Highlights
-
•
Revealing the biodiversity of Red Sea marine invertebrates associated microorganisms by 16S rRNA sequence analysis.
-
•
Isolation of bacteria and actinomycetes with antimicrobial activity from different Red Sea invertebrates.
-
•
Detection of biosynthetic gene clusters (PKS and NRPS) in microorganisms associated with the Red Sea marine invertebrates.
Keywords: Polyketide synthase (PKS), Nonribosomal peptide synthetase (NRPS), Antimicrobial screening, Red Sea, 16s rRNA
Abstract
Marine invertebrates-associated microorganisms were considered to be important sources of marine bioactive products. This study aims to isolate marine invertebrates associated bacteria with antimicrobial activity from the Red Sea and test their biosynthetic potential through the detection of PKS and NRPS gene clusters involved with the production of bioactive secondary metabolites. In this respect, fifty bacterial strains were isolated from eight different Red Sea marine invertebrates and screened for their antimicrobial activity against standard pathogenic bacteria (Staphylococcus aureus ATCC 25923, Escherichia coli ATCC 25922, Bacillus subtilis ATCC 6633) and yeast (Candida albicans ATCC 10231) using the standard well diffusion assay. Five isolates showed antifungal activity against Candida albicans with no activity recorded against other pathogenic bacterial strains. On the other hand when these isolates were screened for the presence of biosynthetic gene clusters (PKS and NRPS) by PCR using five sets of degenerative primers, 60% of the isolates were shown to contain at least one type of PKS and NRPS gene clusters, which indicates the biosynthetic potential of these isolates even if the isolates didn’t express any biological activity in vitro. Moreover the 16S rRNA molecular identification of the isolates reveal the biodiversity of the red sea marine invertebrates associated bacteria as they were found to belong to several bacterial groups present in Alphaproteobacteria, Gammaproteobacteria, Actinobacteria and Firmicutes.
1. Introduction
Antimicrobial resistance (AMR) is a critical global threat which decreases the opportunities for the treatment of infectious diseases caused by viruses, bacteria, and fungi [1,2]. A World Health Organization (WHO) global surveillance report pointed to an increase of morbidity and mortality of infectious diseases due to AMR, which could result in a world-wide economic loss of up to 100 trillion US dollars (USD) in 2050 due to a 2%–3% reduction in the gross domestic product [2,1]. It is estimated that AMR now annually contributes to 700,000 deaths worldwide, with a potential increase to 10 million in 2050 [1].
Therefore, the discovery of new types of antimicrobial compounds with clinical significance is urgent [3,4]. Previously, terrestrial resources have been extensively investigated for the discovery of new bioactive compounds [5]. However, now-a-days, isolation of new sources of antimicrobial and other bioactive compounds from marine environments is increasing rapidly and still under investigation [6].
The marine environment covers more than 70% of planet surface and it harbors high biodiversity as among the 36 known living phyla, 34 of them are found in marine environments with more than 300,000 known species of fauna and flora [7].
Marine macro-organisms such as sponges, tunicates, soft and hard corals, bryozoans, sea slugs and other invertebrates have proven to be important sources of natural bioactive products [8,9]. However, The availability of biomass is a restricting factor for isolating these natural products [10]. In the case of bryostatin, 13,000 kg of Bryozoans neritina is needed to obtain only 18 g of bryostatin for anti cancer clinical trials [11]. On the other hand, mounting evidence suggests that many of the compounds originally associated with the biomass of the marine invertebrates are not produced by the organism itself but are synthesized by symbiotic or associated microorganisms, or derive from a diet of microorganisms [12]. Thus, marine invertebrate-associated microorganisms are considered important sources of marine bioactive products.
It was reported that about 230 marine natural bio-products have been isolated from 2009 to 2011, and most of them (about 102 compounds) have shown significant antimicrobial activities [13].
Moreover, the Red Sea is found to have geochemical and physical parameters which make it a unique marine environment in comparison to other marine ecosystems [14]. It is characterized by a high temperature which varies from about 24 °C in spring and up to 35 °C in summer (at the surface) to 22 °C (from 200 m to the bottom), and high salinity (approximately 41 PSU) [15,16], due to the high rate of evaporation, low level of precipitation and lack of major river inflows [14]. Red sea is also characterized by unique coral reef systems and seasonal fluctuation of air [17]. As a result of these factors, the Red Sea enjoys unique microbial diversity with unique primary and secondary metabolites [18].
It was found that the microorganisms production of many biologically active secondary metabolites require the activation of specific gene clusters encoding multi-modular enzymes. These gene clusters may be in the form of non-ribosomal peptide synthetases (NRPS) and polyketide synthases (PKS). In order to produce these types of secondary metabolites, a set of domains are required within the bacterial biosynthetic pathways; these include ketosynthase (KS), acyltransferase (AT) and acyl carrier proteins in PKSs and adenylation (A), condensation (C) and peptidyl carrier proteins (PCPs) for peptide elongation in NRPSs [19]. Interestingly, antibiotic compounds such as tetracycline and erythromycin, anticancer agent, e.g., bleomycin and the immunosuppressive agent, e.g., rapamycin [20,21] were reported to be produced from PKS and NRPS pathways.
In this article, actinomycetes and bacteria were isolated from different Red Sea marine invertebrates and their metabolic extracts were tested for antimicrobial activity against different pathogens. Further, the isolated microorganisms were tested for the presence of polyketide synthases (PKSs) and non-ribosomal peptide synthetases (NRPSs) coding for multifunctional enzymes which are involved in the synthesis of a wide range of structurally diverse natural compounds, many of which are found to have medical importance [22].
