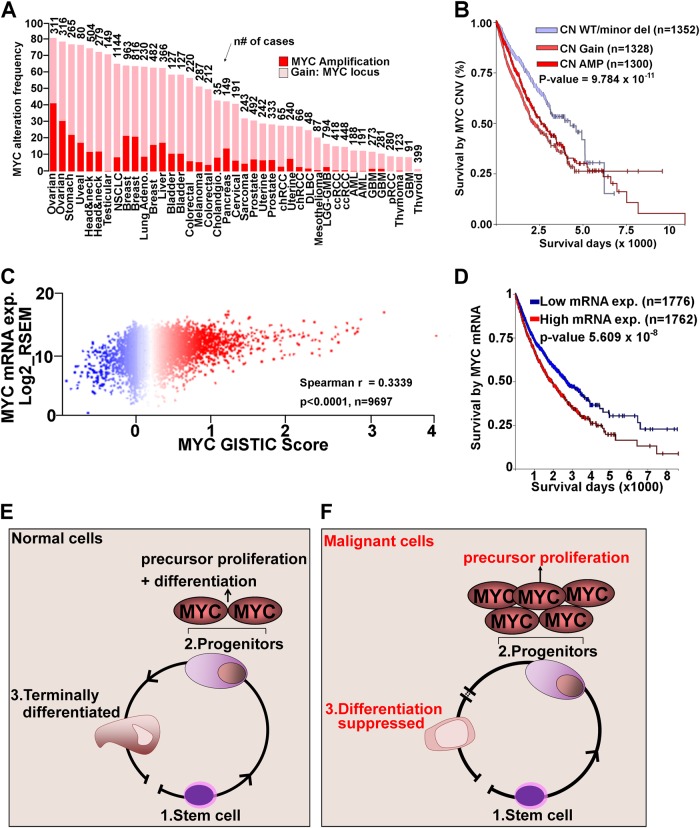
MYC amplification decreases survival across multiple human malignancies
One of the key TFs regulating mammalian cell proliferation is the myelocytomatosis viral oncoprotein (MYC), whose function is conserved across evolutionary hierarchies27–31. Physiologically, MYC regulated proliferation is succeeded by lineage-differentiation programs that antagonize MYC to terminate proliferation20. We analyzed MYC alterations by two approaches. First, we analyzed copy number (CN) alterations at the MYC locus using TCGA and ICGC data available through cBioPortal platform and found frequent amplifications and gains of MYC (Fig. 1a). We then accessed TCGA pan-cancer (PANCAN) data containing 11,000 patients across 33 of the most prevalent tumors and analyzed it through Xena Browser. MYC was highly amplified across these malignancies8,10. In both data sets MYC CN changes were determined using GISTIC score method, where values of −2,−1,0,1,2, represented homozygous deletion, heterozygous deletion, diploid, low-level amplification, or high-level amplification32. We next performed survival analysis using GISTIC scores predicting low level deletion/wild-type MYC, vs. gain/amplification using the PANCAN dataset. MYC amplification correlated with decreased overall survival (p < 9.784 × 10−11, n = 2628) compared to cases with MYC CN WT/low level deletions (n = 1352) (Fig. 1b). We then analyzed the correlation between GISTIC scores at the MYC locus vs. MYC mRNA expression and patient survival. There was a strong correlation (spearman r = 0.3339, p < 0.0001, n = 9697) between MYC GISTIC score and MYC mRNA expression (Fig. 1c). High (n = 1762) vs. low n = 1776) MYC mRNA levels were associated with decreased (p < 5.609 × 10−8) overall survival (Fig. 1d). Thus, MYC is a vital oncogene across many human malignancies and identification of mechanisms to antagonize MYC in cancer could have therapeutic applications. MYC function is conserved across evolutionary hierarchies27–31. The simple life cycle of protozoa requires MYC to generate daughter cells that resemble their parental cells with each cell division27,29. Evolution from single cell organism to multicellular organisms led to intense use of energy to open the chromatin and to expose naked DNA allowing lineage TFs to bind and activate hundreds of terminal differentiation genes that guide cell fate and specialization into various layers of cells. This process does not require actively proliferating cells. Hence, MYC mediated proliferation is potently antagonized at this stage33,34 (Fig. 1e). This form of potent MYC antagonism is also necessary for the existence of multi-cellularity29,35. Convincingly, infection of multicellular organisms with protozoa parasites enhances transformation of infected cells into proliferative cells by complex mechanisms that activate MYC protein and suppress differentiation TFs27,29,36.
Fig. 1. MYC alterations across multiple human malignancies.
a TCGA and IGCG data were analyzed through cBioPortal to determine aberrations at the MYC locus using pre-assigned GISTIC scores in multiple cancers from different tissue types. b We analyzed TCGA PANCAN data sets available through TCGA hub in Xena Browser. Survival analysis of cases with copy number (CN) gains and amplification at the MYC loci vs. those with CN WT/minor deletion of MYC demonstrated a significant overall survival (p-value < 9.784E−11, LogRank test, n = 1352 WT/minor del, 2628 CN gain and amplification). Survival data analyzed in Xena Browser (https://xenabrowser.net/) c Anlysis of MYC GISTIC Score vs. MYC mRNA expression using PANCAN RNA-seq data available in TCGA hub in Xena Browser. There was a strong correlation with spearman r = 0.3339, p < 0.0001, n = 9697. d Survival analysis of patients with increased MYC mRNA compared to those with decreased MYC mRNA expression. Expression levels are normalized relative to expression levels in normal tissues. Increased MYC mRNA was associated with poor survival (n = 1762) compared to decreased MYC mRNA (n = 1776, p = 5.06 × 10−18 e Schematic representation of metazoan differentiation and how differentiation is stalled in malignant cells. Differentiation continuum is initiated through stem cells lineage commitment, followed by exponential proliferation of tissue precursors/progenitors mediated by two copies of the MYC gene. To maintain homeostasis, MYC-mediated proliferation is dominantly antagonized by terminal differentiation pathways. f Human malignancies have impaired differentiation that fails to antagonize the MYC gene allowing for exponential proliferation of tissue precursors
Unlike normal cells, malignant cells undergo proliferation without terminal-differentiation (Fig. 1e, f). This aberrant process is strongly dependent on stabilization of MYC and its co-proteins that modulate cell growth and division17,20,37–39. Genetic and epigenetic alterations ensure that persistent proliferation of lineage committed progenitors occurs without final differentiation in cancer cells (Fig. 1e)7. First, persistent proliferation is achieved by consistent upregulation and chromosome gains of the genetic locus encoding the MYC gene across human malignancies (Fig. 1a). MYC amplification predicts poor overall survival (LogRank p-value = 9.784 × 10−11, n = 3980) (Fig. 1a, b). In studies using genetically engineered mouse models (GEMM) or xenograft models of cancer, antagonizing MYC sustains tumor regression across multiple tumors39–41. For instance, Shachaf et al. developed a transgenic mouse model conditionally expressing MYC in hepatocytes using tetracycline-controlled expression39. Inactivation of Myc induced regression of murine HCC increasing hepatocytes and hepatobiliary cell differentiation, loss of HCC marker α-fetoprotein, and suppressed proliferation39. In a xenograft PDAC model, Zhang et al. targeted MYC-MAX dimerization with a small molecule (10058-F4) that disrupts the MYC transcriptional activity40. Addition of 10058-F4 to gemcitabine led to drastic attenuation of tumorigenesis compared to single agent treatment40. Using a Kras driven mouse model of lung cancer, Soucek et al. targeted MYC using a dominant negative MYC dimerization domain mutant disrupting MYC binding to canonical Myc E-box response element ‘CACGTG’, thereby inhibiting MYC transactivation activity41. Inhibition of MYC transactivation increased mice survival by terminating lung cancer growth41.
From a translational perspective, various challenges exist in the attempt to directly target MYC pharmacologically42. The most important challenge is that proliferation is a feature of normal progenitors and such therapy could have a poor therapeutic index20. Additionally, tumors have heterogeneous genetic backgrounds contributing to sustained MYC activity. Therefore, to understand mechanisms that antagonize excessive MYC actions, it is imperative to define the evolutionary conserved physiological methods by which normal progenitors antagonize MYC to turn off intense proliferation and how these can be restored in cancer.
