Abstract
The objective of this study was to estimate genetic parameters for carcass and meat quality traits, as well as their genetic correlations using pedigree and genomic information. A total of 3,716; 3,702; 3,439; 3,705; and 3,714 records of 12th–13th rib LM area (LMA), backfat thickness (BF), HCW, marbling score (MARB), and Warner–Bratzler peak shear force (WBSF), respectively, were used. Animals were genotyped with BovineHD BeadChip and GeneSeek Genomic Profiler Indicus HD - GGP75Ki panel. The (co)variance components were estimated by Bayesian inference using a multitrait ssGBLUP analysis. The animal model included fixed effects of contemporary group (defined by the combination of farm and year of birth, and management group at yearling) and age of animal at slaughtering as a covariate (linear). Direct additive genetic and residual effects were fitted as random. The posterior means and SD of heritabilities for LMA, BF, HCW, MARB, and WBSF were 0.28 (0.03), 0.21 (0.04), 0.21 (0.04), 0.12 (0.04), and 0.11 (0.03), respectively. The posterior means for genetic correlations between LMA and meat quality were positive and moderate with MARB (0.38 ± 0.12) and negative with WBSF (−0.47 ± 0.12). Low genetic correlations were estimated between BF and WBSF (−0.03 ± 0.16) and between HCW and MARB (−0.04 ± 0.14), indicating that these traits are not controlled by the same set or linked genes. Carcass traits (LMA, BF, and HCW) presented moderate heritability providing quick response to the selection purpose. The estimates of heritability for meat quality traits (MARB and WBSF) were low and indicate that the rate of genetic improvement for these traits would be slow. Genetic correlations indicated that selection for carcass traits would not be strongly antagonistic for improving meat quality.
Keywords: backfat thickness, genetic correlation, heritability, HCW, LM area, marbling score, shear force
INTRODUCTION
Nelore cattle (Bos taurus indicus) is the most representative breed in Brazil raised on extensive production system and pasture-based conditions. Despite of the importance for the Brazilian production system, studies have shown that Nelore cattle have had traditionally inferior carcass and meat quality compared to Bos taurus taurus (O’Connor et al., 1997; Bressan et al., 2011). Therefore, genetic improvement of carcass and meat quality traits in Nelore cattle is essential for the beef industry, once there is a higher demand for such products. The carcass traits LM area (LMA), backfat thickness (BF), and HCW are important for the commercialization of meat products, since they are indicators of the quantitative composition of the carcass. The first trait is related to muscularity and yield of the edible portion. The second trait is used as an indicator of the degree of carcass finishing, while the third is used as a classificatory trait in slaughterhouses and it is directly related to the payment to the producer. Meat quality is defined by the traits of which consumers perceive as desirable. Tenderness and juiciness are one of the most important factors influencing the acceptability of beef and eating quality (Glitsch, 2000; Beermann, 2009).
In order to incorporate carcass and meat quality traits in a genetic evaluation scheme and to design appropriate breeding programs, heritability and genetic correlations for these traits are important to be estimated. Within this context, in their extensive reviews of genetic parameters for carcass and meat quality traits, Burrow et al. (2001) and Warner et al. (2010) reported that these traits are moderately heritable and improvement may be obtained through selection. However, efficient selection programs are restricted, as both carcass and beef quality traits are expensive and difficult to measure on an industry-wide basis. Moreover, due to the small number of animals used in such studies, often only a few hundred, the genetic parameters were estimated with low accuracy (Burrow et al., 2001).
Several authors have estimated genetic parameters for carcass and meat quality traits using only phenotype and pedigree information in Brahman, Angus, Hereford, Simmental, Piemontese, Nelore, and composite breeds (Shanks et al., 2001; Riley et al., 2002; Reverter et al., 2003a, 2003b; Dikeman et al., 2005; Smith et al., 2007; Boukha et al., 2011; Bonfatti et al., 2013; Ferriani et al., 2013). However, few genomic studies have reported heritabilities for these traits in Nelore cattle (Tizioto et al., 2013; Fernandes Júnior et al., 2016; Magalhães et al., 2016). Thus, to learn more about the genetic variation of carcass and meat quality traits in Nelore cattle, the purpose of this paper was to estimate genetic parameters for carcass and meat quality traits as well as their genetic correlations using pedigree and genomic information.
