Abstract
The approval of the first oncolytic virus for the treatment of metastatic melanoma and the compiling evidence that the use of oncolytic viruses can enhance cancer immunotherapies targeted against various immune checkpoint proteins has attracted great interest in the field of cancer virotherapy. We have developed a novel platform for clinically relevant enveloped viruses that can direct the virus-induced immune response against tumor antigens. By physically attaching tumor-specific peptides onto the viral envelope of vaccinia virus and herpes simplex virus 1 (HSV-1), we were able to induce a strong T cell-specific immune response toward these tumor antigens. These therapeutic peptides could be attached onto the viral envelope by using a cell-penetrating peptide sequence derived from human immunodeficiency virus Tat N-terminally fused to the tumor-specific peptides or, alternatively, therapeutic peptides could be conjugated with cholesterol for the attachment of the peptides onto the viral envelope. We used two mouse models of melanoma termed B16.OVA and B16-F10 for testing the efficacy of OVA SIINFEKL-peptide-coated viruses and gp100-Trp2-peptide-coated viruses, respectively, and show that by coating the viral envelope with therapeutic peptides, the anti-tumor immunity and the number of tumor-specific CD8+ T cells in the tumor microenvironment can be significantly enhanced.
Keywords: cancer, immunotherapy, viruses
Ylösmäki et al. developed a method for coating tumor-specific peptides directly onto the viral envelope to effectively enhance tumor-specific T cell responses of clinically relevant enveloped viruses. This personalized cancer vaccine platform has the potential to broaden the number of responders to checkpoint inhibitor therapies.
Introduction
Cancer immunotherapy aims to increase both the activity and the amount of tumor infiltrating tumor-specific T effector cells in order to elicit therapeutic efficacy. The T effector cells, named CD8+ T cells, more specifically cytotoxic T cell lymphocytes (CTLs), are critical components of protective anti-tumor immunity. The tumor-specific CTLs can be found in tumor tissue, and a positive correlation between the amount of tumor-infiltrating CTLs and patient survival has been observed.1, 2, 3, 4, 5 A recent approval of antibodies targeting immune checkpoint molecules, such as programmed death 1 (PD-1), programmed death ligand 1 (PD-L1), and cytotoxic T lymphocyte-associated antigen 4 (CTLA-4), which block the negative feedback systems within the tumor microenvironment to activate the pre-existing anti-tumor immune responses, have met tremendous clinical excitement.6 The use of these immune checkpoint inhibitor (ICI) antibodies can create durable responses in 10%–20% of cancer patients.7 The common feature of the patients responding to ICI therapy is that they have an existing anti-tumor immunity and CTL infiltration in the tumor tissue already prior to ICI therapy.8, 9 However, the remaining 80%–90% of the patients are not responding due to the lack of anti-tumor immune responses or other immune-suppressive aspects of the tumor microenvironment, presenting a strong rationale on finding novel combinational therapies that attract tumor-specific CD8+ T cells into tumors to increase the number of responders to ICI therapies.
Oncolytic viruses are natural pathogens that have been selected or designed to specifically infect and kill cancer cells. They are able to kill cancer cells by two different mechanisms: (1) by direct cell lysis (i.e., oncolysis) of the infected cells and (2) by inducing a strong anti-viral immune response against the therapeutic virus to eliminate the pathogen and in some instances the host cells they infect.10 Virus-induced tumor apoptosis and/or necrosis releases large amounts of tumor-associated proteins that are normally not accessible to antigen-presenting cells. This release of tumor antigens coupled with the virus-associated danger signaling drives antigen cross-presentation by tumor-associated dendritic cells (DCs) in the tumor-draining lymph nodes.11, 12, 13 A major weakness of the oncolytic viruses currently used in the clinics is that while they induce a strong anti-viral immune response, the anti-tumor immune response remains modest at best and thus reduces the therapeutic effect of these viruses.14 To overcome this problem and to significantly increase the anti-tumor immune response of current oncolytic enveloped viruses, we developed a method for coating tumor-specific peptides directly onto the viral envelope (peptide-coated oncolytic-enveloped viruses [PeptiENV]). Our results show that this is an effective strategy to increase the amount of tumor-specific T cells in the tumor microenvironment, and as compared to genetically engineered viruses coding for tumor-specific antigens, this approach is highly suitable for the next generation of personalized approaches that rely on the identification of patient-specific neo-antigens to be used in cancer immunotherapy. The PeptiENV approach is suitable for all enveloped viruses used in the clinics to enhance their tumor-specific T cell responses.
Results
Enveloped Viruses Can Be Coated with Therapeutic Peptides by Using Cell-Penetrating Peptide Sequence or Cholesterol Moiety as an Anchor
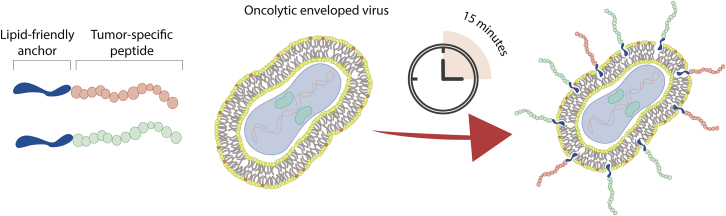
The outer membrane of all enveloped viruses consists of host-derived lipid bilayer.15 We hypothesized that therapeutic peptide sequences could be attached into the viral envelope by using a cell-penetrating peptide (CPP) sequence or peptide-conjugated cholesterol as attachment moieties (see Figure 1 for a schematic presentation of the PeptiENV platform).
Figure 1.
A Schematic Presentation of a PeptiENV-Cancer Vaccine Platform
Anti-tumor immunity-inducing peptides can readily be attached into the envelope of clinically relevant oncolytic enveloped viruses, e.g., HSV-1 and vaccinia viruses. Various different peptides including MHC class I and II epitopes can be delivered by PeptiENV-platform for inducing potent activation of antigen-presenting cells and consequently increased T cell-specific immunological responses.
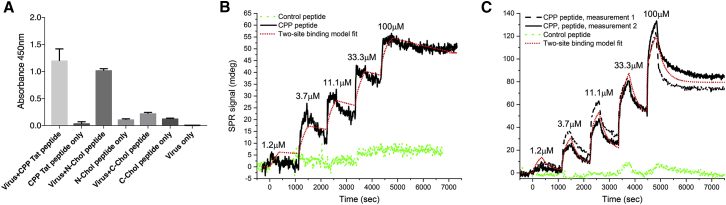
Various CPP sequences tested were able to anchor the therapeutic peptides into the viral envelope (data not shown), and a CPP sequence derived from HIV Tat protein was chosen to be used as a representative of CPP anchoring. Interestingly, cholesterol conjugated to the N terminus but not to the C terminus of the therapeutic peptides was suitable for anchoring the peptides into the viral envelope as assayed by ELISA (Figure 2A). We went on to confirm the complex formation of fluorescein isothiocyanate (FITC)-labeled C-terminally conjugated peptides more quantitatively by using flow cytometry and found that both attachment moieties were capable of forming virus-peptide complexes with envelope-labeling efficiencies of 67.6% and 46.9% for CPP and cholesterol moieties, respectively (Figure S1).
Figure 2.
Physicochemical Characterization of PeptiENV Complexes
(A) ELISA characterization of different C- and N-terminal conjugation strategies to attach FITC-labeled anti-tumor peptides into the viral envelope. An anti-virus antibody was coated to the bottom of 96-well plate, and PeptiENV complexes were incubated in the wells. After washing the unbound fraction, an anti-FITC HRP-conjugated antibody was used for the detection of the PeptiENV complexes. Each bar is the mean ± SEM of technical triplicates. (B) Surface plasmon resonance analysis of the interaction between the CPP Tat peptide and HSV-1 envelope. (C) Surface plasmon resonance analysis of the interaction between the CPP Tat peptide and VACV envelope.
