Abstract
Exosomes (EXOs) are a type of extracellular nanovesicles released from living cells. Accumulating evidence suggests that EXOs are involved in the pathogenesis of human diseases, including lung conditions. In recent years, the potential of EXO-mediated drug delivery has gained increasing interest. In this report, we investigated whether inhaled EXOs serve as an efficient and practical delivery vehicle to activate or inhibit alveolar macrophages (AMs), subsequently modulating pulmonary immune responses. We first identified the recipient cells of the inhaled EXOs, which were labeled with PKH26. We found that only lung macrophages efficiently take up intratracheally instilled EXOs in vivo. Using modified calcium chloride-mediated transformation, we manipulated small RNA molecules in serum-derived EXOs, including siRNAs, microRNA (miRNA) mimics, and miRNA inhibitors. Via intratracheal instillation, we successfully delivered siRNA and miRNA mimics or inhibitors into lung macrophages using the serum-derived EXOs as vehicles. Furthermore, EXO siRNA or miRNA molecules are functional in modulating LPS-induced lung inflammation in vivo. Beneficially, serum-derived EXOs themselves do not trigger lung immune responses, adding more favorable features to serve as drug delivery agents. Collectively, we developed a novel protocol using serum-derived EXOs to deliver designated small RNA molecules into lung macrophages in vivo, potentially shedding light on future gene therapy of human lung diseases.
Keywords: exosome, microRNA, siRNA, macrophage, inflammation
A novel protocol is developed to use serum-derived EXOs as a vehicle to deliver small RNA molecules into lung macrophages in vivo, intratracheally. The loaded exogenous RNA molecules in EXOs exert functional roles in vivo and are non-toxic and non-immunogenic. This method sheds light on novel therapeutic strategies.
Introduction
Extracellular vesicles (EVs) are critical mediators involved in intercellular crosstalk and inter-organ communication.1, 2, 3 They are derived from various cells and potentially transfer functional cellular content between “mother” cells to recipient cells.1, 2, 3 Currently, three categories of EVs are classified based on their size, surface markers, and mode of biogenesis, including apoptotic bodies (ABs), microvesicles (MVs), and exosomes (EXOs).1, 2, 3 Emerging evidence suggests that EXOs serve as vehicles for therapeutic drug delivery, such as small RNA or DNA molecules.4, 5, 6 EXOs are endogenously generated by host cells and may serve as “nature's delivery system.”7, 8 Presumably, EXOs trigger fewer immune responses and/or toxicity compared with exogenous nanoparticles.
Small RNAs, including small interfering RNA (siRNA) and microRNA (miRNA), are enormously promising for the development of therapeutic agents for human diseases.9, 10, 11, 12 siRNA is a chemically synthesized, short, double-stranded RNA (dsRNA) molecule 20–25 bp in length.9, 10 In contrast, miRNAs are endogenously encoded small non-coding RNAs that are crucial regulators of numerous biological processes, such as proliferation, differentiation, development, and cell death.11, 12 To induce post-transcriptional gene silencing, both siRNA and miRNA interact with and activate the RNA-induced silencing complex (RISC).13 These small inhibitory RNA molecules guide the active RISC toward the target mRNAs to silence the designated genes via an endonuclease, Argonaute 2 (AGO2).14, 15 Recently, therapeutic approaches using siRNAs and/or miRNAs have been extensively investigated, and many RNAi-based drugs have entered clinical trials.16, 17, 18 Biopharmaceutical companies are investing a lot of effort to bring small RNA therapeutic agents to the market.
The delivery of a therapeutic siRNA or miRNA to its target tissue or cell is a challenging task that becomes more difficult when delivering siRNA or miRNA into the lungs via inhalation or intratracheal (i.t.) instillation. Inhaled delivery of therapeutic agents is the first-line treatment of various lung diseases. The effectiveness of inhaled therapeutics is closely related to the recipient cells, the amount of drug uptake by the recipient cells, and the kinetics of drug distribution.19, 20 The essential factors affecting the efficacy of an inhaled drug also include respiratory tract geometry, breathing pattern, aerosol properties (such as particle size, shape, density, etc.), and mucociliary clearance.19, 20 As a novel finding, we describe the delivery of inhaled small RNA molecules into the lungs using host-derived EXOs.
In our study, we employed serum-derived EXOs as oligonucleotide delivery vehicles and elucidate fundamental questions regarding the character of EXOs, the recipient cells in lung tissues after i.t. instillation, and the capacity to transport functional small RNAs. We developed an efficient delivery system targeting lung macrophages using serum-derived EXOs. We also demonstrated an effective approach to treating inflammatory lung responses using therapeutic siRNAs or miRNAs delivered in an EXO-mediated manner.
Results
Characterization of EXOs Derived from Mouse Serum
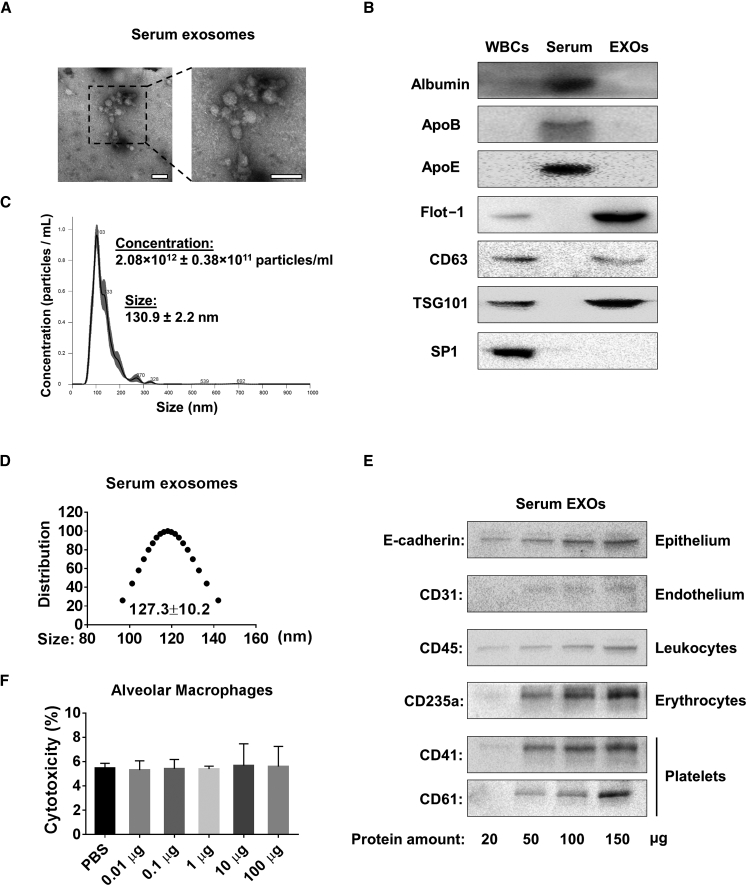
Serum has the potential to provide an unlimited source of EXOs, which are used as vehicles to deliver small RNA molecules. Therefore, we purified EXOs from mouse serum using total EXO isolation reagent as described in Materials and Methods. We first characterized the serum EXOs according to the minimal experimental requirements for extracellular vesicles as defined by the International Society for Extracellular Vesicles.21 The pictures captured using transmission electron microscopy (TEM) show particles with typical serum exosomal morphology, as published before (Figure 1A).22 We also detected traditional positive and negative exosomal markers as well as markers for potential contaminants, such as albumin and lipoproteins (Figure 1B). Nanoparticle tracking analysis (NTA) measurement revealed that the purified serum EXOs were approximately 130 nm in diameter, which is the expected size of EXOs. The concentration of mouse serum EXOs was about 2.08 × 1012 particles/mL (Figure 1C). The size distribution was also confirmed using dynamic light scattering (DLS) (Figure 1D). We next determined the cellular origin of serum EXOs using cell-specific markers as described previously.23, 24 As shown in Figure 1E, we found that serum EXOs were heterogeneous and derived from multiple different types of cells. Using western blot analysis, we detected a variety of cell markers on the isolated serum EXOs, including markers of epithelial cells (E-cadherin), endothelial cell (CD31), leukocytes (CD45), erythrocytes (CD235a), and platelets (CD41 and CD61).