Terminating proliferation by engaging apoptosis is toxic to normal dividing cells
To retain cohesion and integrity between different cell types, multicellular organisms have evolved a system of checks and balances collectively known as apoptosis43,44. The master TFs of apoptosis p53 (TP53) and its cofactor p16 or p14ARF (CDKN2A) play crucial roles by arresting proliferating cells to enable repair of damage, or initiating orderly suicide if such damage cannot be repaired45,46. During embryogenesis, expression of p53 is downregulated perhaps because embryonic stem cells self-renew without exponentially proliferating47–49. Functional studies of differential expression of p53 using reporter assays demonstrated higher expression at later developmental stages, and decreased expression in terminally differentiated cells48. During cell division, p53 pathways potently antagonize MYC pathways to halt proliferation allowing impaired cells to undergo repair; irreparable cells undergo self-destruction through irreversible apoptosis to protect the integrity of entire organism43. Since p53-knockout (KO) mice have normal development and are not enlarged50, this illustrates that apoptosis pathways are not the dominant mechanisms used by lineage-progenitors to terminate exponential proliferation. Thus, mice exhibiting double KO of Trp53, and Phosphatase and tensin homolog (Pten) develop glioma tumors by failing to antagonize MYC, but this phenotype is only observed in the Trp53 and Pten double knockouts45,46. In PDAC the most frequent gene mutation is KRAS (~92%). GEMMs in which mutant KRAS (KC mice) is expressed in pancreas cells develop PDAC in 30 to 40% of cases at ~8–12 months of age51. Adding mutant Trp53 to the above GEMM (KPC mice) increases PDAC penetrance and decreases survival to ~5 months whereas KC mice with Ink4a deletion survive for ~2–3 months52,53. Mice with mutant Trp53 alone without mutant Kras do not develop PDAC53. By contrast, in ovarian cancer mouse models it has been demonstrated that Trp53 inactivation results in invasive tumors but tumor development is accelerated in mice with concomitant inactivation of Brca1 and Trp5354.
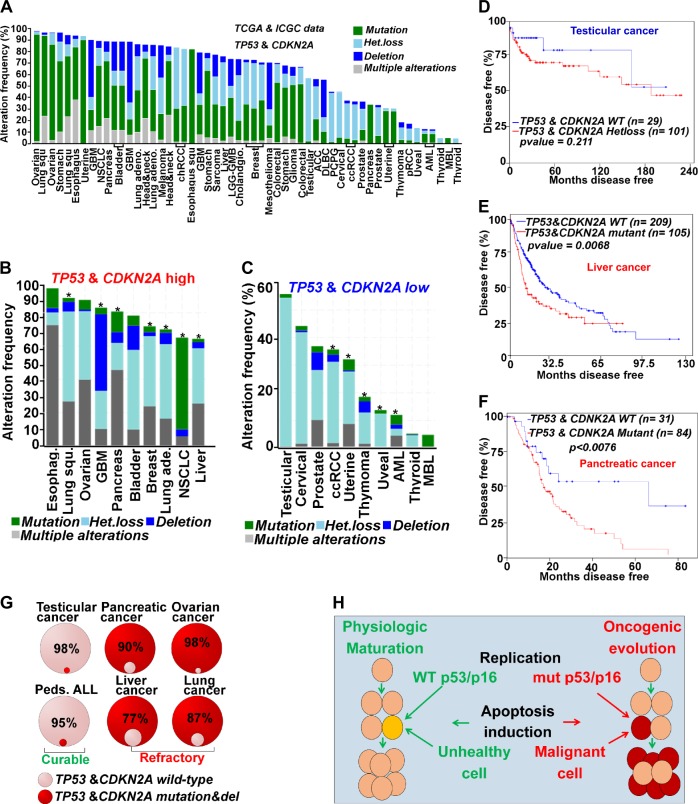
TP53 and CDKN2A are frequently bi-allelically inactivated across human malignancies (Fig. 2a). Such inactivation has major impact on treatment7. To terminate malignant proliferation, conventional chemotherapeutics aim to upregulate p53/p16 by inducing cytotoxic stress that mimics physiological activators of this pathway55. Since malignant cells and normal cells co-exist within the same milieu, such treatment has an unfavorable therapeutic index, as these genes are mutated/physically unavailable in malignant cells, but intact in normal cells. Multiple methods to re-engage apoptosis in cancer therapy have been investigated but it has been difficult to address this fundamental issue of therapeutic index56. Advances in genomic techniques indicate that when TP53/CDKN2A genes are wild-type, as in testicular cancer, treatment with cytotoxic chemotherapy (e.g., cisplatin) produces complete responses that increase overall and disease-free survival57 (Fig. 2a, b). Malignancies with high rates of TP53/CDKN2A inactivation do not exhibit these responses leading to resistance to multiple apoptosis-based treatments (broad chemo-resistance and radio-resistance) (Fig. 2a, b, e, f)7. Even different tumor types originating from the same organ have better responses to therapy if apoptosis genes are intact. For instance, TP53 and CDKN2A mutations occur in ~70 and 90% of PDAC, respectively58 (Fig. 2a). The overall 5 year survival rate in PDAC is ~9% even when including patients treated with chemotherapy or combination therapies and/or surgery59,60. By contrast, pancreatic neuroendocrine tumors (PNETs), generally do not harbor TP53 mutations, exhibit only minimal deletions of CDKN2A61, and have a 5 year survival rate of >50% when treated with apoptosis-inducing therapy62. Similarly, glioblastoma multiform (GBM) exhibits a variety of clinical, histopathologic, and molecular characteristics, and harbor TP53 mutations in ~30% of primary cases and ~65% of secondary GBM63,64. Glioma cells with WT TP53 are responsive to cytotoxic stress induced by clinically available chemotherapeutic agents compared to those with transcriptionally silenced mutant TP5365–67. Additionally, in the Trp53 induced mouse model of PDAC (KPC), genetic inactivation of one allele of Myc sensitizes therapeutic response gemcitabine40. We therefore analyzed genomic data by comparing the top ten malignancies with an elevated frequency of TP53/CDKN2A alterations (TP53/CDKN2A-high) with the bottom ten malignancies with low-frequency TP53/CDKN2A alterations (TP53/CDKN2A-low) (Fig. 2b, c). We found that 7/10 of TP53/CDKN2A-high cancers had a decrease in disease-free and overall survival when these genes were mutated (Fig. 2b; Table S1) (p-values < 0.05). Consistently, even in TP53/CDKN2A-low cases, there was a decrease in disease-free and overall survival when these genes were altered (p-values < 0.05) (Fig 2c; Table S1). Thus, the rate of alterations in apoptosis genes is lower in curable malignancies (testicular cancer/pediatric ALL) compared to high refractory/treatment resistant cancers (PDAC/HCC) (Fig. 2g). During physiologic maturation, WT TP53 induces irreversible apoptosis of unhealthy cells to retain integrity of entire organism (Fig. 2h). By contrast, oncogenic evolution mutates mediators of apoptosis leading to resistance to apoptosis induction (Fig. 2h).
Fig. 2. Apoptosis induction in p53/p16 mutant malignancy remains toxic to normal cells while simultaneous linked to refractory disease.
a Data was downloaded from TCGA and ICGC and analyzed in cBioPortal for mutations in TP53 and CDKN2A genes. b Top 10 malignancies with high TP53/CDKN2A alterations (TP53/CDKN2A high). *Cases where these alterations were linked to poor disease-free or overall survival with a p-value < 0.05 (Table S1). c Bottom 10 cases with least frequency of alterations in TP53/CDKN2A (TP53/CDKN2A low). *Cases where these alterations were linked to poor disease-free or overall survival with a p-value ≤ 0.05 d Disease-free survival of testicular cancer, cases with minor alterations (gains, and heterozygous loss of one allele in TP53 and CDKN2A) vs. cases with wild type TP53 and CDKN2A (p-value = 0.211, LogRank test). e Disease-free survival of pancreatic cancer with mutant TP53 and CDKN2A cases was significantly lower vs. cases with wild-type TP53 and CDKN2A (p-value = 0.0078, LogRank test). f Disease-free survival of liver cancer was also significantly lower in mutant TP53 and CDKN2A cases vs. wild-type (p-value = 0.0068, LogRank test). g Quantitative analysis of TP53 and CDKN2A mutations demonstrated less frequency of alteration of these genes in curable vs. high refractory/treatment resistant human malignancies. h During physiologic maturation, unhealthy cells with WT p53/p16 undergo irreversible apoptosis. Alterations in these proteins sustain oncogenic evolution leading to aberrant proliferation without apoptosis
Genetic and epigenetic alterations of differentiation genes in cancer
The most aggressive human malignancies are poorly differentiated13. While differentiation contributes to poor survival across multiple human malignancies, the mechanisms that underpin differentiation impediment in malignant cells are mostly unclear, but new knowledge is emerging5–7. We identified key lineage master TFs for the development of the ovary, pancreas and liver using published lineage conversion studies, or studies with transgenic mouse models6,68–74 (Table 1). Cellular differentiation and lineage commitment programs are dictated by this handful of master TFs and their cofactors. While multiple cofactors have major roles, the most vital of these are transcriptional coactivators and corepressors that use ATP to remodel chromatin to turn-on or turn-off target genes33,34,75. Accordingly, we analyzed the genetic alterations in lineage TFs, their coactivators and corepressors in OVC, PDAC, and HCC (Table 1).