We further analyzed the CPP anchoring by surface plasmon resonance in order to get an insight of the binding affinities and the number of peptides bound to the viral envelope. Surface plasmon resonance (SPR) analysis showed that the affinity of the CPP moiety toward both the vaccinia virus (VACV) and herpes simplex virus 1 (HSV-1) envelope was very high, and it also seems that the binding is two sided for both envelopes, where most probably, the first interaction is depicting the electrostatic interaction and the second, the penetration of the moiety into the envelope. Interestingly, the Koff was very low, indicating a very strong interaction between the CPP moiety and the viral envelope. The binding was almost completely dependent on the CPP moiety, as the same peptide without the CPP moiety did not have significant affinity toward either of the viral envelopes (Figures 2B and 2C). We also estimated the number of peptides bound to one viral particle, and for HSV-1, it was estimated to be approximately 11,000 peptides and for VACV, which is the bigger of the two viruses, approximately 35,000 peptides (data not shown).
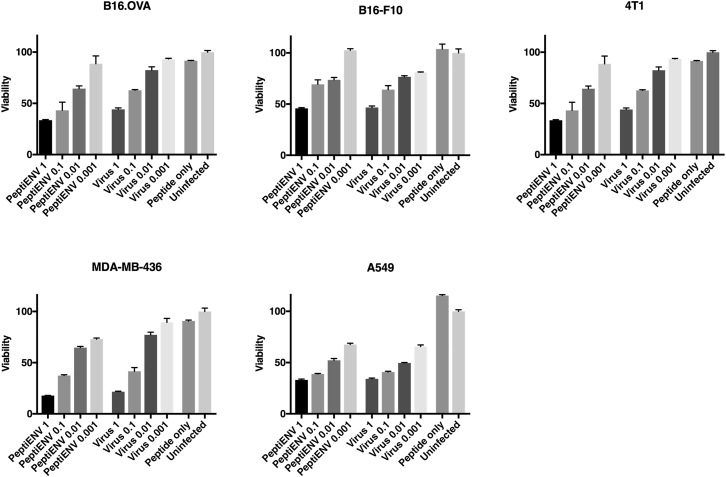
Viruses Coated with Therapeutic Peptides Retain Oncolytic Potential
The degree of lytic cell killing caused by the spread of the infection in a panel of cell lines was monitored using a colorimetric cell-viability assay (Figure 3). The coating of the viral envelope with therapeutic peptides did not have any adverse effect on the oncolytic efficacy on any of the cell lines tested, and the efficacy was found to be similar to the naked virus. The therapeutic peptides alone did not have any toxicity toward the cell lines tested.
Figure 3.
Oncolytic Potency of PeptiENV Is Not Affected by the Attachment of Anti-tumor Peptides into the Viral Envelope
The oncolytic properties of PeptiENV with vaccinia, vaccinia virus, and anti-tumor peptide alone were compared in five cancer cell lines using multiplicities of infection 1, 0.1, 0.01, and 0.001. After 3 days post-infection, the amount of living cells were measured and compared to the viability of uninfected cells. Each bar is the mean ± SEM of technical triplicates.
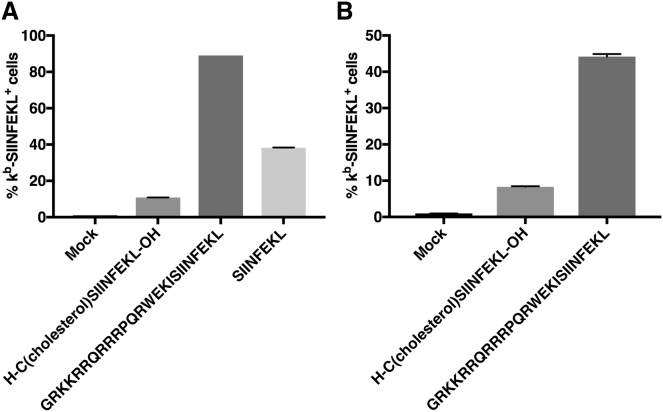
Therapeutic Peptides Are Readily Cross-Presented by Antigen-Presenting Cells
Next, we investigated whether the CPP- or cholesterol-containing therapeutic peptides used in the PeptiENV platform enable the neo-epitope SIINFEKL to be cross-presented on major histocompatibility complex (MHC) class I molecules by the antigen-presenting cells. Mouse DC line Jaws II was pulsed with GRKKRRQRRRPQRWEKISIINFEKL, cholesterol-CSIINFEKL, or SIINFEKL peptide alone, and the efficiency of cross-presentation of the mature form of the neo-epitope (SIINFEKL) from these different peptides was assessed by flow cytometry (Figure 4A). Interestingly, SIINFEKL from the cholesterol-conjugated peptide was cross-presented by only 11% of the Jaws II cells after 4 hr of incubation, likely indicating a compromised processing by the immunoproteasome. In striking contrast, SIINFEKL from the CPP-containing peptide was cross-presented with high efficiency as 89% of Jaws II cells were cross presenting the epitope. SIINFEKL peptide alone was cross-presented by 38% of the Jaws II cells. Since SIINFEKL peptide can be directly loaded to the MHC class I complex without the need for immunoproteasome processing, the cross-presentation kinetics might be faster than with peptides processed by the immunoproteasome and thus the cross-presentation at 4 hr may be too late to see the optimal cross-presentation efficiency of the SIINFEKL peptide.16
Figure 4.
Antigen-Presenting Cells Can Readily Cross-Present Ovalbumin MHC Class I Epitope SIINFEKL from Peptides Used in a PeptiENV Platform
(A) Mouse dendritic cell line Jaws II was pulsed with N-terminal cholesterol-conjugated PeptiENV peptide containing SIINFEKL, N-terminal CPP Tat fusion containing SIINFEKL, or SIINFEKL alone. Cross-presentation was determined by flow cytometry using APC-conjugated anti-H-2Kb bound to SIINFEKL. (B) Mouse bone-marrow-derived dendritic cells were infected with purified PeptiENV viruses complexed with peptides described in (A). Cross-presentation was determined by flow cytometry using APC-conjugated anti-H-2Kb bound to SIINFEKL. Each bar is the mean ± SEM of technical triplicates.
Antigen-Presenting Cells Can Efficiently Present Therapeutic Peptides Delivered by PeptiENV
Next, we tested whether PeptiENV platform can deliver therapeutic peptides to antigen-presenting cells and if they can cross-present the MHC class I epitope from these peptides. PeptiENV viruses coated with GRKKRRQRRRPQRWEKISIINFEKL or cholesterol-CSIINFEKL was used to infect bone marrow-derived DCs (BMDCs) for 4 hr, and the cross-presentation efficacy of the neo-epitope (SIINFEKL) was assessed by flow cytometry (Figure 4B). As expected, the efficacy of SIINFEKL cross-presentation from PeptiENV coated with cholesterol-conjugated SIINFEKL peptide was low, as only 8.3% of BMDCs were shown to cross-present the epitope. In contrast, SIINFEKL was efficiently cross-presented from PeptiENV coated with CPP-conjugated SIINFEKL peptide, as more than 44% of BMDCs were shown to cross-present the SIINFEKL epitope. Since the drastic difference of SIINFEKL cross-presentation efficacy by the PeptiENV coated with cholesterol-conjugated SIINFEKL peptide as compared to the PeptiENV coated with CPP-conjugated SIINFEKL peptide, we chose CPP conjugation as the strategy to anchor therapeutic peptides in the PeptiENV platform.
