Abstract
Gastric cancer (GC) is one of the most frequently diagnosed malignant diseases. The molecular mechanisms of metastasis remain unclear. Recently, studies have shown that long non-coding RNAs (lncRNAs) play critical roles in metastasis. Therefore, deeper understanding of this mechanism could provide potential diagnostic tools and therapeutic targets for metastatic GC. This review focuses on dysregulated lncRNAs in GC metastases. Due to the identification of multiple diverse mechanisms involved in GC metastasis, we classified them into seven categories, including lncRNAs related to epithelial-mesenchymal transition, regulation of degradation of extracellular matrix, angiopoiesis, vasculogenic mimicry, and immunologic escape. As the TNM stage is pivotal for evaluating the severity and prognosis of GC patients, we summarize the lncRNAs relevant to lymphatic metastasis, distant metastasis and TNM classification. This review summarizes the lncRNAs related to metastasis, which may provide insight into the mechanisms, and provide potential markers for prognostic prediction and monitoring the relapse of GC.
Keywords: Long noncoding RNAs, Stomach neoplasms, Metastasis
Core tip: This review summarizes the long noncoding RNAs (lncRNAs) that influence metastasis of gastric cancer. We classified lncRNAs according to their molecular mechanism, which included epithelial-mesenchymal transition, epigenetic regulation, degradation of the extracellular matrix, angiopoiesis, vasculogenic mimicry, and immunologic escape. Finally, we summarized the lncRNAs that have stable expression in serum and describe their clinical value. A table lists the clinical correlation of the lncRNAs in details.
INTRODUCTION
Gastric cancer (GC) is a major public health problem across the life span of human beings and is one of the top two leading causes of cancer-related death worldwide. Eastern Asia has the highest incidence rates of GC, which is particularly prevalent in China[1]. According to statistical analysis, lung cancer is the only cancer with higher rates of incidence and mortality compared to stomach neoplasms[2]. Approximately 28000 cases of gastric neoplasms are expected to be diagnosed in 2017, and 10960 of them are expected to result in death[3]. Patients are usually diagnosed with GC after metastasis has occurred or in an advanced stage due to limitations in early noninvasive detection techniques. Even when diagnosed at an early stage and endoscopic mucosal resection (EMR) or endoscopic submucosal dissection (ESD) are successfully performed, the local recurrence rate is still high, ranging from 2.8%-12.5%[4,5]. Despite multiple post-operative monitoring tools, including endoscopic monitoring, CT, MRI, PET, and serological monitoring (CA19-9, CA153, CA125, and CA724), the sensitivity has not met expectations yet. Recently, circulating tumor DNA (ctDNA) has been studied as GC relapse predictive markers[6,7]. Because of the unsatisfactory prognosis in advanced stage GC patients who have undergone surgery, chemotherapy or radiotherapy, measures should be taken to intensively monitor GC patients[8]. In recent years, significant advances have been made in understanding the molecular mechanisms involved in GC metastasis, however, the overall view of the mechanism map is limited and ambiguous[9,10]. Therefore, clarification of the pathogenesis and corresponding molecular alterations in GC is imperative in seeking diagnostic biomarkers and therapeutic targets.
Noncoding RNAs (ncRNAs) longer than 200 nucleotides are defined as long noncoding RNAs (lncRNAs). ncRNAs are emerging elements that are recognized to play critical roles in cancer development and progression. lncRNAs do not perform transcriptional tasks, but they can affect gene expression at the transcriptional or post-transcriptional levels[11-13].
Increasingly, lncRNAs have been found to participate in GC metastasis. lncRNAs function by impacting embryogenesis, epigenetic regulation, imprinting, angiopoiesis, and vasculogenic mimicry[14-18]. This article reviews the lncRNAs that regulate certain critical steps of GC metastasis, with particular emphasis on epithelial-mesenchymal transition (EMT), vascularization, and vasculogenic mimicry.
LNCRNAS AFFECT EMT
EMT is a vital process involved in embryonic development and cancer metastasis[19]. EMT is the process by which epithelial cells gain increasing migratory potential and mesenchymal characteristics[20]. It has been shown to play an important role in GC metastasis. There are many lncRNAs that facilitate GC metastasis via EMT (Figure 1).
Figure 1.
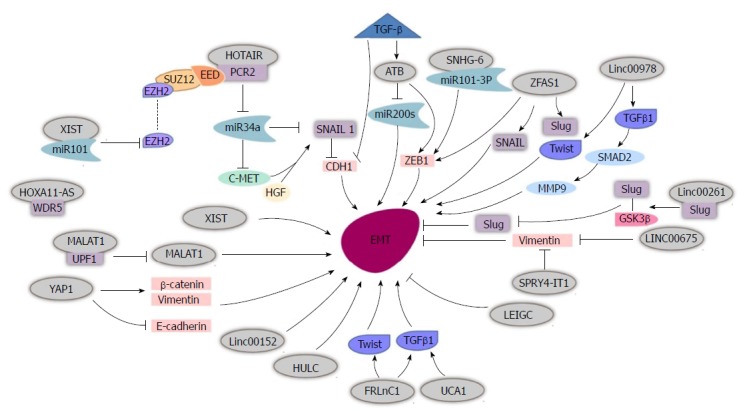
Long non-coding RNAs affect epithelial-mesenchymal transition in gastric cancer cells. HOTAIR recruits PRC2 to silence miR-34a, and then activates the HGF/c-Met/Snail pathway to promote EMT. TGF-β induces lncATB, which inhibits miR-200s and provokes ZEB1 to promote EMT. SNHG6 binds miR-101-3P to activate ZEB1 and then promotes EMT. lncRNA ZFAS1 induces EMT by activating SNAIL, Slug, ZEB1 and Twist. Linc00978 induces Twist and TGFβ1, and TGFβ1 then activates SMAD2 and MMP9 to facilitate EMT. MALAT1 binds UPF1 to reduce its level and activate EMT. FRLnC1 induces EMT by activating Twist and TGFβ1. HUCA1 induces EMT via TGFβ1 activation. YAP1 promotes EMT by increasing vimentin and β-catenin, and decreasing E-cadherin. lncRNA XIST, Linc00152 and HULC promote EMT. Linc00261 binds Slug resulting in reduced Slug levels and decreased EMT. LINC00675 and SPRY4-IT1 restrain EMT by reducing vimentin. LEIGC inhibits EMT. lncRNAs: Long non-coding RNAs; HOTAIR: HOX transcript antisense intergenic RNA; EMT: Epithelial-mesenchymal transition; YAP1: Yes-associated protein 1; MALAT1: Metastasis associated lung adenocarcinoma 1.
Chen et al[14] showed that metastasis associated lung adenocarcinoma 1 (MALAT1) is downregulated in GC cells, and that E-cadherin expression is increased while vimentin expression is decreased at both the mRNA and protein levels. Li et al[21] detected UPF1, a key part of the nonsense-mediated mRNA decay (NMD) pathway, which alters mRNA transcription, and showed that it negatively correlated with MALAT1 expression. Subsequent experiments showed that increased UPF1 expression inhibited migration, invasion and EMT of GC cells. Increased MALAT1 expression decreased the influence of UPF1 in GC cells, including UPF1’s ability to inhibit cell proliferation, EMT and facilitate apoptosis. Taken together, Li et al[22] postulated that UPF1 directly binds MALAT1 to downregulate MALAT1 (UPF1/MALAT1), thus, inhibiting GC progression. Lee et al[23] further confirmed that MALAT1 regulates mesenchymal maker Snail, N-cadherin and ZEB1 to influence EMT.
Another classic lncRNA, HOX transcript antisense intergenic RNA (HOTAIR), has been shown to be elevated in GC cells and promote gastric tumor metastasis via enhancement of EMT. E-cadherin expression was higher in cells with HOTAIR knockdown compared to cells with HOTAIR overexpression, while expression of N-cadherin and vimentin were decreased. The detailed mechanism is believed to involve HOTAIR recruitment and binding of PRC2 to epigenetically silence miR-34a, which activates the HGF/c-Met/Snail pathway, thus facilitating EMT in tumor cells[24].
FRLnc1 is also upregulated in GC cell lines. In vitro functional analysis and a pulmonary metastasis model demonstrated that FRLnc1 enhanced the migration capacity of GC cells. Hui et al[25] discovered that FRLnc1 functions as an EMT promoter to affect the migration of GC cells by upregulating the downstream elements TGFβ-1 and Twist.
lncRNA activated by TGF-β (lncRNA-ATB), also known as lncRNA-AL (ENST00000493038), was overexpressed in TGF-β treated cancer cells, with the cells exhibiting a spindle-like morphology. lncRNA-ATB induced ZEB1 expression and inhibited miR-200s in tumor cells to affect EMT in stomach neoplasm cells. Saito et al[26] uncovered a positive correlation between TGF-β, ZEB1 and lncRNA-ATB, while miR-200c inversely correlated with lncRNA-ATB expression. Saito et al[26] demonstrated that lncRNA-ATB participate in the EMT process in GC via the TGF-β/miR-200/ZEB axis.
It has been reported that the lncRNA X-inactive specific transcript (lncRNA XIST) regulates activation of tumor cell migration and initiates EMT via upregulation of vimentin and fibronectin and downregulation of E-cadherin and α-catenin in stomach cancer cells. lncRNA XIST negatively correlates with miR-101 and decreased lncRNA XIST expression led to downregulation of EZH2 at both the mRNA and protein levels and was reversed with an miR-101 inhibitor. Thus, lncRNA XIST functions by sponging miR-101 and regulating EZH2 in GC cells[27]. The lncRNA small nucleolar RNA host gene 6 (SNHG6) is overexpressed in GC cell lines and facilitates EMT as a competing endogenous (ce) RNA via sponging miR-101-3p, which leads to an increase in ZEB1, thus boosting tumor cell migration at the post-transcriptional level[28].