Figure 1.
Characterization of Serum-Derived EXOs
(A) TEM images of serum EXOs isolated from C57BL/6J wild-type mice. Scale bars, 200 nm. (B) Exosomal positive markers (Flot-1, CD63, and TSG-101) and negative markers (Albumin, ApoB, ApoE, and Sp1) were detected in 100 μg protein from white blood cells (WBCs), total serum, and serum EXOs using western blot. (C) The size and concentration profiles of serum EXOs were measured using NanoSight. (D) The size distribution of serum EXOs was measured using DLS. (E) Purified serum EXOs were subjected to western blot analysis to detect different protein markers. (F) Different amounts of serum EXOs were added to primary alveolar macrophages (1 × 106 cells) and incubated for 12 hr. Cytotoxicity was measured according to the manufacturer’s instructions. Results represent mean ± SD of 3 independent experiments.
Effects of Serum EXOs on Primary AMs
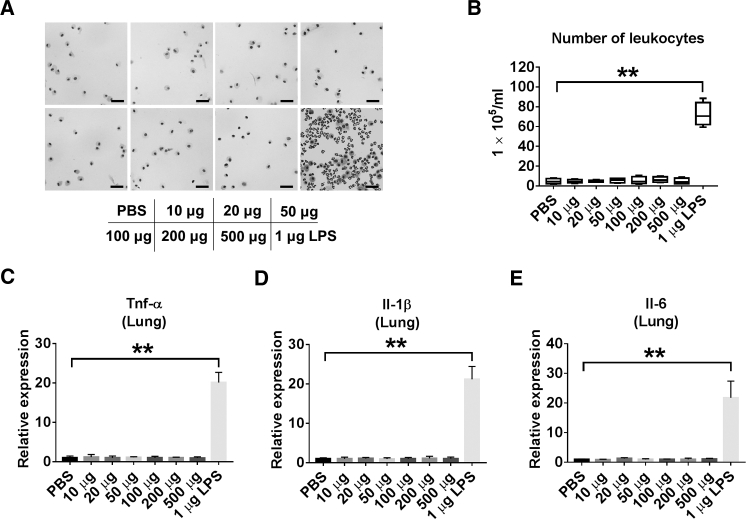
The goal of this study was to develop a practical and efficient method to deliver small RNA molecules into alveolar macrophages (AMs) in vivo. Prior to using serum EXOs as a siRNA/miRNA delivery vehicle, we examined whether the serum EXOs exert cytotoxic or immunogenic effects on primary AMs. No significant cytotoxic responses were observed in primary AMs after exposure to serum EXOs in a dose-dependent manner (Figure 1F). We next evaluated whether serum EXOs trigger inflammatory lung responses in vivo. Purified serum EXOs were given to wild-type (WT) mice via i.t. instillation in a dose-dependent manner. Lipopolysaccharide (LPS) administration was used as a positive control. Inflammatory responses were determined by analyzing broncho-alveolar lavage fluid (BALF) cell counts and differentials as well as the level of secreted cytokines. We observed that BALF cell counts and differentials were not significantly altered in the EXO-treated mice compared with control mice (Figures 2A and 2B). Furthermore, no significant difference was observed in secreted cytokine levels between the EXO-treated group and the control (Figures 2C–2E).
Figure 2.
Instillation of Serum EXOs Has No Pro-inflammatory Effect
(A) C57BL/6J mice were given different amounts of serum EXOs (in 50 μL PBS) or LPS (1 μg in 50 μL PBS), as indicated, via intratracheal instillation. Mice were sacrificed 1 day after treatment, and BALF cells were collected. Shown is H&E staining of BALF cells from each group (n = 4 for each group). Scale bars, 50 μm. (B) Total numbers of leukocytes were counted in the BALF. (C–E) The mRNA expression of TNF-α (C), Il-1β (D), and Il-6 (E) was detected in mouse lungs using real-time RT-PCR. **p < 0.01. Results represent mean ± SD of 3 independent experiments.
AMs Efficiently Take Up Instilled Serum EXOs
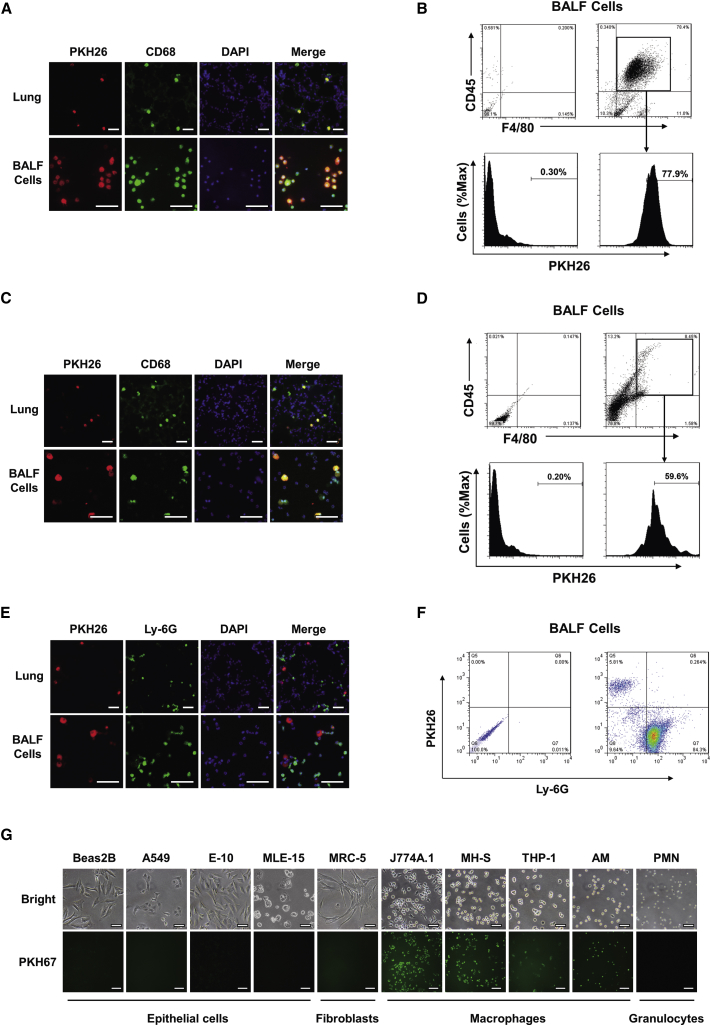
Given that many different types of cells are present in the lung, we first identified the recipient cells of serum EXOs in vivo. Fluorescent PKH26-labeled EXOs were administrated i.t. to untreated WT mice. Co-localization between PKH26 (red) and anti-CD68 (green, a marker of monocyte lineage) were identified, indicating that serum EXOs directly interact with macrophages in the lung (Figure 3A). Consistently, after instillation of PKH26-labeled EXOs i.t., we found that the majority of PKH26-positive cells obtained from BALF were also CD45+F4/80+, which is reported to be an AM marker (Figure 3B).25
Figure 3.