MATERIAL AND METHODS
All animal procedures were approved by the São Paulo State University (Unesp), School of Agricultural and Veterinary Science Ethical Committee (Approval No. 18.340/16).
Description of the Data
A total of 3,742 animals born from 2008 to 2012 belonging to three Nelore breeding programs, DeltaGen, Paint CRV Lagoa, and Nelore Qualitas, were analyzed. The animals were raised on pasture conditions and finished in feedlot systems for around 90 d until slaughter at approximately 2 yr of age. Three carcass traits and two meat quality traits were used in this study: HCW, LMA, BF, marbling score (MARB), and Warner–Bratzler peak shear force (WBSF). At slaughter, HCW was recorded for each animal. After 24 to 48 h chill, the Longissimus thoracis sections between 12/13th ribs were taken from the left side of the carcasses, placed in a labeled bag and immediately frozen at −20 °C for later analyses. Meat samples were then unpacked at the São Paulo State University Meat Science Laboratory and a 2.54 cm thick steak was cut from the collected sample for use in LMA, BF, MARB, and WBSF measurements. Point counting on a plastic grid (where each square corresponds to 1 cm2) was used to measure LMA, in which the grid was placed on the sample and the sum of all squares corresponds to the LMA of the animal. For the determination of BF, the layer of subcutaneous fat located at an angle of 45 degrees from the geometric center of the sample was measured in millimeters with a caliper.
The degree of marbling was scored on a scale from 1 to 10 according to the USDA marbling standards, where: 1 = practically absent; 2 = traces; 3 = slight; 4 = small; 5 = modest; 6 = moderate; 7 = slightly abundant; 8 = moderately abundant; 9 = abundant, and 10 = very abundant. The steaks were then cooked to an internal temperature of 71 °C in an electrical oven at 180 °C. Internal temperatures were monitored with wire thermocouper probes connected to an AKSO temperature recorder. After reaching the endpoint temperature, steaks were cooled at 2 °C for 24 h, and eight cores with half inch of diameter each were removed from the samples and sheared with a V blade attached to a WBSF machine. The WBSF values for the eight cores were averaged and used in the analysis. The contemporary group (CG) for all traits was defined by the effects of year and farm of birth, and management group at yearling. The observations with three SD above or below the mean of their CG were excluded. The description of the data used is shown in Table 1.
Table 1.
Summary statistics for carcass and meat quality traits in Nelore cattle
| Traits | No of observations | No of sires | No of dams | Means (SD) |
|---|---|---|---|---|
| LMA, cm2 | 3,716 | 428 | 3,359 | 68.00 (9.00) |
| BF, mm | 3,702 | 428 | 3,347 | 4.40 (2.25) |
| HCW, kg | 3,439 | 383 | 3,191 | 272 (24.70) |
| MARB | 3,705 | 430 | 3,348 | 2.77 (0.44) |
| WBSF, kg | 3,714 | 429 | 3,358 | 5.98 (1.76) |
Abbreviations: BF = backfat thickness; LMA = LM area; MARB = marbling score; WBSF = Warner–Bratzler peak shear force.
Marker Genotypes
A total of 4,674 animals were genotyped, 2,669 using the BovineHD BeadChip (Illumina, Inc., San Diego, CA) and 1,975 using the GeneSeek Genomic Profiler Indicus HD - GGP75Ki (Neogen Corporation, Lincoln, NE), which contain 777,962 and 74,677 SNP markers distributed across the genome, respectively. Animals genotyped with GGP75Ki were imputed to BovineHD using FImpute software (Sargolzaei et al., 2014) considering pedigree information. For genotype quality control, only autosomal SNPs were considered, and SNPs with minor allele frequency (MAF) <0.05, a Hardy–Weinberg equilibrium P value ≤10−5 and a call rate <0.98 were excluded. Correlation between SNPs within a window of 100 markers also was evaluated and if a pair of markers does not meet the r2 required (r2 ≥ 0.995), the SNP with the lowest MAF was excluded from the analyses. For samples, a call rate of at least 0.90 was required. After quality control, 4,621 genotyped animals and 412,904 SNPs remained.