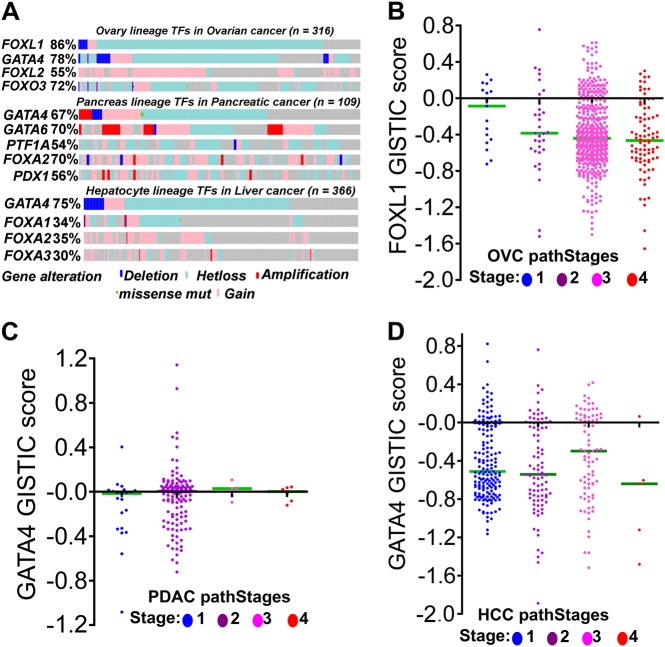
Since malignant cells cannot completely suppress differentiation, as it is a continuum along which all cells exist, master TFs that specify commitment into various lineages are nearly never completely inactivated by mutation but are frequently haploinsufficient (Fig. 3a; Table 1). This dose-reduction is sufficient to stall advances along the differentiation continuum at its most proliferative points5–7. For instance, FOXL1 loss was frequent in OVC (Fig. 3a), and the frequency of FOXL1 loss was highest in poorly differentiated OVC (Fig. 3b). This pattern was similar for GATA4 in PDAC and HCC, even though these malignancies had small numbers of patients surviving beyond stages I and II (Fig. 3b, c). We identified key interacting partners that are coactivators and corepressors of various lineage specific TFs (Table 1) by literature analysis and data deposited in UniProt database (http://www.uniprot.org/). To augment the stalled differentiation, the coactivators were found frequently inactivated and deleted (Table 1; Fig. 4a) favoring repression of downstream genes targeted by key TFs. New lines of evidence now imply that such alterations impair pathways mediating terminal differentiation6,7,76. Early discoveries of the functions of these coactivator enzymes demonstrated that their role in physiology was to utilize ATP to mobilize histone DNA interactions such that naked DNA was exposed, thereby allowing TFs to bind to and activate target genes33,34,75,77. This process is conserved in evolution from yeast78, one of the simplest metazoa, to homo-sapiens77. Inactivation of these genes in cancer could be an attempt to impair the ability of coactivators to expose DNA to master TFs that activate downstream genes. A major clue to this hypothesis is that lineage master TFs are selective in their use of specific coactivators to mediate activation of lineage genes (Table 1). Another clue is that malignant cells tend to lose one allele of lineage specifying TFs, an event that may be sufficient to allow lineage commitment but insufficient for terminal differentiation6,7 (Fig. 3a; Table 1). For instance, liver progenitors require cooperation between GATA4 and FOXA1 to recruit coactivators (e.g., ARID1A) and mediate activation of hepatocyte differentiation genes. In HCC, heterozygous loss of GATA4 is frequent (68%, n = 366, Fig. 3a; Table 1) and inactivating mutations in ARID1A are common (44%, n = 366, Fig. 4a; Table 1)6. Hepatic differentiation is impaired and proliferation enhanced in livers with Gata4 or Arid1a liver-conditional haploinsufficiency6,76,79. Moreover, reintroduction of GATA4 in GATA4 deficient HCC, or ARID1A in ARID1A mutated but GATA4 intact HCC, activates hundreds of hepatocyte epithelial-differentiation genes6. The master TFs of the pancreatic lineage include GATA4 and GATA680,81. Copy number losses of one allele of these factors are seen in PDAC, with loss of function mutations in coactivators also observed (Table 1; Figs. 3a, 4a). However, PDACs also exhibited a high incidence of amplification or gain of GATA4 and GATA6, suggesting that in certain instances these TFs may confer a growth advantage to pancreatic cancer cells. In OVC, one allele of ovarian master TFs FOXL182,83 is frequently lost (80%, Fig. 3a; Table 1 n = 316), while coactivators, such as ARID3A and ARID3B, are often inactivated (Table 1; Fig. 4a). Thus at the core of malignant transformation, differentiation impediment routinely enhances malignant proliferation and is achieved through haploinsufficiency of master TFs and inactivation of the coactivators they use. This understanding could lead to treatments aiming to re-engage forward-differentiation, as an alternative to apoptosis, as the means of terminating malignant proliferation.
Fig. 3. Genetic alterations in lineage specifying master transcription factors in human malignancies.
a Analysis of TCGA data deposited in cBioPortal to determine alterations of master transcription factors of various lineages (Table 1). Key lineage specifying transcriptions factors were mostly haploinsufficient (heterozygous deletion/hetloss) in malignant cells or contained frequent amplification and gains. None of the transcription factors had biallelic frameshift inactivating mutations. Thus stalled differentiation occurs through genetic haploinsuffiency of key lineage specific transcription factors6. b Analysis of FOXL1 deletions across varying degrees of differentiation grades (pathological grades) of ovarian cancer. c Analysis of GATA4 deletions across varying degrees of differentiation grades of pancreatic cancer (PDAC). d Analysis of GATA deletions across varying degrees of differentiation grades liver cancer (HCC)
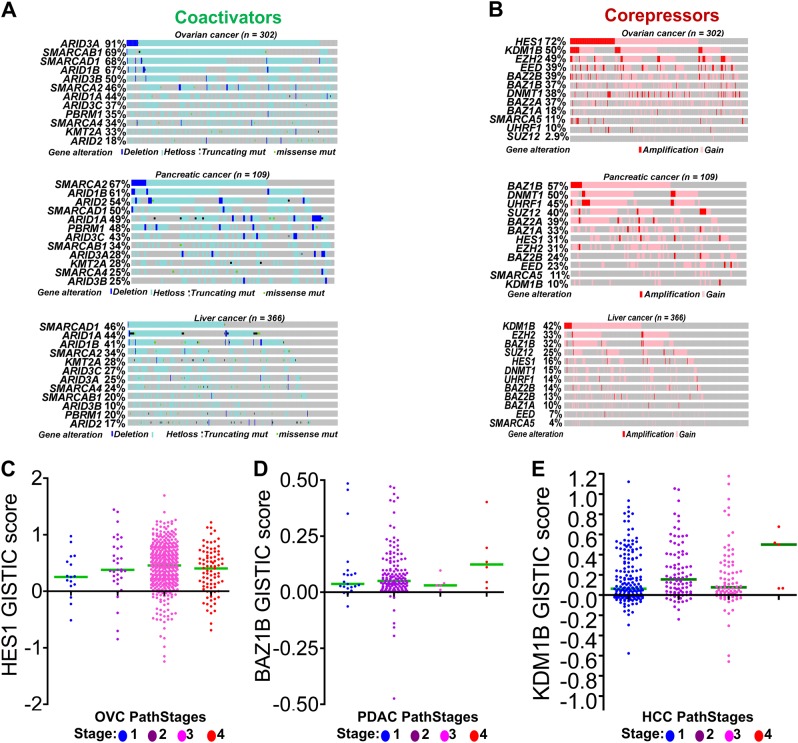
Fig. 4. Frequent inactivating mutations of coactivators and amplification and copy number gains at gene loci of transcriptional corepressors.
