The naphthoquinone unit in 2-hydroxy-3-(prop-2-yn-1-yl)naphthalene-1,4-dione is essentially planar and the linear propargyl group is nearly perpendicular to the naphthalene ring system. In the crystal, O—H⋯O and C—H⋯O hydrogen bonds form an infinite tape structure.
Keywords: crystal structure; naphthoquinones; 2-hydroxy-3-(prop-2-yn-1-yl)naphthalene-1,4-dione
Abstract
The naphthoquinone unit of the title compound, C13H8O3, is essentially planar, with an r.m.s. deviation of 0.013 Å for the non-H atoms. The essentially linear propargyl group is tilted by ca 113° relative to the naphthoquinone plane. In the crystal, molecules are linked via a pair of O—H⋯O hydrogen bonds, forming an inversion dimer. The dimers are further linked via pairs of C—H⋯O hydrogen bonds into a tape structure along [20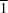 ]. No π–π stacking is observed in the present case as it could be expected for naphthoquinone derivatives.
]. No π–π stacking is observed in the present case as it could be expected for naphthoquinone derivatives.
Chemical context
Lawsone (2-hydroxynaphtalene-1,4-dione), 1, shows promising in the synthesis of analogues of atovaquone, 2, an antimalarial drug (Nixon et al., 2013 ▸) also used in immunosuppressed patients affected by pneumonia caused by Pneumocystis carinii (Cirioni et al., 1995 ▸; Comley et al., 1995 ▸). Recent studies have shown that it can be also useful in the fight against cancer (Fiorillo et al., 2016 ▸; Ashton et al., 2016 ▸). Thus far unknown, 2-hydroxy-3-(prop-2-yn-1-yl)naphthalene-1,4-dione (3) was obtained in a two steps one-pot procedure by reacting 1 with propargyl iodide, prepared in situ from propargyl bromide and potassium iodide. It opens the possibility for the synthesis of triazoles at the C3 position of 1 by [2 + 3] alkyne–azide 1,3-dipolar cycloaddition enabling the preparation of 3-substituted lawsone derivatives with potential pharmacological activity, including atovaquone (2) analogues.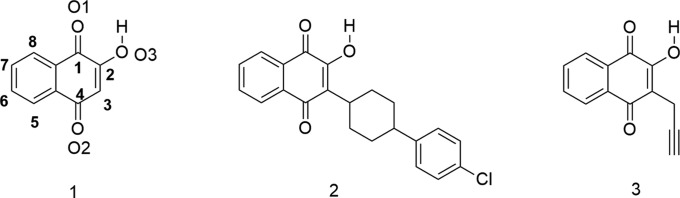
Treatment of 1 with a base leads to the formation of the corresponding enolate that can be O- or C-alkylated depending on the nature of the counter-ion, reaction conditions and nature of the alkyl electrophile (Jordão et al., 2015 ▸). When 1 was reacted with propargyl bromide and sodium carbonate in DMF the 2-O-propargyl derivative was obtained in 20% yield (Valença et al., 2017 ▸). The 3-C-propargyl derivative had not been described thus far. In view of the importance of acetylenic compounds for [2 + 3] alkyne–azide 1,3-dipolar cycloaddition reactions, known as the click reaction, we decided to investigate the 2-O- versus 3-C-propargylation of 1. The 3-C-propargyl derivative is considered to be an interesting intermediate for the synthesis of 3-triazolo analogues of atovaquone, 2, and other bioactive 1,4-naphthoquinones. After evaluating the influence of organic and inorganic bases, protic and aprotic solvents, alkylating agents, temperature and reaction time, we obtained 3 in 28% yield. No product of O-alkylation was observed in the reaction mixture.