Alveolar Macrophages Efficiently Take Up Instilled Serum EXOs
(A and B) Purified serum EXOs (100 μg in 50 μL PBS) labeled with PKH26 were administrated to wild-type mice intratracheally (n = 4 for each group). After 24 hr, mice were sacrificed. Immunofluorescent staining of macrophages was performed in lung sections and BALF cells using an antibody against CD68 (a macrophage marker). The nuclei were stained with DAPI. Cells with red fluorescence indicate the uptake of PKH26 labeled EXOs. Scale bars, 100 μm (A). BALF cells were isolated from EXO-treated wild-type mice as described above. FACS analysis of PKH26 positive cells was performed using an antibody against CD45 and F4/80 (B). (C–F) Mice were given LPS (1 μg in 50 μL PBS) intratracheally 3 hr before administration of PKH26-labeled EXOs (100 μg in 50 μL PBS) (n = 4 for each group). Mice were sacrificed 24 hr after EXO administration. Immunofluorescent staining was performed in lung sections and BALF cells using an antibody against CD68 (C) or Ly-6G (a neutrophil marker) (E). Scale bars, 100 um. BALF cells were subject to FACS analysis, and representative data were obtained using an antibody against CD45 and F4/80 (D) or Ly-6G (F). (G) Serum EXOs were labeled with PKH67 and added to the culture medium of cells as indicated. Pictures were taken using fluorescence microscopy after 2 hr of EXO incubation.
To further determine whether other phagocytes, such as neutrophils, also take up instilled EXOs in the same manner as macrophages, we delivered serum EXOs to both LPS-pretreated mice and PBS-treated mice (control). LPS induces neutrophil recruitment and activation in the lungs.26 Intriguingly, PKH26-labeled EXO uptake was only found in CD68-positive cells (Figure 3C) but not in Ly-6G-positive ones (Figure 3E), suggesting that serum EXOs were primarily taken up by macrophages rather than neutrophils. Using fluorescence-activated cell sorting (FACS), we further analyzed the surface markers of cells that were PKH26-positive and, presumably, had taken up PKH26-labeled EXOs. Consistently, the PKH26-positive cells were CD45+F4/80+ rather than Ly-6G-positive (Figures 3D and 3F). Additionally, to avoid false positive results caused by lipophilic dye labeling of vesicles, we repeated the in vivo experiments using Exo-Red, which stains the single-stranded RNA in EXOs. Similar results were observed in BALF cells and lung sections (Figure S1).
Next, we determined serum EXO uptake using cultured cells in vitro. PKH67-labeled EXOs and controls were added to the cultured cells. As early as 1 hr after incubation, the uptake of PKH67-labeled EXOs by macrophages was visualized using a fluorescence microscope. In contrast, positive green PKH67 fluorescence was not detected in a variety of non-macrophage cells, including many different types of epithelial cells, fibroblasts, and neutrophils (Figure 3G).
Delivery of siRNA Using Serum EXOs
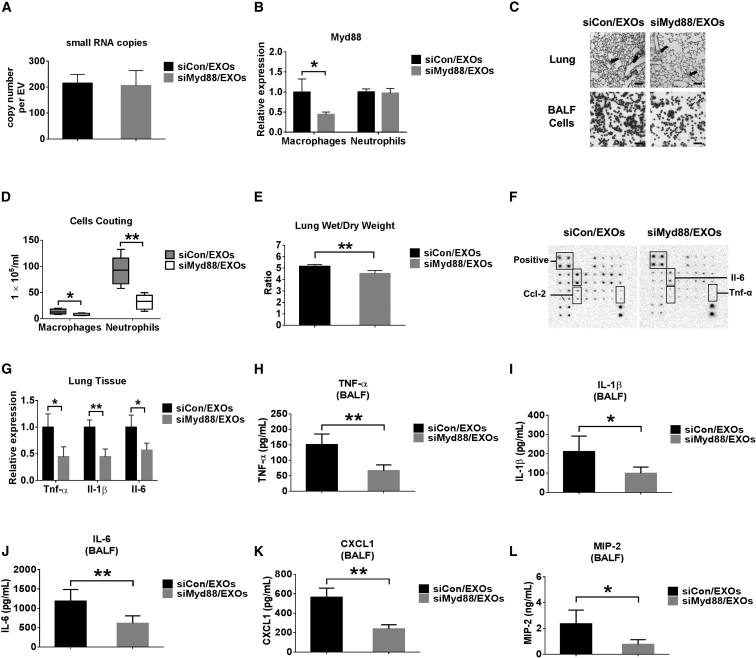
Above, we established the uptake of serum EXOs by lung macrophages and determined whether serum EXOs are safe to be used as delivery vehicles. We next evaluated the efficiency of siRNA delivery in vivo using an EXO-mediated manner. We also determined whether the EXO-delivered Myd88 siRNA exerts functional roles in vivo. Myd88 is a well-known signaling adaptor involved in innate immunity.27 Myd88 siRNA was loaded into serum EXOs using modified calcium-mediated transfection, as described in our previous report.28 We first quantified the copy number of Myd88 siRNA in each EXO after transfection. As shown in Figure 4A, more than 200 copies of control siRNA or Myd88 siRNA per EXO were identified in transfected EXOs. To investigate whether the EXO-delivered Myd88 siRNA has functional activity in vivo, we performed the following experiments. C57BL/6J WT mice were given LPS i.t. Three hours later, control siRNA (siCon)/EXOs or siMyd88/EXOs were administrated i.t. into LPS-pretreated mice. We found that the expression of Myd88 in BALF macrophages was significantly decreased in the mice receiving siMyd88/EXOs. The expression of Myd88 was not altered in the BALF neutrophils (Figure 4B). We further observed a markedly decreased inflammatory cell count in the BALF obtained from siMyd88/EXO-treated mice after exposure to LPS. Using H&E staining, we observed much less cellular infiltration in lung tissue obtained from siMyd88/EXO-treated mice compared with siCon/EXO-treated mice in response to LPS (Figure 4C). Total cell counts of macrophages and neutrophils in siMyd88/EXO-treated mice were reduced significantly compared with the control group (Figure 4D). Moreover, the lung wet to dry weight ratio was robustly reduced in siMyd88/EXO-treated mice (Figure 4E). Using mouse cytokine array analysis, we showed that pro-inflammatory cytokines (tumor necrosis factor alpha [TNF-α], interleukin-2 [IL-2], IL-3, IL-6, and granulocyte-macrophage colony-stimulating factor [GM-CSF]) and chemokines (CCL-2, MCP-5, and CCL-5) were significantly decreased in BALF obtained from siMyd88/EXO-treated mice (Figure 4F). The array data indicated that siMyd88/EXO has anti-inflammatory effects on both recruitment and activation of leukocytes. Consistently, mRNA expression of TNF-α, IL-1β, and IL-6 in the lung tissue was highly inhibited (Figure 4G). In addition, we observed that siMyd88/EXO instillation has inhibitory effects on the secretion of cytokines (TNF-α, IL-1β, and IL-6) and chemokines (CXCL1 and MIP-2) (Figures 4H–4L). These results suggest that the serum EXO-mediated delivery of Myd88 siRNA is successful and functional in recipient lung macrophages and attenuates LPS-induced lung inflammation. It has been reported that EXO membrane integrity is important in the delivery of small RNA.29, 30 Consistent results were observed when siRNA was delivered using lysed EXO (Figure S2).
