Abstract
Purpose
The aim of this study was to detect the genetic defects in a large pedigree of affected individuals with various phenotypes of ocular anomalies including partial aniridia, congenital cataract, and nystagmus.
Methods
The entire coding region of paired box gene 6 (PAX6) was amplified by polymerase chain reaction (PCR), sequenced, and compared with a GenBank database.
Results
A novel mutation (c.1170 C > T; p.Gln297X) was found in the proband and all affected members. This nonsense mutation leads to PAX6 protein truncation.
Conclusions
Our findings suggest that this novel mutation is most likely responsible for the pathogenesis of the congenital aniridia, cataract, and nystagmus in this pedigree. To the best of our knowledge, this is the first report of this mutation of PAX6 gene in a kindred pedigree with various ocular abnormalities.
Keywords: PAX6 nonsense mutation, Polymerase chain reaction, Congenital aniridia, Cataract, Nystagmus
Introduction
The paired box gene 6 (PAX6) is located on chromosome 11p13 and belongs to a family of genes with a highly conserved sequence encoding transcription factors which play essential roles for eye, nose, pancreas, and brain development in both vertebrates and invertebrates.1 PAX6 contains 14 exons that encode a protein containing two DNA binding domains (DBDs); a 128 amino-acid paired domain (PD), a 66 amino-acid-long paired-type homeodomain, and a linker region between them.2 PAX6 is expressed in the optic sulci and surface ectoderm during the early stages of eye development, and its substantial expression in the optic sulci and optic stalk, and subsequently in the anterior surface ectoderm and retina points to its critical role in ocular mesenchymal differentiation.3
The PAX6 gene was originally implicated in human aniridia.2 Studies in humans and mice demonstrated that PAX6 haploinsufficiency leads to aniridia, iris hypoplasia, and other ocular maldevelopment, while homozygous mutations result in the entire absence of eye growth along with central nervous system (CNS) and pancreatic malformations.4, 5 Other ocular malformations linked to PAX6 mutations include corneal opacity and vascularization, band-shape corneal keratitis, eccentric pupil, cataract, ectopia lentis, foveal hypoplasia, or aplasia.6
Approximately 92% of PAX6 mutations are nonsense, producing truncated proteins, and only 2% are missense ones that commonly occurs in the N-terminal region, which can lead to an altered DNA-binding. This difference may be a bias because missense patients have lower likelihood of undergoing genetic analysis, due to their phenotypic difference with nonsense cases outlined in textbooks.
PAX6 seems to have a dose-dependent effect on the target genes; i.e. a critical dose of PAX6 is required to induce the transcription of downstream genes, particularly genes whose products are involved in anterior-segment differentiation.7 The dose-dependent activity of PAX6 products is correlated with the magnitude of abnormal ocular manifestations. Another issue is the dominant-negative characteristic of the mutant-PAX6 which results from the enhanced affinity of truncated PAX6 proteins for DNA-binding compared to wild-type ones.2
In this study, we describe a novel PAX6 nonsense mutation in an Iranian family and report the clinical findings associated with this genotype. Direct sequencing revealed a base substitution in exon 9 of the PAX6 gene that results in a frameshift mutation and protein truncation.
Methods
This study was conducted in accordance with the principles of the Declaration of Helsinki and approved by the Medical Ethics Committee of Farabi Eye Hospital. Informed consents were obtained from participants.
Clinical evaluation
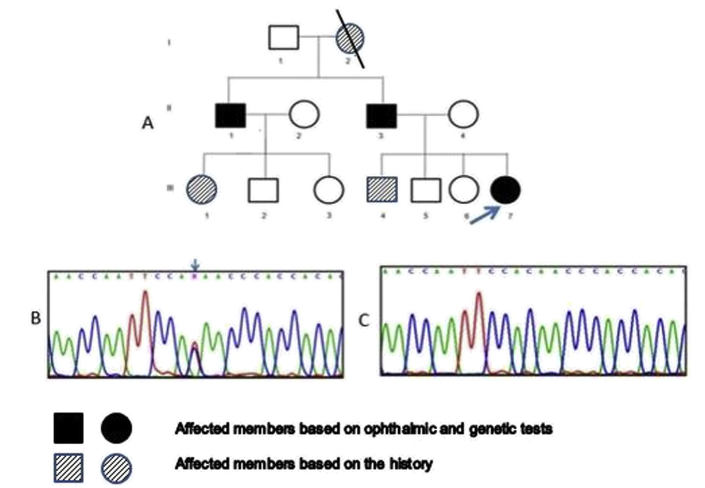
This study concerns 13 members of an Iranian family (Fig. 1A), 6 of which (II-1, 3, 4, and III-5, 6, 7) were available and willing to have a comprehensive ophthalmic examinations at the Anterior Segment Unit of Farabi Eye Hospital, Tehran University of Medical Sciences. After reviewing their history and course of disease, these 6 members underwent examinations including visual acuity tests, slit-lamp examination, funduscopic evaluation, and measurement of the intraocular pressure (IOP) using applanation tonometer, as well as genetic testing. The proband (III-7) also underwent neurological assessment and sampling from the right corneal surface for histopathological studies. The history and clinical information about the remaining 7 family members was obtained from other participants.
