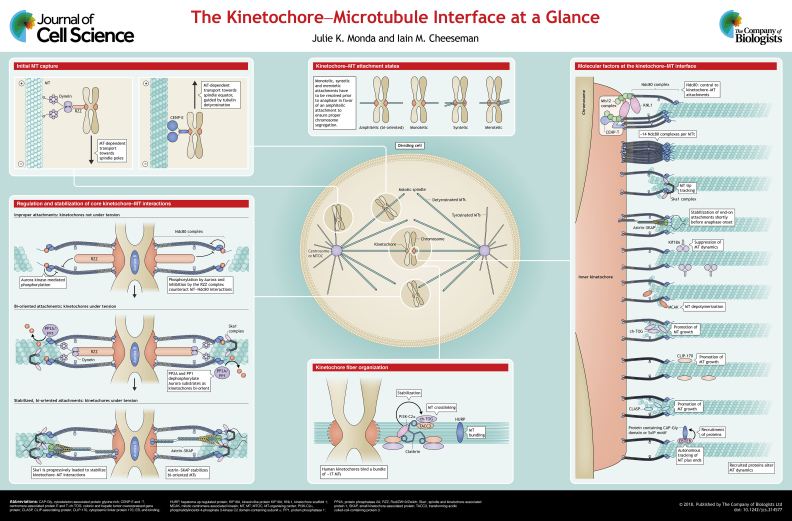
ABSTRACT
Accurate chromosome segregation critically depends on the formation of attachments between microtubule polymers and each sister chromatid. The kinetochore is the macromolecular complex that assembles at the centromere of each chromosome during mitosis and serves as the link between the DNA and the microtubules. In this Cell Science at a Glance article and accompanying poster, we discuss the activities and molecular players that are involved in generating kinetochore–microtubule attachments, including the initial stages of lateral kinetochore–microtubule interactions and maturation to stabilized end-on attachments. We additionally explore the features that contribute to the ability of the kinetochore to track with dynamic microtubules. Finally, we examine the contributions of microtubule-associated proteins to the organization and stabilization of the mitotic spindle and the control of microtubule dynamics.
KEY WORDS: Chromosome, Kinetochore, Microtubule, Mitosis
Summary: Accurate chromosome segregation during cell division relies on attachments between the kinetochores and mitotic spindle microtubules. Here, we discuss the critical players at the kinetochore–microtubule interface.
Introduction
Cell division is a fundamental process that is carried out by all organisms. During mitosis, the genetic material of each cell must be evenly distributed between both resulting daughter cells. The loss or gain of even a single chromosome can result in catastrophic consequences for the organism (Schukken and Foijer, 2018). To ensure the accurate distribution of the DNA in eukaryotes, a large macromolecular complex termed the kinetochore assembles onto the centromere of each chromosome during mitosis. The architecture of the kinetochore will not be extensively discussed in this Cell Science at a Glance article, but has been the topic of several recent review articles (Hara and Fukagawa, 2018; Hinshaw and Harrison, 2018; Joglekar and Kukreja, 2017; Musacchio and Desai, 2017; Nagpal and Fukagawa, 2016; Pesenti et al., 2016). Instead, we will focus on the interface between the kinetochore and microtubules, dynamic polymers of tubulin heterodimers. This kinetochore–microtubule interface is critically important, as depolymerization of kinetochore-associated microtubules ultimately provides the driving force for chromosome segregation (Musacchio and Desai, 2017).
Successfully harnessing the force released by microtubule depolymerization and transducing that force to the DNA requires a sufficiently stable kinetochore–microtubule attachment. The generation of a stable microtubule attachment is not a trivial task. Within the vast expanse of the cytoplasm, the kinetochore must locate and bind to microtubules. If even a single kinetochore lacks microtubule attachments, the spindle assembly checkpoint will prevent anaphase onset (Rieder et al., 1995). Furthermore, the kinetochores of each sister chromatid must bind to microtubules that emanate from opposing poles of the bipolar spindle to achieve a state termed bi-orientation. Kinetochore–microtubule interactions are therefore initially highly dynamic to facilitate correction of erroneous attachments. Once bi-orientation is achieved, the kinetochore–microtubule attachment must be stabilized for force transduction, yet also remain sufficiently dynamic so as to maintain its association even as the microtubule polymerizes and depolymerizes. In this article and accompanying poster, we discuss the features and molecular players that have key roles in overcoming these challenges in order to facilitate the formation of robust kinetochore–microtubule interactions. We focus on the kinetochore-localized proteins that bind microtubules, as well as proteins that alter the dynamics and organization of kinetochore-bound microtubules.
Microtubule capture and lateral-to-end-on conversion
Accurate chromosome segregation depends on the generation of end-on kinetochore–microtubule interactions where the plus-end of the microtubule is embedded within the kinetochore. However, some kinetochores will initially associate with the side of the microtubule, rather than the end (Barisic et al., 2014; Kapoor et al., 2006; Magidson et al., 2011; Tanaka et al., 2005). These lateral associations are mediated by one of two kinetochore-localized, microtubule-based motors – cytoplasmic dynein and centromere-associated protein E (CENP-E). As a minus-end-directed motor, dynein transports chromosomes towards the spindle pole (Li et al., 2007; Vorozhko et al., 2008; Yang et al., 2007) and thereby towards a region of high microtubule density. Chromosome congression is then promoted by the plus-end directed activity of CENP-E (McEwen et al., 2001) (see poster). During mitosis, microtubule plus-ends are located at both the cell cortex and equator (Prosser and Pelletier, 2017). The directionality of CENP-E-driven chromosome transport is guided by tubulin detyrosination (Barisic et al., 2015), a modification that is found to be enriched in the equator-oriented microtubules of the mitotic spindle and depleted in the cortical microtubules (Gundersen and Bulinski, 1986). In this way, the combined actions of dynein and CENP-E help to ensure timely chromosome alignment, and also promote the formation of end-on attachments by ensuring incorporation of the chromosomes into the spindle (Itoh et al., 2018) (see poster).
To generate the initial lateral microtubule interactions, kinetochores expand their reach by forming a structure that is termed the fibrous corona (Jokelainen, 1967; Magidson et al., 2015; McEwen et al., 1993; Rieder, 1982). A subset of outer kinetochore proteins, including dynein (Wordeman et al., 1991), CENP-E (Cooke et al., 1997), CENP-F (Rattner et al., 1993; Zhu et al., 1995) and the Rod–ZW10–Zwilch (RZZ) complex (Rod is also known as kinetochore-associated protein 1; KNTC1) (Basto et al., 2004; Starr et al., 1998), form an extended crescent-shaped structure that surrounds the kinetochore in the absence of microtubules (Dong et al., 2007; Echeverri et al., 1996; Hoffman et al., 2001; Thrower et al., 1996), thereby creating a large platform to capture microtubules. In Xenoups oocytes, an even larger expansion that includes more kinetochore proteins has also been recently described (Wynne and Funabiki, 2015).
