Abstract
Lyophilized keratinocyte-targeted nanocarriers (TLNκ) loaded with locked nucleic acid (LNA) modified anti-miR were developed for topical application to full thickness burn injury. TLNκ were designed to selectively deliver LNA-anti-miR-107 to keratinocytes using the peptide sequence ASKAIQVFLLAG. TLNκ employed DOTAP/DODAP combination pH-responsive lipid components to improve endosomal escape. To minimize interference of clearance by non-targeted cells, especially immune cells in the acute wound microenvironment, surface charge was neutralized. Lyophilization was performed to extend the shelf life of the lipid nanoparticles (LNPs). Encapsulation efficiency of anti-miR in lyophilized TLNκ was estimated to be 96.54%. Cargo stability of lyophilized TLNκ was tested. After 9 days of loading with anti-miR-210, TLNκ was effective in lowering abundance of the hypoxamiR miR-210 in keratinocytes challenged with hypoxia. Keratinocyte uptake of DiD-labeled TLNκ was selective and exceeded 90% within 4 hr. Topical application of hydrogel-dispersed lyophilized TLNκ encapsulating LNA anti-miR-107 twice a week significantly accelerated wound closure and restoration of skin barrier function. TLNκ/anti-miR-107 application depleted miR-107 and upregulated dicer expression, which accelerated differentiation of keratinocytes. Expression of junctional proteins such as claudin-1, loricrin, filaggrin, ZO-1, and ZO-2 were significantly upregulated following TLNκ/anti-miR-107 treatment. These LNPs are promising as topical therapeutic agents in the management of burn injury.
Keywords: microRNAs, drug targeting, nanoparticles, burn wound, wound healing
Graphical Abstract
Sen and colleagues develop a novel cell-targeted nanoparticle capable of delivering miR cargo of choice to the site of tissue injury. Therapeutic efficacy is proven by improved rescue following burn injury by the topical application of lyophilized keratinocyte-targeted nanoparticles loaded with locked nucleic acid (LNA) modified anti-miR-107.
Introduction
Lipid nanoparticles (LNPs) are promising as carriers of nucleic acid therapeutics. They have been used in cancer treatment for delivering targeted agents,1 antisense oligonucleotides,2, 3 small interfering RNA (siRNA),4, 5, 6, 7 mRNA,8 and DNA inhibitors.9 Particle size and surface modifications are important parameters for LNPs.10, 11 Cell targeting can be achieved by conjugating antibodies to the LNP surface.11 Although such approach is effective in vitro, in vivo applications are fraught with the risk of immunogenicity following repeated injections.12 Furthermore, barriers such as high cost, regulatory hurdles, and limited shelf life complicate the translational path.11 Nonetheless, both therapeutic as well as diagnostic nanoparticles are currently undergoing clinical testing.13
Skin is a promising route for drug delivery offering the option to evade the first-pass effect of the liver that can prematurely metabolize drugs.14 Small LNPs (<10 nm) may directly penetrate the stratum corneum of viable human skin.15 Meanwhile, larger LNPs (10–200 nm) access the skin via hair follicle openings, especially after massage.15 These larger particles traverse the epidermis through shunt pathways generated by sweat glands and hair shafts. Such a first-generation delivery system is effective in delivering lipophilic small molecules.14 The low density of hair follicles (32 follicles/cm2) relative to the whole skin surface represents a limitation of the delivery system.16 Current delivery systems rely on chemical enhancers, non-cavitational ultrasound and iontophoresis, as well as additional measures such as microneedles or thermal abrasion to penetrate the stratum corneum.14 Targeted delivery of therapeutic cargo across intact stratum corneum remains a challenge.
In wounds, although skin barrier function is breached, abundance of inflammatory cells at the site of injury poses a different set of challenges. Phagocytic clearance of nanoparticles poses an issue for delivery.17, 18, 19 In this work, we sought to develop a novel delivery platform based on lyophilized keratinocyte targeting LNPs (TLNκ) to facilitate keratinocyte-specific delivery of oligonucleotides such as locked nucleic acid (LNA)-modified anti-microRNAs (miRNAs). Hydrogel-assisted delivery of lyophilized TLNκ was shown to be robust, offering translational advantages and a longer shelf life in wound care.
Results
Preparation and Characterization of Lyophilized TLNκ
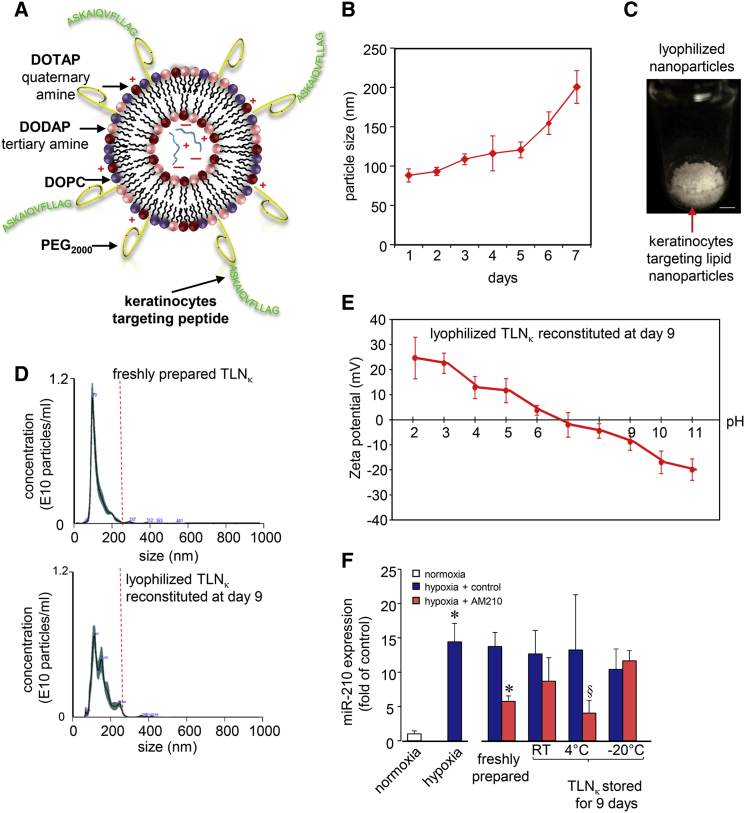
TLNκ was prepared using a keratinocyte-targeting peptide sequence ASKAIQVFLLAG (A5G33).20 The peptide was conjugated to 2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[amino(polyethylene glycol)-2000] (DSPE-PEG2000-Amine) and incorporated onto the surface of LNPs (Figure 1A). Colloidal stability of the LNPs was studied under storage condition. Over a period of 6 days, the particle size doubled from 88 nm to >200 nm (Figure 1B). To address such limitation, TLNκ was lyophilized with addition of a lyoprotectant as described in the Materials and Methods (Figure 1C). The lyophilized keratinocyte-targeted LNPs when stored at 4°C retained their size when reconstituted after 9 days (Figure 1D). Zeta potential of these reconstituted TLNκ ranged from +10 mV to −10 mV in the pH range of 6–8 (Figure 1E). Encapsulation of anti-miRNA in lyophilized TLNκ after 9 days at 4°C was found to be 96.54%. In order to assess cargo stability of lyophilized TLNκ, in vitro transfection was performed with TLNκ loaded with anti-miR-210. Anti-miR-210 was chosen as test cargo based on our previous publications demonstrating that in ischemic wounds, elevated miR-210 compromises wound healing.21, 22 Human immortalized keratinocytes cells, when subjected to hypoxia, showed elevated miR-210 expression. TLNκ loaded with anti-miR-210, stored at 4°C for 9 days, significantly inhibited miR-210 response in cells exposed to hypoxia. Storage of LNPs at 4°C yielded the best result, compared to storage at room temperature or at −20°C (Figure 1F).
Figure 1.
Characterization of the Keratinocytes Targeting Lyophilized Lipid Nanoparticles
(A) Schematic representation of keratinocytes targeting lipid nanoparticles (TLNκ). (B) Change of TLNκ size due to aqueous storage condition under 4°C over a period of 7 days. (C) Representative photograph of the lyophilized keratinocytes targeting lipid nanoparticles. (D) Representative nanoparticle tracking analysis (NanoSight) showing particle size and concentration of freshly prepared TLNκ and reconstituted lyophilized TLNκ after 9 days of storage under 4°C (n = 4). (E) The zeta potential of the TLNκ at different pH. (F) miR-210 expression in HaCaT cells exposed to normoxia and hypoxia 24 hr after delivery of TLNκ/anti-miR-210. Data expressed as mean ± SD (n = 4). §p < 0.05, *p < 0.001 compared to hypoxia, ANOVA.