2. Materials and methods
2.1. Samples collection
On February 2014, specimens of eight different marine invertebrates (from two soft corals, a tunicate, a sponge and four hard corals) were collected from Red Sea at El-Tor, Sinai, Egypt at GPS location (28°13′42.8″N 33°37′19.4″E) in a water depth of 2–3 m. Samples were cut with a sterile dive knife and individual pieces were transferred to sterile plastic sample collection bags, brought to the surface and maintained at ambient seawater temperature until they were transferred to the laboratory within 7 h.
2.2. Sample processing and isolation of associated bacteria
Each sample was rinsed with sterile natural seawater (NSW) to remove transient and loosely attached bacteria. One cm3 of each sample was homogenized with 9 ml of sterile NSW in a sterile mortar then a tenfold serial dilution was carried out using NSW to obtain dilutions varying from 10−1 to 10−6. 100 μl of each dilution was spread onto plates of Marine agar (Difco™, USA), Actinomycetes isolation agar (Difco™, USA), ISP2 agar (4 g/L yeast extract, 10 g/L malt extract, 4 g/L dextrose, 15 g/L agar), Starch casein agar (10 g/L soluble starch, 1 g/L Casein sodium salt, 0.5 g/L K2HPO4, 15 g/L agar) [23], R2A agar plates containing 2% (w/v) NaCl (Difco™) and M1 agar plates (10 g/L soluble starch, 4 g/L yeast, 2 g/L peptone, 15 g/L agar) [24]. All media were prepared by using NSW except Marine and R2A agar, and all except Marine agar were supplemented with 0.2 μm pore size filter sterilized cycloheximide (100 μg/mL), nystatin (25 μg/mL) and nalidixic acid (25 μg/mL). Cycloheximide and nystatin were added to inhibit fungal growth; while nalidixic acid inhibits many fast-growing Gram-negative bacteria [25] then plates were incubated at 30 °C for 6–9 weeks in aerobic condition. The growing colonies were picked and serially streak-plated on agar plates until pure colonies were achieved. For preservation the purified colonies were cultured on liquid media (media on which colonies were initially isolated) and supplemented with 15% (v/v) glycerol then stored in −80 °C [26].
2.3. Metabolic extract preparation and screening for antimicrobial activity
The isolates were cultured in 50 mL of liquid media (the media on which the colonies were initially isolated). Cultures were incubated for 2–10 days depending on their growth rate at 30 °C while shaking at 150 rpm (New Brunswick scientific Innova●43 incubator shaker, USA). The isolates were left in the incubator shaker 3–4 days after their growth to release their metabolites in the culture media. An equal volume of methanol was added to the liquid cultures for cell lysis and shaking was continued (150 rpm, 1 h at room temperature). The broth was centrifuged in 50 mL falcon tubes (5000 rpm, 15 min at room temperature), the supernatant was evaporated and the extract was dissolved in 20 mg/ml of DMSO.
The antimicrobial activity testing was carried out using the standard well diffusion assay as described by Flemer et al. [27]. Briefly, 150 μl of 20 mg/ml metabolic extract of each isolate was tested against 500 μl of standard pathogenic bacteria (OD at ƛ650 nm = 0.45) (Staphylococcus aureus ATCC 25923, Escherichia coli ATCC 25922, Bacillus subtilis ATCC 6633) and yeast (Candida albicans ATCC 10231). Muller-Hinton agar medium was used for testing all pathogenic strains and negative control of DMSO as a solvent of the extract was included. The antimicrobial activity was recorded by measuring the inhibition zone diameter after incubation at 37 °C for 24 h for bacteria and 48 h for Candida albicans.
2.4. Genomic DNA extraction from marine invertebrates associated bacterial isolates
Due to the different natures and characters of the marine bacterial isolates, their genomic DNA were extracted using two different protocols; DNA of some isolates were extracted using QIAamp DNA mini kit (Qiagen, Germany) according to the manufacturer protocol while the other isolates DNA were extracted as described by Maloy [28]. Moreover, DNA of the isolated actinomycetes were extracted using the protocol described by Pospiech and Neumann [29]. DNA quantity and purity were determined using NanoDrop™ 1000 Spectrophotometer V3.7 (Thermo Fisher Scientific Inc, USA).
2.5. Amplification of biosynthetic genes (PKS and NRPS) from extracted DNA
Five sets of degenerative primers (Macrogen Inc, Korea) (listed in Table 1) – A degenerative primer is a mixture of oligonucleotide sequences in which some positions contain a number of possible bases and according to the International Union of Pure and Applied Chemistry (IUPAC) system for degenerate bases we can insert a single-letter code to represent our unknown region as for example Y is code for C or T while V is code for A,C or G - were used for amplification of PKS-I, PKS-II and NRPS. These NRPS and PKS primers target the A and KS domains, respectively. DNA of the reference strain Streptomyces hygroscopicus ATCC 29253 was used as a positive control for the presence of PKS and NRPS gene clusters. PCR cycles with (DKF-MTR) and (MDPQQR f-HGTGT r) primers consisted of: an initial denaturing step of 5 min at 95 °C, 10 cycles of 1 min at 95 °C, 30 s at 60 °C and 1 min at 72 °C, with the annealing temperature reduced by 2 °C per cycle, followed by 30 cycles of 95 °C for 1 min, 40 °C for 30 s and 72 °C for 1 min with a final extension of 10 min at 72 °C [30] with (A3F-A7R) primer set the PCR conditions were 95 °C for 5 min for initial denaturation followed by 35 cycles of 95 °C for 30 s and 56 °C for 2 min and 72 °C for 1.5 min with a final extension of 10 min at 72 °C, with (PF6-PR6) the conditions were 96 °C for 5 min for initial denaturation followed by 30 cycles of 96 °C for 1 min and 58◦C for 1 min and 72 °C for 1.5 min with a final extension of 10 min at 72 °C and with (K1F-M6R) the reaction was done at 95 °C for 1 min for initial denaturation followed by 30 cycles of 94 °C for 35 s and 55 °C for 40 s and 72 °C for 2 min with a final extension of 8 min at 72 °C. All the PCR reactions were performed in (BIO-RAD T100TM Thermal cycler, Singapore) in 25 μl aliquots containing 12.5 μl of PCR Master Mix (BIOLINE, Meridian Life Science Inc, USA), 1 μl of forward primer, 1 μl of reverse primer (each primer of 10 pmol), 1 μl of 100 ng/μl DNA and RNase-free water to final volume of 25 μl. PCR amplicons were analyzed by electrophoresis in 1.2% (w/v) agarose gel with a molecular size marker 100 bp (Cleaver scientific, Ltd, UK).