The eventual conversion from a lateral to an end-on attachment is regulated by the counteracting functions of Aurora B kinase and protein phosphatase 2A (PP2A), and facilitated by the complex between Astrin (also known as SPAG5) and small kinetochore-associated protein (SKAP; also known as KNSTRN) (Shrestha et al., 2017) (Astrin–SKAP complex; see poster). Although geometrically distinct from the final goal of end-on microtubule attachments, the early establishment of kinetochore–microtubule interactions sets the stage for the subsequent mitotic events and helps facilitate chromosome alignment at the metaphase plate.
Core kinetochore–microtubule interactions
Mature end-on microtubule attachments are required for chromosome bi-orientation and satisfaction of the spindle assembly checkpoint (Kuhn and Dumont, 2017). The central player in the formation of these stable attachments is the Ndc80 complex, a component of the kinetochore scaffold 1 (KNL1)/Mis12/Ndc80 (KMN) network. The KMN network serves as the key link between the microtubules and the DNA (see poster), as it also binds the DNA-associated inner kinetochore proteins CENP-C and CENP-T (Gascoigne et al., 2011; Huis In ‘t Veld et al., 2016; Kim and Yu, 2015; Malvezzi et al., 2013; Nishino et al., 2013; Przewloka et al., 2011; Rago et al., 2015; Schleiffer et al., 2012; Screpanti et al., 2011). CENP-T further acts as a platform to expand the microtubule-binding capacity of the kinetochore, as a single CENP-T protein can recruit two Ndc80 complexes in addition to an entire complement of the KMN network (Huis In‘t Veld et al., 2016; Pekgoz Altunkaya et al., 2016; Rago et al., 2015). The precise stoichiometry of Ndc80 and the other components of the kinetochore–microtubule interface are likely to be central to the structure, organization and functionality of these interactions. However, for simplicity, the accompanying poster primarily focuses on the core concepts, rather than attempting to define the relative and absolute molecular numbers.
To ensure proper microtubule interactions, the KMN network is regulated by a variety of mechanisms. For example, Ndc80 binding to microtubules is negatively regulated by the RZZ complex in early mitosis, and that inhibition is relieved by recruitment of dynein to the kinetochore (Cheerambathur et al., 2013). The RZZ complex and dynein show dynamic localization to kinetochores, with RZZ localization being highest at nuclear envelope breakdown, and dynein localization being highest later in prometaphase (Itoh et al., 2018). In this way, RZZ-mediated inhibition of Ndc80 likely prevents the formation of stable end-on attachments early in mitosis when there is a high frequency of incorrect kinetochore–microtubule interactions (Cheerambathur et al., 2013), such as syntelic attachments – where both sister kinetochores are attached to the same pole or merotelic attachments – where a single kinetochore attaches to microtubules from both spindle poles (see poster).
In addition to regulation by RZZ, Ndc80 is also negatively regulated by phosphorylation by mitotic kinases, including Aurora A and Aurora B (Cheeseman et al., 2002; Chmatal et al., 2015; DeLuca et al., 2006; Shrestha et al., 2017; Ye et al., 2015). Aurora-mediated phosphorylation decreases the affinity of Ndc80 for microtubules (Cheeseman et al., 2006), thereby allowing for correction of aberrant kinetochore–microtubule interactions. Consistent with this, Ndc80 phosphorylation is highest in prometaphase and decreases in metaphase through the combined actions of two phosphatases, PP1 and the PP2A-B56 holoenzyme (Liu et al., 2010; Posch et al., 2010; Schleicher et al., 2017). However, recent work has also suggested that a subset of phosphorylation sites are maintained by Aurora A throughout mitosis to ensure proper microtubule dynamics (DeLuca et al., 2017). Thus, the proper segregation of the DNA relies on precise control of the microtubule-binding activity of the KMN network.
Dynamic microtubule tip tracking
During mitosis, the kinetochore needs to not only establish end-on microtubule attachments, but also maintain those attachments while the microtubule grows and shrinks. Indeed, associations with dynamic microtubules contribute to multiple aspects of mitosis. For example, in addition to motor-driven chromosome congression, as discussed above, depolymerization-coupled pulling on kinetochores also contributes to chromosome congression and relies on the ability of the kinetochore to maintain its association with a depolymerizing microtubule (Auckland and McAinsh, 2015). Additionally, chromosomes undergo oscillations during metaphase (Jaqaman et al., 2010; Skibbens et al., 1993), thereby requiring one sister chromatid to associate with depolymerizing microtubules while the other sister chromatid associates with elongating microtubules. Finally, chromosome segregation during anaphase is driven by the association of kinetochores with depolymerizing microtubules (Musacchio and Desai, 2017).
Although critically required for the formation of stable microtubule attachments, the role of Ndc80 in microtubule tip tracking is less clear. In vitro, Ndc80 complexes are unable to track depolymerizing microtubules (Schmidt et al., 2012) unless the complex is artificially oligomerized (McIntosh et al., 2008; Powers et al., 2009; Volkov et al., 2018). It is unclear how well this oligomerization mimics the organization of the ∼14 Ndc80 complexes that bind to each kinetochore microtubule in human cells (Suzuki et al., 2015), but it suggests that Ndc80 contributes to the associations with dynamic microtubules. Indeed, the phosphorylation state of the Ndc80 complex tunes the association of kinetochores with elongating microtubules in vivo (Long et al., 2017).
In fungi, the ring-like Dam1 complex facilitates processive microtubule interactions by binding both Ndc80 and the microtubule, and sliding along the microtubule as protofilaments peel away during depolymerization (Grishchuk et al., 2008; Lampert et al., 2010; Miranda et al., 2005; Tien et al., 2010; Westermann et al., 2005). Interestingly, this mechanism is not widely conserved, as metazoans do not contain homologs of the Dam1 complex (van Hooff et al., 2017). Instead, in species lacking the Dam1 complex, the spindle and kinetochore-associated protein 1 (Ska1) complex likely serves as a functional analog (Gaitanos et al., 2009; Welburn et al., 2009), although the Ska1 complex does not form a ring-like structure (Monda et al., 2017; Schmidt et al., 2012; Welburn et al., 2009).
Ska1 is recruited to the kinetochore by the Ndc80 complex (Cheerambathur et al., 2017; Janczyk et al., 2017), and the phosphatases PP1 and PP2A also promote accumulation of Ska1 at aligned kinetochores (Sivakumar and Gorbsky, 2017) (see poster). Indeed, the levels of kinetochore-localized Ska1 increase throughout congression, thereby enhancing the ability of mature kinetochore–microtubule attachments to sustain load-bearing forces (Auckland et al., 2017). In vitro, Ska1 autonomously tracks both depolymerizing and elongating microtubules (Monda et al., 2017; Schmidt et al., 2012), supporting the model that Ska1 contributes to the ability of the kinetochore to generally associate with dynamic microtubules (Helgeson et al., 2018).
In addition to Ska1, other factors may contribute to dynamic microtubule interactions. For example, CENP-F also tracks depolymerizing microtubules in vitro (Volkov et al., 2015). Taken together, the integrated microtubule-binding activities of Ska1, the Ndc80 complex and perhaps other proteins create the dynamic interface required for persistent association with microtubules (see poster).