The lncRNA zinc finger antisense 1 (ZFAS1) expression level is elevated in GC tissues, serum and exosomes and ZFAS1 also activates ZEB1 to affect EMT. Lei et al[29] showed that ZFAS1 promotes the transformation from mesenchymal-epithelial transition (MET) to EMT by increasing the expression of N-cadherin, Slug, Snail, Twist and ZEB1 and decreasing the expression of E-cadherin. Exosomes that originate from GC cells might promote the GC metastasis by producing ZFAS1.
lncRNA urothelial carcinoma associated 1 (UCA1) is induced by TGFβ-1 and expedites EMT. As UCA1 knockdown partly mitigates the impact of TGFβ-1 on EMT, the specific role of TGFβ-1 in accelerating EMT requires further investigation[30]. Silencing UCA1 inhibits resistance to adriamycin in GC, which suggests that UCA1 may be a novel therapeutic target[31].
LincRNA00978 is reportedly elevated in GC tissues and plasma. It could induce EMT by activating the TGFβ/SMAD2/MMP9 pathway. Another potential pathway is composed of downregulated LincRNA00978, leading to decreased Twist1 and Slug, followed by a decrease in downstream molecules, such as N-cadherin and vimentin and an increase in E-cadherin[32]. Yes-associated protein1 (YAP1) also promotes EMT by upregulating vimentin and β-catenin and downregulating E-cadherin[33]. lncRNAs highly upregulated in liver cancer(HULC) and Linc00152 also increase tumor cell’s migration through acceleration of EMT in GC[34,35].
The lncRNAs mentioned above function by promoting EMT in GC cells, but there are also numerous lncRNAs that function by repressing EMT progression.
Linc00261, which is repressed in GC cells, suppresses E-cadherin and promotes N-cadherin, FN1 and vimentin expression, reverses EMT in gastric tumor cells, and increases the malignant phenotype[36]. Yu et al[37] deduced that Linc00261 reverses EMT by binding Slug. As mass experiments indicated that GSK3β affects the ubiquitin-proteasome pathway to degrade Slug in breast cancer cells[38,39], additional experiments demonstrated that Linc00261 attenuates the stability of Slug proteins through strengthening the interaction between GSK3β and Slug.
Linc00675, also found to be significantly downregulated in GC tissues, suppresses the migration of GC both in vitro and in vivo (pulmonary and hepatic metastases). Mechanistic studies showed that Linc00675 directly interacts with vimentin, resulting in increased phosphorylation of vimentin on Ser83 rather than on Ser39, thereby causing the degradation of vimentin filaments[40,41]. Since vimentin is considered to be a master regulator of EMT, Linc00675 was deduced to be a tumor repressor that inhibits metastasis via reversing EMT[42].
lncRNA SPRY4 intronic transcript 1 (lncRNA SPRY4-IT1), prevents cancer cell migration partly through its role in the regulation of EMT. Xie et al[43] found that SPRY4-IT1 increases the expression of E-cadherin and decreases the expression of vimentin, resulting in EMT inhibition.
After observing significantly decreased lncRNA:chr2:118381039-118383698 levels in GC tissue, Han et al[44] named this lncRNA LEIGC and assessed its role in regulating tumor cell migration. In monolayer cultures, cells with downregulated LEIGC showed a dramatic change in morphology and transitioned from a cobblestone-like-shape to a spindle-like fibroblastic status, whereas LEIGC-overexpressing cells maintained a cobblestone-like morphology. In addition, mRNA and protein levels illustrated that LEIGC could reverse EMT by lowering the expression of vimentin, Snail, Slug, Zeb, and Twist and increasing the expression of E-cadherin. Furthermore, LEIGC overexpression enhances the GC cells sensitivity to 5-fluorouracil, and this characteristic enables LEIGC to be a potential therapeutic target.
LNCRNAS AFFECT EPIGENETIC REGULATION IN GC
Epigenetic processes include the recruitment of histone-modifying enzymes and DNA methyltransferases, and chromatin remodeling. It has been reported that lncRNAs interact with DNA to control gene expression[45]. Given that promoter CpG island hypermethylation, an abnormal DNA modification, is involved in pivotal cellular pathways and is characteristically a hallmark of cancer cells[46], several lncRNAs have been found to play roles in controlling the DNA modification system in GC cells.
Sun et al[15] evaluated the genome-wide expression profile of lncRNAs and discovered BC041951, designating it as gastric cancer-associated lncRNA 1 (GClnc1). Because mice injected with GClnc1 had an increased overall survival time and more metastatic lung nodules than control mice, GClnc1 was determined to enhance the metastatic capability of tumor cells. The mechanism behind GClnc1’s carcinogenesis stems from its ability to function as a molecular scaffold for the WDR5/KAT2A complex, which leads to trimethylation of H3K4 and acetylation of H3K9 in the transcription promoter region of mitochondrial superoxide dismutase (SOD), which upregulates the transcription of mitochondrial superoxide dismutase 2 (SOD2).
LOC100130476, which is dysregulated in gastric cardia adenocarcinoma, is considered to be a tumor suppressor due to the tumor specific hypermethylation of region 1 near the transcription start site. Methylation of region 1 in peripheral white blood cells had a similar effect and may play key roles in gene silencing. Advanced gastric carcinoma patients with low hypermethylation of region 1 preferentially developed metastases, leading to poor prognosis[47].
Xie et al[43] also determined that lncRNA SPRY4-IT1 is downregulated in gastric tumor cells and tissues. Furthermore, Sun et al[48] identified a canonical CpG island in the SPRY4-IT1 loci promoter region. DNMT1 inhibits expression of SPRY4-IT1 in GC cells by altering the DNA methylation level. After treatment with 3.7- and 2.8-fold 5-aza-CdR, the expression of SPRY4-IT1 was significantly higher than in controls. Therefore, SPRY4-IT1 could be a potential therapeutic target[43].
LNCRNAS INVOLVED IN REGULATION OF DEGRADATION OF THE EXTRACELLULAR MATRIX
Tumor cells are exposed to a multitude of abnormal situations due to changes in the ECM that significantly impact cancer cell behavior. Dysregulated ECM cross-linking and repressed stiffness jointly contribute to cancer metastasis and progression[49,50]. Metalloproteases (MMPs) typically participate in adjusting the ECM and vascularization[51].
The lncRNA UCA1 facilitates GC cell migration both in vitro and in vivo via the UCA1/GRK2/ERK/MMP9 axis. Meanwhile, the lncRNA UCA1 increases the degradation of GRK2 via Cbl-c-mediated ubiquitination following the activation of the ERK-MMP9 pathway, which may be involved in vascularization[52]. Xu et al[16] found that FENDRR negatively correlated with FN1 mRNA and that the induction of FENDRR strongly inhibits the activity of MMP2/MMP9, which corroborates FENDRR’s role in preventing GC cell metastasis. Then, Park et al[53] determined that overexpression of BM742401 decreased the B95kDa band, which corresponds to MMP9, via a zymography assay. The reduced concentration of MMP9 in BM74240-induced cells further verified these findings. However, BM742401 did not alter the expression level of intracellular MMP9. Therfore, BM742401 may diminish MMP9 secretion to inhibit cancer metastasis.
lncRNA olfactory receptor, family 3, subfamily A, member 4 (OR3A4), contributes to GC metastasis as it was found to be overexpressed in primary tumor tissue, metastatic tissue and in the peripheral blood. Upregulated OR3A4 induced MMP9, which is involved in the breakdown of the ECM[54]. LINC00052 plays an oncogenous role in GC cells. It promotes GC cell migration and invasion through promoting the SMYD2 related β-catenin methylation to stabilize its expression and activating the Wnt/β-catenin pathway. When upregulating LINC00052 level in GC cells, MMP2, MMP9, and Cyclin D1 expression were upregulated while E-cadherin and P21 were downregulated. The downstream MMP2 and MMP9 are related to the breakdown of the ECM.
Degradation of the extracellular matrix is one way to modulate the tumor microenvironment. Hypoxia is another key change in the tumor microenvironment that promotes tumor metastasis[55]. AK058003, a lncRNA that is induced by hypoxia, is positively associated with γ-synuclein (SNCG) in GC cells. AK058003 and SNCG are both upregulated in hypoxic environments, and SNCG facilitates hypoxia-induced GC cell metastasis, which is regulated by AK058003. Thus, a novel hypoxia/lncRNA-AK058003/SNCG pathway that is related to metastasis was identified[56]. Wang et al[56] found that lncRNA AK058003 is increased in hypoxia-induced GC cells, where it facilitates GC cell migration and invasion in vivo and in vitro. AK058003 positively altered SNCG, a member of the synuclein family, by decreasing methylation of the SNCG gene CpG island. Elevated SNCG expression can also be induced by hypoxia, which in turn induces GC cell metastasis in primary tumor tissue. lncRNA BC005927 is induced by hypoxia and hypoxia inducible factor-1α (HIF-1α), which is a factor involved in hypoxia induced GC metastasis through directly binding the HIF-1 response element to promote GC metastasis and invasion. This hypoxia-induced auxo-action is partially regulated by BPHB4[57].
LNCRNAS INVOLVED IN ANGIOPOIESIS AND VASCULOGENIC MIMICRY
Ample evidence has shown that the development of endothelial vessels (EVs) and vasculogenic mimicry (VM) supply nutrition to tumors and sustain tumor growth. Highly vascular tumors show an increased ability to develop metastases compared to tumors that lack adequate vascularization[58,59]. VM involves the formation of de novo channels by pluripotent embryonic-like and highly invasive tumor cells, mimicking tumor feeding[60]. VM has already been reported in melanoma, soft tissue sarcomas, GIST and hepatocellular carcinoma[61-64].
MALAT1, an oncogenic lncRNA, can increase tumorigenicity and metastasis in GC by facilitating VM and angiogenesis. MALAT1 induces the expression of β-catenin and E-cadherin and increases the p-ERK, p-FAK, and p-paxillin levels. MT1-MMP and MMP2 and MMP9, which are downstream of p-ERK, are consequently altered. MALAT1 functions as an active regulator of VM and EV through the E-cadherin/β-catenin complex and via the ERK/MMP and FAK/paxillin signaling pathways[17]. Another mechanism involving MALAT1 was discovered by which MLAT1 regulates the acetylation level of H3 histone in the EGFL7 promoter region to boost the EGFL7 expression level[65]. An intron of the EGFL7 gene, miR-126, is pivotal in alterations of H3 histone acetylation but not methylation in the EGFL7 promoter in colorectal cancer and non-small cell lung cancer cells and cooperates with MALAT1 to alter angiogenesis[66,67].