Structural commentary
The molecular structure of the title compound, 3, is shown in Fig. 1 ▸. The naphthoquinone unit is essentially planar, with an r.m.s. deviation of 0.013 Å for the non-H atoms. The C—O bond lengths [C1—O1 = 1.2217 (18) Å, C2—O3 = 1.3412 (18) Å and C4—O2 = 1.2488 (19) Å] confirm the presence of 2-hydroxynaphthalene-1,4-dione in the crystalline state and are in agreement with the lengths found by Dekkers et al. (1996 ▸). The 1H and 13C NMR spectra and HMBC experiments confirm atoms C1 and C4 as carbonyls, as well as a hydroxy group at C2. The propargyl group adopts a nearly perpendicular position [C3—C11—C12 = 112.70 (14)°] regarding the naphthalene ring system to avoid hindrance with the O2 and O3 atoms. The naphthoquinone ring system is characterized by the torsion angles C4—C3—C11—C12 = −100.96 (19)° and C2—C3—C11—C12 = 79.9 (2)°.
Figure 1.
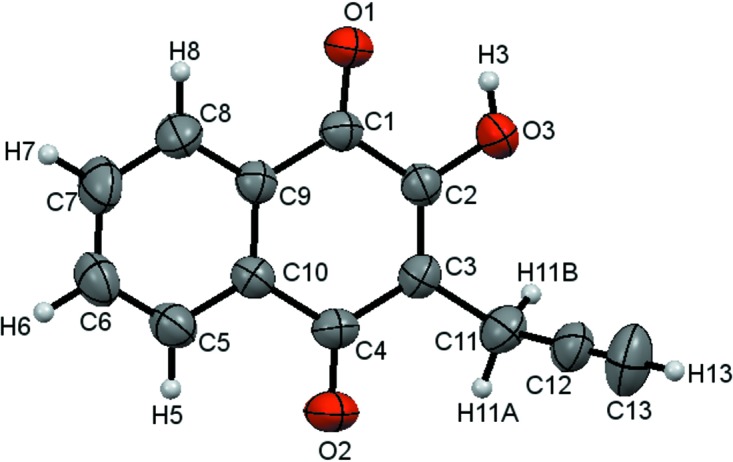
The molecular structure of the title compound 3. Displacement ellipsoids are drawn at the 50% probability level.
Supramolecular features
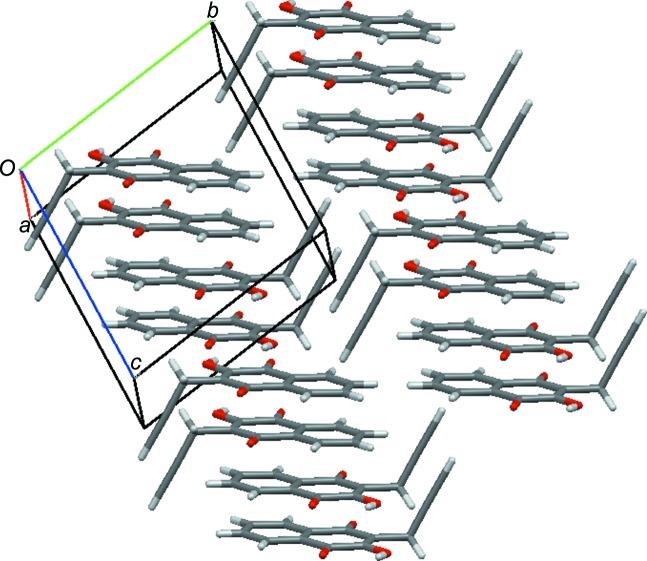
In the crystal, O—H⋯O and C—H⋯O hydrogen bonds (O3—H3⋯O1i and C5—H5⋯O2ii; symmetry codes as in Table 1 ▸) are responsible for an infinite tape structure running along [20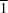 ]. All the naphthoquinone units are arranged in a parallel manner with respect to each other, as shown in Fig. 2 ▸. π–π stacking interactions are expected for naphthoquinone derivatives (Meyer et al., 2003 ▸). However, this type of interaction is not observed here, probably because of the C3 propargyl substituent.
]. All the naphthoquinone units are arranged in a parallel manner with respect to each other, as shown in Fig. 2 ▸. π–π stacking interactions are expected for naphthoquinone derivatives (Meyer et al., 2003 ▸). However, this type of interaction is not observed here, probably because of the C3 propargyl substituent.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O3—H3⋯O1i | 0.89 (3) | 2.06 (3) | 2.8118 (19) | 142 (3) |
| C5—H5⋯O2ii | 0.93 | 2.49 | 3.231 (2) | 137 |
Symmetry codes: (i) 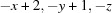 ; (ii)
; (ii) 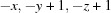 .