Stabilization of end-on attachments
Cycles of microtubule polymerization and depolymerization generate forces that can be transmitted through the kinetochore to drive chromosome movement. Withstanding and harnessing that force requires the kinetochore–microtubule attachment to be sufficiently robust and stable. Although the Ndc80 complex is critical for the formation of microtubule attachments (Cheeseman et al., 2006; DeLuca et al., 2006), other microtubule-binding proteins play key roles in stabilizing and strengthening those microtubule interactions. First, the microtubule-bound Ska1 complex strengthens Ndc80-mediated microtubule interactions and is capable of bearing load (Cheerambathur et al., 2017; Helgeson et al., 2018), in addition to its role in tracking dynamic microtubules (Monda et al., 2017; Schmidt et al., 2012). Second, the Astrin–SKAP complex specifically localizes to microtubule-attached and bi-oriented kinetochores, where it binds to microtubules synergistically with Ndc80 (Kern et al., 2017) (see poster). Thus, the activities of Ndc80, Ska1, Astrin–SKAP and potentially other proteins, create a stable kinetochore–microtubule interface that is capable of withstanding the significant force exerted by the mitotic spindle.
Kinetochore-fiber organization
In most organisms, each kinetochore binds a bundle of multiple microtubules (∼17 in human cells; McEwen et al., 2001; Wendell et al., 1993), collectively referred to as a kinetochore fiber (denoted k-fiber). In contrast to other populations of mitotic microtubules, k-fibers are uniquely stable, as evidenced by their persistence after cold treatment (Salmon and Begg, 1980). This enhanced stability is the result of their plus-ends being embedded within kinetochores, and the cross-linking and bundling of adjacent microtubules. The complex comprising transforming complex acidic coiled-coil-containing protein 3 (TACC3), colonic and hepatic tumor overexpressed gene protein (ch-TOG; also known as CKAP5) and clathrin, stabilized by phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit α (PI3K-C2α; encoded by PIK3C2A) (Gulluni et al., 2017), crosslinks microtubules and contributes to the organization of the mitotic spindle (Booth et al., 2011; Nixon et al., 2017). K-fiber stabilization is also achieved through the microtubule-bundling activity of hepatoma up-regulated protein (HURP; also known as DLGAP5) (Koffa et al., 2006; Silljé et al., 2006; Wong and Fang, 2006) (see poster). Together, the stabilization and organization of k-fibers by these proteins ultimately allows for the generation of sufficient force to drive accurate chromosome segregation.
Control of microtubule dynamics
Chromosome congression, bi-orientation and segregation depend on precise regulation of the stability and dynamics of the microtubules in the mitotic spindle. This balance is achieved though the combined actions of numerous proteins that both positively and negatively regulate microtubule growth through diverse mechanisms. Regulators that localize at or near kinetochores include the kinesin-like protein Kif18A (Mayr et al., 2007), mitotic centromere-associated kinesin (MCAK; also known as KIF2C) (Wordeman and Mitchison, 1995), ch-TOG (Gergely et al., 2003), cytoplasmic linker protein 170 (CLIP-170; also known as CLIP1) (Dujardin et al., 1998), and the CLIP-associating protein (CLASP) (Maiato et al., 2003) and end-binding (EB or MAPRE) (Juwana et al., 1999) families of microtubule-binding proteins (see poster). Metaphase chromosome oscillations are regulated by the kinetochore-localized kinesin Kif18A, which suppresses microtubule dynamics (Du et al., 2010; Stumpff et al., 2008, 2012). At anaphase onset, Kif18A and other factors must be dephosphorylated to allow for a switch from oscillations to robust poleward movement of the separated sister chromatids (Su et al., 2016). MCAK is a kinesin-13 family member (Lawrence et al., 2004) and a microtubule depolymerase (Desai et al., 1999; Hunter et al., 2003). By promoting microtubule depolymerization at kinetochores that are not yet bi-oriented and stably associated with the microtubule, MCAK facilitates the correction of erroneous kinetochore–microtubule attachments and thereby increase the likelihood of achieving bi-orientation (Kline-Smith et al., 2004). The activity of MCAK is counteracted by ch-TOG, a highly processive microtubule polymerase (Brouhard et al., 2008). CLIP-170 and CLASPs also promote microtubule growth, with CLIP-170 promoting the transition from microtubule depolymerization to microtubule polymerization (Komarova et al., 2002) and CLASPs promoting the incorporation of tubulin subunits into k-fibers (Maiato et al., 2005). The EB family of proteins, comprising EB1, EB2 and EB3 (also known as MAPRE1, MAPRE2 and MAPRE3, respectively) in mammals (Su and Qi, 2001), are autonomous plus-end-tracking proteins (Bieling et al., 2007) that recruit a variety of other proteins that contain cytoskeleton-associated proteins (CAP)-Gly domains or SxIP motifs (Kumar and Wittmann, 2012) to affect microtubule growth or organization (Browning et al., 2003; Komarova et al., 2005; Mimori-Kiyosue et al., 2005; Niethammer et al., 2007; Su et al., 1995) (see poster).
The functions of these regulators of microtubule dynamics are also subject to regulatory control themselves. For example, MCAK is phosphorylated by numerous mitotic kinases (Andrews et al., 2004; Lan et al., 2004; Ohi et al., 2004; Sanhaji et al., 2010; Zhang et al., 2007, 2011). Recent work has demonstrated that CDK1-mediated phosphorylation of a single threonine residue in the MCAK motor domain is sufficient to block the ability of MCAK to distinguish the end of the microtubule from the lattice, and thereby reduce microtubule depolymerization (Belsham and Friel, 2017). Additionally, MCAK undergoes significant structural rearrangements upon binding a microtubule that allow for optimal depolymerase activity (Burns et al., 2014; Ems-McClung et al., 2013; Talapatra et al., 2015). Collectively, these diverse microtubule-associated proteins facilitate cell division by ensuring a dynamic mitotic spindle.
Conclusions and perspectives
During every cell division, numerous kinetochore-localized microtubule-binding proteins must act to ensure the accurate segregation of the DNA to the daughter cells. A stable, bipolar spindle must be built, the kinetochore of each sister chromatid must attach to microtubules emanating from one of the spindle poles, and each kinetochore must be capable of maintaining its microtubule attachment, despite elongation and depolymerization of the microtubule. Recent work has defined numerous molecular players that are involved in each of these key aspects of mitosis. Given the complexity of the kinetochore, it is perhaps unsurprising that many of these proteins have opposing activities: dynein and CENP-E transport chromosomes in opposite directions, Astrin–SKAP must stabilize the kinetochore–microtubule attachments yet also allow for dynamic interactions and Ska1-mediated tip-tracking, and MCAK destabilizes microtubules, whereas ch-TOG, CLIP-170 and CLASPs promote microtubule growth. A key goal for future studies will therefore be to uncover how the diverse activities of these molecules are integrated to ensure faithful chromosome segregation.