Table 1.
Primers used in this study.
| Primer ID | Sequence | Target genes | References |
|---|---|---|---|
| 27 F | 5 AGAGTTTGATCCTGGCTCAG 3 | 16S rRNA | [32] |
| 1492R | 5 GGTTACCTTGTTACGACTT 3 | ||
| DKF | 5′GTGCCGGTNCCRTGNGYYTC3′ | NRPS | [33] |
| MTR | 5′GCNGGYGGYGCNTAYGTNCC3′ | ||
| A3F | 5′GCSTACSYSATSTACACSTCSGG 3′ | NRPS | [34] |
| A7R | 5′SASGTCVCCSGTSCGGTAS3′ | ||
| MDPQQR f | 5′RTRGAYCCNCAGCAICG-3′ | PKS-I | [30] |
| HGTGT r | 5′VGTNCCNGTGCCRTG-3′ | ||
| K1F | 5′TSAAGTCSAACATCGGBCA 3′ | PKS-I | [34] |
| M6R | 5′CGCAGGTTSCSGTACCAGTA 3′ | ||
| PF6 | 5′TSGCSTGCTTGGAYGCSATC 3′ | PKS-II | [35] |
| PR6 | 5′TGGAANCCGCCGAABCCGCT 3′ | ||
2.6. Cloning and sequencing
Amplified PKS-II fragments of bacterial isolate (74) were cloned with the TOPO TA cloning kit (Invitrogen, USA, cat.no.450030) according to the manufacturer’s instructions, transformed into chemical competent E. coli DH5α which was prepared as indicated by Sambrook et al. then plasmids were isolated by following the Qiaprep Spin Miniprep kit manufacturer protocol (Qiagen, Germany, cat.no.27104) and checked for inserts by PCR using M13F and M13R primers (Invitrogen,USA). Plasmids with positive inserts were sent to (Solgent co.Ltd, South Korea) for sanger sequencing from both sides using M13F and M13R primers.
2.7. Amplification of partial 16S rRNA gene products
The extracted DNA of 10 isolates found to possess the three types of the biosynthetic genes (NRPS, PKS-I and PKS-II) gene fragments were used as a template for the amplification of approximately 1500 bp of the 16S rRNA gene fragment by polymerase chain reaction (PCR) using the universal bacterial primers 27 F and 1492R (Macrogen, Korea) (Table 1). PCR was carried out under the following conditions: initial denaturation at 94 °C for 5 min, followed by 30 cycles of 94 °C for 45 s, 53 °C for 45 s and 72 °C for 1.5 min, with a final extension 72 °C for 10 min. PCR amplicons were analyzed by electrophoresis in 1.0% (w/v) agarose gel against a molecular size marker 100 bp (Cleaver scientific, Ltd, UK). Where the bands were detected at 1500 bp then the amplicons were sent to (Solgent co.Ltd, South Korea) for purification and Sanger sequencing from both sides using 27 f and 1492r primers.
2.8. Phylogenetic analysis
Vector contamination in the sequencing result of the amplified PKS-II fragment of bacterial isolate (74) was detected using VecScreen tool at the National Center of Biotechnology Information (NCBI) then removed and the full sequence of KS domain and of 16S rRNA fragment were assembled using (DNA baser assembler V4). All Sequences were analyzed using Blast x (for PKS-II) and Blast n (for 16S rRNA) tools at NCBI then phylogenetic trees were constructed using Molecular Evolutionary Genetics Analysis (MEGA) 7 program [31].
3. Results
3.1. Isolation of marine invertebrates associated bacteria and actinomycetes
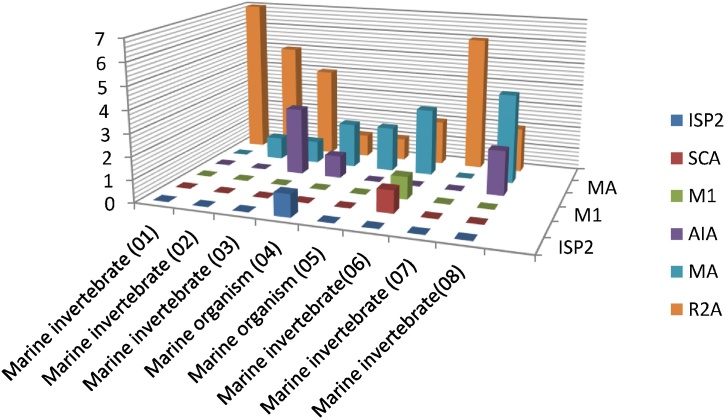
A total of 50 bacterial isolates were obtained from 8 different marine invertebrates. The highest proportion of the strains (56%) were recovered by using R2A agar media (Difco) followed by 26% by Marine agar (Difco) and 12% by Actinomycete isolation agar (Difco) while each of media M1, ISP2 and Starch casein agar recovered only one strain. The data about the isolates and their source were illustrated by Fig. 1.
Fig. 1.