Another lncRNA, C21orF96, which is upregulated in gastric tumor tissues, was found to be significantly higher in metastatic tissues compared to histologically normal lymph node tissues. Yang et al[68] determined that ectopic expression of C21orF96 promotes lymphangiogenesis of stomach neoplasms. With respect to VM, C21orF96 increases the number of tubulars, intersecting nodes, and the length of the tubes in human umbilical vein endothelial cells (HUVECs).
Likewise, OR3A4, an oncogenic lncRNA, was found to facilitate the formation of tubules in HUVECs. Upregulated OR3A4 induces vascular endothelial growth factor C (VEGF-C), which is a known promoter of angiogenesis and vascular permeability[69]. Furthermore, the chicken embryo chorioallantoic membrane (CAM) assay demonstrated that OR3A4 promotes angiogenesis. OR3A4 may exert its effects by inhibiting PDLIM2, promoting MACC1 and GNB2L1, and directly targeting NTN4 to enhance metastasis and tumorigenesis in GC[54].
LNCRNAS RELATED TO IMMUNE ESCAPE OF GC CELLS
Immune escape, the third step of cancer immunoediting[70], reduces the immunogenicity of tumor cells, creating an immunosuppressive tumor microenvironment in which cancer cells can survive and grow[71]. Evading immune destruction has been deemed as a hallmark of cancer[72].
The classical lncRNA, HOTAIR, has been reported to promote GC progression and metastasis[73-76]. Song et al[18] determined that upregulated HOTAIR in GC cells positively correlates with human leukocyte antigen (HLA)-G levels both in tissue and peripheral blood samples. Furthermore, HOTAIR was also found to induce the expression of HLA-G at both the mRNA and protein secretion levels. HOTAIR directly interacts with miR-152 and decreases miR-152 expression level, which reverses the miR-152 induced dysregulated activity of HLA-G 30UTR, while, Mut-HOTAIR fails to have the same effect. Thus, HOTAIR overexpression might play roles in tumor immune escape. Furthermore, polymerase chain reaction-restriction fragment length polymorphism (PCRRFLP) was used to detect three htSNPs of the HOTAIR gene (rs12826786 C > T, rs4759314 A > G, and rs10783618 C > T). In normal and GCA tumor tissues, rs12826786 presented higher HOTAIR expression levels than the CC genotype, and the sore T allele of rs12826786 increased the GCA risk and reduced the five-year survival rates[77].
LNCRNAS DYSREGULATED IN PERIPHERAL BLOOD AND IN GASTRIC ACID
Given that patients are usually asymptomatic and that relapsed GC patients have poor prognosis, many doctors recognize the importance of surveillance in detecting recurrence[78]. Studies have shown that hematogenous metastasis is the most frequent recurrence pattern during the first year following resection[79], and identification of a simple method to monitor patients, for example, using serum lncRNAs, is a top priority. Identification of a noninvasive approach with a high degree of sensitivity and specificity is urgently needed to predict and monitor the prognosis of GC patients and the relapse of patients post-operation.
OR3A4 is upregulated in both metastatic tissue and serum[54], as is ZFAS1 and exosomal ZFAS1. The level of circulating ZFAS1 correlates with lymphatic metastasis and the TNM stage, when the area under the ROC curve is up to 0.792 (95%CI: 0.703-0.881, P < 0.001)[29].
AA174084 is not only ectopically expressed in GC tissue but is also expressed in plasma and in gastric acid. The expression of AA174084 in GC patients’ gastric acid is significantly higher than that in control groups. In addition, the amount of AA174084 in plasma decreases after patients undergo surgery and is positively correlated with invasion and lymphatic metastasis. Thus, AA174084 could serve as a potential biomarker to predict a patient’s prognosis[80].
The lncRNA RNA component of mitochondrial RNA processing endoribonuclease (RMRP) has been reported to be decreased in GC tissues, but increased in the plasma and gastric acid of GC patients. After subtotal gastrectomy, this aberrant expression dramatically declines. Importantly, the RMRP level in gastric acid or in plasma is not only sufficient for clinical detection but that method for RMRP detection are also more sensitive and specific than that for carcinoembryonic antigen (CEA) and carbohydrate antigen19-9 (CA199). These results could provide a new method for GC detection, and the postoperative decline of RMRP implies that this lncRNA has appropriate characteristics for prognostic prediction[81].
Five novel plasma lncRNAs (TINCR, CCAT2, AOC4P, BANCR and LINC00857) demonstrate excellent stability and show little to no change in hostile environments. The diagnostic significance of lncRNA-based Index I, established by logistic regression, is better than that of the CEA-based Index II. Because the lncRNA based index declined dramatically two weeks post-operation, this index is highly effective in monitoring tumor recurrence. The lncRNA based index significantly correlates with tumor size, depth of invasion, lymphatic metastasis and TNM stages[82].
Currently, the majority of GC research focuses on the expression level of lncRNAs in GC tissue, while many of them are stably expressed in plasma. Though systematic evaluation of the lncRNAs mentioned above is lacking, those that are stable in circulation could be useful for predicting metastasis of primary tumors, but this hypothesis must be confirmed. Individual markers, such as a single lncRNA, may not be adequate for determining prognosis in GC, but interested readers could refer to the analysis by Zhang et al[82] and Shao et al[80]. The combination of several lncRNAs known to participate in GC progression may overcome these existing issues.
LNCRNAS AND CLINICAL CORRELATION
Recently, the seventh edition of tumor, node, metastasis (TNM) classification has been widely accepted[83]. Gu et al[84] identified patients diagnosed with GC in the first hospital of the China Medical University and the Liaoning Cancer Hospital from January 1980 to December 2009 and systematically reviewed the data. These authors found that according to the 7th edition of the TNM classification, classification of stage T4b and N0 as stage IIIA had statistical significance in regard to the survival outcome and in predicting prognoses in Chinese GC patients. Given that lymph node and distant metastasis were found to be key factors in the prognosis of GC patients, we identified the lncRNAs that correlated with lymph node, distant metastasis and the TNM stage, as shown in Table 1[14-17,23,25-30,32,33,35,36,40,43,44,53,57,58,69,74-78,80-132].
Table 1.
Mechanistic analysis of long non-coding RNAs involved in gastric cancer metastasis and clinical correlations
| LncRNA ID | Dysregulation | Upstream regulators | Downstream targets | Metastasis processes | Clinical correlation |
Univariate analysis (HR 95%CI) P < 0.05 |
Multivariate analysis (HR 95%CI) P < 0.05 |
Ref. | ||
| OS | DFS | OS | DFS | |||||||
| MALAT1 | Up | JMJD1A | UPF1, Snail, N-cadherin, ZEB1, VE-cadherin/β-catenin complex, ERK/MMP, FAK/paxillin, EGFL7, miR-122 | EMT, Angiopoiesis, VM | Lymphatic metastasis, distant metastasis, TNM stage | 1.38 (1.03-1.85) | 1.40 (1.01-1.94) | [14,17,23,89,90] | ||
| HOTAIR | Up | PCR2, miR-34a, c-MET, SNAIL1, CDH1, miR-152, HLA-G | EMT, immune escape | Lymphatic metastasis, distant metastasis, TNM stage | [73-75,77,91-93] | |||||
| FRLnC | Up | FOXM1 | Twist, TGFβ-1 | EMT | [25] | |||||
| UCA1 | Up | TGFβ-1, GRK2/ERK/MMP9 | EMT, degradation of the ECM | Lymphatic metastasis, TNM stage | 3.909 (1.592-9.599) | 2.917 (1.069-7.962) | [30,73] | |||