Vaccination with PeptiENV Induces a Strong Immune Response against MHC Class I Epitopes Coated into the Viral Envelope
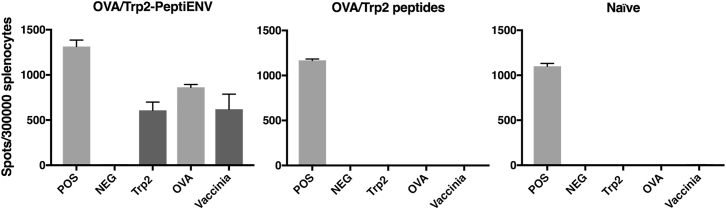
To further validate the PeptiENV platform, we tested whether this platform can induce a robust T cell-specific immune response in naive mice toward the MHC class I restricted epitopes presented by the PeptiENV viruses. To this end, we vaccinated naive C57BL/6JOlaHsd mice with bivalent PeptiENV coated with SIINFEKL- and SVYDFFVWL-containing peptides (OVA/Trp2-PeptiENV). After the vaccination, mice were analyzed for the induction of ovalbumin (OVA)- and tyrosinase-related protein-2 (Trp2)-specific T cell responses by the interferon-gamma enzyme-linked immunospot (ELISPOT) (Figure 5). Vaccination with OVA and Trp2 peptides alone did not induce any SIINFEKL- or SVYDFFVWL-specific T cell response. By striking contrast, bivalent OVA/Trp2-PeptiENV was able to induce a robust SIINFEKL- as well as SVYDFFVWL-specific T cell response. These responses were of similar magnitude as the T cell-specific response toward the VACV used within the platform, indicating a very high immune response toward the attached MHC class I restricted epitopes.
Figure 5.
Bivalent PeptiENV Targeting OVA and Trp2 Can Induce Robust T Cell-Specific Immune Response toward Both Tumor Epitopes
Naive C57BL/6J mice (n = 3/group) were immunized with purified PeptiENV complexed with both OVA- and Trp2-containing peptides or with peptides alone on days 1, 2, 3, and 10. Six days after the last treatment, mice were sacrificed and spleens were collected for the quantification of activated interferon-gamma (IFN-γ) secreting CD8+ cytotoxic T cells specific for the two tumor epitopes (SIINFEKL and SVYDFFVWL) by using a mouse interferon-gamma ELISPOT assay. Each bar is the mean ± SEM of six technical repeats and biological triplicates.
PeptiENV Elicits Potent Anti-tumor Efficacy and Induces Robust Infiltration of Tumor-Specific CD8+ Effector T Cells in a Syngeneic Mouse Model of B16.OVA Melanoma
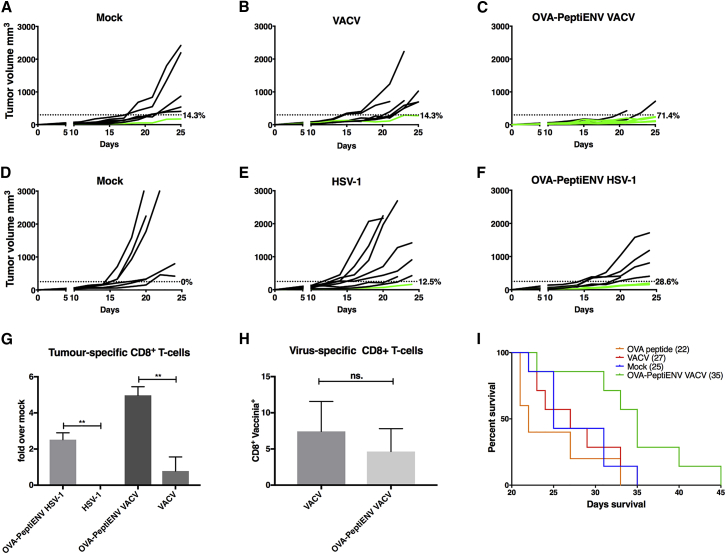
To study the anti-tumor efficacy of PeptiENV platform, we used a well-established syngeneic mouse melanoma model B16 expressing chicken OVA as a model antigen.17 When mice bearing B16.OVA tumors were treated intratumorally with OVA-targeted PeptiENV (OVA-PeptiENV VACV), VACV, peptides alone, or vehicle (mock), we observed significant reduction in tumor growth in OVA-PeptiENV group as compared to all other treatment groups. We set a tumor size threshold of 250 mm3 for defining the responders in each treatment group. Treating mice with SIINFEKL peptide alone did not have any effect on tumor growth, and none of the mice were responsive to the therapy (Figure 6A). In virus- and mock-treated groups, there was one responder in each group accounting for 14.3% of response rate (Figures 6B and 6C). In contrast, OVA-PeptiENV treatment was very efficient in controlling tumor growth, with a response rate of 71.4% for this group (Figure 6D). Although B16 murine melanoma is not permissive for human HSV-1 infection,18, 19 omitting the anti-tumor effect of direct lysis by productive HSV-1 replication in the tumor, we wanted to test whether we could see an indication of direct induction of tumor-specific T cell response by the PeptiENV platform (now used as a cancer vaccine). When mice bearing B16.OVA tumors were treated intratumorally with OVA-PeptiENV HSV-1, HSV-1 alone, or vehicle (mock), we observed a favorable but non-significant trend toward higher reduction of tumor growth in the OVA-PeptiENV HSV-1 group as compared to the HSV-1 or mock groups, with response rates of 0%, 12.5%, and 28.6% for mock, HSV-1, and OVA-PeptiENV HSV-1 groups, respectively (Figures 6E–6G). We went on to analyze whether there were any differences in the infiltration of tumor-specific CD8+ T cells between the treatment groups and whether enhanced infiltration of tumor-specific CD8+ T cells into the tumor microenvironment was accountable for the superior anti-tumor efficacy seen in the OVA-PeptiENV VACV group and for the favorable trend seen in the OVA-PeptiENV HSV-1 group (Figure 6H). As expected, we saw a significantly higher number of tumor-specific T cells infiltrated into the tumors of OVA-PeptiENV VACV-treated mice as compared to the tumors of VACV-, peptide alone-, or mock-treated mice. Indeed, treating mice with VACV did not result in significantly increased tumor-specific CD8+ T cell infiltration to the tumor as compared to mock (0.76-fold increase over mock). In striking contrast, OVA-PeptiENV VACV treatment was able to increase tumor-specific CD8+ T cell infiltration into the tumor by 6-fold as compared to mock. We also saw a significantly higher number of tumor-specific T cells infiltrated into the tumors of OVA-PeptiENV HSV-1-treated mice as compared to the tumors of HSV-1- or mock-treated mice. As expected, HSV-1 treatment alone was not able to induce tumor-specific CD8+ T cell infiltration into the tumor, most likely the reason being the non-permissive nature of B16.OVA for HSV-1 infection and lysis. In contrast, OVA-PeptiENV HSV-1 treatment was able to significantly increase tumor-specific CD8+ T cell infiltration into the tumor as compared to HSV-1 alone or mock. We also analyzed the number of virus-specific T cells in the tumors of VACV- and OVA-PeptiENV VACV-treated groups, and no significant difference between the groups was seen (Figure 6I). The superior anti-tumor efficacy of OVA-PeptiENV VACV treatment was translated into significantly longer survival, with median survival of 35 days as compared to 22, 27, and 25 days with OVA peptide alone, VACV, and mock groups, respectively (Figure 6J). The observed favorable trend toward higher reduction of tumor growth in OVA-PeptiENV HSV-1 group translated into a clear but non-significant trend toward longer survival, with a median survival of 32 days as compared to 25 and 22 days with HSV-1 alone and mock groups, respectively (data not shown).