The number of bacterial strains isolated by different types of media from each marine invertebrate. AIA: Actinomycetes isolation agar, SCA: Starch casein agar, MA: Marine agar, R2A: R2A agar: M1: M1 agar: ISP2: ISP2 agar.
3.2. Anti-microbial activity
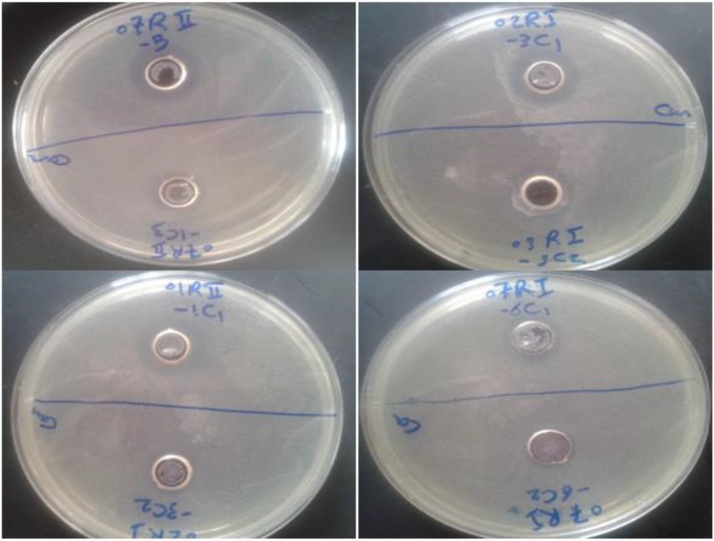
The metabolic extract of the isolated strains were tested against standard pathogenic bacteria (Staphylococcus aureus ATCC 25923, Escherichia coli ATCC 25922, Bacillus subtilis ATCC 6633) and yeast (Candida albicans ATCC 10231) using the standard well diffusion assay. Five isolates showed antifungal activity against Candida albicans ATCC 10231 as shown in Fig. 4 with no activity recorded against other pathogenic bacterial strains. The measurements of the inhibition zone diameters of the 5 bioactive strains were illustrated in Table 2.
Fig. 4.
Isolates with positive antifungal activity against Candida albicans using standard well diffusion assay. The isolates labeled with (07R II -5, 07R II-1C3, 02RI-3C1, 01R II-1C1 and 07RI-6C1) represent isolates (74, 75, 25, 13 and 71) respectively.
Table 2.
Antimicrobial activity of the isolated bacteria.
| Isolate (ID) | Inhibition zone diameter (mm) Candida albicans ATCC 10231 |
|---|---|
| 13 | 16 |
| 25 | 18 |
| 71 | 13 |
| 74 | 17 |
| 75 | 13 |
3.3. Detection of PKS and NRPS biosynthetic genes
All isolated strains were tested for the presence of biosynthetic gene clusters (PKS and NRPS) as the presence of these genes indicates the biosynthetic potential of these isolates, even if the isolates didn’t express any biological activity in the screening test. The KS domain was successfully amplified from 38% (for PKS-1) and 54% for (PKS-II) of the DNA template of the 50 isolates, while 38% of the isolates showed positive amplification of the NRPS A domain. 27% of the strains showing positive PCR amplification had a single type of biosynthetic gene cluster (PKS-I, 2 strains; PKS-II, 5 strains; NRPS, only one strain) while the remaining positive strains had two or more of the biosynthetic gene clusters: 13% possessed both PKS-I and PKS-II; 3% had both PKS-I and NRPS; 17% had both PKS-II and NRPS while 40% of the positive strains had all the three biosynthetic gene clusters. The results of the antimicrobial screening and the PCR amplification of the biosynthetic genes are illustrated in Table 4 (Appendix A. Supplementary data).
3.4. Cloning, sequencing and sequencing analysis of PKS KS domains
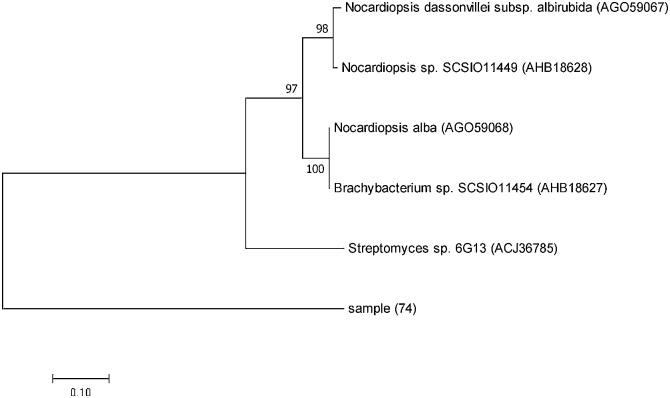
The PKS-II KS domain of isolate 74, which was amplified using primers (PF6-PR6), was chosen to be cloned and sequenced to confirm the result of the PCR amplification. The sequence analysis using BLAST x tool at (NCBI) showed that the amino acid sequence of this gene fragment had 96% identity with type II PKS of Nocardiopsis dassonvillei subsp. Albirubida (accession No. AGO59067.1) and the phylogenetic tree of this KS domain was constructed (see Fig. 2).
Fig. 2.
Phylogenetic analysis of type II PKS KS domain from sample (74) by Maximum Likelihood method. The evolutionary history was inferred by using the Maximum Likelihood method based on the Whelan and Goldman model [36]. The percentage of trees in which the associated taxa clustered together in the bootstrap test (10,000 replicates) is shown next to the branches. Evolutionary analyses were conducted in MEGA7 [31].