| ATB | Up | TGFβ-1 | miR-200s, ZEB1 | EMT | 3.50 (1.73-7.44) | [26] | ||||
| XIST | Up | miR-101 | EMT | Lymphatic metastasis, distant metastasis, TNM stage | [27] | |||||
| SNHG-6 | Up | miR-101-3P, ZEB1 | EMT | Lymphatic metastasis, distant metastasis, TNM stage | [28] | |||||
| ZFAS1 | Up | ZEB1, SNAIL, Slug, Twist | EMT | Lymphatic metastasis, TNM stage | [29] | |||||
| LINC00152 | Up | EMT | Lymphatic metastasis, TNM stage | 2.162 (1.327-3.524) | 1.659 (1.008-2.731) | [94] | ||||
| HULC | Up | EMT | Lymphatic metastasis, distant metastasis, TNM stage | [35] | ||||||
| Linc00978 | Up | TGFβ/SMAD,Twist1, Slug | EMT | Lymphatic metastasis, TNM stage | [32] | |||||
| YAP1 | Up | vimentin, β-catenin, E-cadherin | EMT | Lymphatic metastasis, distant metastasis, TNM stage | [33] | |||||
| Linc00261 | Down | Slug, GSK3β | EMT | Lymphatic, metastasis | 0.494 (0.300-0.812) | 0.551 (0.323-0.940) | [36] | |||
| Linc00675 | Down | Vimentin | EMT | [40] | ||||||
| SPRY4-IT1 | Down | Vimentin | EMT, epigenetic regulation | Lymphatic metastasis, distant metastasis, TNM stage | 1.247 (1473-1.996) | 2.223 (1.806-2.59) | 0.818 (0.314-1.567) | 1.741 (1.324-2477) | [43,95] | |
| LEIGC | Down | EMT | [44] | |||||||
| GClnc1 | Up | WDR5/KAT2, H3K4, H3K9, SOD2 | Epigenetic regulation | 2.21 (1.46-3.33) | 1.93 (1.24-3.00) | [15] | ||||
| LOC100130476 | Down | DNMT1 | Epigenetic regulation | Lymphatic metastasis, distant metastasis, TNM stage | [47] | |||||
| AK058003 | Up | SNCG | Epigenetic regulation, Hypoxia | Lymphatic metastasis, TNM stage | [56] | |||||
| BC005927 | Up | HIF-1α | BPHB4 | Hypoxia | Lymphatic metastasis, TNM stage | [57] | ||||
| SNHG15 | Up | MMP2, MMP9 | Degradation of the ECM | Lymphatic metastasis, TNM stage | [96] | |||||
| FENDRR | Down | MMP2, MMP9 | Degradation of the ECM | Lymphatic metastasis | 0.539 (0.337-0.862) | 0.563 (0.370-0.856) | 0.569 (0.321-0.960) | 0.555 (0.344-0.897) | [16] | |
| BM742401 | Down | MMP9 | Degradation of the ECM | [53] | ||||||
| C21orF96 | Up | Lymphangiogenesis, VM | Lymphatic metastasis, distant metastasis | [68] | ||||||
| LINC00052 | Up | Wnt/β-catenin pathway | TNM stage | [97] | ||||||
| AA174084 | Down | [80] | ||||||||
| RMRP | Down | Lymphatic metastasis | [81] | |||||||
| SNHG1 | Up | Lymphatic metastasis, TNM stage | [98] | |||||||
| SNHG5 | Down | TNM stage | [99] | |||||||
| MSTO2P | Up | miR-335 | Lymphatic metastasis, distant metastasis | [100] | ||||||
| ZEB1-AS1 | Up | miR-335-5p | Lymphatic metastasis, TNM stage | [101] | ||||||
| PTENP1 | Down | miR-106b, miR-93 | Lymphatic metastasis, TNM stage | [102] | ||||||
| RP11-19P22.6-001 | Down | Nitric oxide synthase 2 (NOS2) | Lymphatic metastasis, TNM stage | [103] | ||||||
| PCAT-1 | Up | distant metastasis | [104] | |||||||
| HOXD-ASI | Up | Lymphatic metastasis, distant metastasis, TNM stage | [105] | |||||||
| CARLo-5 | Up | Lymphatic metastasis, distant metastasis | [106] | |||||||
| LINC00673 | Down | Lymphatic metastasis | [107] | |||||||
| LINC00982 | Down | Lymphatic metastasis, TNM stage | [108] | |||||||
| HMlincRNA717 | Down | distant metastasis | [109] | |||||||
| PVT1 | Up | Lymphatic metastasis | [110] | |||||||
| GACAT3 | Up | IL-6/STAT3 | Distant metastasis, TNM stage | [111] | ||||||
| Sox2ot | Down | Distant metastasis | 3.241 (1.239-6.428) | 3.844 (1.873-7.332) | [85] | |||||
| HOTTIP | Up | HOXA13 | Lymphatic metastasis, TNM stage | [112] | ||||||
| NEAT1 | Up | Lymphatic metastasis, distant metastasis | [113,114] | |||||||
| OTUB1-isoform2 | Up | N-cadherin, MMP2, MMP9, E-cadherin | Lymphatic metastasis, TNM stage | 1,538 (1.044-2.265) | 1.615 (1.111-2.348) | 1.498 (1.021-2.200) | [86] | |||
| PANDAR | Lymphatic metastasis, TNM stage | 4.612 (1.59-13.825) | 3.113 (1.591-6.093) | 3.683 (1.125-12.058) | 2.359 (1.153-4.830) | [87] | ||||
| ZMAT1 transcript variant 2 | Down | Lymphatic metastasis, distant metastasis, TNM stage | [115] | |||||||
| JMJD1A | Up | MALAT1, MAPK | Lymphatic metastasis, TNM stage | 8.446 (4.480-15.923) | 3.988 (1.948-8.167) | [116] | ||||
| OR3A4 | Up | PDLIM2, MACC1, NTN4, GNB2L1 | Degradation of the ECM, angiopoiesis, VM | Lymphatic metastasis distant metastasis | [54] | |||||
| HNF1A-AS1 | Down | Lymphatic metastasis | [117] | |||||||
| BANCR | Up | Lymphatic metastasis, distant metastasis | 2.457 (1.715-3.521) | 1.511 (1.02-2.227) | [118] | |||||
| DQ786243 | Up | Lymphatic metastasis, TNM stage | [119] | |||||||
| XLOC_010235 | Up | distant metastasis, TNM stage | [120] | |||||||
| CCAT2 | Up | Lymphatic metastasis, distant metastasis, TNM stage | 2.631 (1.348-5.672) | 2.574 (1.201-5.476) | 2.405 (1.194-5.417) | 2.315 (1.097-5.283) | [121,122] | |||
| Linc-UBC1 | Up | Lymphatic metastasis, TNM stage | [123] | |||||||
| HIF1A-AS2 | Up | Lymphatic metastasis, TNM stage | 2.346 (1.379-3.991) | 1.724 (1.002-2.964) | [124] | |||||
| LET | Down | Lymphatic metastasis, distant metastasis, TNM stage | 2.513 (1.414-5.847) | 2.275 (1.301-5.176) | [125] | |||||
| LSINCT5 | Up | Lymphatic metastasis, TNM stage | 2.501 (1.326-4.719) | 1.081 (1.286-3.564) | [126] | |||||
| AC130710 | Up | distant metastasis, TNM stage | [127] | |||||||
| FER1L4 | Down | Lymphatic metastasis, distant metastasis, TNM stage | [88] | |||||||
| RuPAR | Down | Lymphatic metastasis, distant metastasis, TNM stage | [128] | |||||||
| H19 | Up | miR-675 | Lymphatic metastasis, TNM stage | 1.170 (1.050-1.304) | 1.137 (1.005-1.287) | [129,130] | ||||
| AC096655.1-002 | Down | Lymphatic metastasis, distant metastasis, TNM stage | [131] | |||||||
| SUMO1P3 | Up | Lymphatic metastasis | [132] | |||||||
| IGF2 | Up | Lymphatic metastasis | [133] | |||||||
| CCAT1 | Up | Lymphatic metastasis, TNM stage | [134] | |||||||
EMT: Epithelial-mesenchymal transition; VM: Vasculogenic mimicry; TNM: Tumor, node, metastasis; ECM: Extracellular matrix.
CONCLUSION
Utilizing a variety of techniques, including RT-PCR, computer-assisted microscopic image analysis, bioinformatics methods, ChIP assays, etc., a myriad of lncRNAs have been found to participate in the proliferation, growth, invasion, metastasis, motility, and phenotype of GC cells, with dozens of them correlating with the invasion depth, size, lymph node metastasis, TNM stage, OS and DFS of GC tumors. In this review, we emphasized epithelial-mesenchymal transition, epigenetic regulation, and degradation of the extracellular matrix, angiopoiesis, vasculogenic mimicry, and immune escape in examining the ectopic expression of lncRNAs. lncRNAs involved in specific mechanisms of GC progression could be helpful in GC treatment. Those lncRNAs that are considered as independent prognostic factors by survival analysis such as MALAT1[17], Sox2ot[85], OTUB1-isoform 2[86], PANDAR[87], etc., and those lncRNAs dramatically altered in postoperative GC patients such as FERI4[88], may be utilized as prognosis evaluation markers. Some lncRNAs increased in metastatic tissue compared to primary focus may be beneficial in predicting metastasis.
Footnotes
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
Conflict-of-interest statement: Authors declare no conflict of interests for this article.
Peer-review started: April 25, 2018
First decision: May 16, 2018
Article in press: June 27, 2018
P- Reviewer: Das U, Mavridis K S- Editor: Wang XJ L- Editor: Filipodia E- Editor: Huang Y
Contributor Information
Meng-Ting Lin, Medical School of Ningbo University, Ningbo 315000, Zhejiang Province, China.
Hao-Jun Song, Gastroenterology Department, Ningbo First Hospital, Ningbo 315000, Zhejiang Province, China.
