Abstract
Introduction
Apolipoprotein CII amyloidosis (AApoCII) is a rare form of amyloidosis. Here, we report a novel mutation associated with AApoCII amyloidosis in 5 patients and describe their clinical, renal biopsy, and mass spectrometry findings.
Methods
Five patients with renal AApoCII p.Lys41Thr amyloidosis were identified from our amyloid mass spectrometry cohort. Clinical features, kidney biopsy, and mass spectrometry findings were analyzed in this rare type of amyloidosis.
Results
The patients were older adults (mean age of 71.6 years at diagnosis), presented with nephrotic-range proteinuria, and often had declining renal function. All renal biopsy specimens showed massive mesangial nodules composed of weakly eosinophilic, periodic acid−Schiff negative, Congo red–positive amyloid deposits. There were no interstitial, vascular, or medullary deposits. In all cases, immunofluorescence studies were negative for Igs and electron microscopy showed amyloid fibrils. Proteomic analysis of Congo red−positive amyloid deposits detected large amounts of apolipoprotein CII (APOC2) protein. We also detected APOC2 p.Lys41Thr mutant protein in amyloid deposits of all patients. DNA sequencing in 1 patient confirmed the presence of the mutation. Both mutant and wild-type forms of APOC2 were detected in amyloid deposits of all patients. Molecular dynamic simulations showed the variant mediating a collapse of the native structure of APOC2, thereby destabilizing the protein.
Conclusion
We propose that AApoCII p.Lys41Thr amyloidosis is a new form of amyloidosis seen in elderly individuals, histologically exhibiting massive glomerular involvement, leading to nephrotic-range proteinuria and progressive chronic kidney disease.
Keywords: amyloidosis, apolipoprotein CII, kidney, mutation, p.Lys41Thr, proteomics
Amyloidosis is caused by extracellular deposition of misfolded proteins in an insoluble β pleated physical format. Amyloid deposits are identified by their characteristic apple green−orange birefringence under polarized light on a Congo red stain, as well as the presence of rigid, nonbranching fibrils 7.5 to 10 nm in diameter on electron microscopy.1, 2 Based on the amyloid precursor protein, various types of renal amyloidosis are recognized. Ig light chains are the precursor protein of the most common type of amyloidosis, namely, the light-chain (AL) amyloidosis.3 Renal amyloidosis associated with other precursor proteins include serum amyloid A protein amyloidosis (AA), leukocyte chemotactic factor 2 amyloidosis (ALect2), fibrinogen α-chain amyloidosis (AFib), gelsolin amyloidosis (AGel), lysozyme amyloidosis (ALys), and apolipoproteins I, II, and IV (AApoAI, AApoAII, and AApoAIV).4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14 Many of these are rare, and often have a genetic basis that causes instability and misfolding of the precursor proteins.
Apolipoprotein CII amyloidosis (AApoCII) is a rare form of hereditary amyloidosis that was recently described in a 61-year-old woman with an APOC2 p.Glu69Val mutation.15 We recently found AApoCII amyloidosis associated with a novel APOC2 p.Lys41Thr mutation. This mutation is also more frequently associated with AApoCII amyloidosis than the APOC2 p.Glu69Val mutation. In this report, we present the clinical, renal biopsy, and proteomic findings of 5 patients with AApoCII p.Lys41Thr amyloidosis.
Methods
Proteomic Typing of Amyloid Deposits
Formalin-fixed, paraffin-embedded renal biopsy materials were sent to the Mayo Clinic amyloid typing laboratory for analysis. We used a previously established proteomics method for typing the amyloid deposits.16, 17 For each case, 6-μM-thick, formalin-fixed, paraffin-embedded sections were Congo red stained and examined to confirm the presence of amyloid. Separate 10-μM-thick sections were obtained and mounted on a special laser microdissection slide, and amyloid deposits were visualized using fluorescent light. Congo red–positive deposits were microdissected from an area of 60,000 μM2. Resulting formalin-fixed, paraffin-embedded fragments were collected for mass spectrometry analysis. Multiple independent dissections (replicates) were performed and analyzed for each patient. Data for each patient was processed using a previously described bioinformatics pipeline, and a patient-specific amyloid proteome profile was created. For each patient, a pathologist reviewed the microdissection images to confirm that the amyloid deposits were included in the analyzed tissue. Next, the proteome profile was scrutinized for the presence of universal amyloid tissue biomarkers (apolipoprotein E, serum amyloid P component, and APOAIV).18
Finally, the proteome was searched for potential amyloid precursor proteins. In all 5 cases, previously described canonical amyloid precursor proteins were not present in their corresponding proteomes. However, abundant spectra corresponding to ApoCII protein were detected. An advanced bioinformatics pipeline was used to search for known or unknown mutations in proteomes of all patients. We detected APOC2 p.Lys41Thr mutation in amyloid deposits in all patients.
Immunofluorescence and Electron Microscopy
Routine immunofluorescence microscopy and electron microscopy were performed on frozen biopsy specimens at the referring institutions, and results were obtained for summarization.