Acknowledgements
We thank Leah Bury, Nolan Maier, Lily McKay and Gunter Sissoko for critically reading and providing comments on this manuscript. We apologize to the many authors whose work we could not include due to space restrictions.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
Work in the Cheeseman laboratory is supported by grants from G. Harold and Leila Y. Mathers Charitable Foundation and the National Institute of General Medical Sciences (GM088313 and GM108718). Deposited in PMC for release after 12 months.
Cell science at a glance
A high-resolution version of the poster and individual poster panels are available for downloading at http://jcs.biologists.org/lookup/doi/10.1242/jcs.214577.supplemental
References
- Andrews P. D., Ovechkina Y., Morrice N., Wagenbach M., Duncan K., Wordeman L. and Swedlow J. R. (2004). Aurora B regulates MCAK at the mitotic centromere. Dev. Cell 6, 253-268. 10.1016/S1534-5807(04)00025-5 [DOI] [PubMed] [Google Scholar]
- Auckland P. and McAinsh A. D. (2015). Building an integrated model of chromosome congression. J. Cell Sci. 128, 3363-3374. 10.1242/jcs.169367 [DOI] [PubMed] [Google Scholar]
- Auckland P., Clarke N. I., Royle S. J. and McAinsh A. D. (2017). Congressing kinetochores progressively load Ska complexes to prevent force-dependent detachment. J. Cell Biol. 216, 1623-1639. 10.1083/jcb.201607096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barisic M., Aguiar P., Geley S. and Maiato H. (2014). Kinetochore motors drive congression of peripheral polar chromosomes by overcoming random arm-ejection forces. Nat. Cell Biol. 16, 1249-1256. 10.1038/ncb3060 [DOI] [PubMed] [Google Scholar]
- Barisic M., Silva e Sousa R., Tripathy S. K., Magiera M. M., Zaytsev A. V., Pereira A. L., Janke C., Grishchuk E. L. and Maiato H. (2015). Mitosis. Microtubule detyrosination guides chromosomes during mitosis. Science 348, 799-803. 10.1126/science.aaa5175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R., Scaerou F., Mische S., Wojcik E., Lefebvre C., Gomes R., Hays T. and Karess R. (2004). In vivo dynamics of the rough deal checkpoint protein during Drosophila mitosis. Curr. Biol. 14, 56-61. 10.1016/j.cub.2003.12.025 [DOI] [PubMed] [Google Scholar]
- Belsham H. R. and Friel C. T. (2017). A Cdk1 phosphomimic mutant of MCAK impairs microtubule end recognition. PeerJ 5, e4034 10.7717/peerj.4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling P., Laan L., Schek H., Munteanu E. L., Sandblad L., Dogterom M., Brunner D. and Surrey T. (2007). Reconstitution of a microtubule plus-end tracking system in vitro. Nature 450, 1100-1105. 10.1038/nature06386 [DOI] [PubMed] [Google Scholar]
- Booth D. G., Hood F. E., Prior I. A. and Royle S. J. (2011). A TACC3/ch-TOG/clathrin complex stabilises kinetochore fibres by inter-microtubule bridging. EMBO J. 30, 906-919. 10.1038/emboj.2011.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouhard G. J., Stear J. H., Noetzel T. L., Al-Bassam J., Kinoshita K., Harrison S. C., Howard J. and Hyman A. A. (2008). XMAP215 is a processive microtubule polymerase. Cell 132, 79-88. 10.1016/j.cell.2007.11.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning H., Hackney D. D. and Nurse P. (2003). Targeted movement of cell end factors in fission yeast. Nat. Cell Biol. 5, 812-818. 10.1038/ncb1034 [DOI] [PubMed] [Google Scholar]
- Burns K. M., Wagenbach M., Wordeman L. and Schriemer D. C. (2014). Nucleotide exchange in dimeric MCAK induces longitudinal and lateral stress at microtubule ends to support depolymerization. Structure 22, 1173-1183. 10.1016/j.str.2014.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheerambathur D. K., Gassmann R., Cook B., Oegema K. and Desai A. (2013). Crosstalk between microtubule attachment complexes ensures accurate chromosome segregation. Science 342, 1239-1242. 10.1126/science.1246232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheerambathur D. K., Prevo B., Hattersley N., Lewellyn L., Corbett K. D., Oegema K. and Desai A. (2017). Dephosphorylation of the Ndc80 tail stabilizes kinetochore-microtubule attachments via the Ska complex. Dev. Cell 41, 424-437 e424. 10.1016/j.devcel.2017.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I. M., Anderson S., Jwa M., Green E. M., Kang J., Yates J. R. III, Chan C. S., Drubin D. G. and Barnes G. (2002). Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell 111, 163-172. 10.1016/S0092-8674(02)00973-X [DOI] [PubMed] [Google Scholar]
- Cheeseman I. M., Chappie J. S., Wilson-Kubalek E. M. and Desai A. (2006). The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127, 983-997. 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- Chmatal L., Yang K., Schultz R. M. and Lampson M. A. (2015). Spatial regulation of kinetochore microtubule attachments by destabilization at spindle poles in meiosis I. Curr. Biol. 25, 1835-1841. 10.1016/j.cub.2015.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke C. A., Schaar B., Yen T. J. and Earnshaw W. C. (1997). Localization of CENP-E in the fibrous corona and outer plate of mammalian kinetochores from prometaphase through anaphase. Chromosoma 106, 446-455. 10.1007/s004120050266 [DOI] [PubMed] [Google Scholar]
- DeLuca J. G., Gall W. E., Ciferri C., Cimini D., Musacchio A. and Salmon E. D. (2006). Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell 127, 969-982. 10.1016/j.cell.2006.09.047 [DOI] [PubMed] [Google Scholar]
- DeLuca K. F., Meppelink A., Broad A. J., Mick J. E., Peersen O. B., Pektas S., Lens S. M. A. and DeLuca J. G. (2017). Aurora A kinase phosphorylates Hec1 to regulate metaphase kinetochore-microtubule dynamics. J. Cell Biol. 217, 163-177. 10.1083/jcb.201707160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A., Verma S., Mitchison T. J. and Walczak C. E. (1999). Kin I kinesins are microtubule-destabilizing enzymes. Cell 96, 69-78. 10.1016/S0092-8674(00)80960-5 [DOI] [PubMed] [Google Scholar]
- Dong Y., Vanden Beldt K. J., Meng X., Khodjakov A. and McEwen B. F. (2007). The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions. Nat. Cell Biol. 9, 516-522. 10.1038/ncb1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., English C. A. and Ohi R. (2010). The kinesin-8 Kif18A dampens microtubule plus-end dynamics. Curr. Biol. 20, 374-380. 10.1016/j.cub.2009.12.049 [DOI] [PubMed] [Google Scholar]