TCGA data was analyzed in cBioPortal to determine frequent genetic alterations in transcriptional corepressor and coactivator enzymes (Table 1). a Inactivating mutations, bi-allelic and frameshift mutations and deletions of transcriptional coactivator enzymes in ovarian, pancreatic and liver cancers (Table 1). b Copy number (CN) gain and amplifications of corepressors was frequently observed in various tumors including ovarian cancer (OVC), pancreatic cancer (PDAC) and liver cancer (HCC) (Table 1). c Analysis of HES1 CN gains across varying degrees of differentiation grades (pathological grades) of OVC. d Analysis of CN gains of BAZ1B across varying degrees of differentiation grades PDAC. e Analysis of CN gains KDM1B gains across varying degrees of differentiation grades HCC
Corepressor enzymes: emerging targets for differentiation-restoring oncotherapy
An enhanceosome is composed of multiprotein complexes cooperating to activate genes of a given lineage84,85, e.g., hepatic enhanceosomes activate hepatocyte genes6, whereas pancreas and ovarian enhanceosomes activate pancreatic86 and ovarian genes87, respectively. Genetic disruption of this cooperation can shift the content of these protein hubs away from coactivators to corepressors that repress lineage genes instead76,88,89. Such repression is further enabled by the inherent closed chromatin status of terminal-differentiation genes, contrasting with inherently open chromatin at proliferation and early-differentiation genes6,7,90.
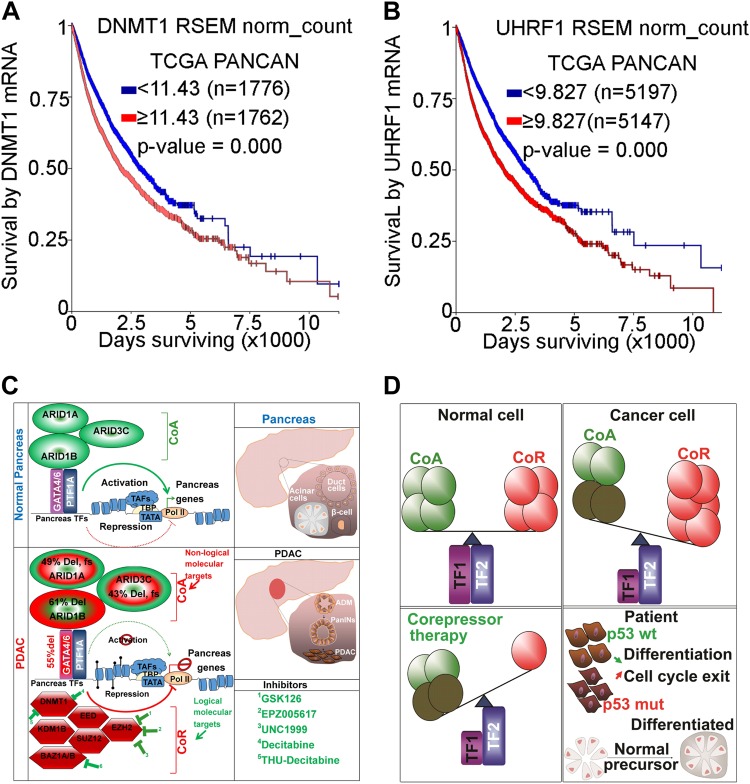
For exponential proliferation to occur decoupled from forward-differentiation, a high degree of corepressor activity is necessary for epigenetic silencing of lineage-differentiation genes. Consequently, aberrant corepressor activity is frequently observed in malignant cells, where hundreds of terminal differentiation genes have accumulation of active corepressors6,89. Unlike coactivators, which are frequently inactivated by genetic mutations/deletions6, corepressors are frequently either wild-type or amplified in malignant cells (Table 1; Fig. 4b). DNA methyl transferase 1 enzyme (DNMT1) is a corepressor for master TF and also the maintenance methyltransferase that recapitulates CpG methylation onto the newly synthesized DNA strand as cells go through cycles of division91–93. In TCGA PANCAN data, high levels of DNMT1 are associated with poor survival (p < 0.00001, n = 5145), compared to cases with low DNMT1 levels (n = 5199) (Fig. 5a). This suggests an important role of this enzyme in numerous human cancers. Therefore, multiple studies have evolved in the last decade attempting to develop therapeutic interventions targeting DNMT1 in cancer therapy94–102. Similarly, Ubiquitin-like, containing PHD and RING finger domains, 1, (UHRF1), closely cooperates with DNMT1 in regulating DNA methylation103,104. We analyzed the expression levels of UHRF1 in the PANCAN dataset and found that high UHRF1 expression levels (p < 0.0001, n = 5150) strongly predicted poor survival rates compared to low levels (n = 5189) (Fig. 5b), illustrating the importance of these methylation genes in human cancers.
Fig. 5. Corepressor upregulation and model for inhibiting corepressors to re-engage forward-differentiation.
a Corepressor DNMT1 mRNA upregulation predicts poor survival across multiple human malignancies in TCGA PANCAN data. b Corepressor UHRF1 (that partners with DNMT1 for epigenetic repression activities) mRNA upregulation predicts poor survival across multiple human malignancies in TCGA PANCAN data. c Model example in PDAC alterations of coactivators and corepressors and candidate small molecules that can be used as corepressor therapy. d Model schematic summary for p-53 independent differentiation-restoring therapy. Non-malignant cells (normal cells) have intact lineage specifying transcription factors of cell fate determination that dynamically recruit coactivators and corepressors enzymes to turn on or turn off differentiation genes. Gene dose reduction by heterozygous deletion of a master transcription factor and inactivating mutations in its coactivators impairs the activation component of differentiation genes epigenetically6. Aberrant amplifications in transcriptional corepressor enzymes facilitate a closed chromatin status and epigenetically silence hundreds of differentiation genes6, 7 (Table 1). This mode of alteration is clinically relevant and can be developed to suppress proliferation even in TP53 mutant malignancies102, 105
DNMT1-depletion without cytotoxicity has therapeutic benefits even in myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML) containing p53-system defects102,105, and multiple clinical trials are ongoing to evaluate DNMT1-depletion more broadly in cancer therapy (although decitabine and 5-azacytidine used to deplete DNMT1 have pharmacologic limitations which can undermine their ability to deplete-DNMT1 from solid tumors) (Table 2). In acute promyelocytic leukemia (APL), complete remissions are achieved by combination of arsenic with retinoic acid to inhibit corepressors recruited on leukemia fusion protein PML-RARA106,107. Since co-repressors are not mutated and have aberrant activity in cancer, they are sufficient and logical molecular targets that may engage terminal differentiation genes for p53 cell cycle exits7,89,99,100,102,105,108–111 (Table 2; Fig. 5c, d).
Table 2.