.
Figure 2.
A packing diagram of the title compound.
Database survey
A search of the Cambridge Structural Database (CSD, Version 5.39, last update May 2018; Groom et al., 2016 ▸) for 2-hydroxy-naphthalene-1,4-dione revealed 40 structures and approximately 787 structures which possess the naphthalene-1,4-dione moiety. 2-Hydroxy-3-(3-oxobutyl)naphthalene-1,4-dione (Nasiri et al., 2006 ▸) and 2-hydroxy-3-(methyl-prop-1-en-1-yl)naphthalene-1,4-dione (Alcantara Emiliano et al., 2016 ▸), compounds with structural similarity to the title compound, were also found. These compounds present a group linked to C3 with an angle nearly perpendicular to the naphthoquinone ring.
Synthesis and crystallization
The synthetic scheme is shown in Fig. 3 ▸. A mixture of propargyl bromide (0.75 ml, 4.47 mmol) and sodium iodide (1.30 g, 5.33 mmol) in dry acetone (3.5 ml) was stirred for 30 min at room temperature in a closed system. Then, a solution of lawsone (0.1 g, 2.4 mmol) and diisopropylethylamine (0.51 ml, 2.93 mmol) in a 2:1 (v/v) mixture of water/tert-butanol (24 ml) was added and the reaction mixture was stirred for a further 24 h at 353 K. The reaction was quenched with dichloromethane (ca 40 ml) and the heterogeneous mixture was transferred to a separatory funnel. The aqueous phase was separated and the organic layer was extracted with 1 mol l−1 hydrochloric acid (3 × 40 ml) and water (3 × 40 ml). The organic layer was dried over anhydrous sodium sulfate and concentrated to dryness. The crude red solid product (0.45 g) was purified by column chromatography (silica) using a 99.5:0.5 (v/v) mixture of hexane/tert-butanol containing 0.1% of acetic acid as eluent. Pure title compound was obtained in 28% yield (0.143 g, m.p. 396.7–397.2 K). Single crystals suitable for X-ray analysis were obtained by slow evaporation of a hexane/tert-butanol solution (ca 0.5 mg ml−1) at room temperature. The infrared and NMR spectral data and corresponding spectra of 3 are available in the supporting information.
Figure 3.
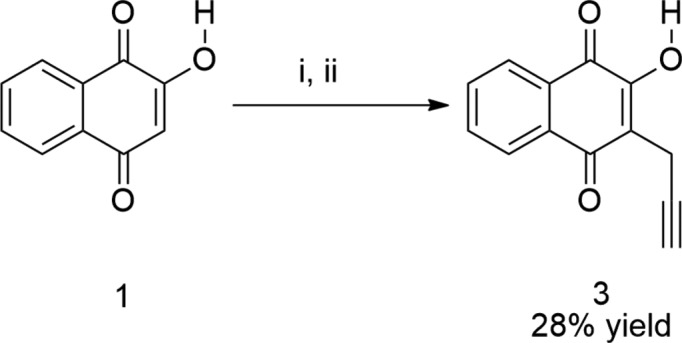
The synthetic scheme of the title compound, 3; (i) propargyl bromide, sodium iodide and dry acetone, 0.5 h; (ii) diisopropylethylamine and t-BuOH/H2O, 353 K, 24 h.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. C-bound H atoms were placed geometrically (C—H = 0.93–0.97 Å) and were refined as riding with U iso(H) = 1.2U eq(C). The O-bound H atom was located in a difference Fourier map and freely refined [O—H = 0.89 (3) Å].