Sanger Sequencing
A DNA sample from patient 1 was extracted from peripheral blood using standard procedures. Direct sequencing of the 3 coding exons and flanking intronic sequences of APOC2 gene was conducted on an ABI 3730 DNA automated sequencer (Applied Biosystems, Frankfurt, Germany) and sequences were analyzed using Sequencher software (Gene Codes). Primer sequences for exon 1 are GGACACCGAGCTCACACAGA (forward) and GGGCTGGGAAGATGCTTGGA (reverse); for exon 2 are CCCAGGCCCTTCTTACCTCT (forward) and GGCCAGACCCCATTTCTCCA (reverse); and for exon 3 are CCCCTCCTCCCTCTAACCATCT (forward) and GGGGAGCTCAGTCTGAACCT (reverse). Conditions for polymerase chain reaction amplification are available upon request. DNA numbering of mutation was based on the APOC2 cDNA sequence (GenBank accession number NM_000483.4).
Molecular Dynamic Simulation of ApoCII Variants
Generalized Born implicit solvent molecular dynamics simulations were carried out using NAMD and the CHARMM22 with CMAP force field.19, 20 The protein data bank structure 1SOH was used for initial conformation. Initial mutant conformations were generated using PyMOL (The PyMOL Molecular Graphics System, Version 1.5.0.3: Schrödinger, LLC).21 We used an interaction cutoff of 15 Å with strength tapering (or switching) beginning at 12 Å, a simulation time step of 1 fs, and conformations recorded every 2 ps. Each initial conformation was used to generate 5 replicates, and each was energy minimized for 20,000 steps, heating to 300 K over 300 ps via Langevin thermostat. A further 50 ns of simulation trajectory was generated and the final 40 ns analyzed. All trajectories were first aligned to the initial WT conformation using Cα atoms. Root-mean-SD and principal component analysis were calculated using Cα atoms. We computed an alignment-free median residue−residue distance matrix within each trajectory.22, 23, 24 The distance difference matrix quantifies the median pairwise proximity of residues to each other. Simulations were analyzed using custom scripts, the Bio3D R package and Visual Molecular Dynamics (VMD).24, 25 Differences between simulations were compared using t tests after subsampling to 100 data points from each condition. The subsampling was repeated 100 times and the median P value reported.
Results
Clinical Features
Five patients with renal AApoCII p.Lys41Thr amyloidosis were identified from our amyloid mass spectrometry cohort. Four patients (80%) were female (Table 1). The mean age at the time of diagnosis was 71.6 years (range 61−86 years). Four patients were white and 1 patient was African American. All patients presented with nephrotic-range proteinuria (mean 4.4 g/24 h, range 3.2−5.9 g/24 h), and 3 of 5 patients also showed an elevated serum creatinine (mean 1.24 mg/dl, range 0.5−1.6 mg/dl). After a mean follow-up of 44.5 months, 1 patient was on dialysis, 1 patient was lost to follow-up, and 3 patients had a rise in serum creatinine level (mean 1.6 mg/dl, range 0.9−3.02 mg/dl). In addition, nephrotic-range proteinuria persisted in all 4 patients with follow-up. Table 1 summarizes clinical findings of all patients.
Table 1.
Clinical and laboratory findings
| Patient | Age/sex/ethnicity | Family history | Monoclonal Ig | Serum creatinine (md/dl) | Urinary protein (g/24 h) | Serum albumin | Follow-up (mo) | Serum creatinine at follow-up | Urinary protein at follow-up | Amyloid location |
|---|---|---|---|---|---|---|---|---|---|---|
| Patient 1 | 74/F/white | No | Negative | 1.6 | 5.4 | 3.6 | 48 | Dialysis | Dialysis | G |
| Patient 2 | 61/M/white | NA | Negative | 0.9 | 3.45 | NA | 54 | 1.01 | 3.8 | G |
| Patient 3a | 68/F/white | Yes (father) | IgG lambda | 0.5 | 3.2 | 2.6 | 52 | 0.9 | 4.8 | G |
| Patient 4 | 86/F/African American | NA | Negative | 1.6 | NA | 2.9 | 24 | 3.02 | NA | G |
| Patient 5 | 69/F/white | Yes (mother) | Negative | 1.6 | 5.9 | 3.8 | N/A | N/A | N/A | G |
F, female; G, glomerular; M, male; N/A, not available (lost to follow-up).
Stem cell transplant.
A brief history of each patient follows: Patient 1 is a 74-year-old white woman who presented with proteinuria and chronic kidney disease. Her serum creatinine at presentation was 1.6 mg/dl and urinary protein was 5.6 g/24 h. Evaluation for a monoclonal gammopathy was negative. There was no known family history of kidney disease. A kidney biopsy showed amyloidosis of unknown type. She had no peripheral or autonomous neuropathy or clinical signs of heart disease. Her lipid profile was normal. Within 2 years of follow-up, the patient’s serum creatinine progressed to 6.1 mg/dl and her urinary protein was 3.9 g/24 h. The patient was put on dialysis. Retyping the amyloid deposits by mass spectrometry studies detected APOC2 protein as the precursor protein along with presence of p.Lys41Thr mutation in the deposit. This patient recently received a kidney transplant.
Patient 2 is a 61-year-old white man who presented with proteinuria of 3.5 g/24 h. His serum creatinine was 0.5 mg/dl. Evaluation for a monoclonal gammopathy was negative. Mass spectrometry of the kidney biopsy detected the APOC2 p.Lys41Thr protein. After 4 years of follow-up, patient continued to have proteinuria of 3.8 g/24 h and his serum creatinine is 1.01 mg/dl.