Key pre-clinical and clinical evaluation of corepressor therapy in cancer
| Drugs | Disease | Dosage | Status | Survival: Ctrl vs. (Tx) | Adverse effects | Ref. |
|---|---|---|---|---|---|---|
| Corepressor therapy in pre-clinical trials | ||||||
| 5-aza-Dc | Ovarian cancer (in-vitro Ovarian cancer cell lines) | 0.1–10 µM | Complete | No survival, decreased cell proliferation | None reported | 119 |
| THU* + Decitabine (Increased decitabine activity in the liver) | AML engrafted in the liver (mouse) | 0.1–0.2 mg/kg | Complete | Median 38 days (61days) | None reported | 109 |
| Guadecitabine | HCC (HCC xenografts) | 2 mg/Kg | Complete | Reduced tumor size | None reported | 120 |
| Decitabine | PDAC (mouse) | 1 μg/g of body weight | Complete | Median 87 days (127.5 days) | None reported | 101 |
| Co-repressor therapy in human clinical trials | ||||||
| Drugs | Disease | Dosage | Status | % ORR (%CR) | Adverse effects | Ref |
| Decitabine | Ovarian cancer | 10–20 mg/m2 | complete | 60(10) | 4 neutropenia at 20 mg/m2, none at 10 mg/m2 | 121 |
| Decitabine | Advanced HCC | 6 mg/m2/d | Complete | 47(6) | Favorable adverse events | 122 |
| Decitabine + THU | Pancreatic cancer | Dec(5 mg/capsule) THU (250 mg/ capsule) | Recruiting | Not available at this time | Not available at this time | Clinicaltrial.gov |
| Decitabine + THU | NSCLC Lung cancer | THU 10mg/kg decitabine 0.2 mg/kg | Recruiting | Not available at this time | Not available at this time | Clinicaltrial.gov |
| Decitabine | MDS | 0.1–0.2 mg/kg/day | Complete | 44(55) | Neutropenic fever | 102 |
Multiple preclinical studies evaluating mechanisms of non-cytotoxic therapy that engages differentiation genes instead of apoptosis genes show the proof principle for corepressor inhibition therapy across multiple tumors by targeting the corepressor DNMT1. Emerging human clinical trials are now evaluating corepressor therapies as p53 independent treatments in cancer therapy
THU tetrahydrouridine, 5-aza-Dc 5-aza-deoxycytadine, ORR overall response rate, CR complete response, TX treatment group, ctrl control group, ref references
Various other corepressors have also been investigated as potential molecular targets for epigenetic therapy of cancer. For instance, histone deacetylase (HDAC) enzymes are key corepressors recruited into TF hubs of many human malignancies and are known epigenetic suppressors of gene expression5,6,88,89. In many pre-clinical studies, HDAC enzymes have been investigated as potential inducers of cell differentiation94,95. One problem with targeting HDACs, however, is their pleiotropic cellular functions—even on-target activity may thus produce unintended side effects. Other common corepressors upregulated in many human malignancies are lysine demethylase enzymes such as KDM1A (Fig. 4b). Various studies have demonstrated differentiation induction by pharmacologic targeting of KDM1A and related clinical trials are currently ongoing112–116. Using a high throughput pan-cancer in vivo screen, Carugo et al. recently demonstrated a link between the corepressor WDR5 and sustained MYC mediated proliferation of PDAC117. Disrupting WDR5 through inhibition assays led to arrested tumor progression and increased survival in PDX mouse models of PDAC117. In this systematic review, we have documented additional corepressors recruited into the master TF hubs of many human malignancies that require additional genetic and pharmacologic validation as candidate molecular targets that enhance differentiation. These include HES1, BAZ1A/B, BAZ2A, EED, SUZ12 and UHRF1 (Figs. 4b, 5b; Table 1). Furthermore, upregulation of these corepressors was found linked to advanced clinical pathological stages suggesting direct effect on differentiation suppression. For example, HES1 was found as the most frequently upregulated corepressor in OVC (Fig. 4b), and stage III and IV OVC had higher HES1 gains compared to stages I and II (Fig. 4c). Thus, HES1 inhibition therapy may be vital for OVC differentiation therapy. This pattern was also seen for BAZ1B in PDAC and KDM1B in HCC (Fig. 4b, d, e). These observations suggest that, in these malignancies, targeting these key enzymes for differentiation induction could provide additional therapeutic strategies that circumvent p53-system defects.
Discussion
Human malignancies upregulate the master regulator of cell proliferation MYC, and this genetic alteration is significantly linked to poor survival rates. Historically, MYC-driven malignant proliferation has been antagonized by induction of apoptosis. Malignant cells, however, often harbor inactivating alterations to the master regulator of apoptosis p53 or its key co-factors, resulting in multi-drug resistance but continued apoptosis-induction in normal dividing cells (poor therapeutic index)7,43,56. To improve on issues of resistance and toxicity, it is thus imperative to find p53-independent strategies for antagonizing MYC6,7. Restoring forward-differentiation is one such potential strategy, and can be guided by an understanding that forward-differentiation is suppressed by partial loss-of-function to master TF that drive lineage-fates and to the coactivators they use to activate lineage-differentiation genes. This results in unbalanced activity of corepressors that repress the lineage-genes instead33,34,76,89. The corepressors are not frequently inactivated in human malignancies but are upregulated by CN gains and amplification of their chromosomal segments. Therefore, targeting these corepressors can provide therapy that engages differentiation instead of apoptosis7,88,89,99,101,102,105. Various clinical trials targeting transcriptional corepressors (e.g., DNMT1) without cytotoxicity have produced meaningful clinical responses102,105.
To sustain a given tissue through daily wear and tear, tissue lineage-progenitors proliferate exponentially2,39. Since cell division involves mechanical processes such as DNA replication, mitosis and meiosis, that are prone to error2,4,9, metazoan cells contain an apoptosis program to ensure that only healthy cells continue through phases of the cell cycle44,45. Therefore, p53/apoptosis potently antagonizes MYC to halt proliferation.
The 2012 Nobel prize awarded to Yamanaka and colleagues demonstrated in spectacular fashion that cell lineage-fates are commanded by handfuls of master TFs68,74. Master TFs combine and collaborate to exchange corepressors (“off enzymes”) for coactivators (“on enzymes”) and activate lineage-programs6,7,118. It is clear from the broadly available genomic data on cancers (TCGA etc.) that lineage master TFs, and the coactivators they use to activate target genes, are very frequently haploinsufficient in cancers. Crucially, however, unlike p53 or p16, these master TF circuits are not completely inactivated, and small molecule drugs that target the transcriptional corepressors aberrantly enriched in these highly expressed master TF hubs can resume forward lineage-differentiation and terminate proliferation, even if p53 or p16 are absent7,89,99,100,102,105,109. Importantly, such treatments simultaneously spare normal tissue stem cells needed for health and life.
Conclusion and perspective
The enduring problems of oncotherapy are resistance and toxicity. A major reason for this situation is that most oncotherapeutics are designed to induce apoptosis, yet, the master regulators of this program are very frequently bi-allelically inactivated in cancers11,43. An alternative pathway for terminating malignant self-replication is to re-engage forward-differentiation in these differentiation-arrested cells. Pre-clinical genetic, epigenetic, biochemical and cell data, and both pre-clinical and clinical in vivo data, suggests that this can be achieved by inhibiting the corepressors aberrantly enriched, at the expense of coactivators, in the lineage master TF hubs highly expressed in replicating cancer cells.
Electronic supplementary material
Disease free and overall survival comparisons in cancers with TP53/CDKN2A high versus low Frequency alterations
Acknowledgements
This work was supported in part by the National Cancer Institute (NCI) of the National Institutes of Health (R01 CA-075059 to M. Korc), and Cancer Center Core Grant P30 (CA043703 to Y. Saunthararajah).
Author contributions
Acquisition and analysis of data: F.O.E., Interpretation of data: F.O.E. and M.K., Drafting the manuscript: F.O.E., Study concept and design: F.O.E., Critical revision of the manuscript for important intellectual content: Y.S., M.K.
Conflict of interest
Y.S.: patents around tetrahydrouridine, decitabine, 5-azacytidine and ISWI corepressor inhibition; consultancy, ownership and royalty eligibility through EpiDestiny. The remaining authors declare that they have no conflict of interest.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by J. Chipuk.
Contributor Information
Francis O. Enane, Phone: +317-274-4837, Email: fenane@iu.edu
Murray Korc, Phone: +317-278-6410, Email: mkorc@iu.edu.
Electronic supplementary material
Supplementary information accompanies this paper at (10.1038/s41419-018-0919-9).