Figure 4.
Delivery of siMyd88 via Serum EXOs
(A) 100 pmol control siRNA (siCon) or Myd88 siRNA (siMyd88) was introduced into 100 μg serum EXOs using modified calcium-mediated transfection. The copy number of siCon or siMyd88 in each EXO was calculated using NanoSight and absolute real-time PCR. Results represent mean ± SD of 3 independent experiments. (B–L) Wild-type mice were pretreated with 1 μg LPS intratracheally. After 3 hr, mice were given siCon- or siMyd88-loaded serum EXOs (n = 4–6 for each group). 24 hr later, the mRNA level of Myd88 in sorted macrophages (F4/80+CD11c+) and neutrophils (Ly-6G+CD11b+) was detected using real-time RT-PCR (B). H&E staining was performed using mouse lung section and BALF cells. Scale bars, 50 μm (C). Differential cell counts were performed (D). Lung wet to dry weight ratios were calculated (E). A mouse cytokine array was used to detect cytokines released from BALF (F). Relative mRNA levels of TNF-α, IL-1β, and IL-6 in lung tissues were measured using real-time PCR (G). The secretion of TNF-α (H), IL-1β (I), IL-6 (J), CXCL1 (K), and MIP-2 (L) was detected using ELISA. Results represent means ± SD. *p < 0.05, **p < 0.01.
Delivery of a miRNA Mimic or Inhibitor via Serum EXOs
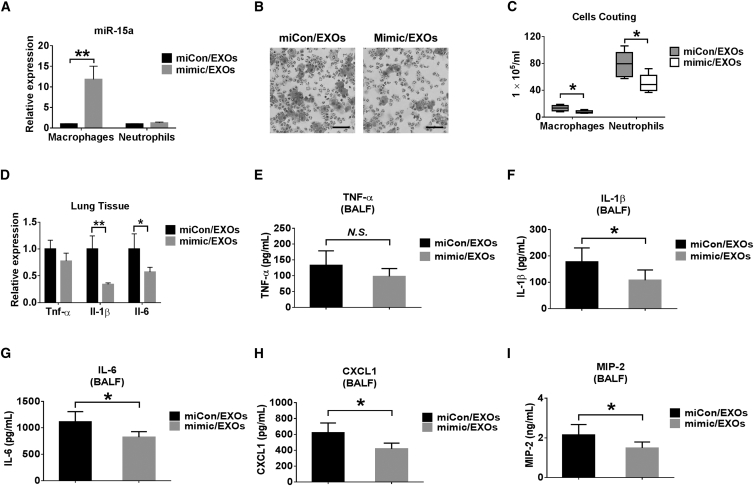
In addition to siRNA, we next evaluated the delivery of a miRNA mimic via EXOs in vivo. We used miR-15a as an example in this study. MiR-15a confers an anti-inflammatory role via the TLR4 signaling pathway.31 Here we evaluated whether EXO-delivered miR-15 attenuates LPS-induced lung inflammation in vivo. C57BL/6J WT mice were pretreated with LPS i.t. After 3 hr, LPS-pretreated mice were treated with an EXO-containing miR-15a mimic (mimic/EXO) or mimic control (miCon/EXO). We confirmed that mimic/EXO administration increased the level of miR-15a in AMs obtained from BALF (Figure 5A). Administration of mimic/EXO resulted in a decreased total amount of macrophages and neutrophils in BALF compared with miCon/EXOs (Figures 5B and 5C). Furthermore, the expression of IL-1β and IL-6 was reduced in the mimic/EXO-treated group (Figure 5D). Using ELISA, we found that mimic/EXO treatment decreases the secretion of cytokines (IL-1β and IL-6) and chemokines (CXCL1 and MIP-2) (Figures 5E–5I). These results indicated that miR-15a-enriched EXOs were successfully delivered and exerted functions in vivo.
Figure 5.
Delivery of miRNA Mimic via Serum EXOs
(A–I) Mice were pretreated with 1 μg LPS intratracheally. After 3 hr, 100 μg serum EXOs transfected with 100 pmol mimic control (miCon/EXOs) or miR-15a mimic (mimic/EXOs) were given to each mouse (n = 4–6 for each group). 24 hr later, the level of miR-15a was detected in sorted macrophages and neutrophils from BALF cells (A). H&E staining was performed using BALF cells (B). Scale bars, 50 μm. The number of macrophages or neutrophils in BALF was counted (C). Relative mRNA levels of TNF-α, IL-1β, and IL-6 in the lung were measured (D). The secretion of TNF-α (E), IL-1β (F), IL-6 (G), CXCL1 (H), and MIP-2 (I) was detected using ELISA. Results represent means ± SD. *p < 0.05, **p < 0.01.
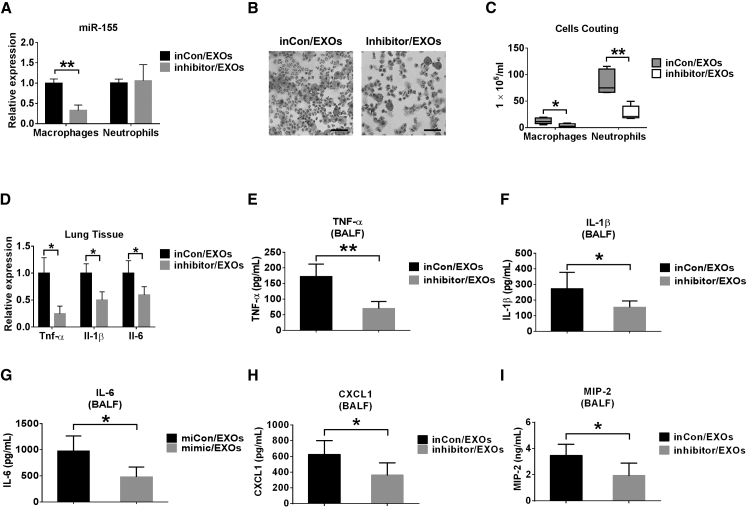
Additionally, we tested a miRNA inhibitor as a cargo of EXO-mediated delivery. miR-155 enhances pro-inflammatory responses in macrophages.32, 33 In our study, the delivery of miR-155 inhibitor-enriched EXOs (inhibitor/EXOs) decreased the level of miR-155 in sorted macrophages obtained from BALF compared with the group treated with inhibitor control-enriched EXOs (inCon/EXO). No significant effect on the level of miR-155 was found in neutrophils (Figure 6A), suggesting that macrophages were the primary target cells of EXO-mediated delivery. Inhibitor/EXO treatment decreased the macrophage and neutrophil counts in BALF and cell infiltration in lung tissue (Figures 6B and 6C). The mRNA expression of TNF-α, IL-1β, and IL-6 was significantly decreased in inhibitor/EXO-treated mice (Figure 6D). TNF-α, IL-1β, IL-6, CXCL1, and MIP-2 in BALF were significantly reduced after miR-155 inhibitor/EXO administration (Figures 6E–6I). These results were consistent with the reported functions of miR-155, suggesting that EXO-mediated delivery of miR-155 inhibitors in vivo was successful.
Figure 6.