In Vitro Targeting Efficiency of TLNκ
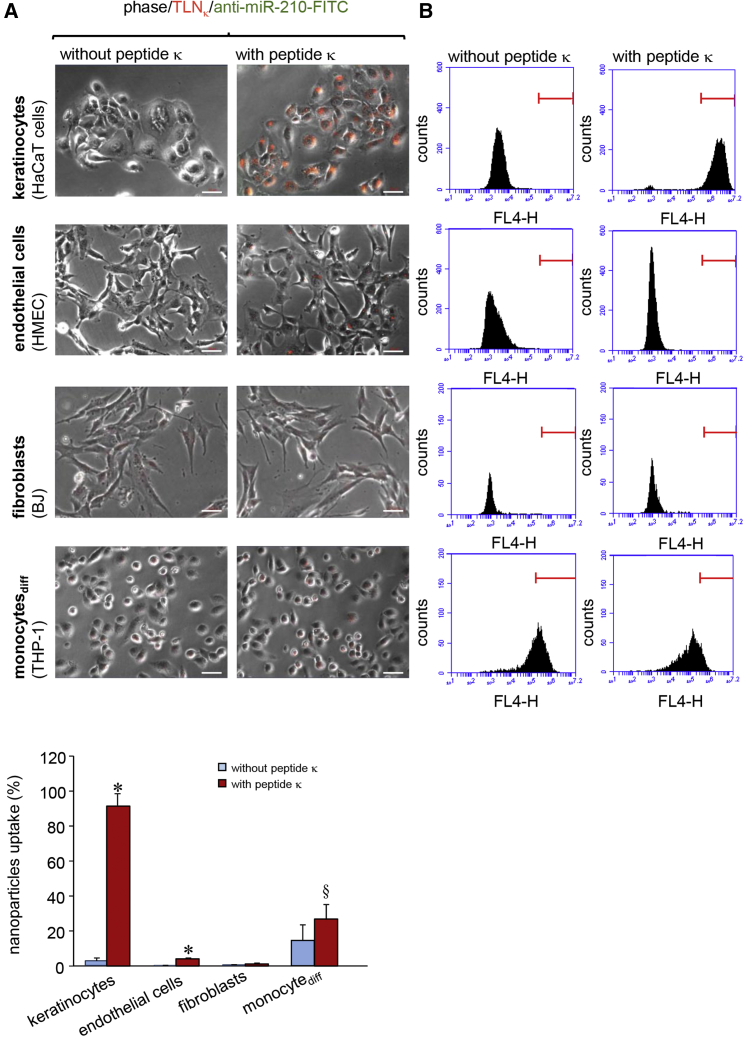
To test specificity of targeted cell delivery of TLNκ, in vitro experiments were carried out using different cell lines, including keratinocytes (HaCaT cells), endothelial cells (HMEC), fibroblasts (BJ), and differentiated monocytes (THP-1). 1,1-Dioctadecyl-3,3,3,3-tetramethylindodicarbocyanine (DiD), a lipophilic fluorescent dye, was incorporated into the LNPs as described in the Materials and Methods.23 Cells were exposed to DiD-labeled TLNκ for 4 hr followed by imaging and flow cytometric analyses. The uptake of DiD-labeled TLNκ was significantly higher in keratinocytes compared to that of non-targeted LNPs (nTLNκ without the targeting peptide) that served as control (Figure 2A). Flow cytometric data showed more than 90% keratinocyte uptake of the DiD-labeled TLNκ, unlike that in endothelial cells (HMEC) or in fibroblasts (BJ) (Figure 2B). Differentiated monocytes (THP-1) appeared promiscuous, demonstrating 14% uptake of even nTLNκ. Nonetheless, delivery of TLNκ to keratinocytes was over 4-fold higher compared to uptake by monocytic cells.
Figure 2.
In Vitro Targeting Efficiency of TLNκ
(A) Overlay of phase contrast and fluorescence microscopic images showing uptake of DiD-labeled (red) TLNκ in keratinocytes (HaCaT cells), endothelial cells (HMEC), fibroblasts (BJ), and differentiated monocytes (THP-1) over a period of 4 hr. DiD-labeled (red) nTLNκ served as control. Scale bars, 50 μm. (B) Flow cytometric analysis showing uptake of DiD-labeled (red) TLNκ in keratinocytes (HaCaT cells), endothelial cells (HMEC), fibroblasts (BJ), and differentiated monocytes (THP-1) over a period of 4 hr. The percentage of cells showing red fluorescence of DiD were plotted graphically. Data expressed as mean ± SD (n = 4). §p < 0.05, *p < 0.001 compared to nTLNκ, ANOVA.
In Vivo Targeting Efficiency of TLNκ
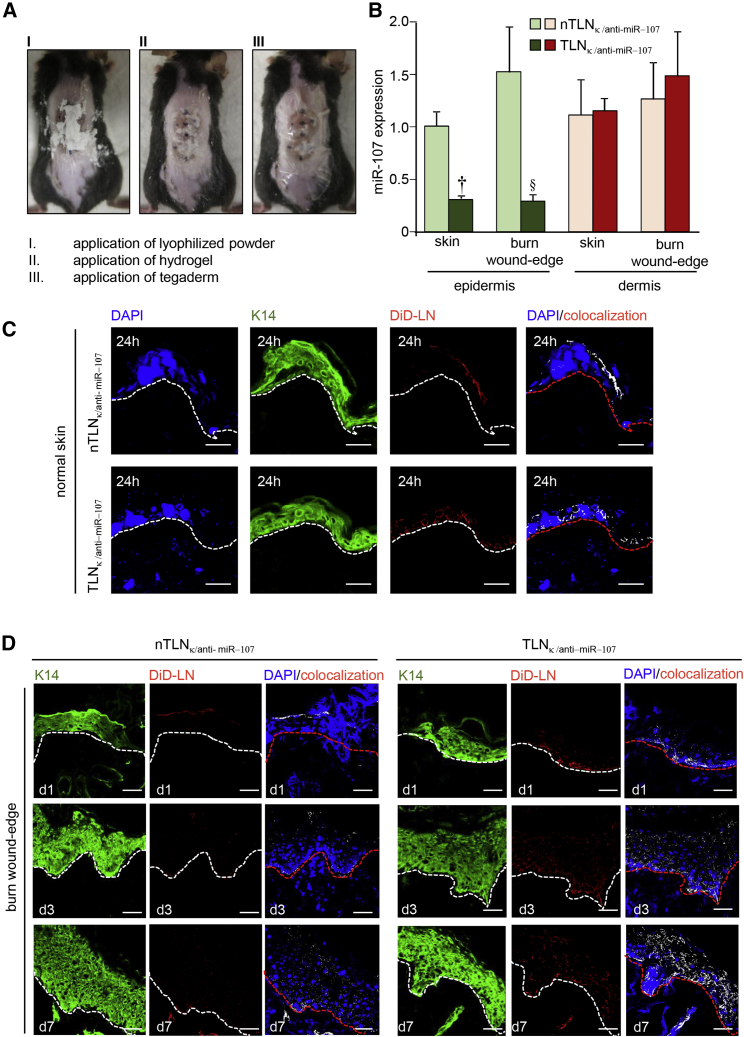
To evaluate targeting efficiency of TLNκ in vivo, full thickness burn wound was developed on the dorsal skin of mice. Full thickness burn wound was confirmed by H&E staining (Figure S1A). Lyophilized DiD-labeled TLNκ was applied with hydrogel on the burn wound (Figure 3A). Lyophilized TLNκ was readily dispersed in hydrogel within 30 s. The burn wound tissue was harvested after 24 hr of injury. To assess anti-miR delivery efficiency of TLNκ in vivo, targeted as well as nTLNκ-containing anti-miR-107 was applied topically on murine skin and burn wound-edge tissue. Tissues were collected after 24 hr of treatment and subjected to laser capture microdissection. qRT-PCR demonstrated significant downregulation of miR-107 in the epidermis in the TLNκ-treated group while the dermis remained unaffected (Figure 3B). Lyophilized TLNκ thus effectively sequestered keratinocyte miR-107 in vivo.
Figure 3.