Xiao-Yun Ding, Gastroenterology Department, Ningbo First Hospital, Ningbo 315000, Zhejiang Province, China. dyyyding@126.com.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 4.Tanabe S, Koizumi W, Mitomi H, Nakai H, Murakami S, Nagaba S, Kida M, Oida M, Saigenji K. Clinical outcome of endoscopic aspiration mucosectomy for early stage gastric cancer. Gastrointest Endosc. 2002;56:708–713. doi: 10.1067/mge.2002.129085. [DOI] [PubMed] [Google Scholar]
- 5.Tanabe S, Ishido K, Higuchi K, Sasaki T, Katada C, Azuma M, Naruke A, Kim M, Koizumi W. Long-term outcomes of endoscopic submucosal dissection for early gastric cancer: a retrospective comparison with conventional endoscopic resection in a single center. Gastric Cancer. 2014;17:130–136. doi: 10.1007/s10120-013-0241-2. [DOI] [PubMed] [Google Scholar]
- 6.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamakawa T, Kukita Y, Kurokawa Y, Miyazaki Y, Takahashi T, Yamasaki M, Miyata H, Nakajima K, Taniguchi K, Takiguchi S, et al. Monitoring gastric cancer progression with circulating tumour DNA. Br J Cancer. 2015;112:352–356. doi: 10.1038/bjc.2014.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferro A, Peleteiro B, Malvezzi M, Bosetti C, Bertuccio P, Levi F, Negri E, La Vecchia C, Lunet N. Worldwide trends in gastric cancer mortality (1980-2011), with predictions to 2015, and incidence by subtype. Eur J Cancer. 2014;50:1330–1344. doi: 10.1016/j.ejca.2014.01.029. [DOI] [PubMed] [Google Scholar]
- 9.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Criscione SW, Zhang Y, Thompson W, Sedivy JM, Neretti N. Transcriptional landscape of repetitive elements in normal and cancer human cells. BMC Genomics. 2014;15:583. doi: 10.1186/1471-2164-15-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Novikova IV, Hennelly SP, Tung CS, Sanbonmatsu KY. Rise of the RNA machines: exploring the structure of long non-coding RNAs. J Mol Biol. 2013;425:3731–3746. doi: 10.1016/j.jmb.2013.02.030. [DOI] [PubMed] [Google Scholar]
- 12.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grammatikakis I, Panda AC, Abdelmohsen K, Gorospe M. Long noncoding RNAs(lncRNAs) and the molecular hallmarks of aging. Aging (Albany NY) 2014;6:992–1009. doi: 10.18632/aging.100710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen D, Liu L, Wang K, Yu H, Wang Y, Liu J, Guo Y, Zhang H. The role of MALAT-1 in the invasion and metastasis of gastric cancer. Scand J Gastroenterol. 2017;52:790–796. doi: 10.1080/00365521.2017.1280531. [DOI] [PubMed] [Google Scholar]
- 15.Sun TT, He J, Liang Q, Ren LL, Yan TT, Yu TC, Tang JY, Bao YJ, Hu Y, Lin Y, et al. LncRNA GClnc1 Promotes Gastric Carcinogenesis and May Act as a Modular Scaffold of WDR5 and KAT2A Complexes to Specify the Histone Modification Pattern. Cancer Discov. 2016;6:784–801. doi: 10.1158/2159-8290.CD-15-0921. [DOI] [PubMed] [Google Scholar]
- 16.Xu TP, Huang MD, Xia R, Liu XX, Sun M, Yin L, Chen WM, Han L, Zhang EB, Kong R, et al. Decreased expression of the long non-coding RNA FENDRR is associated with poor prognosis in gastric cancer and FENDRR regulates gastric cancer cell metastasis by affecting fibronectin1 expression. J Hematol Oncol. 2014;7:63. doi: 10.1186/s13045-014-0063-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, Wu Z, Yuan J, Sun L, Lin L, Huang N, Bin J, Liao Y, Liao W. Long non-coding RNA MALAT1 promotes gastric cancer tumorigenicity and metastasis by regulating vasculogenic mimicry and angiogenesis. Cancer Lett. 2017;395:31–44. doi: 10.1016/j.canlet.2017.02.035. [DOI] [PubMed] [Google Scholar]
- 18.Song B, Guan Z, Liu F, Sun D, Wang K, Qu H. Long non-coding RNA HOTAIR promotes HLA-G expression via inhibiting miR-152 in gastric cancer cells. Biochem Biophys Res Commun. 2015;464:807–813. doi: 10.1016/j.bbrc.2015.07.040. [DOI] [PubMed] [Google Scholar]
- 19.Bonnomet A, Brysse A, Tachsidis A, Waltham M, Thompson EW, Polette M, Gilles C. Epithelial-to-mesenchymal transitions and circulating tumor cells. J Mammary Gland Biol Neoplasia. 2010;15:261–273. doi: 10.1007/s10911-010-9174-0. [DOI] [PubMed] [Google Scholar]
- 20.Guarino M. Epithelial-mesenchymal transition and tumour invasion. Int J Biochem Cell Biol. 2007;39:2153–2160. doi: 10.1016/j.biocel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 21.Hurt JA, Robertson AD, Burge CB. Global analyses of UPF1 binding and function reveal expanded scope of nonsense-mediated mRNA decay. Genome Res. 2013;23:1636–1650. doi: 10.1101/gr.157354.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li L, Geng Y, Feng R, Zhu Q, Miao B, Cao J, Fei S. The Human RNA Surveillance Factor UPF1 Modulates Gastric Cancer Progression by Targeting Long Non-Coding RNA MALAT1. Cell Physiol Biochem. 2017;42:2194–2206. doi: 10.1159/000479994. [DOI] [PubMed] [Google Scholar]
- 23.Lee NK, Lee JH, Ivan C, Ling H, Zhang X, Park CH, Calin GA, Lee SK. MALAT1 promoted invasiveness of gastric adenocarcinoma. BMC Cancer. 2017;17:46. doi: 10.1186/s12885-016-2988-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu YW, Sun M, Xia R, Zhang EB, Liu XH, Zhang ZH, Xu TP, De W, Liu BR, Wang ZX. LincHOTAIR epigenetically silences miR34a by binding to PRC2 to promote the epithelial-to-mesenchymal transition in human gastric cancer. Cell Death Dis. 2015;6:e1802. doi: 10.1038/cddis.2015.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cai H, Chen J, He B, Li Q, Li Y, Gao Y. A FOXM1 related long non-coding RNA contributes to gastric cancer cell migration. Mol Cell Biochem. 2015;406:31–41. doi: 10.1007/s11010-015-2421-3. [DOI] [PubMed] [Google Scholar]
- 26.Saito T, Kurashige J, Nambara S, Komatsu H, Hirata H, Ueda M, Sakimura S, Uchi R, Takano Y, Shinden Y, et al. A Long Non-coding RNA Activated by Transforming Growth Factor-β is an Independent Prognostic Marker of Gastric Cancer. Ann Surg Oncol. 2015;22 Suppl 3:S915–S922. doi: 10.1245/s10434-015-4554-8. [DOI] [PubMed] [Google Scholar]
- 27.Chen DL, Ju HQ, Lu YX, Chen LZ, Zeng ZL, Zhang DS, Luo HY, Wang F, Qiu MZ, Wang DS, et al. Long non-coding RNA XIST regulates gastric cancer progression by acting as a molecular sponge of miR-101 to modulate EZH2 expression. J Exp Clin Cancer Res. 2016;35:142. doi: 10.1186/s13046-016-0420-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan K, Tian J, Shi W, Xia H, Zhu Y. LncRNA SNHG6 is Associated with Poor Prognosis of Gastric Cancer and Promotes Cell Proliferation and EMT through Epigenetically Silencing p27 and Sponging miR-101-3p. Cell Physiol Biochem. 2017;42:999–1012. doi: 10.1159/000478682. [DOI] [PubMed] [Google Scholar]
- 29.Pan L, Liang W, Fu M, Huang ZH, Li X, Zhang W, Zhang P, Qian H, Jiang PC, Xu WR, et al. Exosomes-mediated transfer of long noncoding RNA ZFAS1 promotes gastric cancer progression. J Cancer Res Clin Oncol. 2017;143:991–1004. doi: 10.1007/s00432-017-2361-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zuo ZK, Gong Y, Chen XH, Ye F, Yin ZM, Gong QN, Huang JS. TGFβ1-Induced LncRNA UCA1 Upregulation Promotes Gastric Cancer Invasion and Migration. DNA Cell Biol. 2017;36:159–167. doi: 10.1089/dna.2016.3553. [DOI] [PubMed] [Google Scholar]
- 31.Gao J, Cao R, Mu H. Long non-coding RNA UCA1 may be a novel diagnostic and predictive biomarker in plasma for early gastric cancer. Int J Clin Exp Pathol. 2015;8:12936–12942. [PMC free article] [PubMed] [Google Scholar]
- 32.Fu M, Huang Z, Zang X, Pan L, Liang W, Chen J, Qian H, Xu W, Jiang P, Zhang X. Long noncoding RNA LINC00978 promotes cancer growth and acts as a diagnostic biomarker in gastric cancer. Cell Prolif. 2018:51. doi: 10.1111/cpr.12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun D, Li X, He Y, Li W, Wang Y, Wang H, Jiang S, Xin Y. YAP1 enhances cell proliferation, migration, and invasion of gastric cancer in vitro and in vivo. Oncotarget. 2016;7:81062–81076. doi: 10.18632/oncotarget.13188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao J, Liu Y, Zhang W, Zhou Z, Wu J, Cui P, Zhang Y, Huang G. Long non-coding RNA Linc00152 is involved in cell cycle arrest, apoptosis, epithelial to mesenchymal transition, cell migration and invasion in gastric cancer. Cell Cycle. 2015;14:3112–3123. doi: 10.1080/15384101.2015.1078034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao Y, Guo Q, Chen J, Hu J, Wang S, Sun Y. Role of long non-coding RNA HULC in cell proliferation, apoptosis and tumor metastasis of gastric cancer: a clinical and in vitro investigation. Oncol Rep. 2014;31:358–364. doi: 10.3892/or.2013.2850. [DOI] [PubMed] [Google Scholar]