Delivery of miRNA Inhibitor via Serum EXOs
(A–I) Mice were pretreated with 1 μg LPS intratracheally. After 3 hr, 100 μg serum EXOs transfected with 100 pmol inhibitor control (inCon/EXOs) or miR-155 inhibitor (inhibitor/EXOs) were given to each mouse (n = 4–6 for each group). 24 hr later, the level of miR-155 was detected in sorted macrophages and neutrophils from BALF cells (A). H&E staining was performed using BALF cells (B). Scale bars, 50 μm. The number of macrophages or neutrophils in BALF was counted (C). Relative mRNA levels of TNF-α, IL-1β, and IL-6 in the lung were measured (D). The secretion of TNF-α (E), IL-1β (F), IL-6 (G), CXCL1 (H), and MIP-2 (I) was detected using ELISA. Results represent means ± SD. *p < 0.05, **p < 0.01.
Discussion
Development of an EXO-based drug delivery system has recently attracted increasing attention. EXOs belong to the family of EVs and fall into a similar size range as nanoparticles, with a diameter of around 100 nm.1, 2, 3 Currently, the general consensus is that “EXOs” means vesicles derived from multivesicular endosomes, and “MVs” means vesicles derived from the plasma membrane. In this report, the vesicles we used included both MVs and EXOs. However, based on size, we suspect that the majority of the vesicles probably fell into the category of EXOs. To simplify these terminologies, we used EXO here instead of EXO+MV. EXOs are secreted by the host cells and can be detected in a variety of body fluids, including BALF.1, 2, 3 In the past couple of years, emerging interest has focused on the possibility of using EXOs as a novel delivery agent. Comparing with nanoparticles, liposomes, and viruses, EXO-mediated drug delivery has the following advantages. EXOs are produced endogenously; thus, they are potentially less toxic and less immunogenic compared with exogenous delivery vehicles.34, 35 EXOs have been shown to be a mode of transport across the blood-brain barrier (BBB).34, 36 Additionally, the potential to deliver therapeutic agents via EXOs in a cell type-specific manner is very attractive for gene therapy. Despite EXOs holding great promise as a breakthrough for gene therapy and drug delivery, there are numerous questions to be answered before knowledge-based delivery strategies can be developed. In this report, we addressed several of these unanswered questions. The novel findings in our report included delineating the target cells and the efficiency of EXO-mediated small RNA delivery via the i.t. route in vivo. We identified that macrophages are the main recipient cells when EXO RNA is delivered via inhalation (i.h.) or i.t. instillation. We also determined the copy numbers of the exogenous small RNA molecules loaded into each EXO. Furthermore, we confirmed that EXO-mediated delivery of small RNA molecules, including both miRNA oligos (inhibitors or mimics) and siRNAs, is functional in vivo. Moreover, we tested the probability of using serum-derived EXOs as a vehicle to deliver small RNA molecules to the lungs. The method of using serum-derived EXOs potentially provides sufficient host EXOs to be used in drug delivery in vivo. We confirmed that serum-derived EXOs are not cytotoxic or inflammogenic when delivered into the lungs.
Currently, the majority of reports associated with EXO-mediated drug delivery focus on cancer treatment. The anti-tumor agents are often incorporated into EXOs and delivered intravenously (i.v.). In contrast to the i.v. route, the pulmonary route provides many advantages and is a noninvasive method to deliver therapeutic agents for both local and systemic diseases and disorders. Currently, the delivery of EXO-based therapeutics via i.h. or i.t. remains unexplored. The distribution and target cells of i.t. administrated EXOs are unclear. Therefore, our study potentially provides initial insight into the kinetics and recipient cells of EXO-mediated delivery to the lungs via i.h. or i.t.
EXOs are nanometer-sized membrane vesicles derived from living host cells.1, 2, 3 Prior to the recognition of EXOs, a wide range of nanoparticles or nanoparticulate materials was developed for drug delivery, such as solid lipid nanoparticles, polymeric nanoparticles, and liposomes.37, 38 The application of a colloidal system for successful nebulization and i.t. instillation is challenging because of the difficulties of maintaining their critical physicochemical parameters.39, 40 The deposition of nanoparticles in the lungs depends on multiple physical and chemical factors, including, but not limited to, particle size, velocity, morphology, geometry, surface properties, and the three mechanisms of drug deposition in the respiratory system.39, 40 Impaction, sedimentation, and diffusion are well-established mechanisms of small-particle deposition in the respiratory system.40 Particles with a size larger than 5 μm are commonly deposited in the oropharynx and upper respiratory tract via impaction. Particles with a size between 1 to 5 μm are often deposited in the bronchioles and small airways via sedimentation. Particles smaller than 100 to 500 nm are often deposited via diffusion in the alveoli, where Brownian motion plays an essential role. Nanoparticles and liposomes delivered via the i.t. route usually form aggregates, resulting in a size larger than 1 μm and sediment in the bronchioles. In our studies, we found that i.t. instillation of EXOs led to the deposition of EXOs in alveolar regions (Figure 3) rather than in the bronchioles, where other nanoparticles, such as liposomes, tend to stay (Figure S3). The observed dispersion of EXOs into larger airways, bronchioles, and alveoli is probably caused by Brownian motion, which is consistent with previous reports.41 Currently, liposomes are the prevalent vehicle for drug delivery. Because of their lipid bilayer shell, they can be loaded with nucleic acids and other small molecules. EXOs are also composed of a lipid bilayer with an aqueous inner layer and fall into similar size ranges as liposomes.1, 2, 3, 39, 40 Besides having the advantages of liposomes, EXOs, having generated from host cells, have adapted to avoid detection by the immune system and are less likely to aggregate. Prevention of aggregation has been one of the challenges when liposomes are prepared for drug delivery.42, 43 Furthermore, the surface proteins of EXOs may facilitate uptake by specific cell types, adding a feature for cell type-specific delivery that current nano-sized delivery systems do not allow. Another advantage of EXO-mediated delivery is its high efficiency of intracellular drug delivery. EXOs have been shown to deliver molecules through the hard-to-cross BBB. This feature is probably due to EXOs carrying the same membrane components as the host, which is not a characteristic of synthetic liposomes.1, 2, 3, 39, 40
Despite the potential of delivering EXOs in a cell type-specific manner, we observed that AMs, along with newly recruited macrophages from the circulation, were the primary recipient cells of i.t. delivered EXOs (Figure 3). Curiously, only macrophages, but not other phagocytes, including neutrophils, take up the inhaled EXOs (Figure 3). The serum-derived EXOs we used in this study are heterogeneous and derived from red cells, polynuclear cells, platelets, and other immune cells. No specific cell markers were dominant in the mixture of serum-derived EXOs. Macrophage-mediated phagocytosis is the most important and may be responsible for the clearance of EXOs deposited in the alveolar space in this study. However, in the presence of LPS stimulation, the uptake of EXOs by macrophages, but not by LPS-activated neutrophils, suggests that surface protein-mediated endocytosis plays a pivotal role in this process. In fact, previous reports have revealed that liposomes delivered in vivo are taken up mainly by macrophages, similarly to what we observed with EXO delivery. Lectin receptors, scavenger receptors, Fc receptors, and adhesion molecules that reside on the surface of macrophages potentially facilitate the endocytosis of EXOs and liposomes.44, 45 Apparently, some of these essential surface molecules are missing in phagocytes other than macrophages. This hypothetical explanation will require further exploration.
Our study is an initial investigation of EXO-mediated drug delivery in the lungs via the i.t. route. There are many details that remain to be addressed. First, our study only focused on the delivery of small RNA molecules, including miRNAs and siRNAs. Whether EXO-containing lipid, protein, or other chemical molecules can be delivered and be functional in the lungs in vivo remains unclear. One of the challenges is to load and quantify the desired molecules into EXOs successfully and efficiently. Second, with the emergence of novel technology in the near future, we anticipate that single EXO sorting using FACS will be available and that we will be better able to characterize the distinct components of the serum-derived EXO mixture. Third, to achieve inhaled EXO-mediated drug delivery in other lung cells, such as epithelial cells, we will have to develop a method to avoid uptake of EXOs by macrophages residing in the alveoli.