Patient 3 is a 68-year-old white woman who presented with proteinuria of 3 g/24 h. Her serum creatinine at the time was 0.5 mg/dl. Her family history was positive for kidney disease in her father. Mass spectrometry of the kidney biopsy detected the APOC2 p.Lys41Thr protein (initially reported as unknown type). During evaluation, she was found to have a monoclonal IgM lambda paraprotein, and a bone marrow biopsy showed 4% plasma cells. She underwent an autologous stem cell transplantation without any clinical response in her proteinuria. After 3 years of follow-up, she continues to have significant proteinuria (4.8 g/24 h). Her serum creatinine is 0.9 mg/dl.
Patient 4 is an 86-year-old African American woman who presented with proteinuria and a rise in serum creatinine (1.6 mg/dl at presentation). Urinary protein was not quantitated at presentation but urinalysis showed 3+ proteinuria. Laboratory evaluation revealed positive antinuclear antibodies and elevated double-stranded DNA antibody titers but no monoclonal Igs. Mass spectrometry of the kidney biopsy specimen detected the APOC2 p.Lys41Thr protein (initially reported as unknown type). After a follow-up of 2 years, the patient’s serum creatinine had gone up to 3.0 mg/dl and she continues to have 3+ proteinuria.
Patient 5 is a 69-year-old obese white woman who presented with proteinuria (5.9 g/24 h) and rising serum creatinine (increased to 3.1 mg/dl from a baseline of 1.7 mg/dl). She had a history of diabetes and hypertension for 20 years, although they were controlled. She had a family history of kidney disease. Protein electrophoresis was negative for monoclonal gammopathy. Mass spectrometry of the kidney biopsy specimen detected the APOC2 p.Lys41Thr protein.
Kidney Biopsy Findings
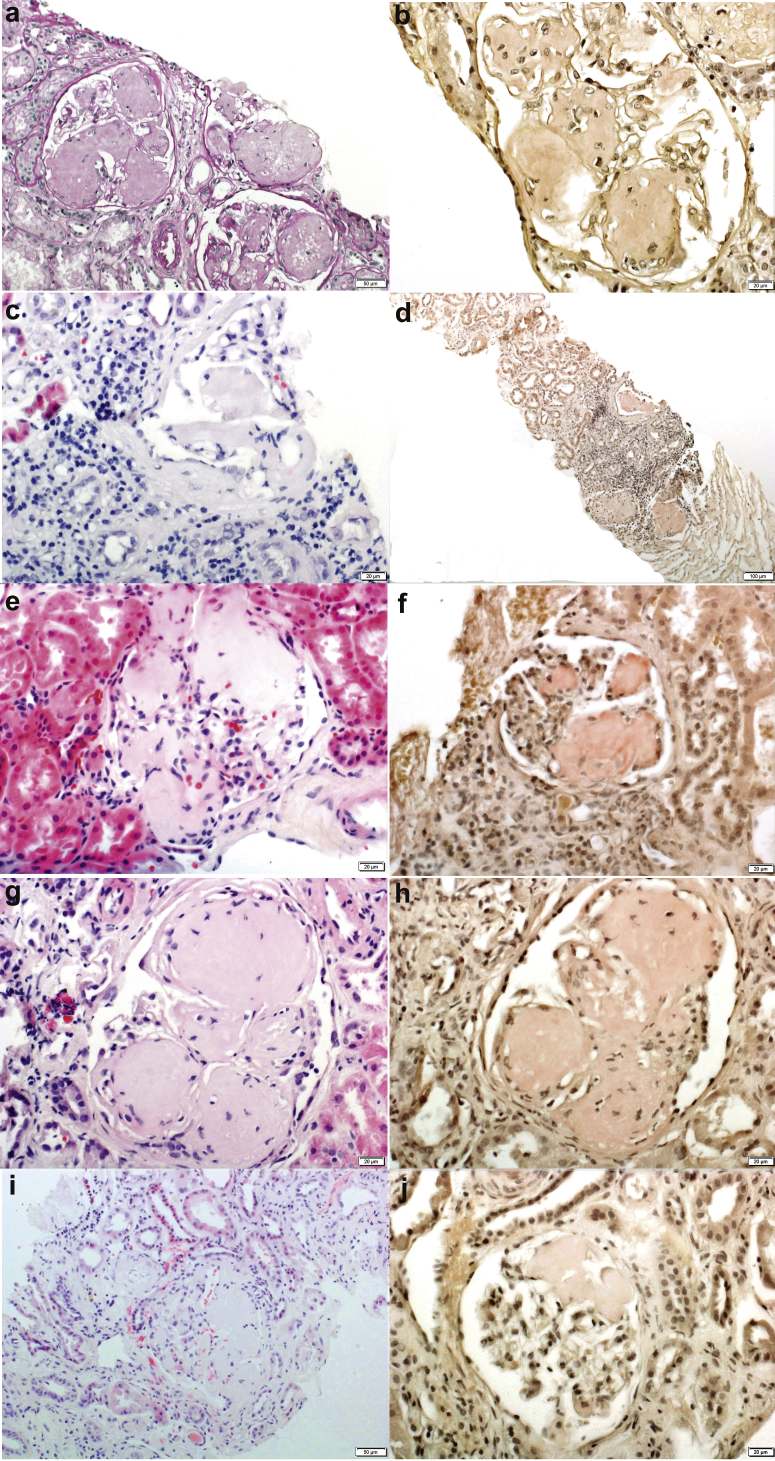
There were an average of 13.8 glomeruli present (range 5−34 glomeruli) per sample, of which an average of 5.4 (37.4%, range 2−13 glomeruli) glomeruli were globally sclerosed. Light microscopy showed mesangial and capillary wall expansion with acellular material, in all cases, forming large mesangial nodules. The nodules had a flaky/fluffy appearance, and although the nodules were large, the glomerular architecture could be appreciated. The nodules were weakly eosinophilic and were silver and periodic acid−Schiff negative. The interstitium showed absent to extensive (range 0%−80%) tubular atrophy and interstitial fibrosis. Arteries showed absent to moderate sclerosis of the intima. Congo red−positive amyloid deposits were present in all cases, although the Congo red was only weakly positive in 2 biopsy specimens. Amyloid was restricted to the glomeruli in all cases. Vascular or cortical/medullary interstitial amyloid deposits were absent. Immunofluorescence microscopy was negative for immune deposits in all 5 kidney biopsies. Electron microscopy showed large aggregates of amyloid fibrils in the mesangium and along capillary walls. Spicule formation was noted along the capillary walls in 1 case. Representative kidney biopsy findings from each patient are shown in Figure 1.