- Dujardin D., Wacker U. I., Moreau A., Schroer T. A., Rickard J. E. and De Mey J. R. (1998). Evidence for a role of CLIP-170 in the establishment of metaphase chromosome alignment. J. Cell Biol. 141, 849-862. 10.1083/jcb.141.4.849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverri C. J., Paschal B. M., Vaughan K. T. and Vallee R. B. (1996). Molecular characterization of the 50-kD subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J. Cell Biol. 132, 617-633. 10.1083/jcb.132.4.617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ems-McClung S. C., Hainline S. G., Devare J., Zong H., Cai S., Carnes S. K., Shaw S. L. and Walczak C. E. (2013). Aurora B inhibits MCAK activity through a phosphoconformational switch that reduces microtubule association. Curr. Biol. 23, 2491-2499. 10.1016/j.cub.2013.10.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaitanos T. N., Santamaria A., Jeyaprakash A. A., Wang B., Conti E. and Nigg E. A. (2009). Stable kinetochore-microtubule interactions depend on the Ska complex and its new component Ska3/C13Orf3. EMBO J. 28, 1442-1452. 10.1038/emboj.2009.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne K. E., Takeuchi K., Suzuki A., Hori T., Fukagawa T. and Cheeseman I. M. (2011). Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell 145, 410-422. 10.1016/j.cell.2011.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergely F., Draviam V. M. and Raff J. W. (2003). The ch-TOG/XMAP215 protein is essential for spindle pole organization in human somatic cells. Genes Dev. 17, 336-341. 10.1101/gad.245603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk E. L., Efremov A. K., Volkov V. A., Spiridonov I. S., Gudimchuk N., Westermann S., Drubin D., Barnes G., McIntosh J. R. and Ataullakhanov F. I. (2008). The Dam1 ring binds microtubules strongly enough to be a processive as well as energy-efficient coupler for chromosome motion. Proc. Natl. Acad. Sci. USA 105, 15423-15428. 10.1073/pnas.0807859105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulluni F., Martini M., De Santis M. C., Campa C. C., Ghigo A., Margaria J. P., Ciraolo E., Franco I., Ala U., Annaratone L. et al. (2017). Mitotic spindle assembly and genomic stability in breast cancer require PI3K-C2alpha scaffolding function. Cancer Cell 32, 444-459 e447. 10.1016/j.ccell.2017.09.002 [DOI] [PubMed] [Google Scholar]
- Gundersen G. G. and Bulinski J. C. (1986). Distribution of tyrosinated and nontyrosinated alpha-tubulin during mitosis. J. Cell Biol. 102, 1118-1126. 10.1083/jcb.102.3.1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara M. and Fukagawa T. (2018). Kinetochore assembly and disassembly during mitotic entry and exit. Curr. Opin. Cell Biol. 52, 73-81. 10.1016/j.ceb.2018.02.005 [DOI] [PubMed] [Google Scholar]
- Helgeson L. A., Zelter A., Riffle M., MacCoss M. J., Asbury C. L. and Davis T. N. (2018). Human Ska complex and Ndc80 complex interact to form a load-bearing assembly that strengthens kinetochore-microtubule attachments. Proc. Natl. Acad. Sci. USA. 115, 2740-2745. 10.1073/pnas.1718553115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw S. M. and Harrison S. C. (2018). Kinetochore function from the bottom up. Trends Cell Biol. 28, 22-33. 10.1016/j.tcb.2017.09.002 [DOI] [PubMed] [Google Scholar]
- Hoffman D. B., Pearson C. G., Yen T. J., Howell B. J. and Salmon E. D. (2001). Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol. Biol. Cell 12, 1995-2009. 10.1091/mbc.12.7.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huis In ‘t Veld P. J., Jeganathan S., Petrovic A., Singh P., John J., Krenn V., Weissmann F., Bange T. and Musacchio A. (2016). Molecular basis of outer kinetochore assembly on CENP-T. eLife 5, e21007 10.7554/eLife.21007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter A. W., Caplow M., Coy D. L., Hancock W. O., Diez S., Wordeman L. and Howard J. (2003). The kinesin-related protein MCAK is a microtubule depolymerase that forms an ATP-hydrolyzing complex at microtubule ends. Mol. Cell 11, 445-457. 10.1016/S1097-2765(03)00049-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh G., Ikeda M., Iemura K., Amin M. A., Kuriyama S., Tanaka M., Mizuno N., Osakada H., Haraguchi T. and Tanaka K. (2018). Lateral attachment of kinetochores to microtubules is enriched in prometaphase rosette and facilitates chromosome alignment and bi-orientation establishment. Sci. Rep. 8, 3888 10.1038/s41598-018-22164-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janczyk P. L., Skorupka K. A., Tooley J. G., Matson D. R., Kestner C. A., West T., Pornillos O. and Stukenberg P. T. (2017). Mechanism of Ska recruitment by Ndc80 complexes to kinetochores. Dev. Cell 41, 438-449 e434. 10.1016/j.devcel.2017.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaqaman K., King E. M., Amaro A. C., Winter J. R., Dorn J. F., Elliott H. L., McHedlishvili N., McClelland S. E., Porter I. M., Posch M. et al. (2010). Kinetochore alignment within the metaphase plate is regulated by centromere stiffness and microtubule depolymerases. J. Cell Biol. 188, 665-679. 10.1083/jcb.200909005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar A. P. and Kukreja A. A. (2017). How kinetochore architecture shapes the mechanisms of its function. Curr. Biol. 27, R816-R824. 10.1016/j.cub.2017.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokelainen P. T. (1967). The ultrastructure and spatial organization of the metaphase kinetochore in mitotic rat cells. J. Ultrastruct. Res. 19, 19-44. 10.1016/S0022-5320(67)80058-3 [DOI] [PubMed] [Google Scholar]
- Juwana J.-P., Henderikx P., Mischo A., Wadle A., Fadle N., Gerlach K., Arends J. W., Hoogenboom H., Pfreundschuh M. and Renner C. (1999). EB/RP gene family encodes tubulin binding proteins. Int. J. Cancer 81, 275-284. [DOI] [PubMed] [Google Scholar]
- Kapoor T. M., Lampson M. A., Hergert P., Cameron L., Cimini D., Salmon E. D., McEwen B. F. and Khodjakov A. (2006). Chromosomes can congress to the metaphase plate before biorientation. Science 311, 388-391. 10.1126/science.1122142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern D. M., Monda J. K., Su K. C., Wilson-Kubalek E. M. and Cheeseman I. M. (2017). Astrin-SKAP complex reconstitution reveals its kinetochore interaction with microtubule-bound Ndc80. eLife 6, e26866 10.7554/eLife.26866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. and Yu H. (2015). Multiple assembly mechanisms anchor the KMN spindle checkpoint platform at human mitotic kinetochores. J. Cell Biol. 208, 181-196. 10.1083/jcb.201407074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline-Smith S. L., Khodjakov A., Hergert P. and Walczak C. E. (2004). Depletion of centromeric MCAK leads to chromosome congression and segregation defects due to improper kinetochore attachments. Mol. Biol. Cell 15, 1146-1159. 10.1091/mbc.e03-08-0581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koffa M. D., Casanova C. M., Santarella R., Köcher T., Wilm M. and Mattaj I. W. (2006). HURP is part of a Ran-dependent complex involved in spindle formation. Curr. Biol. 16, 743-754. 10.1016/j.cub.2006.03.056 [DOI] [PubMed] [Google Scholar]