References
- 1.Kinzler KW, Vogelstein B. Cancer therapy meets p53. N. Engl. J. Med. 1994;331:49–50. doi: 10.1056/NEJM199407073310113. [DOI] [PubMed] [Google Scholar]
- 2.Dominguez-Sola, D. & Gautier, J. MYC and the control of DNA replication. Cold Spring. Harb. Perspect. Med.10.1101/cshperspect.a014423 (2014). [DOI] [PMC free article] [PubMed]
- 3.Lin KI, Lin Y, Calame K. Repression of c-myc is necessary but not sufficient for terminal differentiation of B lymphocytes in vitro. Mol. Cell. Biol. 2000;20:8684–8695. doi: 10.1128/MCB.20.23.8684-8695.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zaytseva, O. & Quinn, L. M. Controlling the master: chromatin dynamics at the MYC promoter integrate developmental signaling. Genes10.3390/genes8040118 (2017). [DOI] [PMC free article] [PubMed]
- 5.Gu, X. et al. Leukemogenic nucleophosmin mutation disrupts the transcription factor hub regulating granulo-monocytic fates. J. Clin. Invest.10.1172/JCI97117 (2018). [DOI] [PMC free article] [PubMed]
- 6.Enane, F. O. et al. GATA4 loss of function in liver cancer impedes precursor to hepatocyte transition. The J. Clin. Invest.10.1172/JCI93488 (2017). [DOI] [PMC free article] [PubMed]
- 7.Velcheti V, Radivoyevitch T, Saunthararajah Y. Higher-level pathway objectives of epigenetic therapy: a solution to the p53 problem in cancer. Am. Soc. Clin. Oncol. Educ. Book. 2017;37:812–824. doi: 10.14694/EDBK_174175. [DOI] [PubMed] [Google Scholar]
- 8.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rahl PB, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gabay, M., Li, Y. & Felsher, D. W. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring. Harb. Perspect. Med. 10.1101/cshperspect.a014241 (2014). [DOI] [PMC free article] [PubMed]
- 11.Vazquez A, Bond EE, Levine AJ, Bond GL. The genetics of the p53 pathway, apoptosis and cancer therapy. Nat. Rev. Drug Discov. 2008;7:979–987. doi: 10.1038/nrd2656. [DOI] [PubMed] [Google Scholar]
- 12.Abegglen LM, et al. Potential mechanisms for cancer resistance in elephants and comparative cellular response to DNA damage in humans. JAMA. 2015;314:1850–1860. doi: 10.1001/jama.2015.13134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jogi A, Vaapil M, Johansson M, Pahlman S. Cancer cell differentiation heterogeneity and aggressive behavior in solid tumors. Ups J. Med. Sci. 2012;117:217–224. doi: 10.3109/03009734.2012.659294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Wath RC, Wilson A, Laurenti E, Trumpp A, Lio P. Estimating dormant and active hematopoietic stem cell kinetics through extensive modeling of bromodeoxyuridine label-retaining cell dynamics. PLoS One. 2009;4:e6972. doi: 10.1371/journal.pone.0006972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwartz GN, et al. Proliferation kinetics of subpopulations of human marrow cells determined by quantifying in vivo incorporation of [2H2]-glucose into DNA of S-phase cells. Blood. 2003;102:2068–2073. doi: 10.1182/blood-2003-01-0139. [DOI] [PubMed] [Google Scholar]
- 16.Nygren JM, Bryder D, Jacobsen SE. Prolonged cell cycle transit is a defining and developmentally conserved hemopoietic stem cell property. J. Immunol. 2006;177:201–208. doi: 10.4049/jimmunol.177.1.201. [DOI] [PubMed] [Google Scholar]
- 17.Arnold I, Watt FM. c-Myc activation in transgenic mouse epidermis results in mobilization of stem cells and differentiation of their progeny. Curr. Biol. 2001;11:558–568. doi: 10.1016/S0960-9822(01)00154-3. [DOI] [PubMed] [Google Scholar]
- 18.Laurenti E, et al. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell. 2008;3:611–624. doi: 10.1016/j.stem.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reavie L, et al. Regulation of hematopoietic stem cell differentiation by a single ubiquitin ligase-substrate complex. Nat. Immunol. 2010;11:207–215. doi: 10.1038/ni.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson A, et al. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 2004;18:2747–2763. doi: 10.1101/gad.313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramaswamy S, Nakamura N, Sansal I, Bergeron L, Sellers WR. A novel mechanism of gene regulation and tumor suppression by the transcription factor FKHR. Cancer Cell. 2002;2:81–91. doi: 10.1016/S1535-6108(02)00086-7. [DOI] [PubMed] [Google Scholar]
- 22.Lucas B, et al. HNF4alpha reduces pr oliferation of kidney cells and affects genes deregulated in renal cell carcinoma. Oncogene. 2005;24:6418–6431. doi: 10.1038/sj.onc.1208794. [DOI] [PubMed] [Google Scholar]
- 23.Green LM, Wagner KJ, Campbell HA, Addison K, Roberts SG. Dynamic interaction between WT1 and BASP1 in transcriptional regulation during differentiation. Nucleic Acids Res. 2009;37:431–440. doi: 10.1093/nar/gkn955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. J. Clin. Epidemiol. 2009;62:1006–1012. doi: 10.1016/j.jclinepi.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 25.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jensen K, et al. The protozoan parasite Theileria annulata alters the differentiation state of the infected macrophage and suppresses musculoaponeurotic fibrosarcoma oncogene (MAF) transcription factors. Int. J. Parasitol. 2009;39:1099–1108. doi: 10.1016/j.ijpara.2009.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dessauge F, Lizundia R, Baumgartner M, Chaussepied M, Langsley G. Taking the Myc is bad for Theileria. Trends. Parasitol. 2005;21:377–385. doi: 10.1016/j.pt.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 29.Young SL, et al. Premetazoan ancestry of the Myc-Max network. Mol. Biol. Evol. 2011;28:2961–2971. doi: 10.1093/molbev/msr132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davis CE, Colmerauer ME, Kim CH, Matthews B, Guiney D. G. myc-related proteins and DNA sequences in Trypanosoma brucei. Microb. Pathog. 1989;7:45–53. doi: 10.1016/0882-4010(89)90110-1. [DOI] [PubMed] [Google Scholar]
- 31.Leyva Leyva M, et al. Expression of sequences related to c-myc in Entamoeba. Parasitol. Res. 1993;79:153–159. doi: 10.1007/BF00932262. [DOI] [PubMed] [Google Scholar]
- 32.Mermel CH, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen L, et al. Subunit organization of the human INO80 chromatin remodeling complex: an evolutionarily conserved core complex catalyzes ATP-dependent nucleosome remodeling. J. Biol. Chem. 2011;286:11283–11289. doi: 10.1074/jbc.M111.222505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- 35.Reinke AW, Baek J, Ashenberg O, Keating AE. Networks of bZIP protein-protein interactions diversified over a billion years of evolution. Science. 2013;340:730–734. doi: 10.1126/science.1233465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dessauge F, et al. c-Myc activation by Theileria parasites promotes survival of infected B-lymphocytes. Oncogene. 2005;24:1075–1083. doi: 10.1038/sj.onc.1208314. [DOI] [PubMed] [Google Scholar]
- 37.Sears R, Leone G, DeGregori J, Nevins JR. Ras enhances Myc protein stability. Mol. Cell. 1999;3:169–179. doi: 10.1016/S1097-2765(00)80308-1. [DOI] [PubMed] [Google Scholar]
- 38.Acosta J. C., Ferrandiz N, Bretones G, et al. Myc inhibits p27-induced erythroid differentiation of leukemia cells by repressing erythroid master genes without reversing p27-mediated cell cycle arrest. Mol. Cell. Biol.28, 7286-95 (2008). [DOI] [PMC free article] [PubMed]
- 39.Shachaf CM, et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431:1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- 40.Zhang M, Fan HY, Li SC. Inhibition of c-Myc by 10058-F4 induces growth arrest and chemosensitivity in pancreatic ductal adenocarcinoma. Biomed. Pharmacother. 2015;73:123–128. doi: 10.1016/j.biopha.2015.05.019. [DOI] [PubMed] [Google Scholar]
- 41.Soucek L, et al. Inhibition of Myc family proteins eradicates KRas-driven lung cancer in mice. Genes Dev. 2013;27:504–513. doi: 10.1101/gad.205542.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soucek L, Evan GI. The ups and downs of Myc biology. Curr. Opin. Genet. Dev. 2010;20:91–95. doi: 10.1016/j.gde.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 44.Molchadsky A, Rivlin N, Brosh R, Rotter V, Sarig R. p53 is balancing development, differentiation and de-differentiation to assure cancer prevention. Carcinogenesis. 2010;31:1501–1508. doi: 10.1093/carcin/bgq101. [DOI] [PubMed] [Google Scholar]
- 45.Zheng H, et al. Pten and p53 converge on c-Myc to control differentiation, self-renewal, and transformation of normal and neoplastic stem cells in glioblastoma. Cold Spring Harb. Symp. Quant. Biol. 2008;73:427–437. doi: 10.1101/sqb.2008.73.047. [DOI] [PubMed] [Google Scholar]