- 36.Fan Y, Wang YF, Su HF, Fang N, Zou C, Li WF, Fei ZH. Decreased expression of the long noncoding RNA LINC00261 indicate poor prognosis in gastric cancer and suppress gastric cancer metastasis by affecting the epithelial-mesenchymal transition. J Hematol Oncol. 2016;9:57. doi: 10.1186/s13045-016-0288-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Yu Y, Li L, Zheng Z, Chen S, Chen E, Hu Y. Long non-coding RNA linc00261 suppresses gastric cancer progression via promoting Slug degradation. J Cell Mol Med. 2017;21:955–967. doi: 10.1111/jcmm.13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu ZQ, Li XY, Hu CY, Ford M, Kleer CG, Weiss SJ. Canonical Wnt signaling regulates Slug activity and links epithelial-mesenchymal transition with epigenetic Breast Cancer 1, Early Onset (BRCA1) repression. Proc Natl Acad Sci U S A. 2012;109:16654–16659. doi: 10.1073/pnas.1205822109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raja, Vidhya G. 2017. GSK3B regulates epithelial-mesenchymal transition and cancer stem cell properties and is a novel drug target for triple-negative breast cancer. [Google Scholar]
- 40.Zeng S, Xie X, Xiao YF, Tang B, Hu CJ, Wang SM, Wu YY, Dong H, Li BS, Yang SM. Long noncoding RNA LINC00675 enhances phosphorylation of vimentin on Ser83 to suppress gastric cancer progression. Cancer letters. 2017;412:179. doi: 10.1016/j.canlet.2017.10.026. [DOI] [PubMed] [Google Scholar]
- 41.Zhang X, Liu W, Yang H, Tan L, Ao L, Liu J, Cao J, Cui Z. Inhibition of PPARα attenuates vimentin phosphorylation on Ser-83 and collapse of vimentin filaments during exposure of rat Sertoli cells in vitro to DBP. Reprod Toxicol. 2014;50:11–18. doi: 10.1016/j.reprotox.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 42.Dave JM, Bayless KJ. Vimentin as an integral regulator of cell adhesion and endothelial sprouting. Microcirculation. 2014;21:333–344. doi: 10.1111/micc.12111. [DOI] [PubMed] [Google Scholar]
- 43.Xie M, Nie FQ, Sun M, Xia R, Liu YW, Zhou P, De W, Liu XH. Decreased long noncoding RNA SPRY4-IT1 contributing to gastric cancer cell metastasis partly via affecting epithelial-mesenchymal transition. J Transl Med. 2015;13:250. doi: 10.1186/s12967-015-0595-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han Y, Ye J, Wu D, Wu P, Chen Z, Chen J, Gao S, Huang J. LEIGC long non-coding RNA acts as a tumor suppressor in gastric carcinoma by inhibiting the epithelial-to-mesenchymal transition. BMC Cancer. 2014;14:932. doi: 10.1186/1471-2407-14-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beckedorff FC, Amaral MS, Deocesanopereira C, Verjovskialmeida S. Long non-coding RNAs and their implications in cancer epigenetics. Bioscience Reports. 2013;33:667–675. doi: 10.1042/BSR20130054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.16 Spec No 1(1): R50; Esteller M. Epigenetic gene silencing in cancer: the DNA hypermethylome. Human Molecular Genetics 2007; [DOI] [PubMed] [Google Scholar]
- 47.Guo W, Dong Z, Shi Y, Liu S, Liang J, Guo Y, Guo X, Shen S, Wang G. Methylation-mediated downregulation of long noncoding RNA LOC100130476 in gastric cardia adenocarcinoma. Clin Exp Metastasis. 2016;33:497–508. doi: 10.1007/s10585-016-9794-x. [DOI] [PubMed] [Google Scholar]
- 48.Sun M, Liu XH, Lu KH, Nie FQ, Xia R, Kong R, Yang JS, Xu TP, Liu YW, Zou YF, et al. EZH2-mediated epigenetic suppression of long noncoding RNA SPRY4-IT1 promotes NSCLC cell proliferation and metastasis by affecting the epithelial-mesenchymal transition. Cell Death Dis. 2014;5:e1298. doi: 10.1038/cddis.2014.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Buchheit CL, Weigel KJ, Schafer ZT. Cancer cell survival during detachment from the ECM: multiple barriers to tumour progression. Nature Reviews Cancer. 2014;14:632. doi: 10.1038/nrc3789. [DOI] [PubMed] [Google Scholar]
- 50.Jodele S, Blavier L, Yoon JM, Declerck YA. Modifying the soil to affect the seed: role of stromal-derived matrix metalloproteinases in cancer progression. Cancer Metasta Revie. 2006;25:35. doi: 10.1007/s10555-006-7887-8. [DOI] [PubMed] [Google Scholar]
- 51.Hadler-Olsen E, Winberg JO, Uhlin-Hansen L. Matrix metalloproteinases in cancer: their value as diagnostic and prognostic markers and therapeutic targets. Bio J of Inter Soci Biol Medic. 2013;34:2041–2051. doi: 10.1007/s13277-013-0842-8. [DOI] [PubMed] [Google Scholar]
- 52.Wang ZQ, He CY, Hu L, Shi HP, Li JF, Gu QL, Su LP, Liu BY, Li C, Zhu Z. Long noncoding RNA UCA1 promotes tumour metastasis by inducing GRK2 degradation in gastric cancer. Cancer Lett. 2017;408:10–21. doi: 10.1016/j.canlet.2017.08.013. [DOI] [PubMed] [Google Scholar]
- 53.Park SM, Park SJ, Kim HJ, Kwon OH, Kang TW, Sohn HA, Kim SK, Moo Noh S, Song KS, Jang SJ, et al. A known expressed sequence tag, BM742401, is a potent lincRNA inhibiting cancer metastasis. Exp Mol Med. 2013;45:e31. doi: 10.1038/emm.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo X, Yang Z, Zhi Q, Wang D, Guo L, Li G, Miao R, Shi Y, Kuang Y. Long noncoding RNA OR3A4 promotes metastasis and tumorigenicity in gastric cancer. Oncotarget. 2016;7:30276–30294. doi: 10.18632/oncotarget.7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Y, Liu X, Zhang H, Sun L, Zhou Y, Jin H, Zhang H, Zhang H, Liu J, Guo H, et al. Hypoxia-inducible lncRNA-AK058003 promotes gastric cancer metastasis by targeting γ-synuclein. Neoplasia. 2014;16:1094–1106. doi: 10.1016/j.neo.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu X, Wang Y, Sun L, Min J, Liu J, Chen D, Zhang H, Zhang H, Zhang H, Zhou Y, et al. Long noncoding RNA BC005927 upregulates EPHB4 and promotes gastric cancer metastasis under hypoxia. Cancer Sci. 2018;109:988–1000. doi: 10.1111/cas.13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koliopanos A, Avgerinos C, Paraskeva C, Touloumis Z, Kelgiorgi D, Dervenis C. Molecu aspe carcinogene pancre cancer. 2008;7:345–356. [PubMed] [Google Scholar]
- 59.Coomber BL, Yu JL, Fathers KE, Plumb C, Rak JW. Angiogenesis and the role of epigenetics in metastasis. Clini experimen metasta. 2003;20:215. doi: 10.1023/a:1022935117193. [DOI] [PubMed] [Google Scholar]
- 60.Chen X, Maniotis AJ, Majumdar D, Pe’Er J, Folberg R. Uveal melanoma cell staining for CD34 and assessment of tumor vascularity. Investigat Ophthalmol Visu Scie. 2002;43:2533–2539. [PubMed] [Google Scholar]
- 61.Sun B, Zhang S, Zhang D, Du J, Guo H, Zhao X, Zhang W, Hao X. Vasculogenic mimicry is associated with high tumor grade, invasion and metastasis, and short survival in patients with hepatocellular carcinoma. Oncol repor. 2006;16:693–698. [PubMed] [Google Scholar]
- 62.Maniotis AJ, Folberg R, Hess A, Seftor EA, Gardner LM, Pe’Er J, Trent JM, Meltzer PS, Hendrix MJ. Vascular channel formation by human melanoma cells in vivo and in vitro: vasculogenic mimicry. Americ J Pathol. 1999;155:739. doi: 10.1016/S0002-9440(10)65173-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun B, Zhang S, Zhao X, Zhang W, Hao X, Sun, B, Zhang, S, Zhao, et al. Vasculogenic mimicry is associated with poor survival in patients with mesothelial sarcomas and alveolar rhabdomyosarcomas. Int J Oncol. 2005;25:1609–1614. [PubMed] [Google Scholar]
- 64.Sun B, Qie S, Zhang S, Sun T, Zhao X, Gao S, Ni C, Wang X, Liu Y, Zhang L. Role and mechanism of vasculogenic mimicry in gastrointestinal stromal tumors. Hum Pathol. 2008;39:444. doi: 10.1016/j.humpath.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 65.Deng QJ, Xie LQ, Li H. Overexpressed MALAT1 promotes invasion and metastasis of gastric cancer cells via increasing EGFL7 expression. Life Sci. 2016;157:38–44. doi: 10.1016/j.lfs.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 66.Hansen TF, Christensen RD, Andersen RF, Sørensen FB, Johnsson A, Jakobsen A. MicroRNA-126 and epidermal growth factor-like domain 7|[ndash]|an angiogenic couple of importance in metastatic colorectal cancer. Results from the Nordic ACT trial. Bri J Cancer. 2013;109:1243. doi: 10.1038/bjc.2013.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sun Y, Bai Y, Zhang F, Wang Y, Guo Y, Guo L. miR-126 inhibits non-small cell lung cancer cells proliferation by targeting EGFL7. Biophyl Resea Communica. 2010;391:1483–1489. doi: 10.1016/j.bbrc.2009.12.098. [DOI] [PubMed] [Google Scholar]
- 68.Yang Z, Zhi Q, Wang D, Zhang L, Preston B, Brandon C, Kuang Y, Miao R, Shi Y, Guo X. Long Noncoding RNA C21orF96 Promotes the Migration, Invasion and Lymph Node Metastasis in Gastric Cancer. Anticancer Agents Med Chem. 2016;16:1101–1108. doi: 10.2174/1871520616666151116122921. [DOI] [PubMed] [Google Scholar]
- 69.Kamei S, Kono K, Amemiya H, Takahashi A, Sugai H, Ichihara F, Fujii H, Matsumoto Y. Evaluation of VEGF and VEGF-C expression in gastric cancer cells producing alpha-fetoprotein. J Gastroenterol. 2003;38:540–547. doi: 10.1007/s00535-002-1099-y. [DOI] [PubMed] [Google Scholar]