Figure 6.
PeptiENV Targeting OVA Elicits Potent Anti-tumor Efficacy and Induces Robust Infiltration of Tumor-Specific CD8+ Effector T Cells into the Tumor in a Syngeneic Mouse Model of B16.OVA Melanoma
(A–F) Tumor growth curves for each mouse/group are shown. C57BL/6 mice were inoculated subcutaneously in right flank with 3.5 × 105 B16.OVA melanoma cells and treated on days 11,13, and 19 with (C) OVA-PeptiENV VACV (n = 7), (B) VACV (n = 7), or (A) injection media alone as mock (n = 7), and in experiments using HSV-1 within the platform, mice were treated on days 10,12, and 18 with (F) OVA-PeptiENV HSV-1 (n = 7), (E) HSV-1 (n = 8), or (D) injection media alone as mock (n = 6). A threshold of 250 mm3 was set to define the percentage of mice responding to the different therapies (dotted line). The percentage of responders in each treatment group is shown on the right side of the dotted line. (G) The number of CD19−CD3+CD8+ tumor-infiltrating lymphocytes were evaluated for tumor antigen specificity (SIINFEKL-pentamer) for each group and plotted as fold increase over the mock group. (H) The number of CD19−CD3+CD8+ tumor-infiltrating lymphocytes were evaluated for virus antigen specificity (vaccinia-pentamer) for the OVA-PeptiENV VACV and VACV groups. (I) Kaplan-Meier survival curve for the OVA-PeptiENV VACV experiment. The median survival for each group is shown in the parentheses. Data shown as mean ± SEM. **p < 0.01; ns., not significant (one-way ANOVA or unpaired t test for H). Flow cytometry was performed with three biological replicates and two technical replicates from each sample.
PeptiENV Induces Enhanced Infiltration of Tumor-Specific CD8+ Effector T Cells in a Syngeneic Mouse Model of B16-F10 Melanoma
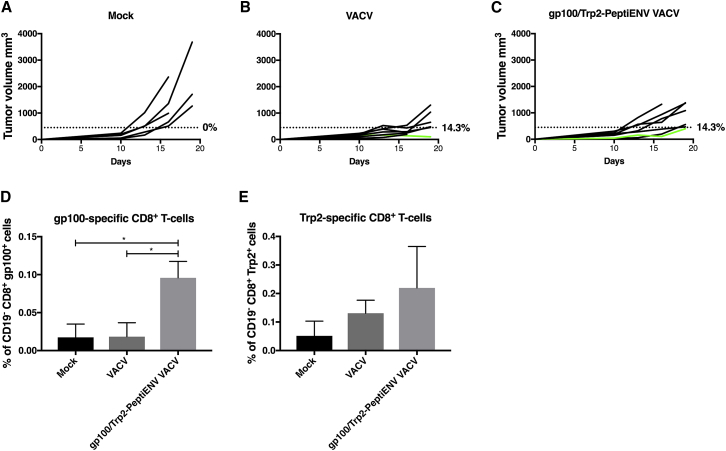
Finally, we tested the PeptiENV platform in B16-F10 melanoma, a representative model of an aggressive and poorly immunogenic tumor.20, 21, 22 B16-F10 cells express endogenous tumor antigens Trp2 and glycoprotein 100 (gp100).23 Viruses were coated with MCH-I-restricted epitopes Trp2180–188 and gp10025–33-containing peptides (Trp2/gp100-PeptiENV). When the tumors reached approximately 150 mm3 is size, mice were treated intratumorally with the bivalent gp100/Trp2-PeptiENV VACV, VACV, or vehicle (mock). We chose to start the treatments when the tumors were considerably big and the immunosuppressive tumor microenvironment already efficiently established (resembling the situation that is found in many human melanomas) to test the PeptiENV platform in very challenging conditions.24 Because of the aggressive nature of the B16-F10 model and the large size of the initial tumors, we set the tumor size threshold to 450 mm3 for defining the responders in each treatment group. The mock-treated group did not respond to therapy, and all mice in this group developed large tumors very rapidly. In contrast, treatment with gp100/Trp2-PeptiENV VACV or VACV alone suppressed the growth of the tumors and, in both groups, 14.3% of mice were defined as responders (Figures 7A–7C). The low number of responders was expected, taking into account the high starting volume of the tumors (150 mm3) and the poorly immunogenic phenotype of the B16-F10 model. We went on to study whether there were any differences in the infiltration of tumor-specific T cells between the groups and whether the PeptiENV platform was able to enhance the infiltration of tumor-specific CD8+ T cells into the tumor microenvironment in these highly immunosuppressive conditions. Treating mice with VACV alone did not result in increased gp100-specific CD8+ T cell infiltration into the tumor as compared to mock. In contrast, gp100/Trp2-PeptiENV VACV treatment significantly increased tumor infiltration of gp100-specific CD8+ T cells over 5-fold when compared to VACV alone or mock groups (Figure 7D). Trp2-specific CD8+ T cells were found more abundantly infiltrated into the tumors in all groups, although similarly to the gp100-specific CD8+ T cells, the highest number of Trp2-specific CD8+ T cells was found in the tumors of gp100/Trp2-PeptiENV VACV-treated mice with over 4-fold increase as compared to mock (Figure 7E).
Figure 7.
Bivalent PeptiENV Targeting gp100 and Trp2 Induces Robust Infiltration of Tumor-Specific CD8+ Effector T Cells into the Tumor in a Syngeneic Mouse Model of B16-F10 Melanoma
(A–C) Tumor growth curves for each mouse/group are shown. C57BL/6 mice were inoculated subcutaneously in right flank with 1 × 105 B16-F10 melanoma cells, and mice were treated on days 10, 11, 12, 13, 17, and 18 with (C) gp100/Trp2-PeptiENV VACV (n = 7), (B) VACV (n = 7), or (A) injection media alone as mock (n = 5). A threshold of 450 mm3 was set to define the percentage of mice responding to the different therapies (dotted line). The percentage of responders in each treatment group is shown on the right side of the dotted line. The percentage of CD19− CD8+ tumor-infiltrating gp100-specific CD8+ T cells (D) and CD19− CD8+ tumor-infiltrating Trp2-specific CD8+ T cells (E) of the total number of CD19− CD8+ cells were assessed for each group. Data shown as mean ± SEM. *p < 0.05 (one-way ANOVA). Flow cytometry was performed with three biological replicates and two technical replicates from each sample.
Discussion
In this study, we show that by coating the viral envelope with immunostimulatory peptides, such as MHC class I-restricted tumor-associated epitopes, we were able to exploit the superior immunostimulatory properties of viruses in eliciting a robust immune response toward the cancer itself. As the attachment moiety for coating the therapeutic peptides onto the viral envelope, we chose the CPP sequence of HIV Tat protein fused to the N terminus of the tumor epitopes. The CPP portion of the therapeutic peptides did not influence the presentation of the tumor epitopes from these peptides by antigen-presenting cells, and when vaccinating naive mice with bivalent PeptiENV targeting both OVA and Trp2 epitopes we saw robust T cell-specific immune response against both epitopes. We also showed using two different mouse models of melanoma termed B16.OVA (expressing chicken OVA as a model antigen) and B16-F10 (a cancer model for an aggressive and poorly immunogenic tumor) that by treating tumor-bearing mice with OVA-PeptiENV or gp100/Trp2-PeptiENV, respectively, we were able induce robust infiltration of tumor-specific T cells into the tumors of treated mice. Although we acknowledge that the OVA is a foreign antigen, in these settings it could be used as model antigen for approximate assessment of neo-antigen immune responses.25, 26 This novel platform could be used with a multitude of enveloped viruses already used in the clinics, such as HSV-1-based talimogene laherparepvec (T-VEC; Imlygic from Amgen), which has already been approved by the US Food and Drug Administration and the European Medicines Agency for the treatment of metastatic melanoma,27 or viruses in ongoing advanced clinical trials, such as an oncolytic VACV, the pexastimogene devacirepvec (Pexa-Vec), currently tested for the treatment of hepatocarcinoma.28 Compared to a variety of other approaches, the clear advantage of this platform is that by introducing the anti-tumor immunity-inducing peptides non-genetically into the clinically approved, good manufacturing practice (GMP)-grade viruses, one can react very quickly to changes in patients’ MHC class I-restricted tumor antigens simply by coating the virus with a new set of tumor-specific peptides.