Fig. 1.
Pedigree chart and molecular study of an Iranian family with various eye anomalies. (A) The pedigree of the family represents an autosomal dominant pattern of inheritance with three affected patients (arrow points to the proband). (B) Direct sequencing revealed a base substitution (c.1170 C > T; p.Gln297X) in exone 9 of PAX6 gene that results in a frameshift mutation and protein truncation (arrow points to the position of the base substitution). (C) Chromatogram of the wild type of exon 9 of PAX6 gene.
Mutation study
Genetic study was performed for the 6 examined participants. Five milliliters of peripheral blood samples were collected into ethylenediaminetetraacetic acid (EDTA)-containing tubes. Genomic DNA was isolated using the QIAamp DNA Blood Mini Kit (Qiagen, Germany) according to the manufacturer's protocol. The quality and quantity of genomic DNA were analyzed using agarose gel electrophoresis and the NanoDrop 1000 Spectrophotometer (Saveen & Werner ApS, Denmark). Finally, 20 ng of DNA was used for amplification of PAX6 coding sequences using specific primers. Amplification was typically performed using 0.2 μl CinnaGen SmarTaq DNA polymerase (SinaClon Inc, Iran), 10 pmol/μl of each primer, 2.5 μl of each dNTPs, 0.7 μl of 50 mM MgCl2, 1 μl DNA, 2.5 μl of polymerase chain reaction (PCR) buffer, and 1 μl dimethyl sulfoxide in 25 μl of PCR. Thermocycling was performed using a touchdown amplification program on an ABI thermocycler. PCR conditions included an initial denaturation step for 5 min at 95 °C (followed by 40 cycles at 95 °C for 30 s), primer annealing 30 s at 68 °C with a 1 °C decrease every second cycle down to 53 °C, then 53 °C for 16 cycles, 45 s at 72 °C for extension, and finally 7 min at 72 °C. PCR products were separated on 1.5% agarose gels and visualized with ethidium bromide. PCR products were purified using the PCR and Gel Purification Kit and protocol (Bioneer, Korea) and used as the template for DNA sequencing which was performed at the DNA sequencing center (Nasle Omid, Iran and Macrogen, Korea). The sequencing data for each fragment was analyzed using Chromas Version 2.32 software and searching the BLAST (http://www.ncbi.nlm.nih.gov/BLAST) and Ensemble (http://www.ensembl.org) databases and finally compared with RefSeq NM_000280.4. (https://www.ncbi.nlm.nih.gov/nuccore/386642908). The nomenclature in this study is according to Human Genome Variation Society recommendation (http://www.hgvs.org/mutomen/).