- Komarova Y. A., Akhmanova A. S., Kojima S., Galjart N. and Borisy G. G. (2002). Cytoplasmic linker proteins promote microtubule rescue in vivo. J. Cell Biol. 159, 589-599. 10.1083/jcb.200208058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova Y., Lansbergen G., Galjart N., Grosveld F., Borisy G. G. and Akhmanova A. (2005). EB1 and EB3 control CLIP dissociation from the ends of growing microtubules. Mol. Biol. Cell 16, 5334-5345. 10.1091/mbc.e05-07-0614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn J. and Dumont S. (2017). Spindle assembly checkpoint satisfaction occurs via end-on but not lateral attachments under tension. J. Cell Biol. 216, 1533-1542. 10.1083/jcb.201611104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P. and Wittmann T. (2012). +TIPs: SxIPping along microtubule ends. Trends Cell Biol. 22, 418-428. 10.1016/j.tcb.2012.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampert F., Hornung P. and Westermann S. (2010). The Dam1 complex confers microtubule plus end-tracking activity to the Ndc80 kinetochore complex. J. Cell Biol. 189, 641-649. 10.1083/jcb.200912021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W., Zhang X., Kline-Smith S. L., Rosasco S. E., Barrett-Wilt G. A., Shabanowitz J., Hunt D. F., Walczak C. E. and Stukenberg P. T. (2004). Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr. Biol. 14, 273-286. 10.1016/j.cub.2004.01.055 [DOI] [PubMed] [Google Scholar]
- Lawrence C. J., Dawe R. K., Christie K. R., Cleveland D. W., Dawson S. C., Endow S. A., Goldstein L. S., Goodson H. V., Hirokawa N., Howard J. et al. (2004). A standardized kinesin nomenclature. J. Cell Biol. 167, 19-22. 10.1083/jcb.200408113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Yu W., Liang Y. and Zhu X. (2007). Kinetochore dynein generates a poleward pulling force to facilitate congression and full chromosome alignment. Cell Res. 17, 701-712. 10.1038/cr.2007.65 [DOI] [PubMed] [Google Scholar]
- Liu D., Vleugel M., Backer C. B., Hori T., Fukagawa T., Cheeseman I. M. and Lampson M. A. (2010). Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J. Cell Biol. 188, 809-820. 10.1083/jcb.201001006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long A. F., Udy D. B. and Dumont S. (2017). Hec1 tail phosphorylation differentially regulates mammalian kinetochore coupling to polymerizing and depolymerizing microtubules. Curr. Biol. 27, 1692-1699 e1693. 10.1016/j.cub.2017.04.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magidson V., O'Connell C. B., Loncarek J., Paul R., Mogilner A. and Khodjakov A. (2011). The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly. Cell 146, 555-567. 10.1016/j.cell.2011.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magidson V., Paul R., Yang N., Ault J. G., O'Connell C. B., Tikhonenko I., McEwen B. F., Mogilner A. and Khodjakov A. (2015). Adaptive changes in the kinetochore architecture facilitate proper spindle assembly. Nat. Cell Biol. 17, 1134-1144. 10.1038/ncb3223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H., Fairley E. A. L., Rieder C. L., Swedlow J. R., Sunkel C. E. and Earnshaw W. C. (2003). Human CLASP1 is an outer kinetochore component that regulates spindle microtubule dynamics. Cell 113, 891-904. 10.1016/S0092-8674(03)00465-3 [DOI] [PubMed] [Google Scholar]
- Maiato H., Khodjakov A. and Rieder C. L. (2005). Drosophila CLASP is required for the incorporation of microtubule subunits into fluxing kinetochore fibres. Nat. Cell Biol. 7, 42-47. 10.1038/ncb1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malvezzi F., Litos G., Schleiffer A., Heuck A., Mechtler K., Clausen T. and Westermann S. (2013). A structural basis for kinetochore recruitment of the Ndc80 complex via two distinct centromere receptors. EMBO J. 32, 409-423. 10.1038/emboj.2012.356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr M. I., Hümmer S., Bormann J., Grüner T., Adio S., Woehlke G. and Mayer T. U. (2007). The human kinesin Kif18A is a motile microtubule depolymerase essential for chromosome congression. Curr. Biol. 17, 488-498. 10.1016/j.cub.2007.02.036 [DOI] [PubMed] [Google Scholar]
- McEwen B. F., Arena J. T., Frank J. and Rieder C. L. (1993). Structure of the colcemid-treated PtK1 kinetochore outer plate as determined by high voltage electron microscopic tomography. J. Cell Biol. 120, 301-312. 10.1083/jcb.120.2.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen B. F., Chan G. K. T., Zubrowski B., Savoian M. S., Sauer M. T. and Yen T. J. (2001). CENP-E is essential for reliable bioriented spindle attachment, but chromosome alignment can be achieved via redundant mechanisms in mammalian cells. Mol. Biol. Cell 12, 2776-2789. 10.1091/mbc.12.9.2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh J. R., Grishchuk E. L., Morphew M. K., Efremov A. K., Zhudenkov K., Volkov V. A., Cheeseman I. M., Desai A., Mastronarde D. N. and Ataullakhanov F. I. (2008). Fibrils connect microtubule tips with kinetochores: a mechanism to couple tubulin dynamics to chromosome motion. Cell 135, 322-333. 10.1016/j.cell.2008.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y., Grigoriev I., Lansbergen G., Sasaki H., Matsui C., Severin F., Galjart N., Grosveld F., Vorobjev I., Tsukita S. et al. (2005). CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J. Cell Biol. 168, 141-153. 10.1083/jcb.200405094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda J. J. L., De Wulf P., Sorger P. K. and Harrison S. C. (2005). The yeast DASH complex forms closed rings on microtubules. Nat. Struct. Mol. Biol. 12, 138-143. 10.1038/nsmb896 [DOI] [PubMed] [Google Scholar]
- Monda J. K., Whitney I. P., Tarasovetc E. V., Wilson-Kubalek E., Milligan R. A., Grishchuk E. L. and Cheeseman I. M. (2017). Microtubule tip tracking by the spindle and kinetochore protein ska1 requires diverse tubulin-interacting surfaces. Curr. Biol. 27, 3666-3675 e3666. 10.1016/j.cub.2017.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A. and Desai A. (2017). A molecular view of kinetochore assembly and function. Biology 6, 5 10.3390/biology6010005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal H. and Fukagawa T. (2016). Kinetochore assembly and function through the cell cycle. Chromosoma 125, 645-659. 10.1007/s00412-016-0608-3 [DOI] [PubMed] [Google Scholar]