In Vivo Targeting Efficiency of TLNκ
(A) Digital photomicrographs showing (I) application of lyophilized TLNκ powder on the dorsal skin of mice post-burn; (II) application of 3M hydrogel to cover and reconstitute the dry powder; (III) application of 3M Tegaderm to cover and strap the dressing from falling off. (B) miR-107 expression from laser microdissected epidermis of murine skin and wound-edge tissue 24 hr after application of lyophilized TLNκ containing anti-miR-107. Data expressed as mean ± SEM (n = 3), §p < 0.05, †p < 0.01, ANOVA compared to nTLNκ/anti-miR-107. (C) Confocal microscopic images showing localization of the DiD-labeled nanoparticles (red) in the epidermis 24 hr after application of nTLNκ/anti-miR-107 and TLNκ/anti-miR-107. The sections were counterstained with K14 (green) and DAPI (blue). (D) Confocal microscopic images showing colocalization (white dots) of the DiD-labeled nanoparticles (red) in the burn wound-edge epidermis stained with K14 (green) at days 1, 3, and 7. The sections were counterstained with DAPI (blue). nTLNκ/anti-miR-107 and TLNκ/anti-miR-107 were applied as mentioned in the Materials and Methods. Scale bars, 50 μm.
Confocal microscopic images of the normal skin demonstrated that TLNκ was successful in penetrating the wound-edge stratum corneum and co-localized in basal keratinocytes (Figure 3C). Non-targeted LNκ primarily localized in the stratum corneum (Figure 3C). Single plane confocal co-localization analysis of wound-edge sections harvested at day 1, day 3, and day 7 was performed. Our data showed that repeated application of TLNκ every third day resulted in cumulative DiD fluorescence in the epidermis, unlike nTLNκ (Figure 3D). At day 7, the TLNκ were found to be more evenly distributed throughout the epidermis.
Functional Wound Closure
miRNA-107 inhibits cell proliferation and migration.24, 25, 26, 27 Delivery of anti-miR-107 using a commercially available transfection reagent (Dharmafect1) accelerates keratinocyte proliferation and migration (Figures S2A and S2B). However, when anti-miR-107 was delivered using TLNκ/anti-miR-107, a marginal change in cell migration was observed, although cell proliferation was increased (Figures S3A and S3B). miRNA-107 also targets the dicer, a key contributor to miR biogenesis.28, 29 Dicer is an RNAase-III enzyme that plays a critical role in re-establishing the barrier function of the skin.30 Delivery of miR-107 silenced dicer expression. Consistently, inhibition of miR-107 upregulated dicer expression in keratinocytes (Figures S2C and S2E). The RNAase-III enzyme dicer is responsible for the biogenesis of key miRs, including miR-20a, miR-93, and miR-106b (Figure S3C). These miRs play pivotal roles in establishing the barrier function of the repairing skin by inhibiting the expression of p21waf1/Cip1 30. TLNκ/anti-miR-107 significantly increased dicer expression (Figure S3D) and downregulated p21Waf1/Cip1 expression (Figure S3E) compared to lyophilized targeted scramble (TLNκ/scramble) or lyophilized nTLNκ/anti-miR-107.
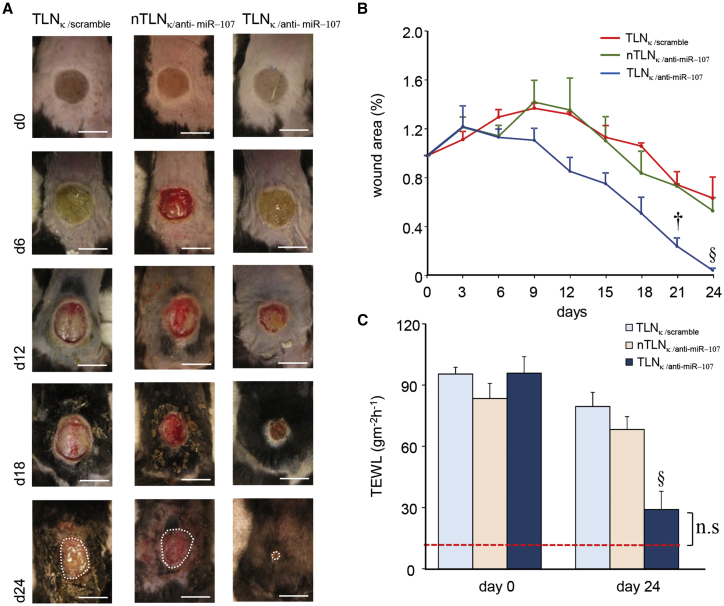
Application of lyophilized TLNκ/anti-miR-107 on the burn wound every third day accelerated wound closure (Figures 4A and 4B). Transepidermal water loss (TEWL) measures skin barrier function.30, 31 TLNκ/anti-miR-107 accelerated re-establishment of successful skin barrier function following burn injury (Figure 4C).
Figure 4.
TLNκ/anti-miR-107 Accelerates Wound Closure and Facilitates in Re-establishing Skin Barrier Function
(A) Digital photographs of the full thickness burn wound at day 0, 6, 12, 18, and 24 days after topical application of TLNκ/scramble, nTLNκ/anti-miR-107, and TLNκ/anti-miR-107. The white dashed lines indicate the wound area. Scale bars, 1 cm. (B) Wound closure after topical application of TLNκ/scramble, nTLNκ/anti-miR-107, and TLNκ/anti-miR-107 was quantified by digital planimetry. Data expressed as mean ± SEM (n = 4), §p < 0.05, †p < 0.01 compared to day 0 (d0), ANOVA. (C) Transepidermal water loss at day 0 and at day 24 after delivery of TLNκ/scramble, nTLNκ/anti-miR-107, and TLNκ/anti-miR-107 was plotted graphically. Data expressed as mean ± SEM (n = 4), §p < 0.05 compared to d0, ANOVA.
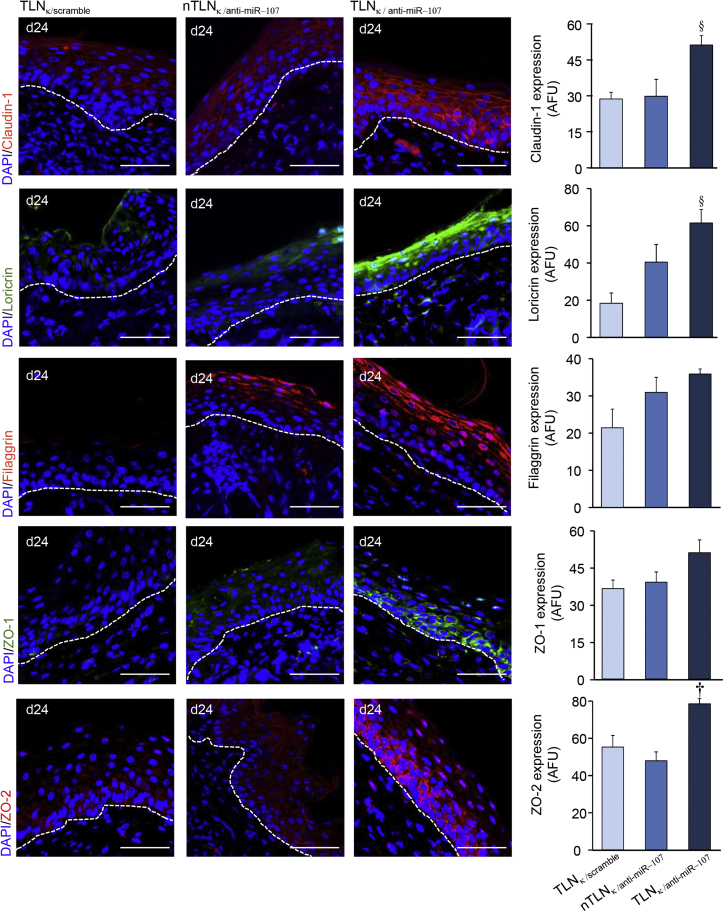
Epidermal Junctional Proteins
Junctional proteins help establish the barrier function of the repaired skin. Topical application of lyophilized TLNκ/anti-miR-107 significantly upregulated the expression of claudin-1, loricrin, filaggrin, ZO-1, and ZO-2 (Figure 5) compared to lyophilized TLNκ/scramble or lyophilized nTLNκ/anti-miR-107.
Figure 5.