Figure 1.
Kidney biopsy findings. Light microscopy shows large acellular mesangial nodules that are periodic acid−Schiff stain negative and Congo red–positive. In some glomeruli, the large mesangial nodules may involve the glomerulus only segmentally. In addition, in some biopsy specimens, the Congo red stain is only weakly positive. The column on the right pertains to the Congo red stain. The column on the left pertains to hematoxylin and eosin except (a), which is a periodic acid−Schiff stain. Each row represents 1 patient: (a,b) patient 1; (c,d) patient 2; (e,f) patient 3; (g,h) patient 4; and (i,j) patient 5. Original magnification for all ×40, except for (a) and (i) (×20) and d (×10).
Mass Spectrometry Findings
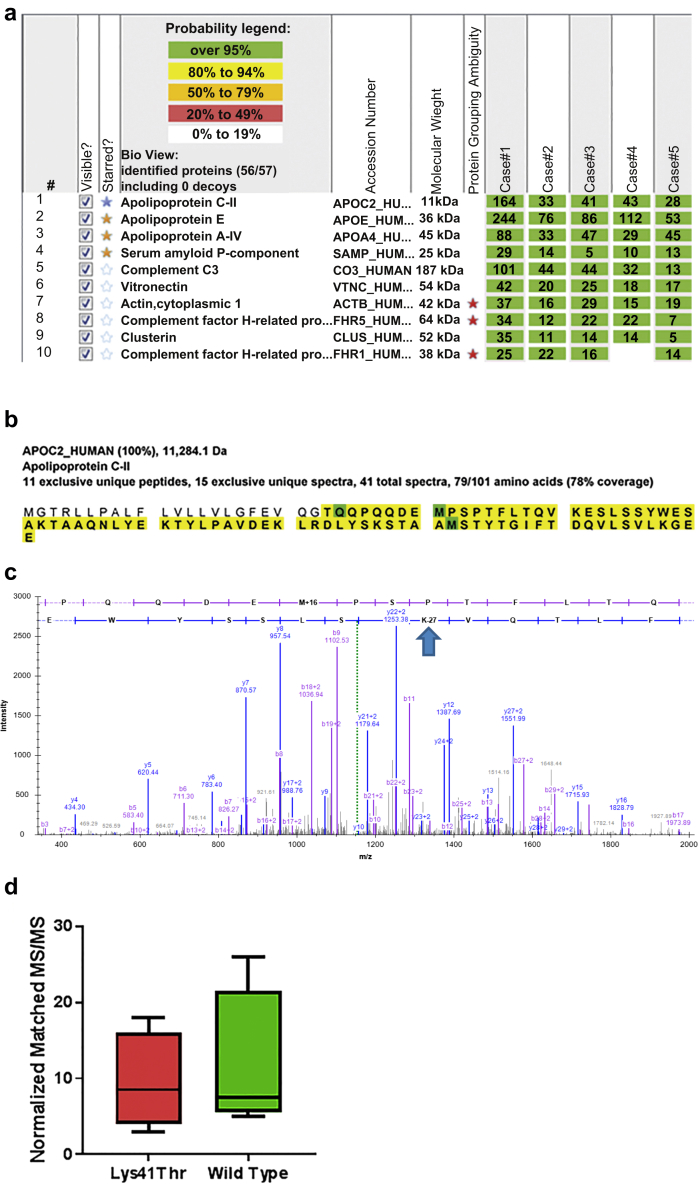
Laser microdissection−assisted mass spectrometry studies were used to make the diagnosis of AApoCII in all cases. The proteomic profiles of the amyloid deposits from all patients are shown in Figure 2a. AApoCII amyloidosis was characterized by the presence of large numbers of APOC2 protein spectra (average spectra number 62, range 28−164). We also detected the universal amyloid tissue biomarkers in all patients: apolipoprotein E (average number of spectra 114, range 53−244), serum amyloid protein (average number of spectra 14, range 5−29), and apolipoprotein AIV (APOA4, average number of spectra 48, range 29−88). Sequence coverage of the deposited APOC2 protein, shown in Figure 2b, suggested that the protein was deposited in its entirety. An unanticipated finding on mutation search detected the presence of APOC2 p.Lys41Thr mutant peptide in the deposits of all 5 patients. Figure 2c shows a sample tandem mass spectrometry spectrum for this mutant peptide. We detected both p.Lys41Thr mutant and corresponding wild-type peptides in deposits of all 5 patients. Figure 2d shows the range of normalized MS/MS counts observed for both forms of APOC2 peptides in the deposits. We did not detect any other amyloid precursor proteins in the deposits of these 5 patients.