Since the recent approval of immunotherapeutic antibodies targeting immune checkpoint molecules such as CTLA-4, PD-1, and PD-L1, there has been a new wave of interest in using oncolytic viruses in combination with ICIs. The first indication of synergistic effects on anti-tumor activity by a combination of oncolytic virus with checkpoint inhibitor antibodies was seen in a phase I study using T-VEC in combination with a checkpoint inhibitor ipilimumab (a therapeutic antibody targeted against CTLA-4) and was later confirmed in a follow-up phase II study showing a significant increase in confirmed objective response rate by the immune-related response criteria (irRC) with the T-VEC + ipilimumab compared with ipilimumab alone (39% versus 18%, respectively; p = 0.002).29, 30 Recently, Ribas et al.31 showed in a phase IB study using T-VEC combined with a checkpoint inhibitor pembrolizumab (a therapeutic antibody targeted against PD-1) high overall and complete response rates of 62% and 33%, respectively, in patients with advanced melanoma. The authors reported that intratumoral injections of T-VEC alone increased CD8+ T cell infiltration in patients responding to combination therapy, indicating a virus-induced beneficial change in the tumor microenvironment, although the number of virus-specific CD8+ T cells was not analyzed. It is intriguing to hypothesize that by using T-VEC in the PeptiENV platform, one could further enhance the number of tumor-specific CD8+ T cells and thus further increase the anti-tumor efficacy of the combination therapy. Our group has earlier shown that treatment of melanoma by oncolytic adenoviruses coated with MHC class I restricted tumor-associated antigen peptides increases the number of tumor-specific CD8+ T cells as compared to naked adenovirus, and the increase of tumor-specific CD8+ T cells also increases the number of responders to the combination therapy with ICIs.16, 32 The PeptiENV platform also enables the use of different MHC class II epitopes coated onto the viral envelope to boost CD4+ T cell responses alone or in combination with MHC class I epitopes. Another important aspect of the platform is that the selected virus needs to go through the rigorous quality control and approval stages only once, reducing time and costs of entering into clinical testing with a new set of therapeutic peptides. In contrast, platforms with genetically modified viruses need to go through every stage of approval every time a new modification or a new peptide or protein is introduced, making it virtually impossible to use these platforms in truly personalized settings.
In summary, by using therapeutic enveloped viruses as active, multifunctional adjuvants for the therapeutic anti-tumor peptides, we can combine the advantages of both approaches; viruses activate efficiently the immune response and induce immunogenic cell death of the tumor cells by lysis, and the therapeutic anti-tumor peptides direct the immune response toward the tumor-associated antigens. As more personalized cancer therapies are needed for enhanced anti-tumor efficacy, there is a rationale to convert the existing immunovirotherapy modalities into more personalized and targeted approaches. The PeptiENV platform can convert existing clinically relevant enveloped viruses into multimodal personalized cancer vaccines without the need for genetic modifications.
Materials and Methods
Cell Lines
Human lung carcinoma cell line A549, human triple-negative breast cancer cell line MDA-MB-436, African Green monkey kidney epithelial cell line Vero and murine melanoma cell line B16-F10 were cultured in RPMI with 10% fetal bovine serum (FBS) (Life Technologies), 1% L-glutamine, and 1% penicillin and streptomycin at 37°C/5% CO2. The cell line B16.OVA,17 a mouse melanoma cell line expressing chicken OVA, was kindly provided by Prof. Richard Vile (Mayo Clinic, Rochester, MN, USA). B16.OVA cells were cultured in DMEM with 10% FBS (Life Technologies), 1% L-glutamine, and 1% penicillin and streptomycin at 37°C/5% CO2. Murine DC line Jaws II was cultured in alpha minimum essential medium with 20% FBS (Life Technologies), ribonucleosides, deoxyribonucleosides, 4 mM L-glutamine (Life Technologies), 1 mM sodium pyruvate (Life Technologies), and 5 ng/mL murine GM-CSF (PeproTech, USA) at 37°C/ 5% CO2. Murine breast cancer cell line 4T1 was cultured in RPMI with 10% FBS (Life Technologies), 1% L-glutamine, and 1% penicillin and streptomycin at 37°C/5% CO2. All cell lines excluding B16.OVA were purchased from ATCC and were not further authenticated. All cells in culture were routinely tested for mycoplasma contamination with commercial detection kit (Lonza).
Peptides
Cholesterol-CSIINFEKL, cholesterol-CRVRRALISLEQLESIINFEKLTEW-FITC, and GRKKRRQRRRPQRVRRALISLEQLESIINFEKLTEW-FITC were purchased from Pepscan (Lelystad, the Netherlands), and an MHC class I-restricted epitope from chicken OVA SIINFEKL (OVA257–264), an MHC class I-restricted epitope from tyrosinase-related protein 2 SVYDFFVWL (TRP2180–188), GRKKRRQRRRPQRWEKISIINFEKL, GRKKRRQRRRPQRWEKISVYDFFVWL, and GRKKRRQRRRPQRWEKIKVPRNQDWL (having human gp100 [gp10025–33] MHC class I-restricted epitope embedded) were purchased from Zhejiang Ontores Biotechnologies (Zhejiang, China).
Viruses
Western reserve strain of VACV coding for mouse DNA-dependent activator of interferon (IFN)-regulatory factors (mDAIs)33 was produced in A549 cells and purified through 36% sucrose cushion ultracentrifugation and eluted in 1 mM Tris (pH 9.0). Herpes simplex virus type 1 (HSV-1) LoxLUC,34 a 17+ derivative, whose genome carried an inserted cassette with a luciferase gene under a human cytomegalovirus promoter (HSV-1(17+) LoxLUC) was produced and tittered in Vero cells as previously described.34
PeptiENV Complex Formation
Peptides were mixed with either VACV or HSV-1 in plain DMEM and incubated at 37°C for 15 to 20 min for peptide-virus complexes to form. After complex formation, PeptiENV was either used directly or unbound peptides were removed by ultracentrifugation (20,000 × g, 50 min) through 36% sucrose cushion in 1 mM Tris (pH 9.0) prior experimentation.
Generation of BMDCs
2 × 106 bone marrow cells isolated from C57BL/6JOlaHsd mouse were seeded in 10 mL complete medium (RPMI-1640) (Sigma) containing 10 ng/mL recombinant granulocyte-macrophage colony-stimulating factor (PeproTech), 10% FBS (Sigma), 2 mM L-glutamine (Gibco), 50 U/mL penicillin, and 50 μg/mL streptomycin (Life Technologies). Cells were cultured at 37°C in a humidified atmosphere of 5% CO2. On day 3, 10 mL of complete medium was added, and on days 6 and 8, 9 mL of media was gently aspirated and replaced with 10 mL of fresh complete medium. Following 10 days of culture, DCs were harvested and used for cross-presentation experiments described below.