- 70.Koebel CM, Vermi W, Swann JB, Zerafa N, Rodig SJ, Old LJ, Smyth MJ, Schreiber RD. Adaptive immunity maintains occult cancer in an equilibrium state. Nature. 2007;450:903–907. doi: 10.1038/nature06309. [DOI] [PubMed] [Google Scholar]
- 71.Khong HT, Restifo NP. Natural selection of tumor variants in the generation of “tumor escape” phenotypes. Nat Immunol. 2002;3:999–1005. doi: 10.1038/ni1102-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 73.Li CY, Liang GY, Yao WZ, Sui J, Shen X, Zhang YQ, Peng H, Hong WW, Ye YC, Zhang ZY, et al. Integrated analysis of long non-coding RNA competing interactions reveals the potential role in progression of human gastric cancer. Int J Oncol. 2016;48:1965–1976. doi: 10.3892/ijo.2016.3407. [DOI] [PubMed] [Google Scholar]
- 74.Zhang ZZ, Shen ZY, Shen YY, Zhao EH, Wang M, Wang CJ, Cao H, Xu J. HOTAIR Long Noncoding RNA Promotes Gastric Cancer Metastasis through Suppression of Poly r(C)-Binding Protein (PCBP) 1. Mol Cancer Ther. 2015;14:1162–1170. doi: 10.1158/1535-7163.MCT-14-0695. [DOI] [PubMed] [Google Scholar]
- 75.Lee NK, Lee JH, Park CH, Yu D, Lee YC, Cheong JH, Noh SH, Lee SK. Long non-coding RNA HOTAIR promotes carcinogenesis and invasion of gastric adenocarcinoma. Biochem Biophys Res Commun. 2014;451:171–178. doi: 10.1016/j.bbrc.2014.07.067. [DOI] [PubMed] [Google Scholar]
- 76.Okugawa Y, Toiyama Y, Hur K, Toden S, Saigusa S, Tanaka K, Inoue Y, Mohri Y, Kusunoki M, Boland CR, et al. Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis. Carcinogenesis. 2014;35:2731–2739. doi: 10.1093/carcin/bgu200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guo W, Dong Z, Bai Y, Guo Y, Shen S, Kuang G, Xu J. Associations between polymorphisms of HOTAIR and risk of gastric cardia adenocarcinoma in a population of north China. Tumour Biol. 2015;36:2845–2854. doi: 10.1007/s13277-014-2912-y. [DOI] [PubMed] [Google Scholar]
- 78.Hondo FY, Kishi H, Safatle-Ribeiro AV, Pessorrusso FCS, Ribeiro U, Maluf-Filho F. Characterization of the mucin phenotype can predict gastric cancer recurrence after endoscopic mucosal resection. Arq Gastroenterol. 2017;54:308–314. doi: 10.1590/S0004-2803.201700000-38. [DOI] [PubMed] [Google Scholar]
- 79.Eom BW, Yoon H, Ryu KW, Lee JH, Cho SJ, Lee JY, Kim CG, Choi IJ, Lee JS, Kook MC, et al. Predictors of timing and patterns of recurrence after curative resection for gastric cancer. Dig Surg. 2010;27:481–486. doi: 10.1159/000320691. [DOI] [PubMed] [Google Scholar]
- 80.Shao Y, Ye M, Jiang X, Sun W, Ding X, Liu Z, Ye G, Zhang X, Xiao B, Guo J. Gastric juice long noncoding RNA used as a tumor marker for screening gastric cancer. Cancer. 2014;120:3320–3328. doi: 10.1002/cncr.28882. [DOI] [PubMed] [Google Scholar]
- 81.Shao Y, Ye M, Li Q, Sun W, Ye G, Zhang X, Yang Y, Xiao B, Guo J. LncRNA-RMRP promotes carcinogenesis by acting as a miR-206 sponge and is used as a novel biomarker for gastric cancer. Oncotarget. 2016;7:37812–37824. doi: 10.18632/oncotarget.9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang K, Shi H, Xi H, Wu X, Cui J, Gao Y, Liang W, Hu C, Liu Y, Li J, et al. Genome-Wide lncRNA Microarray Profiling Identifies Novel Circulating lncRNAs for Detection of Gastric Cancer. Theranostics. 2017;7:213–227. doi: 10.7150/thno.16044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Montes HZ. TNM Classification of Malignant Tumors, 7th edition. Inter J Radia Oncol Biol Phys. 2010;78:1278. [Google Scholar]
- 84.Gu H, Li D, Zhu H, Zhang H, Yu Y, Qin D, Yi M, Li X, Lu P. The prognostic efficacy and improvements of the 7th edition Union for International Cancer Control tumor-node-metastasis classifications for Chinese patients with gastric cancer: Results based on a retrospective three-decade population study. Tumour Biol. 2017;39:1010428317694548. doi: 10.1177/1010428317694548. [DOI] [PubMed] [Google Scholar]
- 85.Zou JH, Li CY, Bao J, Zheng GQ. High expression of long noncoding RNA Sox2ot is associated with the aggressive progression and poor outcome of gastric cancer. Eur Rev Med Pharmacol Sci. 2016;20:4482–4486. [PubMed] [Google Scholar]
- 86.Wang YQ, Zhang QY, Weng WW, Wu Y, Yang YS, Shen C, Chen XC, Wang L, Liu KJ, Xu MD, et al. Upregulation of the Non-Coding RNA OTUB1-isoform 2 Contributes to Gastric Cancer Cell Proliferation and Invasion and Predicts Poor Gastric Cancer Prognosis. Int J Biol Sci. 2016;12:545–557. doi: 10.7150/ijbs.13540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ma P, Xu T, Huang M, Shu Y. Increased expression of LncRNA PANDAR predicts a poor prognosis in gastric cancer. Biomed Pharmacother. 2016;78:172–176. doi: 10.1016/j.biopha.2016.01.025. [DOI] [PubMed] [Google Scholar]
- 88.Liu Z, Shao Y, Tan L, Shi H, Chen S, Guo J. Clinical significance of the low expression of FER1L4 in gastric cancer patients. Tumour Biol. 2014;35:9613–9617. doi: 10.1007/s13277-014-2259-4. [DOI] [PubMed] [Google Scholar]
- 89.Zhang Y, Chen Z, Li MJ, Guo HY, Jing NC. Long non-coding RNA metastasis-associated lung adenocarcinoma transcript 1 regulates the expression of Gli2 by miR-202 to strengthen gastric cancer progression. Biomed Pharmacother. 2017;85:264–271. doi: 10.1016/j.biopha.2016.11.014. [DOI] [PubMed] [Google Scholar]
- 90.Xia H, Chen Q, Chen Y, Ge X, Leng W, Tang Q, Ren M, Chen L, Yuan D, Zhang Y, et al. The lncRNA MALAT1 is a novel biomarker for gastric cancer metastasis. Oncotarget. 2016;7:56209–56218. doi: 10.18632/oncotarget.10941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD, Kong R, Xia R, Lu KH, Li JH, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92. doi: 10.1186/1476-4598-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Endo H, Shiroki T, Nakagawa T, Yokoyama M, Tamai K, Yamanami H, Fujiya T, Sato I, Yamaguchi K, Tanaka N, et al. Enhanced expression of long non-coding RNA HOTAIR is associated with the development of gastric cancer. PLoS One. 2013;8:e77070. doi: 10.1371/journal.pone.0077070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hajjari M, Behmanesh M, Sadeghizadeh M, Zeinoddini M. Up-regulation of HOTAIR long non-coding RNA in human gastric adenocarcinoma tissues. Med Oncol. 2013;30:670. doi: 10.1007/s12032-013-0670-0. [DOI] [PubMed] [Google Scholar]
- 94.Chen WM, Huang MD, Sun DP, Kong R, Xu TP, Xia R, Zhang EB, Shu YQ. Long intergenic non-coding RNA 00152 promotes tumor cell cycle progression by binding to EZH2 and repressing p15 and p21 in gastric cancer. Oncotarget. 2016;7:9773–9787. doi: 10.18632/oncotarget.6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Peng W, Wu G, Fan H, Wu J, Feng J. Long noncoding RNA SPRY4-IT1 predicts poor patient prognosis and promotes tumorigenesis in gastric cancer. Tumour Biol. 2015;36:6751–6758. doi: 10.1007/s13277-015-3376-4. [DOI] [PubMed] [Google Scholar]
- 96.Chen SX, Yin JF, Lin BC, Su HF, Zheng Z, Xie CY, Fei ZH. Upregulated expression of long noncoding RNA SNHG15 promotes cell proliferation and invasion through regulates MMP2/MMP9 in patients with GC. Tumour Biol. 2016;37:6801–6812. doi: 10.1007/s13277-015-4404-0. [DOI] [PubMed] [Google Scholar]
- 97.Shan Y, Ying R, Jia Z, Kong W, Wu Y, Zheng S, Jin H. LINC00052 Promotes Gastric Cancer Cell Proliferation and Metastasis via Activating the Wnt/β-Catenin Signaling Pathway. Oncol Res. 2017;25:1589–1599. doi: 10.3727/096504017X14897896412027. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 98.Hu Y, Ma Z, He Y, Liu W, Su Y, Tang Z. LncRNA-SNHG1 contributes to gastric cancer cell proliferation by regulating DNMT1. Biochem Biophys Res Commun. 2017;491:926–931. doi: 10.1016/j.bbrc.2017.07.137. [DOI] [PubMed] [Google Scholar]
- 99.Zhao L, Guo H, Zhou B, Feng J, Li Y, Han T, Liu L, Li L, Zhang S, Liu Y, et al. Long non-coding RNA SNHG5 suppresses gastric cancer progression by trapping MTA2 in the cytosol. Oncogene. 2016;35:5770–5780. doi: 10.1038/onc.2016.110. [DOI] [PubMed] [Google Scholar]
- 100.Li H, Zhu H, Zhou Y, Wang H, Niu Z, Shen Y, Lv L. Long non-coding RNA MSTO2P promotes the proliferation and colony formation in gastric cancer by indirectly regulating miR-335 expression. Tumour Biol. 2017;39:1010428317705506. doi: 10.1177/1010428317705506. [DOI] [PubMed] [Google Scholar]
- 101.Zhang LL, Zhang LF, Guo XH, Zhang DZ, Yang F, Fan YY. Downregulation of miR-335-5p by Long Noncoding RNA ZEB1-AS1 in Gastric Cancer Promotes Tumor Proliferation and Invasion. DNA Cell Biol. 2018;37:46–52. doi: 10.1089/dna.2017.3926. [DOI] [PubMed] [Google Scholar]