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C13H8O3 |
| M r | 212.19 |
| Crystal system, space group | Triclinic, P
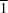
|
| Temperature (K) | 293 |
| a, b, c (Å) | 5.3695 (4), 9.5278 (8), 10.2972 (9) |
| α, β, γ (°) | 96.814 (7), 93.432 (7), 102.977 (7) |
| V (Å3) | 507.68 (8) |
| Z | 2 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.10 |
| Crystal size (mm) | 0.4 × 0.2 × 0.05 |
| Data collection | |
| Diffractometer | Rigaku Xcalibur Atlas Gemini ultra |
| Absorption correction | Multi-scan (CrysAlis PRO; Rigaku OD, 2015 ▸) |
| T min, T max | 0.720, 1.000 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 7946, 2508, 1563 |
| R int | 0.033 |
| (sin θ/λ)max (Å−1) | 0.695 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.051, 0.147, 1.05 |
| No. of reflections | 2508 |
| No. of parameters | 149 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.21, −0.5 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989018011647/is5498sup1.cif
Supporting information file. DOI: 10.1107/S2056989018011647/is5498Isup3.cml
The infrared and NMR spectroscopic data and corresponding spectra of <b>3</b>. DOI: 10.1107/S2056989018011647/is5498sup4.pdf
CCDC reference: 1862442
Additional supporting information: crystallographic information; 3D view; checkCIF report
supplementary crystallographic information
Crystal data
| C13H8O3 | F(000) = 220 |
| Mr = 212.19 | Dx = 1.388 Mg m−3 |
| Triclinic, P1 | Melting point = 396.8–397.5 K |
| a = 5.3695 (4) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 9.5278 (8) Å | Cell parameters from 1721 reflections |
| c = 10.2972 (9) Å | θ = 3.2–28.7° |
| α = 96.814 (7)° | µ = 0.10 mm−1 |
| β = 93.432 (7)° | T = 293 K |
| γ = 102.977 (7)° | Prism, colourless |
| V = 507.68 (8) Å3 | 0.4 × 0.2 × 0.05 mm |
| Z = 2 |
Data collection
| Rigaku Xcalibur Atlas Gemini ultra diffractometer | 2508 independent reflections |
| Radiation source: fine-focus sealed X-ray tube, Enhance (Mo) X-ray Source | 1563 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.033 |
| Detector resolution: 10.4186 pixels mm-1 | θmax = 29.6°, θmin = 2.8° |
| ω scans | h = −7→7 |
| Absorption correction: multi-scan (CrysAlis PRO; Rigaku OD, 2015) | k = −12→12 |
| Tmin = 0.720, Tmax = 1.000 | l = −13→14 |
| 7946 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: dual |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.051 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.147 | w = 1/[σ2(Fo2) + (0.0639P)2 + 0.058P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.05 | (Δ/σ)max < 0.001 |
| 2508 reflections | Δρmax = 0.21 e Å−3 |
| 149 parameters | Δρmin = −0.5 e Å−3 |
| 0 restraints |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O3 | 0.6461 (3) | 0.33269 (14) | 0.01419 (12) | 0.0536 (4) | |