Figure 2.
Proteomic identification of APOC2 Lys41Thr in glomerular amyloid deposits. Congo red−positive glomeruli were microdissected and analyzed using liquid chromatography with mass spectrometry (LC-MS/MS), as described in the Methods. (a) Protein identification report consisting of representative dissections from all 5 cases is shown. Numbers in green boxes represent number of MS/MS matches to the respective protein. Universal amyloid tissue marker proteins (apolipoprotein E [APOE], APOA4, and serum amyloid protein [SAP]) are shown in gold stars. Type-specific marker (APOC2) is shown in the blue star. (b) Sequence coverage map of wild-type APOC2, from the patient 1 specimen, showing that the entire protein is deposited. Amino acids highlighted in bold letters over yellow background were detected in the deposit. The first 22 amino acids correspond to the signal peptide. (c) MS/MS spectrum corresponding to the APOC2 p.Lys41Thr mutant peptide is shown. Blue arrow shows the location of the mutation in the detected peptide. (d) Peptides corresponding to both mutant and wild-type forms of APOC2 were detected in the deposit. Distribution of normalized MS/MS matches to the corresponding APOC2 forms are shown.
Sanger Sequencing
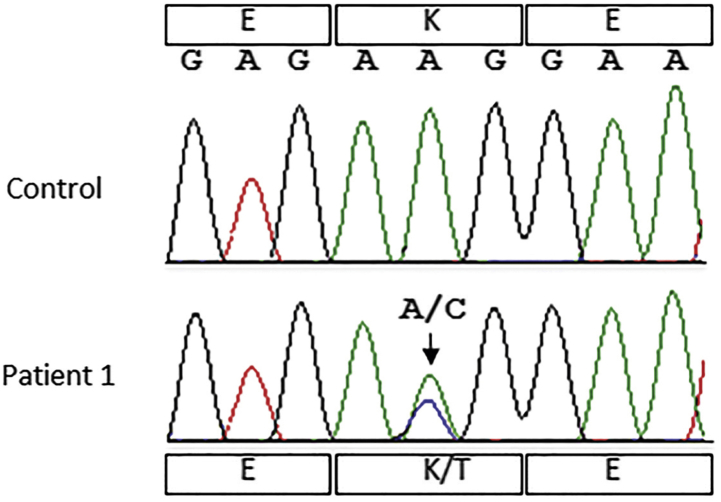
Following the diagnosis of AApoCII in patient 1, Sanger sequencing studies of the APOC2 gene confirmed the p.Lys41Thr (c.122A→C) substitution in the protein (Figure 3). This mutation was reported as extremely rare in Exome Aggregation Consortium (minor allele frequency = 0.00087% in 60,706 individuals), 1000 Genomes (phase 3; minor allele frequency = 0.0004%) human variant data. Genetic studies of TTR, LYZ, FGA, APOA1, APOA2, APOA4, APOC3, APOC2, B2M, LECT2, GSN, PRNP, SAA4, IGKC, and IGLC1 genes did not identify any potential pathogenic variants.
Figure 3.
Sanger sequencing to detect ApoCII variant. Sanger sequencing of blood DNA in patient 1 shows heterozygous APOC2 variant c.122A>C (p.Lys41Thr). The control sequence is shown for comparison.
Molecular Dynamics of APOC2 p.Lys41Thr
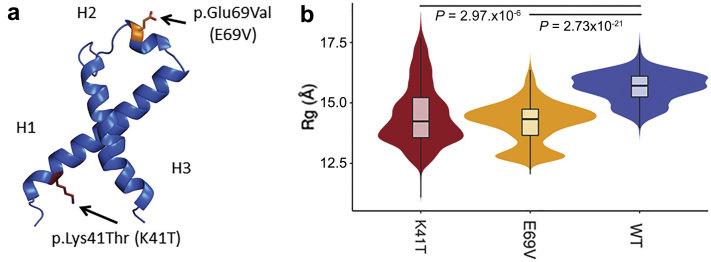
We used molecular dynamics simulations of APOC2 protein to determine whether the new p.Lys41Thr (K41T) variant affected its structure in a way that is similar to the previously described amylogenic variant p.Glu69Val (E69V). Figure 4A shows the location of both variants in the APOC2 3-dimensional structure. APOC2 protein has 3 α-helices and K41T was located in H1 (Figure 4a). Molecular dynamic simulation suggested that both variants were associated with a collapse of the native structure (Figure 4b), as indicated by the radius of gyration (Rg). Rg is a measure of a protein’s compactness. Stably folded proteins have a narrow Rg distribution in molecular dynamics simulations when compared to unstably folded proteins. Both variants of APOC2 protein collapsed the native structure by changing the likelihood of interaction among residues throughout the structure. However, the net effect of these changes on the overall structure was idiosyncratic to each variant. For instance, both K41T and E69V variants decreased the interactions involving the second α-helix (H2) and the rest of the structure and increased interactions between the first and third α-helices (H1 and H3, respectively). This increased interaction between H1 and C-terminal of H3 was noted in our previous work on AApoCII p.Glu69Val amyloidosis.15 However, in this work, we detected that K41T was uniquely associated with the kinking of H3. Furthermore, folding dynamics of both variants are more unstable than those of the wild type (compare Rg distributions in Figure 4b). With this evidence, we hypothesize that APOC2 p.Lys41Thr variant detected in the amyloid deposits of this cohort could be a direct result of the variant destabilizing the H3 APOC2 protein, leading to amyloidogenesis.