TLNκ/anti-miR-210 Upregulates Epidermal Junctional Proteins
TLNκ/anti-miR-107 increased the expression of claudin-1 (red), loricrin (green), filaggrin (red), ZO-1 (green), and ZO-2 (red) in murine skin at day 24. Sections were counterstained with DAPI (blue). Dermal-epidermal junction is indicated by dashed white line. Scale bars, 50 μm. Abundance of junctional proteins were quantified and expressed graphically as mean ± SEM (n = 4). §p < 0.05, †p < 0.01, ANOVA.
Discussion
Lipid nanomaterials with specific biological functions are of therapeutic value. In this work, we report a novel LNP formulation of anti-miR that when applied to the skin topically may penetrate the stratum corneum to specifically target keratinocytes for cargo delivery. The LNPs can be lyophilized to extend the shelf life. This work constitutes the first report on cell-targeting LNPs in a non-cancer health care application.32 According to a recent report from The Center for Drug Evaluation and Research (CDER) within the US Food and Drug Administration (FDA), 234 investigational new drug (IND) applications containing nanomaterials have been submitted, and 34 new drug applications (NDAs) have been approved by the office.33 Liposomes account for 61% of nanomaterials targeting cancer therapeutics having a size less than 300 nm.33 The following targeted nanoparticles are in clinical trial: MCC-465, MBP-426, SGT-53, SGT-94, BIND-014, MM-302, TargomiRs, CALAA-01, Cornell Dot, and ND-L02 s0201.34, 35 Nanoparticles are frequently targeted to cancer cells either through the transferrin receptor or folate receptor.36, 37, 38 In addition, carbohydrates such as lacto bionic acid have been used to target hepatic tumor cells by binding to their asialoglycoprotein receptors.39 Polysaccharides, such as hyaluronic acid or chitosan, have shown promising results in targeting tumor extracellular environment.40 However, the cellular uptake and delivery efficiency are difficult to predict as they depend on the payload-related variables.40 Mannose and galactose derivatives are often used for targeting macrophages through the C-type lectin receptors, which are expressed by all the cells of myeloid origin.41, 42 The targeting ability of functionalized nanoparticles may be lost in a biological environment.43 Even when it does work, efficiency of targeting is highly limited and was estimated to be 30%.44 Some carbohydrate nanocarriers (mono/oligosaccharides and/or polysaccharides) used in biomedical application are reported to reduce unspecific protein adsorption and increases the circulation time in the blood.45 The field of targeted nanoparticle therapy is emergent and at present applications are highly limited. This work underscores the potential of targeted nanoparticles in wound and skin care and offers a new targeting platform.
For topically applied nanoparticles, non-specific uptake was significantly reduced and is mostly limited to follicular (passive) penetration.46 Active penetration of nanoparticles with size >45 nm is an open and controversial topic of discussion. Although the literature reports variable results on porcine and murine skin showing penetration, there are only two publications that report passive permeation of such particles beyond the stratum corneum.46, 47, 48 Toll et al.48 reports penetration of stratum corneum via hair follicle conduits. However, Kohli and Alpar47 attributes penetration of stratum corneum to the strong anionic properties of nanoparticles. If the intent is to target specific cells, high-surface charge of nanoparticles is a confounding factor, as it would be promiscuous and be readily taken up by most cells in proximity.
This work reports a simple and scale-up friendly targeted nanoparticle platform for skin and wound care applications. The design of TLNκ employed 1,2-dioleoyl-3-trimethylammonium-propane/1,2-dioleoyl-3-dimethylammonium-propane (DOTAP/DODAP) combination pH-responsive lipid components to improve endosomal escape. Compared to our previously published nanoparticle formulation,21 we replaced gramicidin with keratinocyte-targeting ligand to promote cell-specific targeting. To minimize interference of clearance by immune cells, surface charge of anti-miR loaded TLNκ was designed to be close to neutral. Additional pH versus zeta-potential analyses were conducted to assess the pH dependence of surface charge. TLNκ possess near-zero surface charges that quickly react to pH drop by boosting surface charges to +15 mV for efficient endosomal release. This design does compromise transfection efficiency but maximizes targeting efficiency, which is desirable in a wound microenvironment complicated by inflammation. By virtue of its design properties, at pH 7 TLNκ is near zero in charge. This poses stability problems due to aggregation, deposition, and sedimentation of the particles.49 Lyophilization of TLNκ not only addresses this limitation but also extends storage life.
Targeted nanoparticles must achieve high uptake efficiency in the desired cell type. The efficiency of uptake is markedly limited in the presence of serum, which may mask targeting ligands by the protein corona.43, 44, 50 In the presence of 10% serum, TLNκ achieved 90% uptake by keratinocytes within the first 4 hr, demonstrating targeting specificity and efficiency. Rapid uptake by intended cells would minimize clearance by immune cells of the wound microenvironment. In its final form, the lyophilized powder and hydrogel may be stored long-term. A clear translational advantage of TLNκ is that all material used for its formulation has prior history of FDA approval for human use. Translational development of TLNκ for human testing is thus warranted.
Materials and Methods
1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), DOTAP, DODAP, and DSPE-PEG2000-Amine were obtained from Avanti Polar Lipids (Alabaster, AL, USA). D-α-Tocopherol polyethylene glycol 1000 succinate (TPGS) and trimethylamine (TEA) were obtained from Sigma-Aldrich (St. Louis, MO, USA). PEGylated bis(sulfosuccinimidyl)suberate (BS(PEG)5) and Vybrant DiD Cell-Labeling Solution were purchased from Thermo Fisher Scientific (Waltham, MA, USA). A5G33 peptide (sequence, ASKAIQVFLLAG) was custom synthesized by Genscript (Township, NJ, USA). 3M Tegaderm hydrogel was obtained from 3M (St. Paul, MN, USA). Anti-miR-107 and anti-miR-210 with LNA modification as well as negative control A-LNA were synthesized by Exiqon (Woburn, MA, USA).
Lipid-Peptide Conjugation
Lipid peptide conjugation was done by dissolving 4 mg of DSPE-PEG2000-Amine in 500 μL of DMSO and combined with 5.7 μL of 250 mM homobifunctional crosslinker (BS(PEG)5) with addition of 4 μL of TEA. The reaction proceeded in an oxygen-free environment (nitrogen purge) at room temperature for overnight. Next, 0.5 mg of A5G33 peptide was added into the solution and incubated for 2 hr at room temperature. The molar ratio between DSPE-PEG2000, BS(PEG)5, and A5G33 peptide was controlled at 1:1.2:1.4. Thin-layer chromatography was used to evaluate the efficiency of conjugation reaction.
Preparation of TLNκ
TLNκ were prepared by an ethanol serial dilution method, as described previously.51, 52 First, lipid components (DOTAP/DODAP/DSPE-PEG2000-A5G33/DOPC/TPGS, 30/24/3/41/2 mol/mol) were dissolved in ethanol. If lipophilic fluorescence dye DiD were chosen to label the LNPs, 0.2% mol/mol amount of dye was added into the above formulation recipe. LNA-based anti-miR was dissolved in equal volume of citric acid buffer (5 mM, pH 4.5) as prepared lipid ethanol solution. The lipid solution was rapidly injected into LNA citric acid buffer at 1:10 anti-miR-107:total lipids weight ratio. This mixture was further diluted by equivalent volumes (1:1) of HEPES buffer (25 mM, 135 mM NaCl, pH 7.4) twice. Non-targeting LNPs were formulated similarly by substituting DSPE-PEG2000-A5G33 with DSPE-PEG2000-Amine and used as controls. The final product was stored at 4°C.
Lyophilization of TLNκ
A primary-secondary drying program was employed.53 1 mL of LNP samples were transferred into 5 mL lyophilization vials (Wheaton, Millville, USA) and diluted with equal volume of 20% sucrose solutions. Slow-freezing processes were chosen: sample vials were partially capped and cooled in a sequence of 2 hr each at 4°C, −20°C, and −80°C. Frozen samples were primary dried at −30°C and 0.12 mbar for 24 hr followed with 25°C and 0.08 mbar secondary drying for an additional 6 hr in a shelf dryer from Labconco (Kansas City, MO, USA). Lyophilized sample stoppers were seated in vacuum and stored at 4°C.
Measurement of TLNκ Size and Zeta Potential
For mean particle diameter and surface charge measurement, aliquotes of TLNκ were diluted in PBS (20 mM, pH 7.4). The particle size was determined by dynamic light scattering method using a Zetasizer Nano ZS90 (Malvern, Worcestershire, UK). TLNκ were dispersed in double-distilled water and tested in volume-weighted size distribution mode. TLN morphology and size distribution were further analyzed by NanoSight NS300 (Malvern, Worcestershire, UK). Aliquots of TLNκ containing anti-miR-107 were diluted in PBS with a series of pHs (50 mM, from 2 to 11) to determine the pH dependency of surface charge. Zeta potential measurement was carried out on a Zetasizer Nano ZS90 (Malvern, Worcestershire, UK).