Statistical Analysis
The (co)variance components were estimated by Bayesian inference using a multitrait ssGBLUP analysis via GIBBSF90 program, and a post-Gibbs analysis was conducted using POSTGIBBSF90 (Misztal et al., 2002). For all the traits, the model included direct additive genetic and residual as random effects, the fixed effects of CG, and the linear effect of age of animal at slaughter. The general model can be written in matrix form as follows:
where y is the vector of the traits observed; β is the vector of fixed effects; α is the vector of genetic additive direct effects of the animal; e is the vector of residual effects; and X and Z are incidence matrices relating β, α, and e to y. In this study, it was assumed that E[y] = Xβ; Var(α) = H ⊗ Sh, Var(e) = I ⊗ Se, where Sh is the additive genetic covariance matrix; Se is the residual covariance matrix; H is the additive genetic relationship matrix based on pedigree and genomic information as proposed by Misztal et al. (2009) and Legarra et al. (2009); I is the identity matrix, and ⊗ is the direct product between matrices.
The inverse of H (H−1) matrix proposed by Aguilar et al. (2010) can be obtained as follows:
where A−1 and G−1 are the inverse of pedigree and genomic-based relationship matrix, respectively and is the inverse of the pedigree-based relationship matrix for genotyped animals. The G matrix proposed by VanRaden (2008) can be written as:
where M is a n × m matrix (n = number of genotyped animals and m = number of markers) and P is a matrix with frequency of the second allele (pj) expressed as 2pj. The markers were coded according to the numbers of copies for the B allele. To facilitate inversion, by default, GIBBSF90 program uses weighted G as proposed by VanRaden (2008): G = 0.95G0 + 0.05A22. Additionally, to make G proportional to A22 the program, by default, scale G based on A22 considering the diagonal mean of G equal to the diagonal mean of A22, and the off-diagonals mean of G equal to the off-diagonals mean of A22.
The vectors β and α are locations parameters from the conditional distribution. Uniform distribution of β was assumed a priori, which reflects a vague prior knowledge about this vector. For (co)variances matrix of random effect, inverted Wishart distributions were defined as a priori. The distribution of y given the parameters of location and scale was assumed (Van Tassell and Van Vleck, 1996): y | β, α, R ∼ N [Xβ + Zα, InR].
A total of 1,000,000 samples were generated, and a burn-in period of 100,000 samples with samples taken each 50 cycles. The convergence was verified through graphical inspection and also using the criteria proposed by Heidelberger and Welch (1983) and Geweke (1992). The package coda of R software was used to calculate Heidelberger and Welch’s and Geweke’s statistics (Plummer et al., 2006). The numerator relationship and genomic matrices contained 64,679 and 4,621 animals, respectively.
RESULTS AND DISCUSSION
The posterior mean, mode, and median of all parameters estimated were similar for all traits (Tables 2 and 3), indicating symmetrical posterior distributions. Thus, the use of the mean will satisfactorily represent the property of the parameter, reflecting the measure of central tendency of the posterior marginal distribution.
Table 2.
Posterior means, SD (PSD), mode, median, and the highest posterior density (HPD) region of heritability estimates for LM area (LMA), backfat thickness (BF), HCW, Warner–Bratzler peak shear force (WBSF), and marbling score (MARB)
| Traits | Means | PSD | Mode | Median | HPD 95(%) |
|---|---|---|---|---|---|
| LMA | 0.28 | 0.03 | 0.27 | 0.28 | 0.21 to 0.35 |
| BF | 0.21 | 0.04 | 0.21 | 0.21 | 0.13 to 0.29 |
| HCW | 0.21 | 0.04 | 0.20 | 0.21 | 0.15 to 0.28 |
| WBSF | 0.12 | 0.04 | 0.12 | 0.12 | 0.06 to 0.18 |
| MARB | 0.11 | 0.03 | 0.10 | 0.11 | 0.06 to 0.16 |
Table 3.