- 102.Zhang R, Guo Y, Ma Z, Ma G, Xue Q, Li F, Liu L. Long non-coding RNA PTENP1 functions as a ceRNA to modulate PTEN level by decoying miR-106b and miR-93 in gastric cancer. Oncotarget. 2017;8:26079–26089. doi: 10.18632/oncotarget.15317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sun W, Mo X, Li T, Xie Y, Guo J. Clinical significance of the long noncoding RNA RP11-19P22.6-001 in gastric cancer. Cancer Biomark. 2017;18:397–403. doi: 10.3233/CBM-160264. [DOI] [PubMed] [Google Scholar]
- 104.Bi M, Yu H, Huang B, Tang C. Long non-coding RNA PCAT-1 over-expression promotes proliferation and metastasis in gastric cancer cells through regulating CDKN1A. Gene. 2017;626:337–343. doi: 10.1016/j.gene.2017.05.049. [DOI] [PubMed] [Google Scholar]
- 105.Zheng L, Chen J, Zhou Z, He Z. Knockdown of long non-coding RNA HOXD-AS1 inhibits gastric cancer cell growth via inactivating the JAK2/STAT3 pathway. Tumour Biol. 2017;39:1010428317705335. doi: 10.1177/1010428317705335. [DOI] [PubMed] [Google Scholar]
- 106.Liu JN, Shangguan YM. Long non-coding RNA CARLo-5 upregulation associates with poor prognosis in patients suffering gastric cancer. Eur Rev Med Pharmacol Sci. 2017;21:530–534. [PubMed] [Google Scholar]
- 107.Huang M, Hou J, Wang Y, Xie M, Wei C, Nie F, Wang Z, Sun M. Long Noncoding RNA LINC00673 Is Activated by SP1 and Exerts Oncogenic Properties by Interacting with LSD1 and EZH2 in Gastric Cancer. Mol Ther. 2017;25:1014–1026. doi: 10.1016/j.ymthe.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 108.Fei ZH, Yu XJ, Zhou M, Su HF, Zheng Z, Xie CY. Upregulated expression of long non-coding RNA LINC00982 regulates cell proliferation and its clinical relevance in patients with gastric cancer. Tumour Biol. 2016;37:1983–1993. doi: 10.1007/s13277-015-3979-9. [DOI] [PubMed] [Google Scholar]
- 109.Shao Y, Chen H, Jiang X, Chen S, Li P, Ye M, Li Q, Sun W, Guo J. Low expression of lncRNA-HMlincRNA717 in human gastric cancer and its clinical significances. Tumour Biol. 2014;35:9591–9595. doi: 10.1007/s13277-014-2243-z. [DOI] [PubMed] [Google Scholar]
- 110.Huang T, Liu HW, Chen JQ, Wang SH, Hao LQ, Liu M, Wang B. The long noncoding RNA PVT1 functions as a competing endogenous RNA by sponging miR-186 in gastric cancer. Biomed Pharmacother. 2017;88:302–308. doi: 10.1016/j.biopha.2017.01.049. [DOI] [PubMed] [Google Scholar]
- 111.Shen W, Yuan Y, Zhao M, Li J, Xu J, Lou G, Zheng J, Bu S, Guo J, Xi Y. Novel long non-coding RNA GACAT3 promotes gastric cancer cell proliferation through the IL-6/STAT3 signaling pathway. Tumour Biol. 2016;37:14895–14902. doi: 10.1007/s13277-016-5372-8. [DOI] [PubMed] [Google Scholar]
- 112.Chang S, Liu J, Guo S, He S, Qiu G, Lu J, Wang J, Fan L, Zhao W, Che X. HOTTIP and HOXA13 are oncogenes associated with gastric cancer progression. Oncol Rep. 2016;35:3577–3585. doi: 10.3892/or.2016.4743. [DOI] [PubMed] [Google Scholar]
- 113.Fu JW, Kong Y, Sun X. Long noncoding RNA NEAT1 is an unfavorable prognostic factor and regulates migration and invasion in gastric cancer. J Cancer Res Clin Oncol. 2016;142:1571–1579. doi: 10.1007/s00432-016-2152-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ma Y, Liu L, Yan F, Wei W, Deng J, Sun J. Enhanced expression of long non-coding RNA NEAT1 is associated with the progression of gastric adenocarcinomas. World J Surg Oncol. 2016;14:41. doi: 10.1186/s12957-016-0799-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lai Y, Xu P, Li Q, Ren D, Wang J, Xu K, Gao W. Downregulation of long noncoding RNA ZMAT1 transcript variant 2 predicts a poor prognosis in patients with gastric cancer. Int J Clin Exp Pathol. 2015;8:5556–5562. [PMC free article] [PubMed] [Google Scholar]
- 116.Yang H, Liu Z, Yuan C, Zhao Y, Wang L, Hu J, Xie D, Wang L, Chen D. Elevated JMJD1A is a novel predictor for prognosis and a potential therapeutic target for gastric cancer. Int J Clin Exp Pathol. 2015;8:11092–11099. [PMC free article] [PubMed] [Google Scholar]
- 117.Dang Y, Lan F, Ouyang X, Wang K, Lin Y, Yu Y, Wang L, Wang Y, Huang Q. Expression and clinical significance of long non-coding RNA HNF1A-AS1 in human gastric cancer. World J Surg Oncol. 2015;13:302. doi: 10.1186/s12957-015-0706-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li L, Zhang L, Zhang Y, Zhou F. Increased expression of LncRNA BANCR is associated with clinical progression and poor prognosis in gastric cancer. Biomed Pharmacother. 2015;72:109–112. doi: 10.1016/j.biopha.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 119.Shan T, Fan J, Zhao Q, Deng K, Xia J. Upregulation of long non-coding RNA DQ786243 promotes the progression of gastric cancer. Mol Med Rep. 2017;16:3761–3768. doi: 10.3892/mmr.2017.7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Song W, Liu YY, Peng JJ, Liang HH, Chen HY, Chen JH, He WL, Xu JB, Cai SR, He YL. Identification of differentially expressed signatures of long non-coding RNAs associated with different metastatic potentials in gastric cancer. J Gastroenterol. 2016;51:119–129. doi: 10.1007/s00535-015-1091-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang CY, Hua L, Yao KH, Chen JT, Zhang JJ, Hu JH. Long non-coding RNA CCAT2 is up-regulated in gastric cancer and associated with poor prognosis. Int J Clin Exp Pathol. 2015;8:779–785. [PMC free article] [PubMed] [Google Scholar]
- 122.Wu SW, Hao YP, Qiu JH, Zhang DB, Yu CG, Li WH. High expression of long non-coding RNA CCAT2 indicates poor prognosis of gastric cancer and promotes cell proliferation and invasion. Minerva Med. 2017;108:317–323. doi: 10.23736/S0026-4806.17.04703-6. [DOI] [PubMed] [Google Scholar]
- 123.Hu Y, Pan J, Wang Y, Li L, Huang Y. Long noncoding RNA linc-UBC1 is negative prognostic factor and exhibits tumor pro-oncogenic activity in gastric cancer. Int J Clin Exp Pathol. 2015;8:594–600. [PMC free article] [PubMed] [Google Scholar]
- 124.Chen WM, Huang MD, Kong R, Xu TP, Zhang EB, Xia R, Sun M, De W, Shu YQ. Antisense Long Noncoding RNA HIF1A-AS2 Is Upregulated in Gastric Cancer and Associated with Poor Prognosis. Dig Dis Sci. 2015;60:1655–1662. doi: 10.1007/s10620-015-3524-0. [DOI] [PubMed] [Google Scholar]
- 125.Zhou B, Jing XY, Wu JQ, Xi HF, Lu GJ. Down-regulation of long non-coding RNA LET is associated with poor prognosis in gastric cancer. Int J Clin Exp Pathol. 2014;7:8893–8898. [PMC free article] [PubMed] [Google Scholar]
- 126.Xu MD, Qi P, Weng WW, Shen XH, Ni SJ, Dong L, Huang D, Tan C, Sheng WQ, Zhou XY, et al. Long non-coding RNA LSINCT5 predicts negative prognosis and exhibits oncogenic activity in gastric cancer. Medicine (Baltimore) 2014;93:e303. doi: 10.1097/MD.0000000000000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Xu C, Shao Y, Xia T, Yang Y, Dai J, Luo L, Zhang X, Sun W, Song H, Xiao B, et al. lncRNA-AC130710 targeting by miR-129-5p is upregulated in gastric cancer and associates with poor prognosis. Tumour Biol. 2014;35:9701–9706. doi: 10.1007/s13277-014-2274-5. [DOI] [PubMed] [Google Scholar]
- 128.Liu L, Yan B, Yang Z, Zhang X, Gu Q, Yue X. ncRuPAR inhibits gastric cancer progression by down-regulating protease-activated receptor-1. Tumour Biol. 2014;35:7821–7829. doi: 10.1007/s13277-014-2042-6. [DOI] [PubMed] [Google Scholar]
- 129.Li H, Yu B, Li J, Su L, Yan M, Zhu Z, Liu B. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5:2318–2329. doi: 10.18632/oncotarget.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhang EB, Han L, Yin DD, Kong R, De W, Chen J. c-Myc-induced, long, noncoding H19 affects cell proliferation and predicts a poor prognosis in patients with gastric cancer. Med Oncol. 2014;31:914. doi: 10.1007/s12032-014-0914-7. [DOI] [PubMed] [Google Scholar]
- 131.Sun W, Wu Y, Yu X, Liu Y, Song H, Xia T, Xiao B, Guo J. Decreased expression of long noncoding RNA AC096655.1-002 in gastric cancer and its clinical significance. Tumour Biol. 2013;34:2697–2701. doi: 10.1007/s13277-013-0821-0. [DOI] [PubMed] [Google Scholar]
- 132.Mei D, Song H, Wang K, Lou Y, Sun W, Liu Z, Ding X, Guo J. Up-regulation of SUMO1 pseudogene 3 (SUMO1P3) in gastric cancer and its clinical association. Med Oncol. 2013;30:709. doi: 10.1007/s12032-013-0709-2. [DOI] [PubMed] [Google Scholar]
- 133.Lu Y, Lu P, Zhu Z, Xu H, Zhu X. Loss of imprinting of insulin-like growth factor 2 is associated with increased risk of lymph node metastasis and gastric corpus cancer. J Exp Clin Cancer Res. 2009;28:125. doi: 10.1186/1756-9966-28-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Li X, Zhou Y, Qian H. CCAT1 expressed in malignant and pre-malignant human gastric tissues. Cell Mol Biol (Noisy-le-grand) 2017;63:89–93. doi: 10.14715/cmb/2017.63.5.16. [DOI] [PubMed] [Google Scholar]