Encapsulation Efficiency of TLNκ
For encapsulation efficiency and stability, TLNκ/anti-miR-107 were first diluted into 1 mL solution by 20 mM HEPES buffer and injected into Slide-A-Lyzer Dialysis Cassette (Thermo Scientific, Waltham, MA, USA). A 1 L beaker was filled with 20 mM HEPES buffer. Dialysis cassette was then dropped into the beaker and incubated at room temperature for 4 hr. Encapsulation efficiency was then performed by Quant-iT RiboGreen RNA Kit (Invitrogen, Grand Island, NY, USA). TLNκ/anti-miR-107 was lysed with Triton X-100 and the fluorescence intensity (FI) was determined using Multi-Mode Microplate Readers (Biotek, Winooski, VT, USA) at 480 nm λex and 520 nm λem. Loading efficiency was calculated with the following formula:
Formulation and lyophilized cake stability was evaluated at −20°C, 4°C, and 25°C over a period of 9 days. The particle size was measured by Zetasizer Nano ZS90.
Cells and Cell Culture
Immortalized human keratinocytes (HaCaT) were grown in Dulbecco’s low-glucose modified Eagle’s medium (Life Technologies) as described previously.54 Human dermal microvascular endothelial cells (HMECs) were cultured in MCDB-131 medium supplemented 10 mM L-glutamine and 100 IU/mL of penicillin and 0.1 mg/mL of streptomycin (Invitrogen), as described previously.55 THP-1 are human (pro-) monocytic cell lines that can be differentiated into either dendritic cells or tissue macrophages.56 THP-1 cells were differentiated to macrophage-like fate using phorbol 12-myristate 13-acetate (PMA; 20 ng/mL, 48 hr) as previously described.57 Differentiated human THP-1 cells were cultured in RPMI complete medium as described previously.57, 58 Human skin fibroblast BJ cells (ATCC CRL-2522) were obtained from ATCC and were cultured in Eagle’s Minimum Essential Medium, (catalog no. 30-2003) as per the instruction provided. All cell cultures media were supplemented with 10% fetal bovine serum (FBS) and 1% antibiotic-antimycotic (AA) (Life Technologies) unless stated otherwise. Cells were maintained in a standard culture incubator with humidified air containing 5% CO2 at 37°C.
Measurement of Targeting Efficiency by Flow Cytometry
The fluorescence of the cells was determined by using an Accuri C6 flow cytometer (Accuri Cytometers, Ann Arbor, MI, USA). Signals from cells labeled with DiD fluorescent dye were collected on channel FL4 (650 ± 20 nm) after excitation with a 488-nm solid-state laser on a gated population of cells. Data were collected from at least 10,000 cells at a flow rate of 250–300 cells/s. A logarithmic scale was used to measure both background and cell fluorescence. Background fluorescence was then subtracted from cell fluorescence, allowing linear comparisons.
Transfection of miRNA Mimic and Inhibitors
HaCaT cells (0.05 × 106 cells/well in 24-well plate) were seeded for 18–24 hr prior to transfection. HaCaT cells were transfected with either miRIDIAN hsa-miR-107 mimic (50 nM) or hsa-miR-107 inhibitor (100 nM) using DharmaFECT 1 transfection reagent (Thermo Scientific Dharmacon RNA Technologies, Lafayette, CO) per the manufacturer’s instructions.59, 60, 61 miRIDIAN miR mimic/inhibitor negative controls (Thermo Scientific Dharmacon RNA Technologies, Lafayette, CO) were used as control. Cells were collected after 48 hr of miR mimic or 72 hr of miR inhibitor transfection for quantification of miRNA or protein expression. In some experiments, the cells were further reseeded in 96-wells plate or 2-well inserts for cell proliferation or cell migration assay.
Determination of Cell Proliferation
Cell proliferation was measured using a Vybrant MTT Cell Proliferation Assay Kit (Thermo Fisher Scientific) per the manufacturer’s instructions as described previously.62 At 24 hr after treatment, cells were incubated in fresh low-glucose DMEM culture medium containing 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) for 4–6 h at 37°C with 5% CO2. After MTT treatment, medium was removed, and DMSO was added (10–20 min at 37°C with 5% CO2) to solubilize formazan produced as a result of MTT metabolism. DMSO extract from each well (100 μL) was collected in a 96-well plate, and formazan content was determined by reading absorbance at 540 nm.
Cell Migration Assay
Cell migration assay was performed using culture inserts (IBIDI, Verona, WI) according to the manufacturer’s instructions as discussed previously.63 In brief, a confluent cellular monolayer was made in the presence of the insert inside a 24-well plate. Removal of the insert generated a gap in the monolayer. Migration of cells across that gap was studied at different time points following withdrawal of the insert. Images were captured using an inverted microscope (Axiovert 200M, Zeiss, Germany) and analyzed using Zen software.
Animal Model
To generate reproducible, full-thickness burn wounds, an electrically heated burn device with controlled pressure delivery was manufactured. A 0.8-cm diameter stainless steel burning stylus was brought to temperature by an electrically heated element (150 W soldering iron, Great American) inserted into the middle of the stylus. Temperature was controlled by an electronic thermostat (eTRON M, JUMO Process Control, NY), which measured the burn stylus temperature using a thermocouple inserted into the center of the stylus and controlled the amount of current sent to the heating element. Applied pressure was regulated through a compression system comprised of upper and lower aluminum guides connected by spring-loaded guide posts. The heating element can be brought into contact with the murine skin and pressure applied. When maximum pressure is reached, the springs collapse fully and prevent further depression of the burner stylus into the skin below.
Male C57BL/6 mice were obtained from Harlan Laboratory (Indianapolis, IN). Experimental thermal injury was caused by 110°C heated stylus for 15 s. Animals were randomly divided into groups based on a computer generated algorithm (https://www.random.org/). Topical application of the LNPs in hydrogel was performed every third day post-burn. All animal studies were performed in accordance with protocols approved by the Laboratory Animal Care and Use Committee of The Ohio State University. During the wounding procedure, mice were anesthetized by low-dose isoflurane inhalation as per standard recommendation. Each wound was digitally photographed at the time point indicated. Wound area was calculated using ImageJ software (NIH, Bethesda, MD).
The animals were euthanized at the indicated post-wounding time point, and wound-edge tissues (2 mm away from the wound) were snap frozen or collected in either optimal cutting temperature compound (OCT) or fixed in formalin for immunohistology.
TEWL
TEWL serves as a reliable index to evaluate the skin barrier function in vivo.31, 64 TEWL was measured from the wounds using DermaLab TEWL Probe (cyberDERM, Broomall, PA).31 The data were expressed in gm−2 h−1.
Laser Capture Microdissection of Epidermis
Epidermal laser capture microdissection (LCM) was performed using the laser microdissection system from PALM Technologies (Bernreid, Germany) as described previously by our group.22, 65 In brief, sections were stained with hematoxylin for 30 s, subsequently washed with DEPC-H2O, and dehydrated in ethanol as described previously.66 Epidermal fractions were identified based on morphology, cut, and captured under a 20× ocular lens. The samples were catapulted into 25 μL of cell direct lysis extraction buffer (Invitrogen). Approximately 1,000,000 μm2 of tissue area was captured into each cap, and the lysate was then stored at −80°C for further processing.22, 65
RNA Extraction and Real-Time qPCR
RNA from mouse wound-edge tissue or cells was isolated using miRVana miRNA Isolation Kit according to the manufacturer’s protocol (Ambion Life Technologies) as described previously.67 The RNA quality was assessed using Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Quantification of mRNA expression was done by real-time qPCR using SYBR Green-I.65, 68 For determination of miR expression, specific TaqMan assays for miRs and the TaqMan miRNA reverse transcription kit were used, followed by real-time PCR using the Universal PCR Master Mix (Applied Biosystems, Foster City, CA).58
Histology, Immunohistochemistry, and Microscopy
Histology of skin was performed from 8-μm-thick paraffin sections after staining with H&E. Immunostaining of Dicer (Abcam, ab13502; 1:200), p21Waf1/Cip1 (Santa Cruz, sc-397; 1:200), Keratin 14 (Covance, PRB-155P; 1:400), Claudin-1 (Invitrogen, RB9209P; 1:200), loricrin (Covance, PRB-145P; 1:400), filaggrin (Covance, PRB-417P; 1:500), ZO-1 (Invitrogen, 617,300; 1:200), ZO-2 (Invitrogen, 38–9,100; 1:200) were performed on paraffin and cryosections of skin sample using specific antibodies as indicated.69 Specificity of the antibodies was validated using rabbit isotype control (Abcam, ab27478; 1:400) (Figure S4). In brief, OCT-embedded tissue was cryosectioned (10 μm), fixed with cold acetone, blocked with 10% normal goat serum, and incubated with specific antibodies overnight at 4°C. Paraffin-embedded tissue were sectioned (8 μm) and deparaffininized, and antigen retrieval was performed prior to blocking. Signal was visualized by subsequent incubation with fluorescence-tagged appropriate secondary antibodies (Alexa 488-tagged α-rabbit, 1:200; Alexa 568-tagged α-rabbit, 1:200).