3.5. 16S rRNA gene analysis
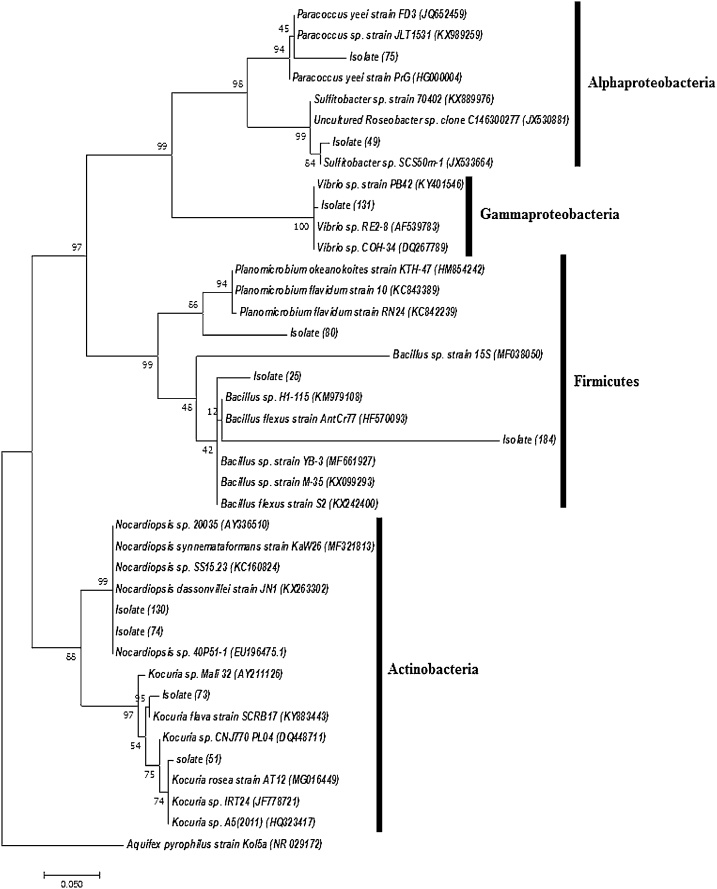
Ten isolates of those showing positive amplification of the three classes of the biosynthetic genes (PKS-1, PKS-II and NRPS) were selected to be identified through the sequencing of the PCR amplified 16S rRNA and analyzing the sequences using BLAST n tool at NCBI to construct a phylogenetic tree (Fig. 3). The results obtained from the sequence analysis are represented in (Table 3). The isolates were found to belong to different bacterial groups as 40% of the isolates were found to belong to Actinobacteria, 20% to Alphaproteobacteria, 10% to Gammaproteobacteria, and 30% to Firmicutes.
Fig. 3.
Molecular rooted phylogenetic 16S rRNA gene tree analysis by Maximum Likelihood method. The tree was rooted using the 16S rRNA partial gene sequence of Aquifex pyrophilus strain kol5a (NR 029172.1). The evolutionary history was inferred by using the Maximum Likelihood method based on the Kimura 2-parameter model [37]. The percentage of trees in which the associated taxa clustered together in the bootstrap test (10,000 replicates) is shown next to the branches. Evolutionary analyses were conducted in MEGA7 [31].
Table 3.
Sequence analysis of 16S rRNA genes of the tested isolates using BLAST n.
| Isolate ID | Name of the most closely related strains | Maximum score | Identity | Accession number |
|---|---|---|---|---|
| 25 | Bacillus sp. strain M-35 | 2174 | 97% | KX099293 |
| 49 | Sulfitobacter sp. strain 70391 | 649 | 98% | MF045096 |
| 51 | Kocuria rosea strain AT12 | 2416 | 98% | MG016449 |
| 73 | Kocuria flava strain SCRB17 | 793 | 98% | KY883443 |
| 74 | Nocardiopsis dassonvillei strain JN1 | 946 | 97% | KX263302 |
| 75 | Paracoccus sp. strain JLT1531 | 532 | 97% | KX989259 |
| 80 | Planomicrobium okeanokoites strain KTH-47 | 2217 | 98% | HM854242 |
| 184 | Geobacillus stearothermophilus strain NB3-8 | 637 | 100% | KR999908 |
| 130 | Nocardiopsis synnemataformans strain Al-H5A-3 | 2410 | 99% | KF384488 |
| 131 | Vibrio splendidus strain GHrC13 | 717 | 99% | GQ375456 |
3.6. Nucleotide sequence accession numbers
The sequences of the 16S rRNA presented in this study were deposited in GenBank and have accession numbers of (MH040868, MH040869, MH040870, MH040871, MH045063, MH045064, MH045263, MH045264, MH046783 and MH046784) for the isolates (51,130, 80 25, 131, 184, 73, 74, 75 and 49) respectively.
4. Discussion
The aim of this study was to isolate marine invertebrate associated bacteria with antimicrobial activity from Red Sea and test their biosynthetic potential through the detection of PKS and NRPS gene clusters involved with the production of bioactive secondary metabolites. In agreement with Palomo et al. [38], high percentage of the isolates (60%) were shown to contain at least one type of PKS or NRPS, while only five isolates, representing 10% showed antimicrobial activity against Candida albicans ATCC 10231. However, this low percentage of the antimicrobial bioactive isolates conflicts with the observations of other previous studies [39,40] which screened Red Sea marine invertebrates associated bacteria and actinomycetes for having antimicrobial activity as Abdelmohsen et al. reported that 35% of the tested actinomycetes isolated from Red Sea sponges were found to have antimicrobial activity [39] while ElAhwany et al. reported that 80% of the tested strains isolated from Red Sea corals showed activity against at least one indicator pathogen used in their screening [40].