- Niethammer P., Kronja I., Kandels-Lewis S., Rybina S., Bastiaens P. and Karsenti E. (2007). Discrete states of a protein interaction network govern interphase and mitotic microtubule dynamics. PLoS Biol. 5, e29 10.1371/journal.pbio.0050029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T., Rago F., Hori T., Tomii K., Cheeseman I. M. and Fukagawa T. (2013). CENP-T provides a structural platform for outer kinetochore assembly. EMBO J. 32, 424-436. 10.1038/emboj.2012.348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon F. M., Honnor T. R., Clarke N. I., Starling G. P., Beckett A. J., Johansen A. M., Brettschneider J. A., Prior I. A. and Royle S. J. (2017). Microtubule organization within mitotic spindles revealed by serial block face scanning electron microscopy and image analysis. J. Cell Sci. 130, 1845-1855. 10.1242/jcs.203877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi R., Sapra T., Howard J. and Mitchison T. J. (2004). Differentiation of cytoplasmic and meiotic spindle assembly MCAK functions by Aurora B-dependent phosphorylation. Mol. Biol. Cell 15, 2895-2906. 10.1091/mbc.e04-02-0082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekgoz Altunkaya G., Malvezzi F., Demianova Z., Zimniak T., Litos G., Weissmann F., Mechtler K., Herzog F. and Westermann S. (2016). CCAN assembly configures composite binding interfaces to promote cross-linking of Ndc80 complexes at the kinetochore. Curr. Biol. 26, 2370-2378. 10.1016/j.cub.2016.07.005 [DOI] [PubMed] [Google Scholar]
- Pesenti M. E., Weir J. R. and Musacchio A. (2016). Progress in the structural and functional characterization of kinetochores. Curr. Opin. Struct. Biol. 37, 152-163. 10.1016/j.sbi.2016.03.003 [DOI] [PubMed] [Google Scholar]
- Posch M., Khoudoli G. A., Swift S., King E. M., Deluca J. G. and Swedlow J. R. (2010). Sds22 regulates aurora B activity and microtubule-kinetochore interactions at mitosis. J. Cell Biol. 191, 61-74. 10.1083/jcb.200912046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers A. F., Franck A. D., Gestaut D. R., Cooper J., Gracyzk B., Wei R. R., Wordeman L., Davis T. N. and Asbury C. L. (2009). The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell 136, 865-875. 10.1016/j.cell.2008.12.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser S. L. and Pelletier L. (2017). Mitotic spindle assembly in animal cells: a fine balancing act. Nat. Rev. Mol. Cell Biol. 18, 187-201. 10.1038/nrm.2016.162 [DOI] [PubMed] [Google Scholar]
- Przewloka M. R., Venkei Z., Bolanos-Garcia V. M., Debski J., Dadlez M. and Glover D. M. (2011). CENP-C is a structural platform for kinetochore assembly. Curr. Biol. 21, 399-405. 10.1016/j.cub.2011.02.005 [DOI] [PubMed] [Google Scholar]
- Rago F., Gascoigne K. E. and Cheeseman I. M. (2015). Distinct organization and regulation of the outer kinetochore KMN network downstream of CENP-C and CENP-T. Curr. Biol. 25, 671-677. 10.1016/j.cub.2015.01.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattner J. B., Rao A., Fritzler M. J., Valencia D. W. and Yen T. J. (1993). CENP-F is a.ca 400 kDa kinetochore protein that exhibits a cell-cycle dependent localization. Cell Motil. Cytoskelet. 26, 214-226. 10.1002/cm.970260305 [DOI] [PubMed] [Google Scholar]
- Rieder C. L. (1982). The formation, structure, and composition of the mammalian kinetochore and kinetochore fiber. Int. Rev. Cytol. 79, 1-58. 10.1016/S0074-7696(08)61672-1 [DOI] [PubMed] [Google Scholar]
- Rieder C. L., Cole R. W., Khodjakov A. and Sluder G. (1995). The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J. Cell Biol. 130, 941-948. 10.1083/jcb.130.4.941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon E. D. and Begg D. A. (1980). Functional implications of cold-stable microtubules in kinetochore fibers of insect spermatocytes during anaphase. J. Cell Biol. 85, 853-865. 10.1083/jcb.85.3.853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanhaji M., Friel C. T., Kreis N.-N., Krämer A., Martin C., Howard J., Strebhardt K. and Yuan J. (2010). Functional and spatial regulation of mitotic centromere-associated kinesin by cyclin-dependent kinase 1. Mol. Cell. Biol. 30, 2594-2607. 10.1128/MCB.00098-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleicher K., Porter M., Ten Have S., Sundaramoorthy R., Porter I. M. and Swedlow J. R. (2017). The Ndc80 complex targets Bod1 to human mitotic kinetochores. Open Biol. 7, 170099 10.1098/rsob.170099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiffer A., Maier M., Litos G., Lampert F., Hornung P., Mechtler K. and Westermann S. (2012). CENP-T proteins are conserved centromere receptors of the Ndc80 complex. Nat. Cell Biol. 14, 604-613. 10.1038/ncb2493 [DOI] [PubMed] [Google Scholar]
- Schmidt J. C., Arthanari H., Boeszoermenyi A., Dashkevich N. M., Wilson-Kubalek E. M., Monnier N., Markus M., Oberer M., Milligan R. A., Bathe M. et al. (2012). The kinetochore-bound Ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments. Dev. Cell 23, 968-980. 10.1016/j.devcel.2012.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schukken K. M. and Foijer F. (2018). CIN and aneuploidy: different concepts, different consequences. BioEssays 40, 1700147 10.1002/bies.201700147 [DOI] [PubMed] [Google Scholar]
- Screpanti E., De Antoni A., Alushin G. M., Petrovic A., Melis T., Nogales E. and Musacchio A. (2011). Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr. Biol. 21, 391-398. 10.1016/j.cub.2010.12.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha R. L., Conti D., Tamura N., Braun D., Ramalingam R. A., Cieslinski K., Ries J. and Draviam V. M. (2017). Aurora-B kinase pathway controls the lateral to end-on conversion of kinetochore-microtubule attachments in human cells. Nat. Commun. 8, 150 10.1038/s41467-017-00209-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silljé H. H., Nagel S., Körner R. and Nigg E. A. (2006). HURP is a Ran-importin beta-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr. Biol. 16, 731-742. 10.1016/j.cub.2006.02.070 [DOI] [PubMed] [Google Scholar]
- Sivakumar S. and Gorbsky G. J. (2017). Phosphatase-regulated recruitment of the spindle- and kinetochore-associated (Ska) complex to kinetochores. Biol. Open 6, 1672-1679. 10.1242/bio.026930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens R. V., Skeen V. P. and Salmon E. D. (1993). Directional instability of kinetochore motility during chromosome congression and segregation in mitotic newt lung cells: a push-pull mechanism. J. Cell Biol. 122, 859-875. 10.1083/jcb.122.4.859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D. A., Williams B. C., Hays T. S. and Goldberg M. L. (1998). ZW10 helps recruit dynactin and dynein to the kinetochore. J. Cell Biol. 142, 763-774. 10.1083/jcb.142.3.763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J., von Dassow G., Wagenbach M., Asbury C. and Wordeman L. (2008). The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev. Cell 14, 252-262. 10.1016/j.devcel.2007.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J., Wagenbach M., Franck A., Asbury C. L. and Wordeman L. (2012). Kif18A and chromokinesins confine centromere movements via microtubule growth suppression and spatial control of kinetochore tension. Dev. Cell 22, 1017-1029. 10.1016/j.devcel.2012.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L.-K. and Qi Y. (2001). Characterization of human MAPRE genes and their proteins. Genomics 71, 142-149. 10.1006/geno.2000.6428 [DOI] [PubMed] [Google Scholar]