Data Collection and Statistical Analyses
Samples were coded and data analysis was performed in a blinded fashion. For animal studies, data are reported as mean ± SD of at least 4–8 animals as indicated. In vitro data are reported as mean ± SD of 3–6 experiments, as indicated in respective figure legends. Student’s t test (two-tailed) was used to determine significant differences. Comparisons among multiple groups were tested using ANOVA. p < 0.05 was considered statistically significant.
Author Contributions
Conceptualization, J.L., S.G., R.J.L., C.K.S.; Methodology, J.L., S.G., Y.L., R.J.L., C.K.S.; Investigation and Validation, J.L., S.G., M.S.E.M., A.D., Y.L.; Formal Analysis, J.L., S.G., M.S.E.M., S.R.; Writing – Original Draft, J.L., S.G., M.S.E.M., S.R., R.J.L., C.K.S.; Writing – Review and Editing, J.L, S.G., M.S.E.M., A.D., S.R., R.J.L, C.K.S.; Visualization, J.L., S.G.; Funding Acquisition, C.K.S.; Resources, R.J.L., S.R., C.K.S.; Supervision, R.J.L., S.R., C.K.S.
Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgments
We acknowledge Xin Xin from the Department of Chemical and Biomolecular Engineering, College of Engineering, The Ohio State University, Columbus for helping with the design of the nanoparticles and refining the synthesis procedures to achieve better conjugation efficiency, and MALDI-TOF mass spectrometry to identify the product. We thank The Ohio State University Laboratory Animal Resources for care of mice in accordance with NIH guidelines. This study was supported by NIH grants RO1 GM069589, GM077185, GM108014 NR015676, and NR013898to C.K.S. and in part by NIH grant DK076566 to S.R.
Footnotes
Supplemental Information includes four figures and can be found with this article online at https://doi.org/10.1016/j.ymthe.2018.04.021.
Supplemental Information
References
- 1.Ashton S., Song Y.H., Nolan J., Cadogan E., Murray J., Odedra R., Foster J., Hall P.A., Low S., Taylor P. Aurora kinase inhibitor nanoparticles target tumors with favorable therapeutic index in vivo. Sci. Transl. Med. 2016;8:325ra17. doi: 10.1126/scitranslmed.aad2355. [DOI] [PubMed] [Google Scholar]
- 2.Dritschilo A., Huang C.H., Rudin C.M., Marshall J., Collins B., Dul J.L., Zhang C., Kumar D., Gokhale P.C., Ahmad A. Phase I study of liposome-encapsulated c-raf antisense oligodeoxyribonucleotide infusion in combination with radiation therapy in patients with advanced malignancies. Clin. Cancer Res. 2006;12:1251–1259. doi: 10.1158/1078-0432.CCR-05-1260. [DOI] [PubMed] [Google Scholar]
- 3.Elazar V., Adwan H., Bäuerle T., Rohekar K., Golomb G., Berger M.R. Sustained delivery and efficacy of polymeric nanoparticles containing osteopontin and bone sialoprotein antisenses in rats with breast cancer bone metastasis. Int. J. Cancer. 2010;126:1749–1760. doi: 10.1002/ijc.24890. [DOI] [PubMed] [Google Scholar]
- 4.Davis M.E., Zuckerman J.E., Choi C.H., Seligson D., Tolcher A., Alabi C.A., Yen Y., Heidel J.D., Ribas A. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tabernero J., Shapiro G.I., LoRusso P.M., Cervantes A., Schwartz G.K., Weiss G.J., Paz-Ares L., Cho D.C., Infante J.R., Alsina M. First-in-humans trial of an RNA interference therapeutic targeting VEGF and KSP in cancer patients with liver involvement. Cancer Discov. 2013;3:406–417. doi: 10.1158/2159-8290.CD-12-0429. [DOI] [PubMed] [Google Scholar]
- 6.Schultheis B., Strumberg D., Santel A., Vank C., Gebhardt F., Keil O., Lange C., Giese K., Kaufmann J., Khan M., Drevs J. First-in-human phase I study of the liposomal RNA interference therapeutic Atu027 in patients with advanced solid tumors. J. Clin. Oncol. 2014;32:4141–4148. doi: 10.1200/JCO.2013.55.0376. [DOI] [PubMed] [Google Scholar]
- 7.Jensen S.A., Day E.S., Ko C.H., Hurley L.A., Luciano J.P., Kouri F.M., Merkel T.J., Luthi A.J., Patel P.C., Cutler J.I. Spherical nucleic acid nanoparticle conjugates as an RNAi-based therapy for glioblastoma. Sci. Transl. Med. 2013;5:209ra152. doi: 10.1126/scitranslmed.3006839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Islam M.A., Reesor E.K., Xu Y., Zope H.R., Zetter B.R., Shi J. Biomaterials for mRNA delivery. Biomater. Sci. 2015;3:1519–1533. doi: 10.1039/c5bm00198f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tolcher A.W., Rodrigueza W.V., Rasco D.W., Patnaik A., Papadopoulos K.P., Amaya A., Moore T.D., Gaylor S.K., Bisgaier C.L., Sooch M.P. A phase 1 study of the BCL2-targeted deoxyribonucleic acid inhibitor (DNAi) PNT2258 in patients with advanced solid tumors. Cancer Chemother. Pharmacol. 2014;73:363–371. doi: 10.1007/s00280-013-2361-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uchechi O., Ogbonna J.D.N., Attama A.A. Nanoparticles for dermal and transdermal drug delivery. In: Sezer A.D., editor. Application of Nanotechnology in Drug Delivery. ed. InTech Open; 2014. pp. 193–235. [Google Scholar]
- 11.Weissleder R., Kelly K., Sun E.Y., Shtatland T., Josephson L. Cell-specific targeting of nanoparticles by multivalent attachment of small molecules. Nat. Biotechnol. 2005;23:1418–1423. doi: 10.1038/nbt1159. [DOI] [PubMed] [Google Scholar]
- 12.Högemann D., Ntziachristos V., Josephson L., Weissleder R. High throughput magnetic resonance imaging for evaluating targeted nanoparticle probes. Bioconjug. Chem. 2002;13:116–121. doi: 10.1021/bc015549h. [DOI] [PubMed] [Google Scholar]
- 13.Bobo D., Robinson K.J., Islam J., Thurecht K.J., Corrie S.R. Nanoparticle-based medicines: a review of FDA-approved materials and clinical trials to date. Pharm. Res. 2016;33:2373–2387. doi: 10.1007/s11095-016-1958-5. [DOI] [PubMed] [Google Scholar]
- 14.Prausnitz M.R., Langer R. Transdermal drug delivery. Nat. Biotechnol. 2008;26:1261–1268. doi: 10.1038/nbt.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prow T.W., Grice J.E., Lin L.L., Faye R., Butler M., Becker W., Wurm E.M., Yoong C., Robertson T.A., Soyer H.P., Roberts M.S. Nanoparticles and microparticles for skin drug delivery. Adv. Drug Deliv. Rev. 2011;63:470–491. doi: 10.1016/j.addr.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 16.Lauterbach A., Müller-Goymann C.C. Applications and limitations of lipid nanoparticles in dermal and transdermal drug delivery via the follicular route. Eur. J. Pharm. Biopharm. 2015;97(Pt A):152–163. doi: 10.1016/j.ejpb.2015.06.020. [DOI] [PubMed] [Google Scholar]
- 17.Desai N. Challenges in development of nanoparticle-based therapeutics. AAPS J. 2012;14:282–295. doi: 10.1208/s12248-012-9339-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi J., Kantoff P.W., Wooster R., Farokhzad O.C. Cancer nanomedicine: progress, challenges and opportunities. Nat. Rev. Cancer. 2017;17:20–37. doi: 10.1038/nrc.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miele E., Spinelli G.P., Miele E., Di Fabrizio E., Ferretti E., Tomao S., Gulino A. Nanoparticle-based delivery of small interfering RNA: challenges for cancer therapy. Int. J. Nanomedicine. 2012;7:3637–3657. doi: 10.2147/IJN.S23696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masuda R., Mochizuki M., Hozumi K., Takeda A., Uchinuma E., Yamashina S., Nomizu M., Kadoya Y. A novel cell-adhesive scaffold material for delivering keratinocytes reduces granulation tissue in dermal wounds. Wound Repair Regen. 2009;17:127–135. doi: 10.1111/j.1524-475X.2008.00450.x. [DOI] [PubMed] [Google Scholar]