Depending on the studies which considered that the presence of the biosynthetic gene clusters (PKS and NRPS) in the genomic DNA of an organism is an indication for the biosynthetic potential of that organism to produce bioactive metabolites [[41], [42], [43]], there may be two explanations for the previous results of our study. The first one is that the products may have another activity rather than the antimicrobial such as antitumor activity as discussed in the study of El-Moneam et al. which reported a relation between cytotoxic bioactivity of the Red Sea Sponge, Hyrtios aff. Erectus associated bacteria and the presence of associated bioactive metabolic pathways genes related to polyketides [44]. As well, Sagar et al. and Abdelfattah et al. support this previous probability as both of them reported a potent activity against cancer cell lines of a red sea derived strains of Sulfitobacter sp. and Nocardiopsis sp. respectively while, in our study isolate (49) which was molecularly identified as a strain of Sulfitobacter sp. and isolates (74) and (130) which were identified as strains of Nocadiopsis sp., found to possess three different types of biosynthetic genes (PKS-1, II and NRPS) [45,46].
The second explanation may be due to the poor expression or repression of the biosynthetic gene clusters, many biosynthetic genes remain silent and are not expressed in vitro until they are activated by stimuli [47], such as providing proper cultivation conditions as reported by Dashti et al. [48].
As a result of these explanations, it is recommended to screen for further targets to reveal other biological activities of these isolates or use appropriate cultivation conditions to activate the expression of the silent gene clusters. In terms of the phylogenetic analyses of the 16S rRNA sequences of the isolates, it revealed the biodiversity of the Red Sea marine invertebrate associated bacteria as they were found to belong to several bacterial groups represented in Alphaproteobacteria, Gammaproteobacteria, Actinobacteria and Firmicutes. Additionally, isolates belong to species of Vibrio, Sulfitobacter, Kocuria and Nocardioposis were found to possess the three different types of biosynthetic gene clusters (PKS1, II and NRPS) and this was in agreement with being well known for their production of bioactive secondary metabolites [38,45,[49], [50], [51], [52]].
5. Conclusion
The presence of the biosynthetic gene clusters (PKS or NRPS or both) in 60% of the strains isolated from Red Sea invertebrates indicates the high biosynthetic potential of these isolates to produce bioactive metabolites with either antimicrobial activity or any other biological activities. This was supported by the presence of these genes in isolates (74, 75 and 25) which showed antimicrobial activity in the standard well diffusion assay and this is considered as an indication for the high value of these isolates in further studies. As well, the Red Sea marine invertebrates showed high biodiversity revealed by the 16S rRNA molecular identification of a number of the isolated strains associated with these animals.
Declarations of interests
The authors have no competing interests.
Acknowledgement
The research was supported by Swedish International Development Cooperation Agency (SIDA) Project ID (2009-6420), Styrelsen för internationellt utvecklingssamarbete.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.btre.2018.e00275.
Contributor Information
Samar M. Solyman, Email: samar.solyman@gmail.com.
Amro Hanora, Email: a.hanora@pharm.suez.ed.eg.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Indraningrat A., Smidt H., Sipkema D. Bioprospecting sponge-associated microbes for antimicrobial compounds. Mar. Drugs. 2016;14:87. doi: 10.3390/md14050087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization, editor. Antimicrobial Resistance: Global Report on Surveillance. World Health Organization; Geneva, Switzerland: 2014. [Google Scholar]
- 3.Aminov R.I. A brief history of the antibiotic era: lessons learned and challenges for the future. Front. Microbiol. 2010;1 doi: 10.3389/fmicb.2010.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moellering R.C. Discovering new antimicrobial agents. Int. J. Antimicrob. Agents. 2011;37:2–9. doi: 10.1016/j.ijantimicag.2010.08.018. [DOI] [PubMed] [Google Scholar]
- 5.Bibi F., Faheem M., Azhar E.I., Yasir M., Alvi S.A., Kamal M.A., Ullah I., Nasser M.I. Bacteria from marine sponges: a source of new drugs. Curr. Drug Metab. 2016;18(1):11–15. doi: 10.2174/1389200217666161013090610. [DOI] [PubMed] [Google Scholar]
- 6.Biswas K., Paul D., Sinha S.N. Marine bacteria: a potential tool for antibacterial activity. J. Appl. Environ. Microbiol. 2016;4:25–29. [Google Scholar]
- 7.Jimeno J., Faircloth G., Sousa-Faro J.F., Scheuer P., Rinehart K. New marine derived anticancer therapeutics—a journey from the sea to clinical trials. Mar. Drugs. 2004;2:14–29. [Google Scholar]
- 8.Blunt J.W., Copp B.R., Hu W.-P., Munro M.H.G., Northcote P.T., Prinsep M.R. Marine natural products. Nat. Prod. Rep. 2007;24:31–86. doi: 10.1039/b603047p. [DOI] [PubMed] [Google Scholar]
- 9.Hill R.A. Marine natural products. Annu. Rep. Sect. “B” (Org. Chem.) 2006;102:123. [Google Scholar]
- 10.Zhang W., Zhang F., Li Z., Miao X., Meng Q., Zhang X. Investigation of bacteria with polyketide synthase genes and antimicrobial activity isolated from South China Sea sponges. J. Appl. Microbiol. 2009;107:567–575. doi: 10.1111/j.1365-2672.2009.04241.x. [DOI] [PubMed] [Google Scholar]