ELISAs
2.5 × 107 plaque-forming units (PFU) of VACV particles were complexed with GRKKRRQRRRPQRVRRALISLEQLESIINFEKLTEW-FITC (CPP-peptide-FITC) or cholesterol-CRVRRALISLEQLESIINFEKLTEW-FITC (cholesterol-conjugated peptide-FITC) in 100 μL of DMEM for 15 min at 37°C. After complexation, unbound peptides were removed by ultracentrifugation (20,000 × g, 50 min) through 36% sucrose cushion in 1 mM Tris (pH 9.0). For ELISA, anti-vaccinia polyclonal antibody (ab21039, Abcam) was coated overnight at 4°C into Nunc maxisorb 96-well immunoplates (Thermo Fisher) at the concentration of 2 μg/mL. VACV-peptide complexes were incubated for 30–60 min at 37°C or room temperature (RT) and washed with 1× PBS three times. Complexes were detected with anti-FITC antibody conjugated to horseradish peroxidase (ab19224, Abcam) (1:5,000 dilution in 2% BSA-PBS).
2.5 × 107 PFU of HSV-1 particles were complexed with CPP-peptide-FITC or cholesterol-conjugated peptide-FITC in 100 μL of DMEM for 15 min at 37°C. For ELISA, anti-HSV-1 polyclonal antibody (ab19946, Abcam) was coated overnight at 4°C into maxisorb 96-well immunoplates at the concentration of 2 μg/mL. HSV-1-peptide complexes were incubated for 30–60 min at 37°C or RT and washed with 1× PBS three times. Complexes were detected with anti-FITC antibody conjugated to horseradish peroxidase (ab19224, Abcam) (1:5,000 dilution in 2% BSA-PBS).
Cross-Presentation Experiments
For peptide cross-presentation experiments, 1.6 × 106 Jaws II cells were incubated with 50 nmol of GRKKRRQRRRPQRWEKISIINFEKL, cholesterol-CSIINFEKL, or SIINFEKL peptide. After 4 hr of incubation, cells were washed and stained with either allophycocyanin (APC)-conjugated anti-mouse H-2Kb bound to SIINFEKL (141606, BioLegend) or APC-conjugated mouse IgG κ isotype Ctrl (400119, BioLegend), and the samples were analyzed by flow cytometry. For PeptiENV cross-presentation experiments, 15 nmol of either GRKKRRQRRRPQRWEKISIINFEKL or cholesterol-CSIINFEKL were complexed with 4.8 × 107 PFU of VACV, and after the complexation, unbound peptides were removed by ultracentrifugation (20,000 × g, 50 min) through 36% sucrose cushion in 1 mM Tris (pH 9.0), and the purified viral pellets were suspended in 100 μL of complete RPMI-1640. 2 × 106 BMDCs were plated in 2 mL of complete RPMI-1640 media and were infected with the purified PeptiENV viruses. After 4 hr of incubation, cells were washed and stained with either APC-conjugated anti-mouse H-2Kb bound to SIINFEKL (141606, BioLegend) or APC-conjugated mouse IgG κ isotype Ctrl (400119, BioLegend), and the samples were analyzed by flow cytometry.
Cell Viability Assay
50,000 cells were plated in a 96-well plate 1 day prior to infections. Three days post-infection cell viability was measured using the The CellTiter-Fluor Cell Viability Assay (Promega), and a multi-well plate reader (Varioscan; ThermoLabsystems) was used to determine the fluorescence of the samples.
Surface Plasmon Resonance
Measurements were performed using a multi-parametric SPR Navi 220A instrument (Bionavis, Tampere, Finland). PBS (pH 7.4) was used as a running buffer. A constant flow rate of 20 μL/min was used throughout the experiments, and temperature was set to +20°C. Laser light with a wavelength of 670 nm was used for surface plasmon excitation. A sensor slide with a silicon dioxide surface was activated by 5 min of plasma treatment followed by coating with APTES ((3-aminopropyl)triethoxysilane) by incubating the sensor in 50 mM APTES in isopropanol for 4 hr. The sensor was then washed and placed into the SPR device, and viruses were immobilized in situ on the sensor surface of the two test channels by injecting approximately 1.1 × 107 PFU of VACV or 5.5 × 107 PFU of HSV-1 in PBS (pH 7.4) for 12 min, followed by a 3-min wash with PBS. CPP-containing anti-tumor peptide or peptide without CPP sequence (a non-interacting control) was then injected into both flow channels of the flow cell in parallel, with increasing concentrations ranging from 1.2 μM to 100 μM.
Animal Experiments
All animal experiments were reviewed and approved by the Experimental Animal Committee of the University of Helsinki and the Provincial Government of Southern Finland (license number ESAVI/9817/04.10.07/2016).
For vaccination experiments, 8- to 9-week-old immune competent female naive C57BL/6JOlaHsd mice were injected on days 0, 1, 2, and 9 with 2 × 106 PFU of purified bivalent OVA/Trp2-PeptiENV VACV-containing tumor antigens SIINFEKL (OVA257–264) and SVYDFFVWL (TRP2180–188) or SIINFEKL and SVYDFFVWL-containing peptides alone. Untreated mice were used as a mock group. Six days after the last treatment, mice were sacrificed and spleens were collected for immunological analysis. C57BL/6JOlaHsd mice were obtained from Scanbur (Karlslunde, Denmark). For PeptiENV melanoma experiments with VACV as a viral backbone, 8- to 9-week-old immune-competent female C57BL/6JOlaHsd mice were injected in the right flank with 350,000 B16.OVA melanoma cells, and when the tumor size reached approximately 50 mm3 (11 days after injection), mice were treated with 2 × 106 PFU of VACV, 2 × 106 PFU of OVA-PeptiENV VACV, peptides only, or injection media only (mock). Mice were treated on days 11 and 13, and a booster treatment was given on day 19. For PeptiENV experiments with HSV-1 as a viral backbone, 8- to 9-week-old immune-competent female C57BL/6JOlaHsd mice were injected in the right flank with 350,000 B16.OVA melanoma cells, and when the tumor size reached approximately 50 mm3 (10 days after injection), mice were treated with 5 × 106 PFU of HSV-1, 5 × 106 PFU of OVA-PeptiENV HSV-1, or injection media only (mock). Mice were treated on days 10 and 12 and a booster treatment was given on day 18. In B16.OVA experiments, mice were considered as responders when the tumor volume at the time of final measurement remained less than five times the initial volume. For PeptiENV B16-F10 melanoma experiments, 8- to 9-week-old immune-competent female C57BL/6JOlaHsd mice were injected in the right flank with 100,000 B16-F10 melanoma cells, and when the tumor size reached approximately 150 mm3 (11 days after injection), mice were treated with 2 × 106 PFU of sucrose cushion-purified VACV, 2 × 106 PFU of sucrose cushion-purified gp100/Trp2-PeptiENV VACV, or injection media only (mock). Since the initial tumors were of considerably large in size, mice were treated on days 10, 11, 12, and 13, and booster treatments were given on days 17 and 18. In B16-F10 experiments, mice were considered as responders when the tumor volume at the time of final measurement remained less than three times the initial volume. In all experiments, tumors were measured every second or third day until the tumor size reached the maximum allowed, and mice were then sacrificed and tumors collected. For all mouse models, n ≥ 7 was considered adequate for performing statistical analysis. Animals were excluded from the experiments only if tumors failed to form or they were too big before starting the experiment or if health concerns were reported. For all animal experiments, mice were randomly grouped and during the experiments, and the measurements for tumor size were done blinded for the groups.