- 21.Ghatak S., Li J., Chan Y.C., Gnyawali S.C., Steen E., Yung B.C., Khanna S., Roy S., Lee R.J., Sen C.K. AntihypoxamiR functionalized gramicidin lipid nanoparticles rescue against ischemic memory improving cutaneous wound healing. Nanomedicine (Lond.) 2016;12:1827–1831. doi: 10.1016/j.nano.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biswas S., Roy S., Banerjee J., Hussain S.R., Khanna S., Meenakshisundaram G., Kuppusamy P., Friedman A., Sen C.K. Hypoxia inducible microRNA 210 attenuates keratinocyte proliferation and impairs closure in a murine model of ischemic wounds. Proc. Natl. Acad. Sci. USA. 2010;107:6976–6981. doi: 10.1073/pnas.1001653107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Texier I., Goutayer M., Da Silva A., Guyon L., Djaker N., Josserand V., Neumann E., Bibette J., Vinet F. Cyanine-loaded lipid nanoparticles for improved in vivo fluorescence imaging. J. Biomed. Opt. 2009;14:054005. doi: 10.1117/1.3213606. 054005–054011. [DOI] [PubMed] [Google Scholar]
- 24.Wang B., Zuo Z., Lv F., Zhao L., Du M., Gao Y. MiR-107 inhibits proliferation of lung cancer cells through regulating TP53 regulated inhibitor of apoptosis 1 (TRIAP1) Open Life Sci. 2017;12:200–205. [Google Scholar]
- 25.Xia H., Li Y., Lv X. MicroRNA-107 inhibits tumor growth and metastasis by targeting the BDNF-mediated PI3K/AKT pathway in human non-small lung cancer. Int. J. Oncol. 2016;49:1325–1333. doi: 10.3892/ijo.2016.3628. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Li X.-Y., Luo Q.-F., Wei C.-K., Li D.-F., Li J., Fang L. MiRNA-107 inhibits proliferation and migration by targeting CDK8 in breast cancer. Int. J. Clin. Exp. Med. 2014;7:32–40. [PMC free article] [PubMed] [Google Scholar]
- 27.Piao L., Zhang M., Datta J., Xie X., Su T., Li H., Teknos T.N., Pan Q. Lipid-based nanoparticle delivery of Pre-miR-107 inhibits the tumorigenicity of head and neck squamous cell carcinoma. Mol. Ther. 2012;20:1261–1269. doi: 10.1038/mt.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y., Mao L., Gao Y., Baral S., Zhou Y., Hu B. MicroRNA-107 contributes to post-stroke angiogenesis by targeting Dicer-1. 2015;5:13316. doi: 10.1038/srep13316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ristori E., Lopez-Ramirez M.A., Narayanan A., Hill-Teran G., Moro A., Calvo C.F., Thomas J.L., Nicoli S. A dicer-miR-107 interaction regulates biogenesis of specific miRNAs crucial for neurogenesis. Dev. Cell. 2015;32:546–560. doi: 10.1016/j.devcel.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghatak S., Chan Y.C., Khanna S., Banerjee J., Weist J., Roy S., Sen C.K. Barrier function of the repaired skin is disrupted following arrest of dicer in keratinocytes. Mol. Ther. 2015;23:1201–1210. doi: 10.1038/mt.2015.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roy S., Elgharably H., Sinha M., Ganesh K., Chaney S., Mann E., Miller C., Khanna S., Bergdall V.K., Powell H.M. Mixed-species biofilm compromises wound healing by disrupting epidermal barrier function. J. Pathol. 2014;233:331–343. doi: 10.1002/path.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zatsepin T.S., Kotelevtsev Y.V., Koteliansky V. Lipid nanoparticles for targeted siRNA delivery—going from bench to bedside. Int. J. Nanomedicine. 2016;11:3077–3086. doi: 10.2147/IJN.S106625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.D’Mello S.R., Cruz C.N., Chen M.L., Kapoor M., Lee S.L., Tyner K.M. The evolving landscape of drug products containing nanomaterials in the United States. Nat. Nanotechnol. 2017;12:523–529. doi: 10.1038/nnano.2017.67. [DOI] [PubMed] [Google Scholar]
- 34.Davis M.E., Chen Z.G., Shin D.M. Nanoparticle therapeutics: an emerging treatment modality for cancer. Nat. Rev. Drug Discov. 2008;7:771–782. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]
- 35.Anselmo A.C., Mitragotri S. Nanoparticles in the clinic. Bioeng. Transl. Med. 2016;1:10–29. doi: 10.1002/btm2.10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gatter K.C., Brown G., Trowbridge I.S., Woolston R.E., Mason D.Y. Transferrin receptors in human tissues: their distribution and possible clinical relevance. J. Clin. Pathol. 1983;36:539–545. doi: 10.1136/jcp.36.5.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stevens P.J., Sekido M., Lee R.J. A folate receptor-targeted lipid nanoparticle formulation for a lipophilic paclitaxel prodrug. Pharm. Res. 2004;21:2153–2157. doi: 10.1007/s11095-004-7667-5. [DOI] [PubMed] [Google Scholar]
- 38.Li H., Liu Y., Chen L., Liu Q., Qi S., Cheng X., Lee Y.B., Ahn C.H., Kim D.J., Lee R.J. Folate receptor-targeted lipid-albumin nanoparticles (F-LAN) for therapeutic delivery of an Akt1 antisense oligonucleotide. J. Drug Target. 2018;26:466–473. doi: 10.1080/1061186X.2018.1433678. [DOI] [PubMed] [Google Scholar]
- 39.Chen W., Zou Y., Meng F., Cheng R., Deng C., Feijen J., Zhong Z. Glyco-nanoparticles with sheddable saccharide shells: a unique and potent platform for hepatoma-targeting delivery of anticancer drugs. Biomacromolecules. 2014;15:900–907. doi: 10.1021/bm401749t. [DOI] [PubMed] [Google Scholar]
- 40.Lallana E., Rios de la Rosa J.M., Tirella A., Pelliccia M., Gennari A., Stratford I.J., Puri S., Ashford M., Tirelli N. Chitosan/hyaluronic acid nanoparticles: rational design revisited for RNA delivery. Mol. Pharm. 2017;14:2422–2436. doi: 10.1021/acs.molpharmaceut.7b00320. [DOI] [PubMed] [Google Scholar]
- 41.Martínez-Avila O., Hijazi K., Marradi M., Clavel C., Campion C., Kelly C., Penadés S. Gold manno-glyconanoparticles: multivalent systems to block HIV-1 gp120 binding to the lectin DC-SIGN. Chemistry. 2009;15:9874–9888. doi: 10.1002/chem.200900923. [DOI] [PubMed] [Google Scholar]
- 42.Chavez-Santoscoy A.V., Roychoudhury R., Pohl N.L., Wannemuehler M.J., Narasimhan B., Ramer-Tait A.E. Tailoring the immune response by targeting C-type lectin receptors on alveolar macrophages using “pathogen-like” amphiphilic polyanhydride nanoparticles. Biomaterials. 2012;33:4762–4772. doi: 10.1016/j.biomaterials.2012.03.027. [DOI] [PubMed] [Google Scholar]
- 43.Salvati A., Pitek A.S., Monopoli M.P., Prapainop K., Bombelli F.B., Hristov D.R., Kelly P.M., Åberg C., Mahon E., Dawson K.A. Transferrin-functionalized nanoparticles lose their targeting capabilities when a biomolecule corona adsorbs on the surface. Nat. Nanotechnol. 2013;8:137–143. doi: 10.1038/nnano.2012.237. [DOI] [PubMed] [Google Scholar]
- 44.Harrison C. Nanotechnology: biological proteins knock nanoparticles off target. Nat. Rev. Drug Discov. 2013;12:264. doi: 10.1038/nrd3983. [DOI] [PubMed] [Google Scholar]
- 45.Kang B., Opatz T., Landfester K., Wurm F.R. Carbohydrate nanocarriers in biomedical applications: functionalization and construction. Chem. Soc. Rev. 2015;44:8301–8325. doi: 10.1039/c5cs00092k. [DOI] [PubMed] [Google Scholar]