- 11.Taylor M.W., Radax R., Steger D., Wagner M. Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 2007;71:295–347. doi: 10.1128/MMBR.00040-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anjum Komal, Abbas Syed Qamar, Shah Sayed Asmat Ali, Akhter Najeeb, Batool Sundas, Hassan Syed Shamsul. Marine sponges as a drug treasure. Biomol. Ther. 2016;24:347–362. doi: 10.4062/biomolther.2016.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayer A.M.S., Rodríguez A.D., Taglialatela-Scafati O., Fusetani N. Marine pharmacology in 2009–2011: marine compounds with antibacterial, antidiabetic, antifungal, anti-inflammatory, antiprotozoal, antituberculosis, and antiviral activities; affecting the immune and nervous systems, and other miscellaneous mechanisms of action. Mar. Drugs. 2013;11:2510–2573. doi: 10.3390/md11072510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qian P.-Y., Wang Y., Lee O.O., Lau S.C.K., Yang J., Lafi F.F., Al-Suwailem A., Wong T.Y. Vertical stratification of microbial communities in the Red Sea revealed by 16S rDNA pyrosequencing. ISME J. 2011;5:507–518. doi: 10.1038/ismej.2010.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ngugi D.K., Antunes A., Brune A., Stingl U. Biogeography of pelagic bacterioplankton across an antagonistic temperature-salinity gradient in the Red Sea: biogeography of pelagic bacteria in the Red Sea. Mol. Ecol. 2012;21:388–405. doi: 10.1111/j.1365-294X.2011.05378.x. [DOI] [PubMed] [Google Scholar]
- 16.Ngugi D.K., Stingl U. Correction: combined analyses of the ITS loci and the corresponding 16S rRNA genes reveal high micro- and macrodiversity of SAR11 populations in the Red Sea. PLoS One. 2013;8 doi: 10.1371/journal.pone.0050274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Temraz T.A., Houssen W.E., Jaspars M., Woolley D.R., Wease K.N., Davies S.N., Scott R.H. A pyridinium derivative from Red Sea soft corals inhibited voltage-activated potassium conductances and increased excitability of rat cultured sensory neurones. BMC Pharmacol. 2006;6:10. doi: 10.1186/1471-2210-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nadeem F., Oves M., Qari H.A., Ismail I.M.I. Red Sea microbial diversity for antimicrobial and anticancer agents. J. Mol. Biomark. Diagn. 2016;07 [Google Scholar]
- 19.Graça A.P., Viana F., Bondoso J., Correia M.I., Gomes L., Humanes M., Reis A., Xavier J.R., Gaspar H., Lage O.M. The antimicrobial activity of heterotrophic bacteria isolated from the marine sponge Erylus deficiens (Astrophorida, Geodiidae) Front. Microbiol. 2015;6 doi: 10.3389/fmicb.2015.00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santos O.C.S., Soares A.R., Machado F.L.S., Romanos M.T.V., Muricy G., Giambiagi-deMarval M., Laport M.S. Investigation of biotechnological potential of sponge-associated bacteria collected in Brazilian coast. Lett. Appl. Microbiol. 2015;60:140–147. doi: 10.1111/lam.12347. [DOI] [PubMed] [Google Scholar]
- 21.Williams G.J. Engineering polyketide synthases and nonribosomal peptide synthetases. Curr. Opin. Struct. Biol. 2013;23:603–612. doi: 10.1016/j.sbi.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hutchinson C.R. Polyketide and non-ribosomal peptide synthases: falling together by coming apart. Proc. Natl. Acad. Sci. U. S. A. 2003;100:3010–3012. doi: 10.1073/pnas.0730689100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Webster N.S., Wilson K.J., Blackall L.L., Hill R.T. Phylogenetic diversity of bacteria associated with the marine sponge Rhopaloeides odorabile. Appl. Environ. Microbiol. 2001;67:434–444. doi: 10.1128/AEM.67.1.434-444.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mincer T.J., Jensen P.R., Kauffman C.A., Fenical W. Widespread and persistent populations of a major new marine actinomycete taxon in ocean sediments. Appl. Environ. Microbiol. 2002;68:5005–5011. doi: 10.1128/AEM.68.10.5005-5011.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Webster N.S., Watts J.E.M., Hill R.T. Detection and phylogenetic analysis of novel Crenarchaeote and Euryarchaeote 16S ribosomal RNA gene sequences from a Great Barrier Reef Sponge. Mar. Biotechnol. 2001;3:600–608. doi: 10.1007/s10126-001-0065-7. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J., Fritsch E., Maniatis T. 2nd ed. vol. 2. Cold Spring Harbor, S.l.; 1989. (Molecular Cloning: A Laboratory Manual). [Google Scholar]
- 27.Flemer B., Kennedy J., Margassery L.M., Morrissey J.P., O’Gara F., Dobson A.D.W. Diversity and antimicrobial activities of microbes from two Irish marine sponges, Suberites carnosus and Leucosolenia sp.: antimicrobial activities of sponge microbes. J. Appl. Microbiol. 2012;112:289–301. doi: 10.1111/j.1365-2672.2011.05211.x. [DOI] [PubMed] [Google Scholar]
- 28.Maloy S.R. Jones & Bartlett Learning; 1990. Experimental Techniques in Bacterial Genetics. [Google Scholar]
- 29.Pospiech A., Neumann B. A versatile quick-prep of genomic DNA from gram-positive bacteria. Trends Genet. 1995;11:217–218. doi: 10.1016/s0168-9525(00)89052-6. [DOI] [PubMed] [Google Scholar]
- 30.Kim T.K., Garson M.J., Fuerst J.A. Marine actinomycetes related to the “Salinospora” group from the Great Barrier Reef sponge Pseudoceratina clavata. Environ. Microbiol. 2005;7:509–518. doi: 10.1111/j.1462-2920.2005.00716.x. [DOI] [PubMed] [Google Scholar]