Posterior means, SD (PSD), mode, median, and the highest posterior density (HPD) region of genetic correlations estimates for carcass traits with meat quality traits
| Trait pairs | Means | PSD | Mode | Median | HPD 95(%) |
|---|---|---|---|---|---|
| LMA–WBSF | −0.47 | 0.12 | −0.46 | −0.48 | −0.70 to −0.23 |
| LMA–MARB | 0.38 | 0.12 | 0.37 | 0.38 | 0.12 to 0.62 |
| BF–WBSF | −0.03 | 0.16 | −0.04 | −0.04 | −0.30 to 0.25 |
| BF–MARB | 0.14 | 0.16 | 0.13 | 0.14 | −0.18 to 0.46 |
| HCW–WBSF | −0.27 | 0.14 | −0.26 | −0.27 | −0.49 to −0.06 |
| HCW–MARB | −0.04 | 0.14 | −0.03 | −0.04 | −0.31 to 0.24 |
Abbreviations: BF = backfat thickness; LMA = LM area; MARB = marbling score; WBSF = Warner–Bratzler peak shear force.
Heritability Estimates
The mean heritabilities for LMA, BF, and HCW were of moderate magnitudes, varying from 0.20 to 0.31 (Table 2), indicating that part of the variation in these traits are attributed to genes with additive effects. Although the heritability estimated for LMA here had the highest magnitude, it was lower than those reported in the literature for Zebu animals using relationship matrix based on pedigree information (A), which ranged from 0.35 to 0.63 (Riley et al., 2002; Smith et al., 2007), and similar to that reported by Tizioto et al. (2013) using the genomic relationship matrix (0.27). The heritability estimated for BF was lower than those reported by Riley et al. (2002) and Rezende et al. (2009) (0.63 and 0.52, respectively) but similar to that reported by Tizioto et al. (2013) for Nelore animals (0.21). For HCW, the heritability estimate was lower than those obtained by Riley et al. (2002) for Brahman animals (0.55). Studying Nelore animals, Rezende et al. (2009) reported a higher heritability (0.38) estimate than that observed in our study. On the other hand, Ferriani et al. (2013) obtained a similar heritability (0.20) for animals of the same breed.
Heritability estimates for meat quality traits were of low magnitude (Table 2) and indicate that these traits are affected mostly by environmental effects, and consequently, selection for meat quality traits in Nelore cattle would result in a slow rate of genetic improvement. The low magnitude of the heritability estimated for WBSF was similar to those related by Riley et al. (2003) in Brahman cattle—0.14, 0.14, and 0.06 for shear force at 7, 14, and 21 d of maturation, respectively—and by Boukha et al. (2011) in Piemontese cattle (0.14). In Nelore cattle, Tizioto et al. (2013) reported heritability for shear force close (0.16) to that obtained in this study. However, in comparison with our results, larger heritability estimates for shear force measurements have been obtained by Johnston et al. (2003) in tropically adapted breed, Dikeman et al. (2005) in three beef cattle breeds, and have been summarized in a literature review by Burrow et al. (2001). Regarding the heritability estimated for MARB, higher estimates have been reported in taurine and composite cattle by several authors (Shackelford et al., 1994; Wheeler et al., 2001; Dikeman et al., 2005), which ranged from 0.57 to 0.93. Utrera and Van Vleck (2004) reported a mean heritability estimate of 0.45 on 29 estimates. For zebuine cattle, Riley et al. (2002) and Smith et al. (2007) reported heritability estimates for Brahman cattle of 0.44 and 0.37, respectively. The heritability estimates for carcass and meat quality traits presented in this study were generally lower than those reported in the literature. This may be due to genetic aspects of the breed, but also to the commercial animals studied, which were probably submitted to greater variability of environmental conditions in comparison to those raised in research stations.