In summary, we developed a novel protocol to use serum-derived EXOs as a vehicle to deliver small RNA molecules into the lung macrophages in vivo, i.t. We quantified the loaded exogenous RNA molecules in each EXO. We also confirmed with our current method that the EXO-containing small RNA molecules exerted functional roles in the lungs in vivo. The serum-derived EXOs are non-toxic and do not trigger an immune response; thus, they are safe to be used in mice in vivo. This report potentially sheds light on future gene therapy of human lung diseases via inhalational delivery using EXOs.
Materials and Methods
Animals, Cell Culture, and Isolation of AMs and Polymorphonuclear Leukocytes
WT C57BL/6 mice of both genders (8 weeks of age) were obtained from The Jackson Laboratory (Maine, USA). To induce inflammation in the lung, mice were given 1 μg LPS (Sigma, St. Louis, MO) i.t. 3 hr before EXO administration. All the protocols involving animals in this study were approved by the institutional animal care and use committee (IACUC) of Boston University.
Cell lines Beas2B, A549, MRC-5, J774A.1, MH-S, and THP-1 were obtained from the ATCC (Manassas, VA) and cultured according to the standard protocol provided by the ATCC. E-10 and MLE-15 cells were a gift from Dr. Mizgerd at Boston University and maintained as described previously.46, 47 Isolation of mouse polymorphonuclear leukocytes (PMNs) from blood was performed as described previously47 using Percoll density gradient centrifugation (Sigma, St. Louis, MO). The method used for isolation of murine AMs was described previously.28 Briefly, after tracheostomy, 2 mL (1 mL × 2) of PBS was used to lavage total mouse lungs, and BALF was obtained. BALF cells were collected after centrifugation at 400 × g for 10 min. All cells were cultured at 37°C in a humidified atmosphere of 5% CO2 and 95% air.
RNA Preparation, Reverse Transcription, and Real-Time qPCR
MiRNeasy Mini Kits (QIAGEN, Valencia, CA) were used for purification of total RNA from tissues and cells. Single-stranded cDNA was generated according to the manuals of the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, Waltham, MA). For miR-15a and miR-155 detection, real-time PCR was performed using a TaqMan PCR kit (Thermo Fisher Scientific, Waltham, MA) and the Applied Biosystems StepOnePlus real-time PCR system. Mouse Hprt (Thermo Fisher Scientific, Waltham, MA) was used as a normalization control.
Copy Number Analysis
The copy numbers of siRNA molecules were calculated using real-time PCR based on the absolute quantification method, as described previously.48, 49 To make a standard curve, 1 ng of synthetic control siRNA or Myd88 siRNA was used for the reaction of poly(A)-tailed reverse transcription, as described previously.50, 51 Standard curves were made with these cDNA samples via serial dilutions and then used to determine the copy number in each sample. siRNA-specific primers and the universal reverse primer used in the real-time PCR were as follows:
Control siRNA primer: 5′-GAACACAGAATACGUCTGAATTAAC-3′
Myd88 siRNA primer: 5′-UGUAGAUAAUCGUCAGAAACAACCA-3′
Universal reverse primer: 5′-CGAATTCTAGAGCTCGAGGCAGG-3′.
Serum EXO Isolation and Labeling
Serum EXOs were isolated according to the manufacturer’s protocol (Thermo Fisher Scientific, Waltham, MA). PKH26 and PKH67 Fluorescent Cell Linker Kits for General Cell Membrane Labeling (Sigma, St. Louis, MO) were purchased and used according to the manufacturer’s protocol. The EXO-GLOW Exosome Labeling Kit (System Biosciences, Palo Alto, CA) was also used for serum EXO labeling, following the manufacturer’s protocol.
TEM, DLS, and NTA
For TEM, an EXO preparation kit for TEM imaging was obtained from 101Bio (Palo Alto, CA). The TEM images were taken using a Philips CM120 electron microscope. DLS (Brookhaven 90plus Nano-particle Sizer) was performed to determine the average EXO size. NTA was performed to determine the size and concentration of serum EXOs. NTA data were obtained using NanoSight NS500 at the Nanomedicines Characterization Core Facility (University of North Carolina at Chapel Hill, Chapel Hill, NC).
Preparation of Small RNA-Loaded EXOs and Delivery of Serum EXOs In Vivo and In Vitro
Both the control siRNA (5′-GAACACAGAAUACGUCUGAAUUAAC-3′) and Myd88 siRNA (5′-UGUAGAUAAUCGUCAGAAACAACCA-3′) used in the study were ordered from Integrated DNA Technologies (Coralville, IA). All siRNAs were synthesized with a 3′-UU overhang on each strand. The mimic control (catalog no. HMC0002), miR-15a mimic (catalog no. HMI0256), inhibitor control (catalog no. NCSTUD001), and miR-155 inhibitor (catalog no. HSTUD0254) were purchased from Sigma-Aldrich (St. Louis, MO). Modified calcium-mediated transfection was used to introduce small RNAs, including siMyd88, the miR-15a mimic, and the miR-155 inhibitor, and their respective controls into serum EXOs.28 To deliver small RNAs into serum EXOs, 100 pmol small RNA was loaded into 100 μg serum EXOs quantified by protein content (about 6.0 × 1010 particles). The final volume was adjusted to 200 μL using sterile PBS. To wash the serum EXOs, ExoQuick (System Biosciences, Palo Alto, CA) was used according to the user manual provided.
For in vivo experiments, different numbers of EXOs in 50 μL PBS were i.t. instilled into the mouse lung. One day after instillation, mice were sacrificed, and inflammation in the lung was evaluated. For in vitro experiments, serum EXOs were added to a cell culture dish containing 1.0 × 106 cells with 10% EXO-depleted fetal bovine serum (FBS) (System Biosciences, Palo Alto, CA). The time and dose are indicated in each figure.
Cytotoxicity and Cell Sorting Using Flow Cytometry
Cytotoxicity was detected 12 hr after EXO treatment using a cytotoxicity detection kit (Sigma, St. Louis, MO). Macrophages (CD11c+ F4/80+) and neutrophils (CD11b+ Ly6G+) were sorted from BALF cells at the Boston University Medical Campus (BUMC) Flow Cytometry Core Facility using antibodies purchased from BioLegend (San Diego, CA).
Immunofluorescence and H&E Staining
For cytospin preparations, the cell suspension was cytocentrifuged at 300 × g for 5 min using a Shandon Cytospin 4 (Thermo Fisher Scientific, Waltham, MA). Total inflammatory cell counts in the BALF were determined using a hemocytometer as described previously.52 BALF cells and lung sections were air-dried and stained with PROTOCOL Hema 3 fixative and solutions (Thermo Fisher Scientific, Waltham, MA).
To identify cell types in the lung that take up EXOs labeled with PKH26, immunofluorescence staining was performed as described previously.28 BALF cells or lung sections were briefly incubated overnight at 4°C with an antibody against mouse CD68 (Santa Cruz Biotechnology, Dallas, TX) as a marker of macrophages or the Ly-6G antibody (BD Biosciences, San Jose, CA) as a marker of granulocytes. After washing, Alexa 488-conjugated secondary antibodies (Thermo Fisher Scientific, Waltham, MA) were applied. Nuclei were counterstained with DAPI. Images of the stained BALF cells and lung sections were visualized and captured using a fluorescence microscope (Axioplan-2, Zeiss, Oberkochen, Germany), a high-speed 5-megapixel microscope camera (AxioCam, Zeiss, Oberkochen, Germany), and a software package (AxioVision, Zeiss, Oberkochen, Germany) with an N-Achroplan 20×/0.45 (Zeiss, Oberkochen, Germany) objective lens.