| H3 | 0.796 (5) | 0.389 (3) | 0.002 (3) | 0.104 (10)* | |
| O1 | 0.9195 (2) | 0.59305 (13) | 0.12532 (12) | 0.0518 (3) | |
| O2 | 0.0447 (3) | 0.36386 (14) | 0.31887 (13) | 0.0647 (4) | |
| C1 | 0.7232 (3) | 0.54469 (17) | 0.17533 (15) | 0.0388 (4) | |
| C2 | 0.5615 (3) | 0.39989 (17) | 0.11881 (15) | 0.0403 (4) | |
| C3 | 0.3415 (3) | 0.33844 (17) | 0.16560 (15) | 0.0419 (4) | |
| C4 | 0.2483 (3) | 0.41556 (18) | 0.27733 (16) | 0.0431 (4) | |
| C5 | 0.3230 (4) | 0.6358 (2) | 0.44404 (17) | 0.0511 (4) | |
| H5 | 0.1693 | 0.5951 | 0.4767 | 0.061* | |
| C6 | 0.4664 (4) | 0.7695 (2) | 0.50175 (19) | 0.0586 (5) | |
| H6 | 0.4109 | 0.8182 | 0.5740 | 0.070* | |
| C7 | 0.6917 (4) | 0.8311 (2) | 0.4525 (2) | 0.0642 (6) | |
| H7 | 0.7871 | 0.9218 | 0.4915 | 0.077* | |
| C8 | 0.7782 (3) | 0.7597 (2) | 0.34557 (18) | 0.0530 (5) | |
| H8 | 0.9302 | 0.8021 | 0.3123 | 0.064* | |
| C9 | 0.6345 (3) | 0.62361 (16) | 0.28851 (15) | 0.0389 (4) | |
| C10 | 0.4061 (3) | 0.56178 (17) | 0.33796 (15) | 0.0397 (4) | |
| C11 | 0.1798 (4) | 0.19019 (18) | 0.10516 (18) | 0.0536 (5) | |
| H11A | 0.0054 | 0.1811 | 0.1290 | 0.064* | |
| H11B | 0.1751 | 0.1833 | 0.0103 | 0.064* | |
| C12 | 0.2779 (4) | 0.07067 (19) | 0.14761 (17) | 0.0552 (5) | |
| C13 | 0.3566 (5) | −0.0258 (2) | 0.1781 (2) | 0.0829 (7) | |
| H13 | 0.4196 | −0.1030 | 0.2026 | 0.099* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O3 | 0.0570 (8) | 0.0492 (7) | 0.0513 (7) | 0.0084 (6) | 0.0167 (6) | −0.0053 (5) |
| O1 | 0.0456 (7) | 0.0531 (7) | 0.0548 (7) | 0.0048 (6) | 0.0176 (6) | 0.0060 (5) |
| O2 | 0.0540 (8) | 0.0635 (9) | 0.0699 (9) | −0.0031 (6) | 0.0256 (7) | 0.0043 (7) |
| C1 | 0.0372 (9) | 0.0400 (8) | 0.0405 (8) | 0.0091 (7) | 0.0057 (7) | 0.0094 (6) |
| C2 | 0.0433 (9) | 0.0397 (9) | 0.0391 (9) | 0.0119 (7) | 0.0057 (7) | 0.0047 (6) |
| C3 | 0.0437 (9) | 0.0374 (8) | 0.0430 (9) | 0.0075 (7) | 0.0018 (7) | 0.0040 (7) |
| C4 | 0.0395 (9) | 0.0443 (9) | 0.0453 (9) | 0.0061 (7) | 0.0073 (7) | 0.0110 (7) |
| C5 | 0.0499 (10) | 0.0575 (11) | 0.0468 (10) | 0.0140 (8) | 0.0121 (8) | 0.0047 (8) |
| C6 | 0.0615 (12) | 0.0620 (12) | 0.0503 (10) | 0.0185 (10) | 0.0071 (9) | −0.0096 (8) |
| C7 | 0.0615 (13) | 0.0533 (11) | 0.0678 (13) | 0.0076 (10) | 0.0001 (10) | −0.0172 (9) |
| C8 | 0.0441 (10) | 0.0488 (10) | 0.0611 (11) | 0.0041 (8) | 0.0066 (8) | −0.0004 (8) |
| C9 | 0.0377 (9) | 0.0390 (8) | 0.0402 (8) | 0.0101 (7) | 0.0019 (7) | 0.0046 (6) |
| C10 | 0.0389 (9) | 0.0430 (9) | 0.0380 (8) | 0.0106 (7) | 0.0049 (7) | 0.0055 (7) |
| C11 | 0.0497 (10) | 0.0458 (10) | 0.0582 (11) | 0.0004 (8) | 0.0023 (9) | 0.0006 (8) |
| C12 | 0.0671 (12) | 0.0395 (10) | 0.0515 (10) | 0.0016 (9) | 0.0070 (9) | −0.0039 (8) |