Genetic Correlations
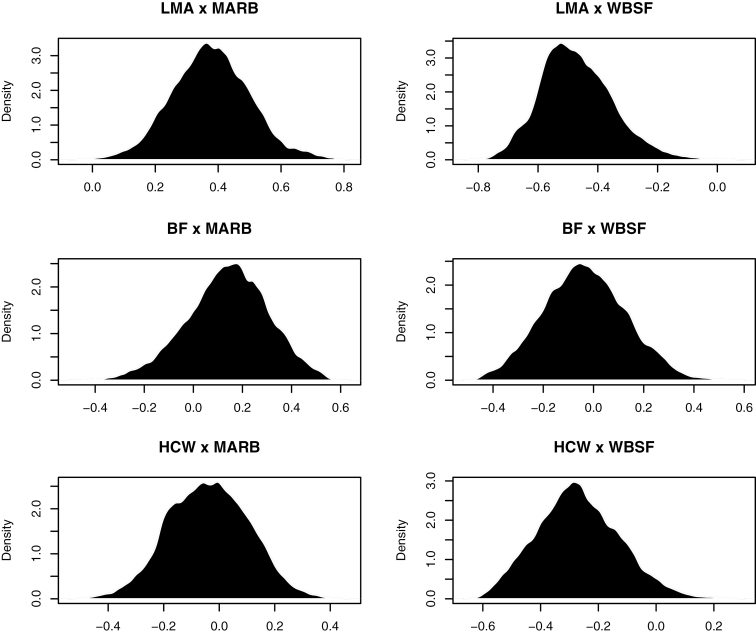
Genetic correlations of carcass traits with meat quality traits are given in Table 3. For presenting these results in more detail, Figure 1 shows the posterior distributions of the genetic correlations between these traits. There was a positive genetic correlation between LMA and MARB, suggesting that the genes with the potential to produce larger LMA are the same set of genes as, or closely linked to, those with the potential to produce marbling meat. On the other hand, lower genetic correlation (0.17) was found by Smith et al. (2007). The genetic relationship between LMA and WBSF was moderate and negative, indicating a lack of association between these traits. However, due to the wide amplitude of the highest posterior density region, this genetic correlation should be treated with caution. In agreement with the presented findings, Reverter et al. (2003b) and Bonfatti et al. (2013) reported the existence of low genetic correlation between LMA and shear force (0.15 and −0.08, respectively). The low genetic correlation (−0.03) between BF and WBSF and the value of zero within the 95% posterior density interval suggest that selection for BF should not affect WBSF. In contrast to our result, Wheeler et al. (2001) and Smith et al. (2007) reported that BF is negatively genetically associated with shear force, with values ranging from −0.41 to −0.50. The posterior mean for genetic correlation between BF and MARB was low. Our finding was similar to the low values reported by Crews et al. (2004) and Smith et al. (2007), of −0.10 and 0.04, respectively; but different (−0.47) to that reported by Shanks et al. (2001) the Simmental breed.
Figure 1.
Marginal posterior density of the genetic correlations among carcass and meat quality traits.
The HCW was negatively genetically correlated with WBSF (Table 3). This suggests that genetically heavier animals tend to produce more tender meat. This estimate is in agreement with the genetic correlation between HCW and shear force reported by Reverter et al. (2003b) in tropically adapted beef (−0.21).There was a low genetic correlation between HCW and MARB (−0.04), indicating that selection for higher HCW will not result in a significant increase in MARB. Near-zero genetic estimates between carcass weight and intramuscular fat were also reported by Marshall (1994) and Reverter et al. (2003b) of 0.18 and −0.03, respectively. Although some genetic correlation estimates among carcass and meat quality traits suggest an association between traits, the interpretation presented in our study should be tempered due to the low heritability estimates for meat quality traits. Furthermore, the 95% posterior density intervals for some estimates were relatively large, albeit they were in agreement with several studies cited above. Because of the scarce literature reporting genetic parameters for carcass and meat quality traits in Nelore cattle, our study provided estimates that could be useful in the design of breeding programs for improving such traits in this breed.