- 31.Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ehrenreich I.M., Waterbury J.B., Webb E.A. Distribution and diversity of natural product genes in marine and freshwater cyanobacterial cultures and genomes. Appl. Environ. Microbiol. 2005;71:7401–7413. doi: 10.1128/AEM.71.11.7401-7413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neilan B.A., Dittmann E., Rouhiainen L., Bass R.A., Schaub V., Sivonen K., Börner T. Nonribosomal peptide synthesis and toxigenicity of cyanobacteria. J. Bacteriol. 1999;181:4089–4097. doi: 10.1128/jb.181.13.4089-4097.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ayuso-Sacido A., Genilloud O. New PCR primers for the screening of NRPS and PKS-I systems in actinomycetes: detection and distribution of these biosynthetic gene sequences in major taxonomic groups. Microb. Ecol. 2005;49:10–24. doi: 10.1007/s00248-004-0249-6. [DOI] [PubMed] [Google Scholar]
- 35.Metsä-Ketelä M., Salo V., Halo L., Hautala A., Hakala J., Mäntsälä P., Ylihonko K. An efficient approach for screening minimal PKS genes from Streptomyces. FEMS Microbiol. Lett. 1999;180:1–6. doi: 10.1111/j.1574-6968.1999.tb08770.x. [DOI] [PubMed] [Google Scholar]
- 36.Whelan S., Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 37.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 38.Palomo S., González I., de la Cruz M., Martín J., Tormo J.R., Anderson M., Hill R.T., Vicente F., Reyes F., Genilloud O. Sponge-derived Kocuria and Micrococcus spp. as sources of the new thiazolyl peptide antibiotic kocurin. Mar. Drugs. 2013;11:1071–1086. doi: 10.3390/md11041071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abdelmohsen U.R., Pimentel-Elardo S.M., Hanora A., Radwan M., Abou-El-Ela S.H., Ahmed S., Hentschel U. Isolation, phylogenetic analysis and anti-infective activity screening of marine sponge-associated actinomycetes. Mar. Drugs. 2010;8:399–412. doi: 10.3390/md8030399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.ElAhwany A.M.D., Ghozlan H.A., ElSharif H.A., Sabry S.A. Phylogenetic diversity and antimicrobial activity of marine bacteria associated with the soft coral Sarcophyton glaucum. J. Basic Microbiol. 2015;55:2–10. doi: 10.1002/jobm.201300195. [DOI] [PubMed] [Google Scholar]
- 41.Machado H., Sonnenschein E.C., Melchiorsen J., Gram L. Genome mining reveals unlocked bioactive potential of marine gram-negative bacteria. BMC Genom. 2015;16:158. doi: 10.1186/s12864-015-1365-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schneemann I., Nagel K., Kajahn I., Labes A., Wiese J., Imhoff J.F. Comprehensive investigation of marine Actinobacteria associated with the sponge Halichondria panicea. Appl. Environ. Microbiol. 2010;76:3702–3714. doi: 10.1128/AEM.00780-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zerikly M., Challis G.L. Strategies for the discovery of new natural products by genome mining. ChemBioChem. 2009;10:625–633. doi: 10.1002/cbic.200800389. [DOI] [PubMed] [Google Scholar]
- 44.El-Moneam N., El-Assar S., Shreadah M., Adam A. Isolation, identification and molecular screening of Pseudomonas sp. metabolic pathways NRPs and PKS associated with the Red sea Sponge, Hyrtios aff. Erectus, Egypt. J. Pure Appl. Microbiol. 2017;11:1299–1311. [Google Scholar]
- 45.Sagar S., Esau L., Hikmawan T., Antunes A., Holtermann K., Stingl U., Bajic V.B., Kaur M. Cytotoxic and apoptotic evaluations of marine bacteria isolated from brine-seawater interface of the Red Sea. BMC Complement. Altern. Med. 2013;13 doi: 10.1186/1472-6882-13-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abdelfattah M.S., Elmallah M.I.Y., Hawas U.W., Abou El-Kassema L.T., Eid M.A.G. Isolation and characterization of marine-derived actinomycetes with cytotoxic activity from the Red Sea coast. Asian Pac. J. Trop. Biomed. 2016;6:651–657. [Google Scholar]
- 47.Ayuso A., Clark D., González I., Salazar O., Anderson A., Genilloud O. A novel actinomycete strain de-replication approach based on the diversity of polyketide synthase and nonribosomal peptide synthetase biosynthetic pathways. Appl. Microbiol. Biotechnol. 2005;67:795–806. doi: 10.1007/s00253-004-1828-7. [DOI] [PubMed] [Google Scholar]
- 48.Dashti Y., Grkovic T., Abdelmohsen U., Hentschel U., Quinn R. Production of induced secondary metabolites by a co-culture of sponge-associated actinomycetes, Actinokineospora sp. EG49 and Nocardiopsis sp. RV163. Mar. Drugs. 2014;12:3046–3059. doi: 10.3390/md12053046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Al-Zereini W., Fotso Fondja Yao C.B., Laatsch H., Anke H. Aqabamycins A–G: novel nitro maleimides from a marine Vibrio species. I. Taxonomy, fermentation, isolation and biological activities. J. Antibiot. 2010;63:297–301. doi: 10.1038/ja.2010.34. [DOI] [PubMed] [Google Scholar]
- 50.Bennur T., Kumar A.R., Zinjarde S., Javdekar V. Nocardiopsis species: incidence, ecological roles and adaptations. Microbiol. Res. 2015;174:33–47. doi: 10.1016/j.micres.2015.03.010. [DOI] [PubMed] [Google Scholar]
- 51.Mansson M., Gram L., Larsen T.O. Production of bioactive secondary metabolites by marine Vibrionaceae. Mar. Drugs. 2011;9:1440–1468. doi: 10.3390/md9091440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu Z., Xie L., Xia G., Zhang J., Nie Y., Hu J., Wang S., Zhang R. A new tetrodotoxin-producing actinomycete, Nocardiopsis dassonvillei, isolated from the ovaries of puffer fish Fugu rubripes. Toxicon. 2005;45:851–859. doi: 10.1016/j.toxicon.2005.02.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.