ELISPOT Assays
The amount of SIINFEKL (OVA257–264), SVYDFFVWL (TRP2180–188) and VACV-specific activated, interferon-γ secreting T cells were measured by ELISPOT assay (CTL, Ohio USA) according to the manufacturer’s instructions. Briefly, 2 μg of SIINFEKL peptide or SVYDFFVWL peptide was used to stimulate the antigen-presenting cells (NB. these peptides contained only the MHC class I epitope in order to be able to rule out any unspecific stimulation which could derive from CPP Tat sequence or immunoproteasome processing sequence used in the PeptiENV platform). After 3 days of stimulation, plates where stained and sent to CTL-Europe GmbH for counting of the spots.
Flow Cytometry
The following antibodies were used in the experiments: TruStain Fc block anti-mouse and anti-human CD16/32 (101320, BioLegend), FITC anti-mouse CD8 (A502-3B-E, ProImmune), phycoerythrin-cyanin 7 (PE/Cy7) anti-mouse CD3e (100320, BioLegend), PE/Cy7 anti-mouse CD19 (115520, BioLegend), FITC anti-mouse CD11c (553801, BD Bioscience), APC anti-mouse H-2Kb bound to SIINFEKL (141606, BioLegend), APC mouse IgG (usually mouse IgG) κ isotype Ctrl (400119, BioLegend). SIINFEKL epitope-specific T cells were studied using R-PE-labeled H-2Kb/SIINFEKL pentamer (F093-84A-E, ProImmune). Trp2(180–188) epitope-specific T cells were studied using R-PE-labeled H-2Kb/SVYDFFVWL pentamer (F185-2B-E, ProImmune), and gp100(25–33) epitope-specific T cells were studied using APC-labeled H-2Kb/KVPRNQDWL pentamer (F1333-4B-E, ProImmune).
Flow cytometric analyses were performed using a Gallios flow cytometer (Beckman Coulter) or BD Accuri 6C plus flow cytometer (BD Biosciences). FlowJo software v10 (FlowJo) was used for the data analysis.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 6.0 software (GraphPad Software, USA). For tumor growth curve analysis, two-way ANOVA was used, and p < 0.05 was considered statistically significant. For flow cytometry data analysis, one-way ANOVA or unpaired t test were used. All results are expressed as the mean ± SEM.
Data Availability
All data supporting the findings of this study are available within the article and the Supplemental Information files and/or from the corresponding author upon reasonable request.
Author Contributions
E.Y. and V.C. conceived and planned the experiments. E.Y., C.M., C.C., O.H., M.F., B.M., L.Y., A.L., S.F., K.P., and T.V. carried out the experiments. H.P. contributed to sample preparation. E.Y., C.C., L.Y., S.F., K.P., V.H., T.V., and V.C. contributed to the interpretation of the results. E.Y. took the lead in writing the manuscript. All authors provided critical feedback and helped shape the research, analysis, and manuscript.
Conflicts of Interest
V.C. is a co-founder and shareholder of VALO therapeutics. V.C., E.Y., and C.C. are co-inventors in a patent application based on the present work.
Acknowledgments
The research leading to these results has received funding from the European Research Council under an ERC-consolidator grant (agreement no. 681219). Moreover, this research was supported by the Helsinki Institute of Life Science (HiLIFE) and the Jane and Aatos Erkko Foundation (grant number 170046 to V.C. and V.H.).
Footnotes
Supplemental Information includes Supplemental Materials and Methods and one figure and can be found with this article online at https://doi.org/10.1016/j.ymthe.2018.06.008.
Supplemental Information
References
- 1.Clemente C.G., Mihm M.C., Jr., Bufalino R., Zurrida S., Collini P., Cascinelli N. Prognostic value of tumor infiltrating lymphocytes in the vertical growth phase of primary cutaneous melanoma. Cancer. 1996;77:1303–1310. doi: 10.1002/(SICI)1097-0142(19960401)77:7<1303::AID-CNCR12>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 2.Galon J., Costes A., Sanchez-Cabo F., Kirilovsky A., Mlecnik B., Lagorce-Pagès C., Tosolini M., Camus M., Berger A., Wind P. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 3.Loi S., Sirtaine N., Piette F., Salgado R., Viale G., Van Eenoo F., Rouas G., Francis P., Crown J.P., Hitre E. Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02-98. J. Clin. Oncol. 2013;31:860–867. doi: 10.1200/JCO.2011.41.0902. [DOI] [PubMed] [Google Scholar]
- 4.Pagès F., Galon J., Dieu-Nosjean M.C., Tartour E., Sautès-Fridman C., Fridman W.H. Immune infiltration in human tumors: a prognostic factor that should not be ignored. Oncogene. 2010;29:1093–1102. doi: 10.1038/onc.2009.416. [DOI] [PubMed] [Google Scholar]
- 5.Sato E., Olson S.H., Ahn J., Bundy B., Nishikawa H., Qian F., Jungbluth A.A., Frosina D., Gnjatic S., Ambrosone C. Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc. Natl. Acad. Sci. USA. 2005;102:18538–18543. doi: 10.1073/pnas.0509182102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ott P.A., Hodi F.S., Robert C. CTLA-4 and PD-1/PD-L1 blockade: new immunotherapeutic modalities with durable clinical benefit in melanoma patients. Clin. Cancer Res. 2013;19:5300–5309. doi: 10.1158/1078-0432.CCR-13-0143. [DOI] [PubMed] [Google Scholar]
- 7.Topalian S.L., Drake C.G., Pardoll D.M. Targeting the PD-1/B7-H1(PD-L1) pathway to activate anti-tumor immunity. Curr. Opin. Immunol. 2012;24:207–212. doi: 10.1016/j.coi.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ku G.Y., Yuan J., Page D.B., Schroeder S.E., Panageas K.S., Carvajal R.D., Chapman P.B., Schwartz G.K., Allison J.P., Wolchok J.D. Single-institution experience with ipilimumab in advanced melanoma patients in the compassionate use setting: lymphocyte count after 2 doses correlates with survival. Cancer. 2010;116:1767–1775. doi: 10.1002/cncr.24951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan J., Adamow M., Ginsberg B.A., Rasalan T.S., Ritter E., Gallardo H.F., Xu Y., Pogoriler E., Terzulli S.L., Kuk D. Integrated NY-ESO-1 antibody and CD8+ T-cell responses correlate with clinical. Proc. Natl. Acad. Sci. USA. 2011;108:16723–16728. doi: 10.1073/pnas.1110814108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breitbach C.J., Lichty B.D., Bell J.C. Oncolytic Viruses: Therapeutics With an Identity Crisis. EBioMedicine. 2016;9:31–36. doi: 10.1016/j.ebiom.2016.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nowak A.K., Lake R.A., Marzo A.L., Scott B., Heath W.R., Collins E.J., Frelinger J.A., Robinson B.W. Induction of tumor cell apoptosis in vivo increases tumor antigen cross-presentation, cross-priming rather than cross-tolerizing host tumor-specific CD8 T cells. J. Immunol. 2003;170:4905–4913. doi: 10.4049/jimmunol.170.10.4905. [DOI] [PubMed] [Google Scholar]
- 12.van der Most R.G., Currie A.J., Robinson B.W., Lake R.A. Decoding dangerous death: how cytotoxic chemotherapy invokes inflammation, immunity or nothing at all. Cell Death Differ. 2008;15:13–20. doi: 10.1038/sj.cdd.4402255. [DOI] [PubMed] [Google Scholar]