CONCLUSIONS
Nelore cattle has been constantly critiqued for reduced carcass quality. This paper has shown that carcass traits have moderate heritabilities, which may respond to the selection purpose quickly. On the other hand, the low estimates of heritability for meat quality traits indicate that the rate of genetic improvement for these traits would be slow. Genetic correlations indicated that selection for carcass traits would not be strongly antagonistic to improvement in meat quality.
Footnotes
This research was funded by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), grant #2009/16118-5.
LITERATURE CITED
- Aguilar I., Misztal I., Johnson D. L., Legarra A., Tsuruta S., and Lawlor T. J.. 2010. A unified approach to utilize phenotypic, full pedigree, and genomic information for genetic evaluation of Holstein final score. J. Dairy Sci. 93:743–752. doi: 10.3168/jds.2009-2730 [DOI] [PubMed] [Google Scholar]
- Beermann D. H. 2009. ASAS centennial paper: a century of pioneers and progress in meat science in the United States leads to new frontiers. J. Anim. Sci. 87:1192–1198. doi: 10.2527/jas.2008-1542 [DOI] [PubMed] [Google Scholar]
- Bonfatti V., Albera A., and Carnier P.. 2013. Genetic associations between daily BW gain and live fleshiness of station-tested young bulls and carcass and meat quality traits of commercial intact males in Piemontese cattle. J. Anim. Sci. 91:2057–2066. doi: 10.2527/jas.2012-5386 [DOI] [PubMed] [Google Scholar]
- Boukha A., Bonfatti V., Cecchinato A., Albera A., Gallo L., Carnier P., and Bittante G.. 2011. Genetic parameters of carcass and meat quality traits of double muscled Piemontese cattle. Meat Sci. 89:84–90. doi: 10.1016/j.meatsci.2011.03.024 [DOI] [PubMed] [Google Scholar]
- Burrow H., Moore S., Johnston D., Barendse W., and Bindon B.. 2001. Quantitative and molecular genetic influences on properties of beef: a review. Aust. J. Exp. Agric. 41:893–919. doi: 10.1071/EA00015 [DOI] [Google Scholar]
- Bressan M. C., Rodrigues E. C., Rossato L. V., Ramos E. M., Gama L. T.. 2011. Physicochemical properties of meat from Bos taurus and Bos indicus. R. Bras. Zootec. 40:1250–1259. doi: 10.1590/S1516-35982011000600013 [DOI] [Google Scholar]
- Crews D. H. Jr. Lowerison M., Caron N., and Kemp R. A.. 2004. Genetic parameters among growth and carcass traits of Canadian Charolais cattle. Can. J. Anim. Sci. 84:589–597. doi: 10.4141/A04-019 [DOI] [Google Scholar]
- Dikeman M. E., Pollak E. J., Zhang Z., Moser D. W., Gill C. A., and Dressler E. A.. 2005. Phenotypic ranges and relationships among carcass and meat palatability traits for fourteen cattle breeds, and heritabilities and expected progeny differences for Warner-Bratzler shear force in three beef cattle breeds. J. Anim. Sci. 83:2461–2467. doi: 10.2527/2005.83102461x [DOI] [PubMed] [Google Scholar]
- Ferriani L., Albuquerque L. G., Baldi F., Venturini G., Bignardi A., Silva J. I., Chud T., Munari D., and Oliveira J.. 2013. Parâmetros genéticos de características de carcaça e de crescimento de bovinos da raça Nelore. Arch. Zootec. 62:123–129. doi: 10.4321/S0004-05922013000100013 [DOI] [Google Scholar]