- 46.Schneider M., Stracke F., Hansen S., Schaefer U.F. Nanoparticles and their interactions with the dermal barrier. Dermatoendocrinol. 2009;1:197–206. doi: 10.4161/derm.1.4.9501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kohli A.K., Alpar H.O. Potential use of nanoparticles for transcutaneous vaccine delivery: effect of particle size and charge. Int. J. Pharm. 2004;275:13–17. doi: 10.1016/j.ijpharm.2003.10.038. [DOI] [PubMed] [Google Scholar]
- 48.Toll R., Jacobi U., Richter H., Lademann J., Schaefer H., Blume-Peytavi U. Penetration profile of microspheres in follicular targeting of terminal hair follicles. J. Invest. Dermatol. 2004;123:168–176. doi: 10.1111/j.0022-202X.2004.22717.x. [DOI] [PubMed] [Google Scholar]
- 49.Jiang J., Oberdörster G., Biswas P. Characterization of size, surface charge, and agglomeration state of nanoparticle dispersions for toxicological studies. J. Nanopart. Res. 2009;11:77–89. [Google Scholar]
- 50.Mirshafiee V., Mahmoudi M., Lou K., Cheng J., Kraft M.L. Protein corona significantly reduces active targeting yield. Chem. Commun. (Camb.) 2013;49:2557–2559. doi: 10.1039/c3cc37307j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang X., Koh C.G., Liu S., Pan X., Santhanam R., Yu B., Peng Y., Pang J., Golan S., Talmon Y. Transferrin receptor-targeted lipid nanoparticles for delivery of an antisense oligodeoxyribonucleotide against Bcl-2. Mol. Pharm. 2009;6:221–230. doi: 10.1021/mp800149s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yung B.C., Li J., Zhang M., Cheng X., Li H., Yung E.M., Kang C., Cosby L.E., Liu Y., Teng L., Lee R.J. Lipid nanoparticles composed of quaternary amine-tertiary amine cationic lipid combination (QTsome) for therapeutic delivery of AntimiR-21 for lung cancer. Mol. Pharm. 2016;13:653–662. doi: 10.1021/acs.molpharmaceut.5b00878. [DOI] [PubMed] [Google Scholar]
- 53.Abdelwahed W., Degobert G., Stainmesse S., Fessi H. Freeze-drying of nanoparticles: formulation, process and storage considerations. Adv. Drug Deliv. Rev. 2006;58:1688–1713. doi: 10.1016/j.addr.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 54.Sen C.K., Khanna S., Babior B.M., Hunt T.K., Ellison E.C., Roy S. Oxidant-induced vascular endothelial growth factor expression in human keratinocytes and cutaneous wound healing. J. Biol. Chem. 2002;277:33284–33290. doi: 10.1074/jbc.M203391200. [DOI] [PubMed] [Google Scholar]
- 55.Chan Y.C., Khanna S., Roy S., Sen C.K. miR-200b targets Ets-1 and is down-regulated by hypoxia to induce angiogenic response of endothelial cells. J. Biol. Chem. 2011;286:2047–2056. doi: 10.1074/jbc.M110.158790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bosshart H., Heinzelmann M. THP-1 cells as a model for human monocytes. Ann. Transl. Med. 2016;4:438. doi: 10.21037/atm.2016.08.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ganesh K., Das A., Dickerson R., Khanna S., Parinandi N.L., Gordillo G.M., Sen C.K., Roy S. Prostaglandin E2 induces oncostatin M expression in human chronic wound macrophages through Axl receptor tyrosine kinase pathway. J. Immunol. 2012;189:2563–2573. doi: 10.4049/jimmunol.1102762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Das A., Ganesh K., Khanna S., Sen C.K., Roy S. Engulfment of apoptotic cells by macrophages: a role of microRNA-21 in the resolution of wound inflammation. J. Immunol. 2014;192:1120–1129. doi: 10.4049/jimmunol.1300613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chan Y.C., Roy S., Huang Y., Khanna S., Sen C.K. The microRNA miR-199a-5p down-regulation switches on wound angiogenesis by derepressing the v-ets erythroblastosis virus E26 oncogene homolog 1-matrix metalloproteinase-1 pathway. J. Biol. Chem. 2012;287:41032–41043. doi: 10.1074/jbc.M112.413294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Park H.A., Kubicki N., Gnyawali S., Chan Y.C., Roy S., Khanna S., Sen C.K. Natural vitamin E α-tocotrienol protects against ischemic stroke by induction of multidrug resistance-associated protein 1. Stroke. 2011;42:2308–2314. doi: 10.1161/STROKEAHA.110.608547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gordillo G.M., Biswas A., Khanna S., Pan X., Sinha M., Roy S., Sen C.K. Dicer knockdown inhibits endothelial cell tumor growth via microRNA 21a-3p targeting of Nox-4. J. Biol. Chem. 2014;289:9027–9038. doi: 10.1074/jbc.M113.519264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rink C., Gnyawali S., Stewart R., Teplitsky S., Harris H., Roy S., Sen C.K., Khanna S. Glutamate oxaloacetate transaminase enables anaplerotic refilling of TCA cycle intermediates in stroke-affected brain. FASEB J. 2017;31:1709–1718. doi: 10.1096/fj.201601033R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Banerjee J., Das Ghatak P., Roy S., Khanna S., Sequin E.K., Bellman K., Dickinson B.C., Suri P., Subramaniam V.V., Chang C.J., Sen C.K. Improvement of human keratinocyte migration by a redox active bioelectric dressing. PLoS ONE. 2014;9:e89239. doi: 10.1371/journal.pone.0089239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pinnagoda J., Tupker R.A., Agner T., Serup J. Guidelines for transepidermal water loss (TEWL) measurement. A report from the Standardization Group of the European Society of Contact Dermatitis. Contact Dermat. 1990;22:164–178. doi: 10.1111/j.1600-0536.1990.tb01553.x. [DOI] [PubMed] [Google Scholar]
- 65.Roy S., Patel D., Khanna S., Gordillo G.M., Biswas S., Friedman A., Sen C.K. Transcriptome-wide analysis of blood vessels laser captured from human skin and chronic wound-edge tissue. Proc. Natl. Acad. Sci. USA. 2007;104:14472–14477. doi: 10.1073/pnas.0706793104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kuhn D.E., Roy S., Radtke J., Khanna S., Sen C.K. Laser microdissection and capture of pure cardiomyocytes and fibroblasts from infarcted heart regions: perceived hyperoxia induces p21 in peri-infarct myocytes. Am. J. Physiol. Heart Circ. Physiol. 2007;292:H1245–H1253. doi: 10.1152/ajpheart.01069.2006. [DOI] [PubMed] [Google Scholar]
- 67.Das A., Ghatak S., Sinha M., Chaffee S., Ahmed N.S., Parinandi N.L., Wohleb E.S., Sheridan J.F., Sen C.K., Roy S. Correction of MFG-E8 resolves inflammation and promotes cutaneous wound healing in diabetes. J. Immunol. 2016;196:5089–5100. doi: 10.4049/jimmunol.1502270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roy S., Khanna S., Rink C., Biswas S., Sen C.K. Characterization of the acute temporal changes in excisional murine cutaneous wound inflammation by screening of the wound-edge transcriptome. Physiol. Genomics. 2008;34:162–184. doi: 10.1152/physiolgenomics.00045.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ahmed N.S., Ghatak S., El Masry M.S., Gnyawali S.C., Roy S., Amer M., Everts H., Sen C.K., Khanna S. Epidermal E-cadherin dependent β-catenin pathway is phytochemical inducible and accelerates anagen hair cycling. Mol. Ther. 2017;25:2502–2512. doi: 10.1016/j.ymthe.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.