- 46.Zheng H, et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455:1129–1133. doi: 10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gottlieb E, et al. Transgenic mouse model for studying the transcriptional activity of the p53 protein: age- and tissue-dependent changes in radiation-induced activation during embryogenesis. EMBO J. 1997;16:1381–1390. doi: 10.1093/emboj/16.6.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Komarova EA, et al. Transgenic mice with p53-responsive lacZ: p53 activity varies dramatically during normal development and determines radiation and drug sensitivity in vivo. EMBO J. 1997;16:1391–1400. doi: 10.1093/emboj/16.6.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.MacCallum DE, et al. Thep53 response to ionising radiation in adult and developing murine tissues. Oncogene. 1996;13:2575–2587. [PubMed] [Google Scholar]
- 50.Donehower LA, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 51.Hingorani SR, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/S1535-6108(03)00309-X. [DOI] [PubMed] [Google Scholar]
- 52.Westphalen CB, Olive KP. Genetically engineered mouse models of pancreatic cancer. Cancer J. 2012;18:502–510. doi: 10.1097/PPO.0b013e31827ab4c4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morton JP, et al. Mutant p53 drives metastasis and overcomes growth arrest/senescence in pancreatic cancer. Proc. Natl Acad. Sci. USA. 2010;107:246–251. doi: 10.1073/pnas.0908428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clark-Knowles KV, Senterman MK, Collins O, Vanderhyden BC. Conditional inactivation of Brca1, p53 and Rb in mouse ovaries results in the development of leiomyosarcomas. PLoS One. 2009;4:e8534. doi: 10.1371/journal.pone.0008534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sellers WR, Fisher DE. Apoptosis and cancer drug targeting. J. Clin. Invest. 1999;104:1655–1661. doi: 10.1172/JCI9053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Loehrer PJ, et al. Vinblastine plus ifosfamide plus cisplatin as initial salvage therapy in recurrent germ cell tumor. J. Clin. Oncol. 1998;16:2500–2504. doi: 10.1200/JCO.1998.16.7.2500. [DOI] [PubMed] [Google Scholar]
- 58.Kleeff, J. et al. Pancreatic cancer. Nat. Rev. Dis. Primers2, 16022 (2016). [DOI] [PubMed]
- 59.Picozzi VJ, et al. Multicenter phase II trial of adjuvant therapy for resected pancreatic cancer using cisplatin, 5-fluorouracil, and interferon-alfa-2b-based chemoradiation: ACOSOG Trial Z05031. Ann. Oncol. 2011;22:348–354. doi: 10.1093/annonc/mdq384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Katz MH, et al. Standardization of surgical and pathologic variables is needed in multicenter trials of adjuvant therapy for pancreatic cancer: results from the ACOSOG Z5031 trial. Ann. Surg. Oncol. 2011;18:337–344. doi: 10.1245/s10434-010-1282-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scarpa A, et al. Whole-genome landscape of pancreatic neuroendocrine tumours. Nature. 2017;543:65–71. doi: 10.1038/nature21063. [DOI] [PubMed] [Google Scholar]
- 62.Fjallskog ML, et al. Treatment with cisplatin and etoposide in patients with neuroendocrine tumors. Cancer. 2001;92:1101–1107. doi: 10.1002/1097-0142(20010901)92:5<1101::AID-CNCR1426>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 63.Ohgaki H, et al. Genetic pathways to glioblastoma: a population-based study. Cancer Res. 2004;64:6892–6899. doi: 10.1158/0008-5472.CAN-04-1337. [DOI] [PubMed] [Google Scholar]
- 64.Ichimura K, Ohgaki H, Kleihues P, Collins VP. Molecular pathogenesis of astrocytic tumours. J. Neurooncol. 2004;70:137–160. doi: 10.1007/s11060-004-2747-2. [DOI] [PubMed] [Google Scholar]
- 65.Batista LF, Roos WP, Christmann M, Menck CF, Kaina B. Differential sensitivity of malignant glioma cells to methylating and chloroethylating anticancer drugs: p53 determines the switch by regulating xpc, ddb2, and DNA double-strand breaks. Cancer Res. 2007;67:11886–11895. doi: 10.1158/0008-5472.CAN-07-2964. [DOI] [PubMed] [Google Scholar]
- 66.Roos WP, et al. Apoptosis in malignant glioma cells triggered by the temozolomide-induced DNA lesion O6-methylguanine. Oncogene. 2007;26:186–197. doi: 10.1038/sj.onc.1209785. [DOI] [PubMed] [Google Scholar]
- 67.Hermisson M, et al. O6-methylguanine DNA methyltransferase and p53 status predict temozolomide sensitivity in human malignant glioma cells. J. Neurochem. 2006;96:766–776. doi: 10.1111/j.1471-4159.2005.03583.x. [DOI] [PubMed] [Google Scholar]
- 68.Bar-Nur O, et al. Lineage conversion induced by pluripotency factors involves transient passage through an iPSC stage. Nat. Biotechnol. 2015;33:761–768. doi: 10.1038/nbt.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vierbuchen T, Wernig M. Molecular roadblocks for cellular reprogramming. Mol. Cell. 2012;47:827–838. doi: 10.1016/j.molcel.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Efe JA, et al. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat. Cell Biol. 2011;13:215–222. doi: 10.1038/ncb2164. [DOI] [PubMed] [Google Scholar]
- 71.Kim J, et al. Direct reprogramming of mouse fibroblasts to neural progenitors. Proc. Natl Acad. Sci. USA. 2011;108:7838–7843. doi: 10.1073/pnas.1103113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li K, et al. Small molecules facilitate the reprogramming of mouse fibroblasts into pancreatic lineages. Cell Stem Cell. 2014;14:228–236. doi: 10.1016/j.stem.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu S, et al. Mouse liver repopulation with hepatocytes generated from human fibroblasts. Nature. 2014;508:93–97. doi: 10.1038/nature13020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 75.Saha A, Wittmeyer J, Cairns BR. Mechanisms for nucleosome movement by ATP-dependent chromatin remodeling complexes. Results Probl. Cell Differ. 2006;41:127–148. doi: 10.1007/400_005. [DOI] [PubMed] [Google Scholar]
- 76.Enane, F. O. Hepatocyte Differentiation and Hepatocellular Carcinoma: Rationale for p53 Independent Therapy. PhD thesis, (Case Western Reserve University, 2017).
- 77.Saha A, Wittmeyer J, Cairns BR. Chromatin remodelling: the industrial revolution of DNA around histones. Nat. Rev. Mol. Cell Biol. 2006;7:437–447. doi: 10.1038/nrm1945. [DOI] [PubMed] [Google Scholar]
- 78.Tran HG, Steger DJ, Iyer VR, Johnson AD. The chromo domain protein chd1p from budding yeast is an ATP-dependent chromatin-modifying factor. EMBO J. 2000;19:2323–2331. doi: 10.1093/emboj/19.10.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sun X, et al. Suppression of the SWI/SNF component Arid1a promotes mammalian regeneration. Cell Stem Cell. 2016;18:456–466. doi: 10.1016/j.stem.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xuan S, et al. Pancreas-specific deletion of mouse Gata4 and Gata6 causes pancreatic agenesis. J. Clin. Invest. 2012;122:3516–3528. doi: 10.1172/JCI63352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shi, Z. D. et al. Genome Editing in hPSCs Reveals GATA6 Haploinsufficiency and a Genetic Interaction with GATA4 in human pancreatic development. Cell Stem Cell20, 675–688 e676 (2017). [DOI] [PMC free article] [PubMed]
- 82.Chan AL, et al. Germline stem cell activity is sustained by SALL4-dependent silencing of distinct tumor suppressor genes. Stem Cell Rep. 2017;9:956–971. doi: 10.1016/j.stemcr.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Efimenko E, et al. The transcription factor GATA4 is required for follicular development and normal ovarian function. Dev. Biol. 2013;381:144–158. doi: 10.1016/j.ydbio.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Merika M, Thanos D. Enhanceosomes. Curr. Opin. Genet. Dev. 2001;11:205–208. doi: 10.1016/S0959-437X(00)00180-5. [DOI] [PubMed] [Google Scholar]
- 85.Long HK, Prescott SL, Wysocka J. Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell. 2016;167:1170–1187. doi: 10.1016/j.cell.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lomberk G, et al. Distinct epigenetic landscapes underlie the pathobiology of pancreatic cancer subtypes. Nat. Commun. 2018;9:1978. doi: 10.1038/s41467-018-04383-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Garcia-Ortiz JE, et al. Foxl2 functions in sex determination and histogenesis throughout mouse ovary development. Bmc. Dev. Biol. 2009;9:36. doi: 10.1186/1471-213X-9-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hu Z, et al. RUNX1 regulates corepressor interactions of PU.1. Blood. 2011;117:6498–6508. doi: 10.1182/blood-2010-10-312512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gu X, et al. Runx1 regulation of Pu.1 corepressor/coactivator exchange identifies specific molecular targets for leukemia differentiation therapy. J. Biol. Chem. 2014;289:14881–14895. doi: 10.1074/jbc.M114.562447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schaniel C, et al. Smarcc1/Baf155 couples self-renewal gene repression with changes in chromatin structure in mouse embryonic stem cells. Stem Cells. 2009;27:2979–2991. doi: 10.1002/stem.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.De Marzo AM, et al. Abnormal regulation of DNA methyltransferase expression during colorectal carcinogenesis. Cancer Res. 1999;59:3855–3860. [PubMed] [Google Scholar]