Western Blot Analysis, ELISA, and Mouse Cytokine Array
Western blot analysis was performed as described previously.50 In brief, cells were homogenized in radioimmunoprecipitation analysis (RIPA) lysis buffer supplemented with protease inhibitor cocktail and phosphatase inhibitor cocktail (Sigma, St. Louis, MO). Protein lysates were resolved on SDS-PAGE gels before being transferred to a polyvinylidene difluoride (PVDF) membrane (EMD Millipore, Darmstadt, Germany). CD41 and CD235a antibodies were purchased from Santa Cruz Biotechnology (Dallas, TX). CD31 and CD45 antibodies were ordered from BD Biosciences (San Jose, CA). Anti-E-cadherin and anti-CD61 were ordered from Cell Signaling Technology (Danvers, MA).
To quantify the cytokine and chemokine amounts in BALF, TNF-α, IL-1β, IL-6, CXCL1, and MIP-2, ELISA kits (R&D Systems) were used according to the manufacturer’s protocol.
The Mouse Cytokine Array C1 Kit (RayBiotech, Norcross, GA) was obtained to determine the levels of cytokines in BALF collected from mice according to the standard protocol.
Statistical Analysis
All data were presented as means ± SD. All the data from three independent experiments were averaged before normalization. For real-time qPCR, the same amount of cDNA was used, and all data were analyzed at the same time. Comparisons between 2 groups were performed using a two-tailed unpaired Student’s t test. Multiple groups were compared using a one-way ANOVA with the Tukey method. p < 0.05 was considered statistically significant.
Author Contributions
D.Z., H.L., X.W., and Y.J. designed the research. D.Z. performed the research. D.Z., A.R., M.G., and Y.J. analyzed data and wrote the paper.
Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgments
This work was supported by the NIH (R01GM127596 to Y.J., R21AI121644 and R33AI121644 to Y.J., R01GM111313 to Y.J., and K99HL141685 to D.Z.). Funding for the open access charge was provided by the NIH.
Footnotes
Supplemental Information includes three figures and can be found with this article online at https://doi.org/10.1016/j.ymthe.2018.06.007.
Supplemental Information
References
- 1.Tkach M., Théry C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell. 2016;164:1226–1232. doi: 10.1016/j.cell.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 2.Pitt J.M., Kroemer G., Zitvogel L. Extracellular vesicles: masters of intercellular communication and potential clinical interventions. J. Clin. Invest. 2016;126:1139–1143. doi: 10.1172/JCI87316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raposo G., Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andalussi S. El, Mäger I., Breakefield X.O., Wood M.J. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013;12:347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 5.Johnsen K.B., Gudbergsson J.M., Skov M.N., Pilgaard L., Moos T., Duroux M. A comprehensive overview of exosomes as drug delivery vehicles - endogenous nanocarriers for targeted cancer therapy. Biochim. Biophys. Acta. 2014;1846:75–87. doi: 10.1016/j.bbcan.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 6.Lamichhane T.N., Raiker R.S., Jay S.M. Exogenous DNA Loading into Extracellular Vesicles via Electroporation is Size-Dependent and Enables Limited Gene Delivery. Mol. Pharm. 2015;12:3650–3657. doi: 10.1021/acs.molpharmaceut.5b00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hessvik N.P., Llorente A. Current knowledge on exosome biogenesis and release. Cell. Mol. Life Sci. 2018;75:193–208. doi: 10.1007/s00018-017-2595-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ha D., Yang N., Nadithe V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: current perspectives and future challenges. Acta Pharm. Sin. B. 2016;6:287–296. doi: 10.1016/j.apsb.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lares M.R., Rossi J.J., Ouellet D.L. RNAi and small interfering RNAs in human disease therapeutic applications. Trends Biotechnol. 2010;28:570–579. doi: 10.1016/j.tibtech.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wittrup A., Lieberman J. Knocking down disease: a progress report on siRNA therapeutics. Nat. Rev. Genet. 2015;16:543–552. doi: 10.1038/nrg3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Broderick J.A., Zamore P.D. MicroRNA therapeutics. Gene Ther. 2011;18:1104–1110. doi: 10.1038/gt.2011.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christopher A.F., Kaur R.P., Kaur G., Kaur A., Gupta V., Bansal P. MicroRNA therapeutics: Discovering novel targets and developing specific therapy. Perspect. Clin. Res. 2016;7:68–74. doi: 10.4103/2229-3485.179431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carthew R.W., Sontheimer E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agrawal N., Dasaradhi P.V., Mohmmed A., Malhotra P., Bhatnagar R.K., Mukherjee S.K. RNA interference: biology, mechanism, and applications. Microbiol. Mol. Biol. Rev. 2003;67:657–685. doi: 10.1128/MMBR.67.4.657-685.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rand T.A., Petersen S., Du F., Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 16.Lam J.K., Chow M.Y., Zhang Y., Leung S.W. siRNA Versus miRNA as Therapeutics for Gene Silencing. Mol. Ther. Nucleic Acids. 2015;4:e252. doi: 10.1038/mtna.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zuckerman J.E., Davis M.E. Clinical experiences with systemically administered siRNA-based therapeutics in cancer. Nat. Rev. Drug Discov. 2015;14:843–856. doi: 10.1038/nrd4685. [DOI] [PubMed] [Google Scholar]
- 18.Rupaimoole R., Slack F.J. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 19.Sakagami M. In vivo, in vitro and ex vivo models to assess pulmonary absorption and disposition of inhaled therapeutics for systemic delivery. Adv. Drug Deliv. Rev. 2006;58:1030–1060. doi: 10.1016/j.addr.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 20.Patil J.S., Sarasija S. Pulmonary drug delivery strategies: A concise, systematic review. Lung India. 2012;29:44–49. doi: 10.4103/0970-2113.92361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lötvall J., Hill A.F., Hochberg F., Buzás E.I., Di Vizio D., Gardiner C., Gho Y.S., Kurochkin I.V., Mathivanan S., Quesenberry P. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles. 2014;3:26913. doi: 10.3402/jev.v3.26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Helwa I., Cai J., Drewry M.D., Zimmerman A., Dinkins M.B., Khaled M.L., Seremwe M., Dismuke W.M., Bieberich E., Stamer W.D. A Comparative Study of Serum Exosome Isolation Using Differential Ultracentrifugation and Three Commercial Reagents. PLoS ONE. 2017;12:e0170628. doi: 10.1371/journal.pone.0170628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L., Bennett S.A., Wang L. Role of E-cadherin and other cell adhesion molecules in survival and differentiation of human pluripotent stem cells. Cell Adhes. Migr. 2012;6:59–70. doi: 10.4161/cam.19583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pelikan Z. Expression of surface markers on the blood cells during the delayed asthmatic response to allergen challenge. Allergy Rhinol. (Providence) 2014;5:96–109. doi: 10.2500/ar.2014.5.0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sabatel C., Radermecker C., Fievez L., Paulissen G., Chakarov S., Fernandes C., Olivier S., Toussaint M., Pirottin D., Xiao X. Exposure to Bacterial CpG DNA Protects from Airway Allergic Inflammation by Expanding Regulatory Lung Interstitial Macrophages. Immunity. 2017;46:457–473. doi: 10.1016/j.immuni.2017.02.016. [DOI] [PubMed] [Google Scholar]