Figure 4.
Molecular dynamics (MD) simulation of APOC2 p.Lys41Thr. (a) Variant p.Lys41Thr was located in the third helix (H1) in the APOC2 native structure, whereas the previously known amyloidogenic variant p.Gly69Val was located in the second α-helix (H2). (b) Molecular dynamics simulations were performed for both variants, and radius of gyration (Rg) was monitored during simulations (Rg is a measure of the protein’s compactness). Furthermore, stable proteins have a narrower Rg in MD simulations when compared to unstable proteins. Distribution of Rg observed for wild-type (WT), K41T, and E69V variants. Like E69V, the new K41T was associated with a collapse of the protein, as indicated by significantly lower Rg measures. K41T folding, like that of E69V, is also more unstable than in WT, as indicated by a wider Rg distribution.
Discussion
Apolipoproteins, including APOC2, are small peptides that play an important part in lipid transport and form components of chylomicrons, as well as very-low-density and high-density lipoproteins.26, 27 Excess ApoCII has been associated with increased triglyceride-rich particles and alterations in high-density lipoprotein particle distribution, factors that may increase the risk of cardiovascular disease.27 However, in the absence of lipid binding, ApoCII becomes unstable and exhibits a propensity to form amyloid fibrils.28, 29, 30 It has been proposed that genetic alterations to APOC2 could alter the lipid-binding capabilities, resulting in amyloid fibril formation. We recently reported the first case of renal amyloidosis associated with deposition of APOC2 with a p.Glu69Val mutation.15 In this study, we identified AApoCII amyloidosis, which is associated with deposition of APOC2 protein with a c.122A→C transition, resulting in a p.Lys41Thr substitution. Based on the frequency of APOC2 mutations seen in our amyloid typing practice, we find APOC2 p.Lys41Thr mutant to be the most common form associated with renal AApoCII amyloidosis.
AApoCII p.Lys41Thr amyloidosis was seen in elderly patients who presented with progressive nephrotic-range proteinuria either with or without progressive chronic kidney disease. Laboratory evaluation showed monoclonal gammopathy in 1 patient at the time of the initial diagnosis of amyloidosis. As ApoCII amyloidosis type was unknown at that time, this patient received a stem cell transplant, with no improvement in renal function. AApoCII can occur with concurrent monoclonal gammopathy, and there is no specific treatment available for AApoCII. Hence, it is imperative to correctly type the AApoCII to avoid unnecessary treatment of presumed amyloid light chain amyloidosis based solely on the presence of monoclonal gammopathy.
The kidney biopsy specimen in AApoCII p.Lys41Thr amyloidosis shows distinctive features characterized by very large mesangial amyloid nodules, often pushing into and obliterating the capillary lumen. The nodules have an almost papery or flaky appearance, are periodic acid−Schiff negative, and are only very weakly eosinophilic. Despite the massive mesangial nodules, some of the glomeruli show only segmental involvement (Figure 1, lowest panel). In some cases, the Congo red stain is weakly positive. In all our 5 cases, there was no interstitial or vascular involvement. Electron microscopy of representative vessels and tubules did not reveal amyloid material. Unlike other amyloidoses associated with apolipoproteins AI and AIV, and also in contrast to the single case of AApoCII p.Glu69Val amyloidosis, which had both glomerular and medullary interstitial amyloid deposits, AApoCII p.Lys41Thr amyloidosis cases lacked both cortical and medullary interstitial involvement. All 5 cases were also negative for vascular involvement. The glomerular restriction is similar to that in AGel and AFib amyloidosis, which also show predominant glomerular involvement.31, 32, 33
Proteomic analysis of amyloid deposits showed an overabundance of APOC2 protein along with the presence of universal amyloid tissue signature proteins (apolipoprotein E and serum amyloid P-component).34 Interestingly, large numbers of apolipoprotein A-IV spectra were present in all cases. However, the abundance of APOAIV protein in these deposits was not typical of true renal AApoAIV amyloidosis, which is characterized by massive medullary interstitial amyloid deposits and absence of detectable ApoCII.12, 35 Furthermore, the clinical presentation and biopsy findings of AApoAIV amyloidosis are quite distinctive from those of AApoCII amyloidosis, in that the patients present with gradual loss of renal function and no proteinuria.