Results
Ocular findings
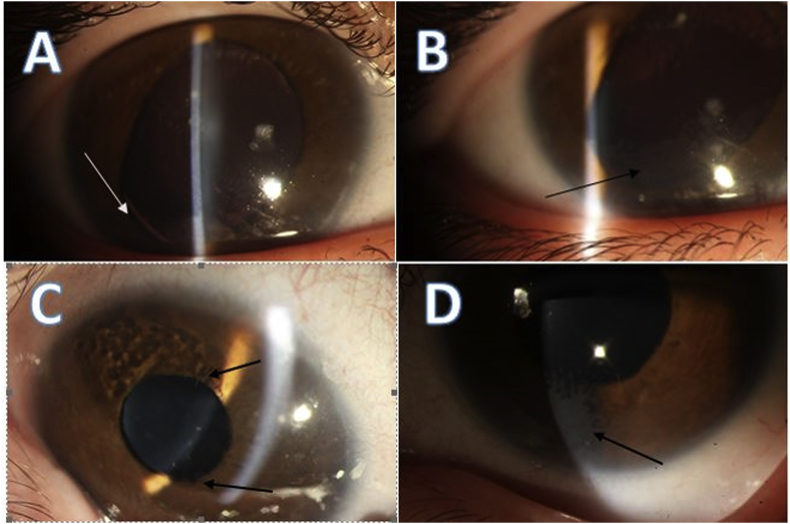
Of the six family members examined in this study (II-1, 3, 4, and III-5, 6, 7), three had abnormal findings in their examinations (II-1, II-3, and III-7). The proband (III-7) presented with normal developmental milestones but had a history of nystagmus and visual inattention from the first few months of the life. Ophthalmological examination of the right eye revealed corneal opacity with a fibrovascular component extending centripetally from the limbus suggesting limbal stem-cell deficiency (LSCD), iris hypoplasia, lens, and iris coloboma. Similar findings and coloboma was observed in the left eye, accompanied by posterior subcapsular opacity (Fig. 2). IOP was 18 mmHg in both eyes as measured with a Schiǿtz tonometer under general anesthesia, and the funduscopic examination was unremarkable. The histopathological study revealed the presence of conjunctival epithelium and goblet cells over the corneal surface, confirming the diagnosis of LSCD. The proband also underwent a thorough neurological work-up and neuroimaging, and no prominent neurological impairment was found.
Fig. 2.
Ophthalmological examinations of the proband (III-7) revealed variable expression of ocular conditions. (A) Incomplete iris and lens coloboma (white arrow). (B) Fibrovascular pannus extending from the limbus (black arrow) of the right eye. (C) Partial iris hypoplasia and crypt-less feature. (D) Superficial fibrovascular pannus extension over the cornea.
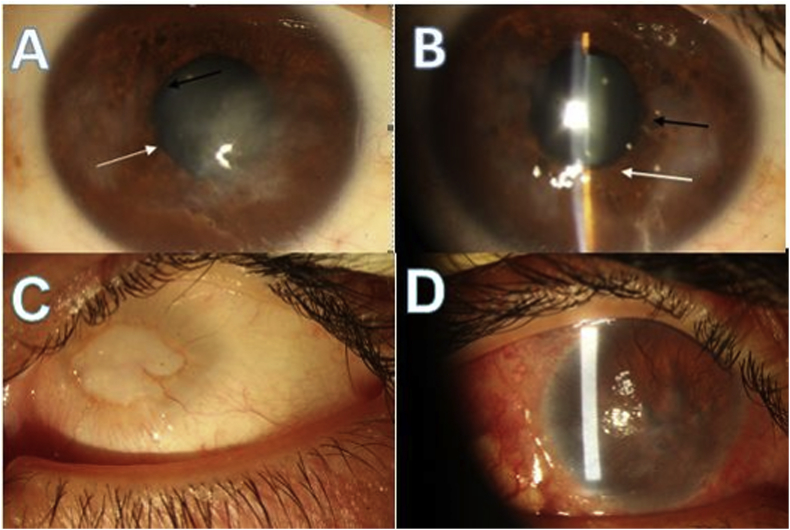
The 46-year-old paternal uncle (II-1) had a history of visual inattention from early infancy, which gradually progressed to severe vision loss and abnormal ocular movement. The best corrected visual acuity of the right and left eyes was light perception (LP) and no light perception (NLP), respectively. He had a history of a glaucoma filtering procedure for advanced glaucoma and cataract surgery in the right eye several years ago. Slit-lamp examination of the right eye revealed extensive conjunctival hyperemia, vascularization, and subconjunctival fibrosis probably due to impaired ocular surface integrity, topical medications, and prior surgical interventions. The right cornea also showed extensive fibrovascular opacity and surface irregularity presumably due to LSCD (Fig. 3C). The left eye was a disorganized small globe with the characteristic features of sclerocornea (Fig. 3D).
Fig. 3.
Ophthalmological features of other affected family members. (A, B) Bilateral fibrovascular pannus, uveal ectropion (black arrow), and iris stromal atrophy adjacent to pupillary border (white arrow) in II-3. (C) Sclerocornea and total corneal opacity of the left eye. (D) Prominent subconjunctival fibrosis and extensive corneal fibrovascular opacity of the right eye in II-1.