- 13.Zitvogel L., Apetoh L., Ghiringhelli F., Kroemer G. Immunological aspects of cancer chemotherapy. Nat. Rev. Immunol. 2008;8:59–73. doi: 10.1038/nri2216. [DOI] [PubMed] [Google Scholar]
- 14.Ranki T., Pesonen S., Hemminki A., Partanen K., Kairemo K., Alanko T., Lundin J., Linder N., Turkki R., Ristimäki A. Phase I study with ONCOS-102 for the treatment of solid tumors - an evaluation of clinical response and exploratory analyses of immune markers. J. Immunother. Cancer. 2016;4:17. doi: 10.1186/s40425-016-0121-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuhn R.J., Strauss J.H. Enveloped viruses. Adv. Protein Chem. 2003;64:363–377. doi: 10.1016/s0065-3233(03)01010-6. [DOI] [PubMed] [Google Scholar]
- 16.Capasso C., Hirvinen M., Garofalo M., Romaniuk D., Kuryk L., Sarvela T., Vitale A., Antopolsky M., Magarkar A., Viitala T. Oncolytic adenoviruses coated with MHC-I tumor epitopes increase the antitumor immunity and efficacy against melanoma. OncoImmunology. 2015;5:e1105429. doi: 10.1080/2162402X.2015.1105429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore M.W., Carbone F.R., Bevan M.J. Introduction of soluble protein into the class I pathway of antigen processing and presentation. Cell. 1988;54:777–785. doi: 10.1016/s0092-8674(88)91043-4. [DOI] [PubMed] [Google Scholar]
- 18.Miller C.G., Krummenacher C., Eisenberg R.J., Cohen G.H., Fraser N.W. Development of a syngenic murine B16 cell line-derived melanoma susceptible to destruction by neuroattenuated HSV-1. Mol. Ther. 2001;3:160–168. doi: 10.1006/mthe.2000.0240. [DOI] [PubMed] [Google Scholar]
- 19.Speranza M.C., Kasai K., Lawler S.E. Preclinical Mouse Models for Analysis of the Therapeutic Potential of Engineered Oncolytic Herpes Viruses. ILAR J. 2016;57:63–72. doi: 10.1093/ilar/ilw002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baird J.R., Byrne K.T., Lizotte P.H., Toraya-Brown S., Scarlett U.K., Alexander M.P., Sheen M.R., Fox B.A., Bzik D.J., Bosenberg M. Immune-mediated regression of established B16F10 melanoma by intratumoral injection of attenuated Toxoplasma gondii protects against rechallenge. J. Immunol. 2013;190:469–478. doi: 10.4049/jimmunol.1201209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chien C.H., Lee M.J., Liou H.C., Liou H.H., Fu W.M. Local immunosuppressive microenvironment enhances migration of melanoma cells to lungs in DJ-1 knockout mice. PLoS ONE. 2015;10:e0115827. doi: 10.1371/journal.pone.0115827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J., Saffold S., Cao X., Krauss J., Chen W. Eliciting T cell immunity against poorly immunogenic tumors by immunization with dendritic cell-tumor fusion vaccines. J. Immunol. 1998;161:5516–5524. [PubMed] [Google Scholar]
- 23.Sorensen M.R., Pedersen S.R., Lindkvist A., Christensen J.P., Thomsen A.R. Quantification of B16 melanoma cells in lungs using triplex Q-PCR—a new approach to evaluate melanoma cell metastasis and tumor control. PLoS ONE. 2014;9:e87831. doi: 10.1371/journal.pone.0087831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Umansky V., Sevko A. Overcoming immunosuppression in the melanoma microenvironment induced by chronic inflammation. Cancer Immunol. Immunother. 2012;61:275–282. doi: 10.1007/s00262-011-1164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Evans R.A., Diamond M.S., Rech A.J., Chao T., Richardson M.W., Lin J.H., Bajor D.L., Byrne K.T., Stanger B.Z., Riley J.L. Lack of immunoediting in murine pancreatic cancer reversed with neoantigen. JCI Insight. 2016;1:e88328. doi: 10.1172/jci.insight.88328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knocke S., Fleischmann-Mundt B., Saborowski M., Manns M.P., Kuhnel F., Wirth T.C., Woller N. Tailored Tumor Immunogenicity Reveals Regulation of CD4 and CD8 T Cell Responses. Cell Rep. 2016;17:2234–2246. doi: 10.1016/j.celrep.2016.10.086. [DOI] [PubMed] [Google Scholar]
- 27.Andtbacka R.H., Ross M., Puzanov I., Milhem M., Collichio F., Delman K.A., Amatruda T., Zager J.S., Cranmer L., Hsueh E. Patterns of Clinical Response with Talimogene Laherparepvec (T-VEC) in Patients with Melanoma Treated in the OPTiM Phase III Clinical Trial. Ann. Surg. Oncol. 2016;23:4169–4177. doi: 10.1245/s10434-016-5286-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heo J., Reid T., Ruo L., Breitbach C.J., Rose S., Bloomston M., Cho M., Lim H.Y., Chung H.C., Kim C.W. Randomized dose-finding clinical trial of oncolytic immunotherapeutic vaccinia JX-594 in liver cancer. Nat. Med. 2013;19:329–336. doi: 10.1038/nm.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chesney J., Puzanov I., Collichio F., Singh P., Milhem M.M., Glaspy J., Hamid O., Ross M., Friedlander P., Garbe C. Randomized, Open-Label Phase II Study Evaluating the Efficacy and Safety of Talimogene Laherparepvec in Combination With Ipilimumab Versus Ipilimumab Alone in Patients With Advanced, Unresectable Melanoma. J. Clin. Oncol. 2018;36:1658–1667. doi: 10.1200/JCO.2017.73.7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Puzanov I., Milhem M.M., Minor D., Hamid O., Li A., Chen L., Chastain M., Gorski K.S., Anderson A., Chou J. Talimogene Laherparepvec in Combination With Ipilimumab in Previously Untreated, Unresectable Stage IIIB-IV Melanoma. J. Clin. Oncol. 2016;34:2619–2626. doi: 10.1200/JCO.2016.67.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ribas A., Dummer R., Puzanov I., VanderWalde A., Andtbacka R.H.I., Michielin O., Olszanski A.J., Malvehy J., Cebon J., Fernandez E. Oncolytic Virotherapy Promotes Intratumoral T Cell Infiltration and Improves. Cell. 2017;170:1109–1119.e10. doi: 10.1016/j.cell.2017.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feola S., Capasso C., Fusciello M., Martins B., Tähtinen S., Medeot M., Carpi S., Frascaro F., Ylosmäki E., Peltonen K. Oncolytic vaccines increase the response to PD-L1 blockade in immunogenic and poorly immunogenic tumors. OncoImmunology. 2018 doi: 10.1080/2162402X.2018.1457596. Published online May 7, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirvinen M., Capasso C., Guse K., Garofalo M., Vitale A., Ahonen M., Kuryk L., Vähä-Koskela M., Hemminki A., Fortino V. Expression of DAI by an oncolytic vaccinia virus boosts the immunogenicity of the virus and enhances antitumor immunity. Mol. Ther. Oncolytics. 2016;3:16002. doi: 10.1038/mto.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nygårdas M., Paavilainen H., Müther N., Nagel C.H., Röyttä M., Sodeik B., Hukkanen V. A herpes simplex virus-derived replicative vector expressing LIF limits experimental demyelinating disease and modulates autoimmunity. PLoS ONE. 2013;8:e64200. doi: 10.1371/journal.pone.0064200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the article and the Supplemental Information files and/or from the corresponding author upon reasonable request.