- 92.Nicolle R, et al. Pancreatic adenocarcinoma therapeutic targets revealed by tumor-stroma cross-talk analyses in patient-derived xenografts. Cell Rep. 2017;21:2458–2470. doi: 10.1016/j.celrep.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Furst AL, Barton JK. DNA electrochemistry shows DNMT1 methyltransferase hyperactivity in colorectal tumors. Chem. Biol. 2015;22:938–945. doi: 10.1016/j.chembiol.2015.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Belinsky SA, et al. Inhibition of DNA methylation and histone deacetylation prevents murine lung cancer. Cancer Res. 2003;63:7089–7093. [PubMed] [Google Scholar]
- 95.Kumagai T, et al. Histone deacetylase inhibitor, suberoylanilide hydroxamic acid (Vorinostat, SAHA) profoundly inhibits the growth of human pancreatic cancer cells. Int. J. Cancer. 2007;121:656–665. doi: 10.1002/ijc.22558. [DOI] [PubMed] [Google Scholar]
- 96.Cecconi D, et al. Proteomic analysis of pancreatic ductal carcinoma cells treated with 5-aza-2’-deoxycytidine. Electrophoresis. 2003;24:4291–4303. doi: 10.1002/elps.200305724. [DOI] [PubMed] [Google Scholar]
- 97.Ng KP, et al. p53 independent epigenetic-differentiation treatment in xenotransplant models of acute myeloid leukemia. Leukemia. 2011;25:1739–1750. doi: 10.1038/leu.2011.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Negrotto S, et al. Noncytotoxic differentiation treatment of renal cell cancer. Cancer Res. 2011;71:1431–1441. doi: 10.1158/0008-5472.CAN-10-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Negrotto S, et al. CpG methylation patterns and decitabine treatment response in acute myeloid leukemia cells and normal hematopoietic precursors. Leukemia. 2012;26:244–254. doi: 10.1038/leu.2011.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Alcazar O, et al. Epigenetic regulation by decitabine of melanoma differentiation in vitro and in vivo. Int. J. Cancer. 2012;131:18–29. doi: 10.1002/ijc.26320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shakya R, et al. Hypomethylating therapy in an aggressive stroma-rich model of pancreatic carcinoma. Cancer Res. 2013;73:885–896. doi: 10.1158/0008-5472.CAN-12-1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Saunthararajah Y, et al. Evaluation of noncytotoxic DNMT1-depleting therapy in patients with myelodysplastic syndromes. J. Clin. Invest. 2015;125:1043–1055. doi: 10.1172/JCI78789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu X, et al. UHRF1 targets DNMT1 for DNA methylation through cooperative binding of hemi-methylated DNA and methylated H3K9. Nat. Commun. 2013;4:1563. doi: 10.1038/ncomms2562. [DOI] [PubMed] [Google Scholar]
- 104.Guan D, Factor D, Liu Y, Wang Z, Kao HY. The epigenetic regulator UHRF1 promotes ubiquitination-mediated degradation of the tumor-suppressor protein promyelocytic leukemia protein. Oncogene. 2013;32:3819–3828. doi: 10.1038/onc.2012.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Welch JS, et al. TP53 and decitabine in acute myeloid leukemia and myelodysplastic syndromes. N. Engl. J. Med. 2016;375:2023–2036. doi: 10.1056/NEJMoa1605949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lo-Coco F, et al. Retinoic acid and arsenic trioxide for acute promyelocytic leukemia. N. Engl. J. Med. 2013;369:111–121. doi: 10.1056/NEJMoa1300874. [DOI] [PubMed] [Google Scholar]
- 107.Huang ME, et al. All-trans retinoic acid with or without low dose cytosine arabinoside in acute promyelocytic leukemia. Report of 6 cases. Chin. Med. J. 1987;100:949–953. [PubMed] [Google Scholar]
- 108.Bejanyan N, et al. A phase 2 trial of combination therapy with thalidomide, arsenic trioxide, dexamethasone, and ascorbic acid (TADA) in patients with overlap myelodysplastic/myeloproliferative neoplasms (MDS/MPN) or primary myelofibrosis (PMF) Cancer. 2012;118:3968–3976. doi: 10.1002/cncr.26741. [DOI] [PubMed] [Google Scholar]
- 109.Ebrahem Q, Mahfouz RZ, Ng KP, Saunthararajah Y. High cytidine deaminase expression in the liver provides sanctuary for cancer cells from decitabine treatment effects. Oncotarget. 2012;3:1137–1145. doi: 10.18632/oncotarget.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lavelle D, et al. Effects of tetrahydrouridine on pharmacokinetics and pharmacodynamics of oral decitabine. Blood. 2012;119:1240–1247. doi: 10.1182/blood-2011-08-371690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Smith ML, Hills RK, Grimwade D. Independent prognostic variables in acute myeloid leukaemia. Blood Rev. 2011;25:39–51. doi: 10.1016/j.blre.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 112.Harris WJ, et al. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell. 2012;21:473–487. doi: 10.1016/j.ccr.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 113.Schenk T, et al. Inhibition of the LSD1 (KDM1A) demethylase reactivates the all-trans-retinoic acid differentiation pathway in acute myeloid leukemia. Nat. Med. 2012;18:605–611. doi: 10.1038/nm.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mould DP, McGonagle AE, Wiseman DH, Williams EL, Jordan AM. Reversible inhibitors of LSD1 as therapeutic agents in acute myeloid leukemia: clinical significance and progress to date. Med. Res. Rev. 2015;35:586–618. doi: 10.1002/med.21334. [DOI] [PubMed] [Google Scholar]
- 115.Mohammad HsssP, et al. A DNA hypomethylation signature predicts antitumor activity of LSD1 inhibitors in SCLC. Cancer Cell. 2015;28:57–69. doi: 10.1016/j.ccell.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 116.Komura S, et al. An EWS-FLI1-induced osteosarcoma model unveiled a crucial role of impaired osteogenic differentiation on osteosarcomadevelopment. Stem Cell Rep. 2016;6:592–606. doi: 10.1016/j.stemcr.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Carugo A, et al. In vivo functional platform targeting patient-derived xenografts identifies WDR5-Myc association as a critical determinant of pancreatic cancer. Cell Rep. 2016;16:133–147. doi: 10.1016/j.celrep.2016.05.063. [DOI] [PubMed] [Google Scholar]
- 118.Ahmed K, et al. Global chromatin architecture reflects pluripotency and lineage commitment in the early mouse embryo. PLoS One. 2010;5:e10531. doi: 10.1371/journal.pone.0010531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Balch C, et al. Antimitogenic and chemosensitizing effects of the methylation inhibitor zebularine in ovarian cancer. Mol. Cancer Ther. 2005;4:1505–1514. doi: 10.1158/1535-7163.MCT-05-0216. [DOI] [PubMed] [Google Scholar]
- 120.Lee, C. S., Friedman, J. R., Fulmer, J. T. & Kaestner, K. H. The initiation of liver development is dependent on Foxa transcription factors. Nature 435, 944-947, 10.1038/nature03649 (2005). [DOI] [PubMed]
- 121.Fang F, et al. A phase 1 and pharmacodynamic study of decitabine in combination with carboplatin in patients with recurrent, platinum-resistant, epithelial ovarian cancer. Cancer. 2010;116:4043–4053. doi: 10.1002/cncr.25204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rodolosse, A., Campos, M. L., Rooman, I., Lichtenstein, M. & Real, F. X. p/CAF modulates the activity of the transcription factor p48/Ptf1a involved in pancreatic acinar differentiation. Biochem J 418, 463-473, 10.1042/BJ20080293 (2009). [DOI] [PubMed]
- 123.Lee, C. S., Sund, N. J., Behr, R., Herrera, P. L. & Kaestner, K. H. Foxa2 is required for the differentiation of pancreatic alpha-cells. Dev Biol 278, 484-495, 10.1016/j.ydbio.2004.10.012 (2005). [DOI] [PubMed]
- 124.Hosaka, T. et al. Disruption of forkhead transcription factor (FOXO) family members in mice reveals their functional diversification. Proc Natl Acad Sci USA 101, 2975-2980, 10.1073/pnas.0400093101 (2004). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Disease free and overall survival comparisons in cancers with TP53/CDKN2A high versus low Frequency alterations