Similarly, the proband's 42-year-old father (II-3) had a history of progressive visual loss from early childhood, and visual acuity had gradually declined to counting finger in both eyes, ensued by a high frequency pendular nystagmus. The signs of LSCD were also evident during slit-lamp examination (Fig. 3A–B). The fundus examination was unremarkable, and the IOP was within normal values.
The other 3 examined family members had no complaints of visual impairment, and we found no ocular anomaly during ophthalmologic examinations. Of the remaining 7 members who were not available for ophthalmic or genetic testing, 3 appeared to be affected according to their medical history.
Mutation screening
The coding region of the PAX6 gene was successfully amplified through PCR in all studied DNA samples. Direct sequencing of the PCR product of exon 9 revealed a heterozygous substitution of cytosine by thymine at nucleotide position 1170 of the open reading frame of the PAX6 gene in the proband. This genetic alteration was detected in all three affected family members, but not the others participants (Fig. 1). The point mutation generated a stop codon (TAA) for glutamine (CAA) at amino-acid position 297, resulting in a truncated protein NM_000280.4 (PAX6): c.1170C > T; p. Q297X. Cosegregation of this mutation among affected individuals suggested that it represents an inheritable mutation causing aniridia with unique phenotypes.
Discussion
The PAX6 gene is essential for ocular development and many ocular abnormalities have been reported due to the mutations in this gene. This study reported an Iranian family with various eye anomalies because of PAX6 mutation.
Six persons underwent full ophthalmic examinations, and ocular anomalies were detected in three of the cases. Interestingly, each patient had a unique manifestation, ranging from a subtle change to extensive abnormality. The proband (III-7) presented with infantile nystagmus, iris- and lens-coloboma, unilateral cataract, and centripetal fibrovascular extension across superficial cornea that suggested LSCD confirmed by histopathologic study. The proband's uncle (II-1) shows complete aniridia, and fibrovascular pannus of the cornea in right eye and microphthalmic–sclerocornea in left one, and the proband's father (II-2) presented nystagmus and corneal fibrovascular pannus. In this study we found great phenotypical variability between cases, but fibrovascular extension over the superficial cornea and probably LSCD was the most consistent finding in these subjects.
Literature suggest that approximately 92% of PAX6 mutations are nonsense, and frameshift, generating premature termination codon (PTC) that consequently lead to the production of truncated transcription factors.8
The mechanism by which PAX6 mutations induce variable phenotypic abnormality is complicated and not well understood yet.9, 10, 11 Previous reports postulated nonsense mediated decay (NMD) as a probable mechanism that degrades M-RNAs containing PTC, which leads to significant reduction in the level of truncated proteins.12 Other investigators claimed the specific level of PAX6 protein could be necessary to activate the PAX6 target gene. It might present the haploinsufficiency as a causative mechanism.2 Since the truncated transcripts have no effective activity, and the other normal allele is not capable of producing a high enough dose of transcriptional proteins, the ultimate effect is abolished.2, 4
Another mechanism is the competition of truncated proteins with the wild-type ones, producing a dominant-negative effect. Surprisingly, the truncated protein may have a higher DNA binding affinity and attenuates the normal transcriptional activity of the wild-type one.2 Additionally, various PAX6 transcripts, due to multiple promoters and splice sites, might promote variable phenotypes formation.13, 14, 15
In this study, genetic analyses revealed a novel nonsense mutation c.1170 C > T (p. Gln297X) in the C-terminal of PAX6 which encoding homeodomain part of the transcription factor and resulted in a frameshift and protein truncation. Cross-segregation among members and the mutation that was detected only in the affected people suggested that PAX6 truncated proteins were responsible for the ocular anomalies found in this family.
Although exon 9 of PAX6 is one of the most known sites of mutation with variable phenotypes, in this study, we identified c.1170 C > T (p. Gln297X), a novel nonsense mutation in an Iranian family with variable phenotypes that has not been reported according to human PAX6 databases (http://www.hgu.mrc.ac.uk/Softdata/PAX6/). This study enriches our knowledge about PAX6 mutations and the related phenotypes.