- Su L.-K., Burrell M., Hill D. E., Gyuris J., Brent R., Wiltshire R., Trent J., Vogelstein B. and Kinzler K. W. (1995). APC binds to the novel protein EB1. Cancer Res. 55, 2972-2977. [PubMed] [Google Scholar]
- Su K.-C., Barry Z., Schweizer N., Maiato H., Bathe M. and Cheeseman I. M. P. (2016). A regulatory switch alters chromosome motions at the metaphase-to-anaphase transition. Cell Reports 17, 1728-1738. 10.1016/j.celrep.2016.10.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A., Badger B. L. and Salmon E. D. (2015). A quantitative description of Ndc80 complex linkage to human kinetochores. Nat. Commun. 6, 8161 10.1038/ncomms9161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talapatra S. K., Harker B. and Welburn J. P. (2015). The C-terminal region of the motor protein MCAK controls its structure and activity through a conformational switch. eLife 4, e06421 10.7554/eLife.06421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Mukae N., Dewar H., van Breugel M., James E. K., Prescott A. R., Antony C. and Tanaka T. U. (2005). Molecular mechanisms of kinetochore capture by spindle microtubules. Nature 434, 987-994. 10.1038/nature03483 [DOI] [PubMed] [Google Scholar]
- Thrower D. A., Jordan M. A. and Wilson L. (1996). Modulation of CENP-E organization at kinetochores by spindle microtubule attachment. Cell Motil. Cytoskelet. 35, 121-133. [DOI] [PubMed] [Google Scholar]
- Tien J. F., Umbreit N. T., Gestaut D. R., Franck A. D., Cooper J., Wordeman L., Gonen T., Asbury C. L. and Davis T. N. (2010). Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. J. Cell Biol. 189, 713-723. 10.1083/jcb.200910142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hooff J. J. E., Snel B. and Kops G. J. P. L. (2017). Unique phylogenetic distributions of the Ska and Dam1 complexes support functional analogy and suggest multiple parallel displacements of Ska by Dam1. Genome Biol. Evol. 9, 1295-1303. 10.1093/gbe/evx088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov V. A., Grissom P. M., Arzhanik V. K., Zaytsev A. V., Renganathan K., McClure-Begley T., Old W. M., Ahn N. and McIntosh J. R. (2015). Centromere protein F includes two sites that couple efficiently to depolymerizing microtubules. J. Cell Biol. 209, 813-828. 10.1083/jcb.201408083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov V. A., Huis In ‘t Veld P. J., Dogterom M. and Musacchio A. (2018). Multivalency of NDC80 in the outer kinetochore is essential to track shortening microtubules and generate forces. eLife 7, e36764 10.7554/eLife.36764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorozhko V. V., Emanuele M. J., Kallio M. J., Stukenberg P. T. and Gorbsky G. J. (2008). Multiple mechanisms of chromosome movement in vertebrate cells mediated through the Ndc80 complex and dynein/dynactin. Chromosoma 117, 169-179. 10.1007/s00412-007-0135-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn J. P. I., Grishchuk E. L., Backer C. B., Wilson-Kubalek E. M., Yates J. R. III and Cheeseman I. M. (2009). The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Dev. Cell 16, 374-385. 10.1016/j.devcel.2009.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendell K. L., Wilson L. and Jordan M. A. (1993). Mitotic block in HeLa cells by vinblastine: ultrastructural changes in kinetochore-microtubule attachment and in centrosomes. J. Cell Sci. 104, 261-274. [DOI] [PubMed] [Google Scholar]
- Westermann S., Avila-Sakar A., Wang H.-W., Niederstrasser H., Wong J., Drubin D. G., Nogales E. and Barnes G. (2005). Formation of a dynamic kinetochore- microtubule interface through assembly of the Dam1 ring complex. Mol. Cell 17, 277-290. 10.1016/j.molcel.2004.12.019 [DOI] [PubMed] [Google Scholar]
- Wong J. and Fang G. (2006). HURP controls spindle dynamics to promote proper interkinetochore tension and efficient kinetochore capture. J. Cell Biol. 173, 879-891. 10.1083/jcb.200511132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wordeman L. and Mitchison T. J. (1995). Identification and partial characterization of mitotic centromere-associated kinesin, a kinesin-related protein that associates with centromeres during mitosis. J. Cell Biol. 128, 95-104. 10.1083/jcb.128.1.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wordeman L., Steuer E. R., Sheetz M. P. and Mitchison T. (1991). Chemical subdomains within the kinetochore domain of isolated CHO mitotic chromosomes. J. Cell Biol. 114, 285-294. 10.1083/jcb.114.2.285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynne D. J. and Funabiki H. (2015). Kinetochore function is controlled by a phospho-dependent coexpansion of inner and outer components. J. Cell Biol. 210, 899-916. 10.1083/jcb.201506020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Tulu U. S., Wadsworth P. and Rieder C. L. (2007). Kinetochore dynein is required for chromosome motion and congression independent of the spindle checkpoint. Curr. Biol. 17, 973-980. 10.1016/j.cub.2007.04.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye A. A., Deretic J., Hoel C. M., Hinman A. W., Cimini D., Welburn J. P. and Maresca T. J. (2015). Aurora a kinase contributes to a pole-based error correction pathway. Curr. Biol. 25, 1842-1851. 10.1016/j.cub.2015.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Lan W., Ems-McClung S. C., Stukenberg P. T. and Walczak C. E. (2007). Aurora B phosphorylates multiple sites on mitotic centromere-associated kinesin to spatially and temporally regulate its function. Mol. Biol. Cell 18, 3264-3276. 10.1091/mbc.e07-01-0086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Shao H., Huang Y., Yan F., Chu Y., Hou H., Zhu M., Fu C., Aikhionbare F., Fang G. et al. (2011). PLK1 phosphorylates mitotic centromere-associated kinesin and promotes its depolymerase activity. J. Biol. Chem. 286, 3033-3046. 10.1074/jbc.M110.165340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Mancini M. A., Chang K. H., Liu C. Y., Chen C. F., Shan B., Jones D., Yang-Feng T. L. and Lee W. H. (1995). Characterization of a novel 350-kilodalton nuclear phosphoprotein that is specifically involved in mitotic-phase progression. Mol. Cell. Biol. 15, 5017-5029. 10.1128/MCB.15.9.5017 [DOI] [PMC free article] [PubMed] [Google Scholar]