| C13 | 0.113 (2) | 0.0483 (12) | 0.0862 (16) | 0.0257 (13) | −0.0048 (14) | 0.0002 (11) |
Geometric parameters (Å, º)
| O3—H3 | 0.88 (3) | C6—H6 | 0.9300 |
| O3—C2 | 1.3426 (19) | C6—C7 | 1.375 (3) |
| O1—C1 | 1.2217 (18) | C7—H7 | 0.9300 |
| O2—C4 | 1.2189 (19) | C7—C8 | 1.384 (3) |
| C1—C2 | 1.485 (2) | C8—H8 | 0.9300 |
| C1—C9 | 1.473 (2) | C8—C9 | 1.392 (2) |
| C2—C3 | 1.340 (2) | C9—C10 | 1.390 (2) |
| C3—C4 | 1.465 (2) | C11—H11A | 0.9700 |
| C3—C11 | 1.521 (2) | C11—H11B | 0.9700 |
| C4—C10 | 1.499 (2) | C11—C12 | 1.458 (3) |
| C5—H5 | 0.9300 | C12—C13 | 1.161 (3) |
| C5—C6 | 1.376 (3) | C13—H13 | 0.9300 |
| C5—C10 | 1.381 (2) | ||
| C2—O3—H3 | 106.5 (17) | C6—C7—C8 | 120.80 (18) |
| O1—C1—C2 | 118.90 (14) | C8—C7—H7 | 119.6 |
| O1—C1—C9 | 123.40 (15) | C7—C8—H8 | 120.5 |
| C9—C1—C2 | 117.69 (14) | C7—C8—C9 | 119.08 (17) |
| O3—C2—C1 | 115.90 (14) | C9—C8—H8 | 120.5 |
| C3—C2—O3 | 120.71 (15) | C8—C9—C1 | 120.23 (15) |
| C3—C2—C1 | 123.38 (14) | C10—C9—C1 | 119.71 (14) |
| C2—C3—C4 | 119.88 (15) | C10—C9—C8 | 120.07 (15) |
| C2—C3—C11 | 122.10 (15) | C5—C10—C4 | 119.53 (15) |
| C4—C3—C11 | 118.01 (15) | C5—C10—C9 | 119.69 (15) |
| O2—C4—C3 | 121.03 (16) | C9—C10—C4 | 120.78 (14) |
| O2—C4—C10 | 120.40 (15) | C3—C11—H11A | 109.1 |
| C3—C4—C10 | 118.56 (14) | C3—C11—H11B | 109.1 |
| C6—C5—H5 | 119.8 | H11A—C11—H11B | 107.8 |
| C6—C5—C10 | 120.37 (17) | C12—C11—C3 | 112.70 (14) |
| C10—C5—H5 | 119.8 | C12—C11—H11A | 109.1 |
| C5—C6—H6 | 120.0 | C12—C11—H11B | 109.1 |
| C7—C6—C5 | 119.98 (17) | C13—C12—C11 | 178.3 (2) |
| C7—C6—H6 | 120.0 | C12—C13—H13 | 180.0 |
| C6—C7—H7 | 119.6 | ||
| O1—C1—C2—O3 | −0.4 (2) | C4—C3—C11—C12 | −100.96 (19) |
| O1—C1—C2—C3 | −179.15 (16) | O2—C4—C10—C5 | 2.0 (3) |
| C9—C1—C2—O3 | 178.72 (14) | O2—C4—C10—C9 | −177.93 (16) |
| C9—C1—C2—C3 | −0.1 (2) | C3—C4—C10—C5 | −179.45 (16) |
| O1—C1—C9—C8 | −1.0 (2) | C3—C4—C10—C9 | 0.7 (2) |
| O1—C1—C9—C10 | 178.81 (15) | C10—C5—C6—C7 | 1.0 (3) |
| C2—C1—C9—C8 | 179.98 (14) | C6—C5—C10—C4 | 179.40 (17) |
| C2—C1—C9—C10 | −0.2 (2) | C6—C5—C10—C9 | −0.7 (3) |
| O3—C2—C3—C4 | −178.09 (15) | C5—C6—C7—C8 | −0.5 (3) |
| O3—C2—C3—C11 | 1.0 (2) | C6—C7—C8—C9 | −0.4 (3) |
| C1—C2—C3—C4 | 0.7 (2) | C7—C8—C9—C1 | −179.48 (16) |
| C1—C2—C3—C11 | 179.77 (15) | C7—C8—C9—C10 | 0.7 (3) |
| C2—C3—C4—O2 | 177.64 (16) | C1—C9—C10—C4 | −0.1 (2) |
| C2—C3—C4—C10 | −0.9 (2) | C1—C9—C10—C5 | −180.00 (16) |
| C11—C3—C4—O2 | −1.5 (2) | C8—C9—C10—C4 | 179.73 (15) |
| C11—C3—C4—C10 | 179.92 (15) | C8—C9—C10—C5 | −0.2 (2) |
| C2—C3—C11—C12 | 79.9 (2) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O3—H3···O1i | 0.89 (3) | 2.06 (3) | 2.8118 (19) | 142 (3) |
| C5—H5···O2ii | 0.93 | 2.49 | 3.231 (2) | 137 |
Symmetry codes: (i) −x+2, −y+1, −z; (ii) −x, −y+1, −z+1.