Footnotes
Conflict of Interest: All authors certify that they have no affiliations with or involvement in any organization or entity with any financial interest or non-financial interest in the subject matter or materials discussed in this manuscript.
Peer review under responsibility of the Iranian Society of Ophthalmology.
References
- 1.Chow R.L., Lang R.A. Early eye development in vertebrates. Annu Rev Cell Dev Biol. 2001;17:255–296. doi: 10.1146/annurev.cellbio.17.1.255. [DOI] [PubMed] [Google Scholar]
- 2.Kokotas H., Petersen M. Clinical and molecular aspects of aniridia. Clin Genet. 2010;77(5):409–420. doi: 10.1111/j.1399-0004.2010.01372.x. [DOI] [PubMed] [Google Scholar]
- 3.Walther C., Gruss P. Pax-6, a murine paired box gene, is expressed in the developing CNS. Development. 1991;113(4):1435–1449. doi: 10.1242/dev.113.4.1435. [DOI] [PubMed] [Google Scholar]
- 4.Glaser T., Jepeal L., Edwards J.G., Young S.R., Favor J., Maas R.L. PAX6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous system defects. Nat Genet. 1994;7(4):463–471. doi: 10.1038/ng0894-463. [DOI] [PubMed] [Google Scholar]
- 5.Sander M., Neubüser A., Kalamaras J., Ee H.C., Martin G.R., German M.S. Genetic analysis reveals that PAX6 is required for normal transcription of pancreatic hormone genes and islet development. Genes Dev. 1997;11(13):1662–1673. doi: 10.1101/gad.11.13.1662. [DOI] [PubMed] [Google Scholar]
- 6.St-Onge L., Sosa-Pineda B., Chowdhury K., Mansouri A., Gruss P. Pax6 is required for differentiation of glucagon-producing α-cells in mouse pancreas. Nature. 1997;387(6631):406–409. doi: 10.1038/387406a0. [DOI] [PubMed] [Google Scholar]
- 7.Cvekl A., Tamm E.R. Anterior eye development and ocular mesenchyme: new insights from mouse models and human diseases. Bioessays. 2004;26(4):374–386. doi: 10.1002/bies.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanson I., Churchill A., Love J. Missense mutations in the most ancient residues of the PAX6 paired domain underlie a spectrum of human congenital eye malformations. Hum Mol Genet. 1999;8(2):165–172. doi: 10.1093/hmg/8.2.165. [DOI] [PubMed] [Google Scholar]
- 9.Park S.H., Kim M.S., Chae H., Kim Y., Kim M. Molecular analysis of the PAX6 gene for congenital aniridia in the Korean population: identification of four novel mutations. Mol Vis. 2012;18:488. [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson D.S., Guenther B., Desplan C., Kuriyan J. High resolution crystal structure of a paired (Pax) class cooperative homeodomain dimer on DNA. Cell. 1995;82(5):709–719. doi: 10.1016/0092-8674(95)90468-9. [DOI] [PubMed] [Google Scholar]
- 11.Wolf M.T.F., Lorenz B., Winterpacht A. Ten novel mutations found in aniridia. Hum Mutat. 1998;12(5):304–313. doi: 10.1002/(SICI)1098-1004(1998)12:5<304::AID-HUMU3>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 12.Culbertson M.R. RNA surveillance: unforeseen consequences for gene expression, inherited genetic disorders and cancer. Trends Genet. 1999;15(2):74–80. doi: 10.1016/s0168-9525(98)01658-8. [DOI] [PubMed] [Google Scholar]
- 13.Vincent M.C., Gallai R., Olivier D. Variable phenotype related to a novel PAX 6 mutation (IVS4+5G>C) in a family presenting congenital nystagmus and foveal hypoplasia. Am J Ophthalmol. 2004;138(6):1016–1021. doi: 10.1016/j.ajo.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 14.Kim J., Lauderdale J.D. Analysis of Pax6 expression using a BAC transgene reveals the presence of a paired-less isoform of Pax6 in the eye and olfactory bulb. Dev Biol. 2006;292(2):486–505. doi: 10.1016/j.ydbio.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 15.Bandah D., Rosenmann A., Blumenfeld A., Averbukh E., Banin E., Sharon D. A novel de novo PAX6 mutation in an Ashkenazi-Jewish family with aniridia. Mol Vis. 2008;14:142. [PMC free article] [PubMed] [Google Scholar]