Mass spectrometry played a critical role in detecting the presence of APOC2 protein and the p.Lys41Thr mutation in the amyloid deposits of all patients. Sanger sequencing of the APOC2 gene in 1 patient confirmed the mutation at the DNA level in 1 patient. This mutation is likely to disrupt the native APOC2 protein stability in 2 ways. This mutation is located in the lipid-binding domain, but not the lipoprotein lipase−binding domain of the APOC2. Previous studies have indicated that mutations in the lipid-binding domain alter the protein’s lipid-binding ability, making the APOC2 protein unstable.36, 37 In addition, molecular dynamics simulations suggested that the p.Lys41Thr mutation led to a collapse of the protein’s native structure as well as kinking the third helix (H3) of native protein. Mature APOC2 protein has a molecular weight of 8.9 kDa and is a byproduct of very-low-density lipoprotein clearance, and small amounts are likely filtered into the mesangium. On the other hand, the mutant APOC2 protein, which is an unstable protein, is not completely filtered but likely binds to the chaperone proteins such as apolipoprotein E/IV and serum amyloid protein, resulting in the formation, accumulation, and glomerular localization of the AApoCII amyloid fibrils.
To summarize, AApoCII p.Lys41Thr amyloidosis is a novel but rare type of amyloidosis that occurs in elderly individuals, presents with nephrotic-range proteinuria and progressive loss of renal function, and involves the glomeruli and spares the interstitium and vessels. Laser microdissection of the amyloid deposits and mass spectrometry and, if feasible, confirmation by Sanger sequencing studies of the APOC2 gene are required to confirm the diagnosis. Although rare, correct diagnosis is of paramount importance to avoid unnecessary treatment regimens that may be directed to other forms of amyloidosis.
Disclosure
All the authors declared no competing interests.
Acknowledgments
We acknowledge Dr. Sophie Valleix for the sequencing of the other genes involved in hereditary amyloidosis in patient 1.
Contributor Information
Sanjeev Sethi, Email: sethi.sanjeev@mayo.edu.
Ellen D. McPhail, Email: McPhail.ellen@mayo.edu.
Paul J. Kurtin, Email: Kurtin.Paul@mayo.edu.
References
- 1.Dember L.M. Amyloidosis-associated kidney disease. J Am Soc Nephrol. 2006;17:3458–3471. doi: 10.1681/ASN.2006050460. [DOI] [PubMed] [Google Scholar]
- 2.Merlini G., Bellotti V. Molecular mechanisms of amyloidosis. N Engl J Med. 2003;349:583–596. doi: 10.1056/NEJMra023144. [DOI] [PubMed] [Google Scholar]
- 3.Said S.M., Sethi S., Valeri A.M. Renal amyloidosis: origin and clinicopathologic correlations of 474 recent cases. Clin J Am Soc Nephrol. 2013;8:1515–1523. doi: 10.2215/CJN.10491012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Picken M.M. New insights into systemic amyloidosis: the importance of diagnosis of specific type. Curr Opin Nephrol Hypertens. 2007;16:196–203. doi: 10.1097/MNH.0b013e3280bdc0db. [DOI] [PubMed] [Google Scholar]
- 5.Vigushin D.M., Gough J., Allan D. Familial nephropathic systemic amyloidosis caused by apolipoprotein Al variant Arg26. Q J Med. 1994;87:149–154. [PubMed] [Google Scholar]
- 6.Yazaki M., Liepnieks J.J., Barats M.S. Hereditary systemic amyloidosis associated with a new apolipoprotein AII stop codon mutation Stop78Arg. Kidney Int. 2003;64:11–16. doi: 10.1046/j.1523-1755.2003.00047.x. [DOI] [PubMed] [Google Scholar]
- 7.Yazaki M., Liepnieks J.J., Yamashita T. Renal amyloidosis caused by a novel stop-codon mutation in the apolipoprotein A-II gene. Kidney Int. 2001;60:1658–1665. doi: 10.1046/j.1523-1755.2001.00024.x. [DOI] [PubMed] [Google Scholar]
- 8.Benson M., Liepnieks J., Uemichi T. Hereditary renal amyloidosis associated with a mutant fibrinogen [alpha]-chain. Blood. 1993;3:252–255. doi: 10.1038/ng0393-252. [DOI] [PubMed] [Google Scholar]
- 9.Benson M.D. Ostertag revisited: the inherited systemic amyloidoses without neuropathy. Amyloid. 2005;12:75–87. doi: 10.1080/13506120500106925. [DOI] [PubMed] [Google Scholar]
- 10.Joy T., Wang J., Hahn A. Apoa1 related amyloidosis: a case report and literature review. Clin Biochem. 2003;36:641–645. doi: 10.1016/s0009-9120(03)00110-3. [DOI] [PubMed] [Google Scholar]
- 11.Gillmore J.D., Booth D.R., Madhoo S. Hereditary renal amyloidosis associated with variant lysozyme in a large English family. Nephrol Dial Transplant. 1999;14:2639–2644. doi: 10.1093/ndt/14.11.2639. [DOI] [PubMed] [Google Scholar]
- 12.Dasari S, Amin MS, Kurtin PJ, et al. Clinical, biopsy, and mass spectrometry characteristics of renal apolipoprotein A-IV amyloidosis. Kidney Int. 90:658–664. [DOI] [PubMed]
- 13.Sethi S., Dasari S., Amin M.S. Clinical, biopsy, and mass spectrometry findings of renal gelsolin amyloidosis. Kidney Int. 2017;91:964–971. doi: 10.1016/j.kint.2016.11.017. [DOI] [PubMed] [Google Scholar]
- 14.Sethi S., Theis J.D. Pathology and diagnosis of renal non-AL amyloidosis. J Nephrol. 2017 doi: 10.1007/s40620-017-0426-6. [DOI] [PubMed] [Google Scholar]