Funding Statement
This work was funded by Conselho Nacional de Desenvolvimento Científico e Tecnológico grant 303901/2017-9. Fundação de Amparo à Pesquisa do Estado de Minas Gerais grant CDS-APQ-02541-15.
References
- Alcantara Emiliano, S., Welma Duarte Silva, S., Alves Pereira, M., R. dos Santos Malta, V. & Luciano Balliano, T. (2016). Acta Cryst. E72, 188–190. [DOI] [PMC free article] [PubMed]
- Ashton, T. M., Fokas, E., Kunz-Schughart, L. A., Folkes, L. K., Anbalagan, S., Huether, M., Kelly, C. J., Pirovano, G., Buffa, F. M., Hammond, E. M., Stratford, M., Muschel, R. J., Higgins, G. S. & Mckenna, W. G. (2016). Nat. Commun. 7, 12308. [DOI] [PMC free article] [PubMed]
- Cirioni, O., Giacometti, A., Balducci, M., Burzacchini, F. & Scalise, G. (1995). J. Antimicrob. Chemother. 36, 740–741. [DOI] [PubMed]
- Comley, J. C. W., Yeates, C. L. & Frend, T. J. (1995). J. Antimicrob. Chemother. 39, 806–809. [DOI] [PMC free article] [PubMed]
- Dekkers, J., Kooijman, H., Kroon, J. & Grech, E. (1996). Acta Cryst. C52, 2896–2899.
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- Fiorillo, M., Lamb, R., Tanowitz, H. B., Mutti, L., Krstic-Demonacos, M., Cappello, A. R., Martinez-Outschoorn, U. E., Sotgia, F. & Lisanti, M. F. (2016). Oncotarget, 7, 34084–34099. [DOI] [PMC free article] [PubMed]
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Jordão, A. K., Vargas, M. D., Pinto, A. C., da Silva, F. de C. & Ferreira, V. F. (2015). RSC Adv. 5, 67909–67943.
- Meyer, E. A., Castellano, R. K. & Diederich, F. (2003). Angew. Chem. 42, 1210–1250. [DOI] [PubMed]
- Nasiri, H. R., Madej, M. G., Lancaster, C. R. D., Schwalbe, H. & Bolte, M. (2006). Acta Cryst. C62, o671–o673. [DOI] [PubMed]
- Nixon, G. L., Moss, D. M., Shone, A. E., Lalloo, D. G., Fisher, N., O’Neill, P. M., Ward, S. A. & Biagini, G. A. (2013). J. Antimicrob. Chemother. 68, 977–985. [DOI] [PMC free article] [PubMed]
- Rigaku OD (2015). CrysAlis PRO. Rigaku Oxford Diffaction, Yarnton, Oxfordshire, England.
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Valença, W. O., Baiju, T. V., Brito, F. G., Araujo, M. H., Pessoa, C., Cavalcanti, B. C., Simore, C. A., Jacob, C., Namboothiri, I. N. N. & da Silva Júnior, E. N. (2017). ChemistrySelect, 2, 4301–4308.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989018011647/is5498sup1.cif
Supporting information file. DOI: 10.1107/S2056989018011647/is5498Isup3.cml
The infrared and NMR spectroscopic data and corresponding spectra of <b>3</b>. DOI: 10.1107/S2056989018011647/is5498sup4.pdf
CCDC reference: 1862442
Additional supporting information: crystallographic information; 3D view; checkCIF report