- 26.Reutershan J., Morris M.A., Burcin T.L., Smith D.F., Chang D., Saprito M.S., Ley K. Critical role of endothelial CXCR2 in LPS-induced neutrophil migration into the lung. J. Clin. Invest. 2006;116:695–702. doi: 10.1172/JCI27009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawasaki T., Kawai T. Toll-like receptor signaling pathways. Front. Immunol. 2014;5:461. doi: 10.3389/fimmu.2014.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang D., Lee H., Zhu Z., Minhas J.K., Jin Y. Enrichment of selective miRNAs in exosomes and delivery of exosomal miRNAs in vitro and in vivo. Am. J. Physiol. Lung Cell. Mol. Physiol. 2017;312:L110–L121. doi: 10.1152/ajplung.00423.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haney M.J., Klyachko N.L., Zhao Y., Gupta R., Plotnikova E.G., He Z., Patel T., Piroyan A., Sokolsky M., Kabanov A.V., Batrakova E.V. Exosomes as drug delivery vehicles for Parkinson’s disease therapy. J. Control. Release. 2015;207:18–30. doi: 10.1016/j.jconrel.2015.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luan X., Sansanaphongpricha K., Myers I., Chen H., Yuan H., Sun D. Engineering exosomes as refined biological nanoplatforms for drug delivery. Acta Pharmacol. Sin. 2017;38:754–763. doi: 10.1038/aps.2017.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moon H.G., Yang J., Zheng Y., Jin Y. miR-15a/16 regulates macrophage phagocytosis after bacterial infection. J. Immunol. 2014;193:4558–4567. doi: 10.4049/jimmunol.1401372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu F., Kang Y., Zhang H., Piao Z., Yin H., Diao R., Xia J., Shi L. Akt1-mediated regulation of macrophage polarization in a murine model of Staphylococcus aureus pulmonary infection. J. Infect. Dis. 2013;208:528–538. doi: 10.1093/infdis/jit177. [DOI] [PubMed] [Google Scholar]
- 33.O’Connell R.M., Chaudhuri A.A., Rao D.S., Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc. Natl. Acad. Sci. USA. 2009;106:7113–7118. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wood M.J., O’Loughlin A.J., Samira L. Exosomes and the blood-brain barrier: implications for neurological diseases. Ther. Deliv. 2011;2:1095–1099. doi: 10.4155/tde.11.83. [DOI] [PubMed] [Google Scholar]
- 35.Tan S., Wu T., Zhang D., Zhang Z. Cell or cell membrane-based drug delivery systems. Theranostics. 2015;5:863–881. doi: 10.7150/thno.11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsumoto J., Stewart T., Banks W.A., Zhang J. The transport mechanism of extracellular vesicles at the blood-brain barrier. Curr. Pharm. Des. 2017;23:6206–6214. doi: 10.2174/1381612823666170913164738. [DOI] [PubMed] [Google Scholar]
- 37.Mudshinge S.R., Deore A.B., Patil S., Bhalgat C.M. Nanoparticles: Emerging carriers for drug delivery. Saudi Pharm. J. 2011;19:129–141. doi: 10.1016/j.jsps.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naseri N., Valizadeh H., Zakeri-Milani P. Solid Lipid Nanoparticles and Nanostructured Lipid Carriers: Structure, Preparation and Application. Adv. Pharm. Bull. 2015;5:305–313. doi: 10.15171/apb.2015.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paranjpe M., Müller-Goymann C.C. Nanoparticle-mediated pulmonary drug delivery: a review. Int. J. Mol. Sci. 2014;15:5852–5873. doi: 10.3390/ijms15045852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fernández Tena A., Casan Clarà P. Deposition of inhaled particles in the lungs. Arch. Bronconeumol. 2012;48:240–246. doi: 10.1016/j.arbres.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 41.Sercombe L., Veerati T., Moheimani F., Wu S.Y., Sood A.K., Hua S. Advances and Challenges of Liposome Assisted Drug Delivery. Front. Pharmacol. 2015;6:286. doi: 10.3389/fphar.2015.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bozzuto G., Molinari A. Liposomes as nanomedical devices. Int. J. Nanomedicine. 2015;10:975–999. doi: 10.2147/IJN.S68861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kelly C., Jefferies C., Cryan S.A. Targeted liposomal drug delivery to monocytes and macrophages. J. Drug Deliv. 2011;2011:727241. doi: 10.1155/2011/727241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Imai T., Takahashi Y., Nishikawa M., Kato K., Morishita M., Yamashita T., Matsumoto A., Charoenviriyakul C., Takakura Y. Macrophage-dependent clearance of systemically administered B16BL6-derived exosomes from the blood circulation in mice. J. Extracell. Vesicles. 2015;4:26238. doi: 10.3402/jev.v4.26238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamamoto K., Ferrari J.D., Cao Y., Ramirez M.I., Jones M.R., Quinton L.J., Mizgerd J.P. Type I alveolar epithelial cells mount innate immune responses during pneumococcal pneumonia. J. Immunol. 2012;189:2450–2459. doi: 10.4049/jimmunol.1200634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wikenheiser K.A., Vorbroker D.K., Rice W.R., Clark J.C., Bachurski C.J., Oie H.K., Whitsett J.A. Production of immortalized distal respiratory epithelial cell lines from surfactant protein C/simian virus 40 large tumor antigen transgenic mice. Proc. Natl. Acad. Sci. USA. 1993;90:11029–11033. doi: 10.1073/pnas.90.23.11029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swamydas, M., Luo, Y., Dorf, M.E., and Lionakis, M.S. (20153.20.1). Isolation of mouse neutrophils. Curr. Protoc. Immunol. 110, 3.20.1–3.20.15. [DOI] [PMC free article] [PubMed]
- 48.Chevillet J.R., Kang Q., Ruf I.K., Briggs H.A., Vojtech L.N., Hughes S.M., Cheng H.H., Arroyo J.D., Meredith E.K., Gallichotte E.N. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc. Natl. Acad. Sci. USA. 2014;111:14888–14893. doi: 10.1073/pnas.1408301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alexander M., Hu R., Runtsch M.C., Kagele D.A., Mosbruger T.L., Tolmachova T., Seabra M.C., Round J.L., Ward D.M., O’Connell R.M. Exosome-delivered microRNAs modulate the inflammatory response to endotoxin. Nat. Commun. 2015;6:7321. doi: 10.1038/ncomms8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang D., Li X., Chen C., Li Y., Zhao L., Jing Y., Liu W., Wang X., Zhang Y., Xia H. Attenuation of p38-mediated miR-1/133 expression facilitates myoblast proliferation during the early stage of muscle regeneration. PLoS ONE. 2012;7:e41478. doi: 10.1371/journal.pone.0041478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee H., Zhang D., Wu J., Otterbein L.E., Jin Y. Lung Epithelial Cell-Derived Microvesicles Regulate Macrophage Migration via MicroRNA-17/221-Induced Integrin β1 Recycling. J. Immunol. 2017;199:1453–1464. doi: 10.4049/jimmunol.1700165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang D., Lee H., Haspel J.A., Jin Y. Long noncoding RNA FOXD3-AS1 regulates oxidative stress-induced apoptosis via sponging microRNA-150. FASEB J. 2017;31:4472–4481. doi: 10.1096/fj.201700091R. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.