- 15.Nasr S.H., Dasari S., Hasadsri L. Novel type of renal amyloidosis derived from apolipoprotein-CII. J Am Soc Nephrol. 2017;28:439–445. doi: 10.1681/ASN.2015111228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vrana J.A., Gamez J.D., Madden B.J. Classification of amyloidosis by laser microdissection and mass spectrometry based proteomic analysis in clinical biopsy specimens. Blood. 2009;114:4957–4959. doi: 10.1182/blood-2009-07-230722. [DOI] [PubMed] [Google Scholar]
- 17.Dasari S., Theis J.D., Vrana J.A. Clinical proteome informatics workbench detects pathogenic mutations in hereditary amyloidoses. J Proteome Res. 2014;13:2352–2358. doi: 10.1021/pr4011475. [DOI] [PubMed] [Google Scholar]
- 18.Sethi S., Vrana J.A., Theis J.D. Laser microdissection and mass spectrometry-based proteomics aids the diagnosis and typing of renal amyloidosis. Kidney Int. 2012;82:226–234. doi: 10.1038/ki.2012.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phillips J.C., Braun R., Wang W. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mackerell A.D., Feig M., Brooks C.L. Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 21.MacRaild C.A., Howlett G.J., Gooley P.R. The structure and interactions of human apolipoprotein C-II in dodecyl phosphocholine. Biochemistry. 2004;43:8084–8093. doi: 10.1021/bi049817l. [DOI] [PubMed] [Google Scholar]
- 22.Rashin A.A., Domagalski M.J., Zimmermann M.T. Factors correlating with significant differences between X-ray structures of myoglobin. Acta Crystallog Sect D. 2014;70:481–491. doi: 10.1107/S1399004713028812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holm L., Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 24.Grant B.J., Rodrigues A.P.C., El Sawy K.M. Bio3d: an R package for the comparative analysis of protein structures. Bioinformatics. 2006;22:2695–2696. doi: 10.1093/bioinformatics/btl461. [DOI] [PubMed] [Google Scholar]
- 25.Humphrey W., Dalke A., Schulten K.V.M.D. visual molecular dynamics. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–38. [DOI] [PubMed] [Google Scholar]
- 26.Ryan T.M., Mok Y.-F., Howlett G.J. The role of lipid in misfolding and amyloid fibril formation by apolipoprotein C-II. In: Gursky O., editor. Lipids in Protein Misfolding. Springer International Publishing; Cham: 2015. pp. 157–174. [DOI] [PubMed] [Google Scholar]
- 27.Kei A.A., Filippatos T.D., Tsimihodimos V. A review of the role of apolipoprotein C-II in lipoprotein metabolism and cardiovascular disease. Metabolism. 2012;61:906–921. doi: 10.1016/j.metabol.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 28.Teoh C.L., Pham C.L.L., Todorova N. A structural model for apolipoprotein C-II amyloid fibrils: experimental characterization and molecular dynamics simulations. J Mol Biol. 2011;405:1246–1266. doi: 10.1016/j.jmb.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 29.Hanson C.L., Ilag L.L., Malo J. Phospholipid complexation and association with apolipoprotein C-II: insights from mass spectrometry. Biophys J. 2003;85:3802–3812. doi: 10.1016/S0006-3495(03)74795-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hatters D., Howlett G. The structural basis for amyloid formation by plasma apolipoproteins: a review. Eur Biophys J. 2002;31:2–8. doi: 10.1007/s002490100172. [DOI] [PubMed] [Google Scholar]
- 31.Sethi S., Theis J.D., Quint P. Renal amyloidosis associated with a novel sequence variant of gelsolin. Am J Kidney Dis. 2013;61:161–166. doi: 10.1053/j.ajkd.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 32.Sethi S, Dasari S, Amin MS, et al. Clinical, biopsy, and mass spectrometry findings of renal gelsolin amyloidosis. Kidney Int. 91:964–971. [DOI] [PubMed]
- 33.Gillmore J.D., Lachmann H.J., Rowczenio D. Diagnosis, pathogenesis, treatment, and prognosis of hereditary fibrinogen A{alpha}-chain amyloidosis. J Am Soc Nephrol. 2009;20:444–451. doi: 10.1681/ASN.2008060614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vrana J.A., Theis J.D., Dasari S. Clinical diagnosis and typing of systemic amyloidosis in subcutaneous fat aspirates by mass spectrometry-based proteomics. Haematologica. 2014;99:1239–1247. doi: 10.3324/haematol.2013.102764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sethi S., Theis J.D., Shiller S.M. Medullary amyloidosis associated with apolipoprotein A-IV deposition. Kidney Int. 2012;81:201–206. doi: 10.1038/ki.2011.316. [DOI] [PubMed] [Google Scholar]
- 36.Legge F.S., Binger K.J., Griffin M.D.W. Effect of oxidation and mutation on the conformational dynamics and fibril assembly of amyloidogenic peptides derived from apolipoprotein C-II. J Phys Chem B. 2009;113:14006–14014. doi: 10.1021/jp903842u. [DOI] [PubMed] [Google Scholar]
- 37.Legge F.S., Treutlein H., Howlett G.J. Molecular dynamics simulations of a fibrillogenic peptide derived from apolipoprotein C-II. Biophys Chem. 2007;130:102–113. doi: 10.1016/j.bpc.2007.08.002. [DOI] [PubMed] [Google Scholar]