- Fernandes Júnior G. A., Rosa G. J., Valente B. D., Carvalheiro R., Baldi F., Garcia D. A., Gordo D. G., Espigolan R., Takada L., Tonussi R. L., et al. 2016. Genomic prediction of breeding values for carcass traits in Nellore cattle. Genet. Sel. Evol. 48:7. doi: 10.1186/s12711-016-0188-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geweke J. 1992. Evaluating the accuracy of sampling-based approaches to the calculation of posterior moments. In: Bernardo J. M., Berger J., Dawid A. P., and Smith J. F. M., editor, Bayesian Statistics 4. Oxford University Press, Oxford: p. 169–193. [Google Scholar]
- Glitsch K. 2000. Consumer perceptions of fresh meat quality: cross-national comparison. Brit. Food J. 102:177–194. doi: 10.1108/00070700010332278 [DOI] [Google Scholar]
- Heidelberger P. and Welch P. D.. 1983. Simulation run length control in the presence of an initial transient. Oper. Res. 31:1109–1144. doi: 10.1287/opre.31.6.1109. [DOI] [Google Scholar]
- Johnston D. J. A., Reverter A. A., Ferguson D. M. B., Thompson J. M. C., and Burrow H. M. D.. 2003. Genetic and phenotypic characterisation of animal, carcass, and meat quality traits from temperate and tropically adapted beef breeds. 3. Meat quality traits. Aust. J. Exp. Agric. 54:135–147. doi: 10.1071/AR02085.0004-9409/03/020107 [DOI] [Google Scholar]
- Legarra A., Aguilar I., and Misztal I.. 2009. A relationship matrix including full pedigree and genomic information. J. Dairy Sci. 92:4656–4663. doi: 10.3168/jds.2009-2061 [DOI] [PubMed] [Google Scholar]
- Magalhães A. F. B., Camargo G. M. F., Fernandes Junior G. A., Gordo D. G. M., Tonussi R. L., Costa R. B., Espigolan R., Silva R. M. O., Bresolin T., Andrade W. B. F., Takada L.,. et al. 2016. Genome-Wide Association Study of meat quality traits in Nellore cattle. PLoS ONE. 11:1–12. doi: 10.1371/journal.pone.0157845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall D. M. 1994. Breed differences and genetic parameters for body composition traits in beef cattle. J. Anim. Sci. 72:2745–2755. doi: 10.2527/1994.72102745x [DOI] [PubMed] [Google Scholar]
- Misztal I., Legarra A., and Aguilar I.. 2009. Computing procedures for genetic evaluation including phenotypic, full pedigree, and genomic information. J. Dairy Sci. 92:4648–4655. doi: 10.3168/jds.2009-2064 [DOI] [PubMed] [Google Scholar]
- Misztal I., Tsuruta S., Strabel T., Auvray B., Druet T., and Lee D. H.. 2002. BLUPF90 and related programs (BGF90). Proceedings from the 7th World Congress on Genetics Applied to Livestock Production; August 2002;Montpellier, France 28:21–22. [Google Scholar]
- O’Connor S. F., Tatum J. D., Wulf D. M., Green R. D., and Smith G. C.. 1997. Genetic effects on beef tenderness in Bos indicus composite and Bos taurus cattle. J. Anim. Sci. 75:1822–1830. [DOI] [PubMed] [Google Scholar]
- Plummer M., Best N., Cowles K., and Vines K.. 2006. CODA: convergence diagnosis and output analysis for MCMC. R News. 6:7–11. http://oro.open.ac.uk/id/eprint/22547 [Google Scholar]
- Reverter A. A., Johnston D. J. A., Ferguson D. M. B., Perry D. C., Goddard M. E. A., Burrow H. M. E., Oddy V. H. F., Thompson J. M. G., and Bindon B. M. H.. 2003a. Genetic and phenotypic characterisation of animal, carcass, and meat quality traits from temperate and tropically adapted beef breeds. 4. Correlations among animal carcass and meat quality traits. Aust. J. Exp. Agric. 54:149–158. doi: 10.1071/AR02088.0004-9409/03/020149 [DOI] [Google Scholar]
- Reverter A. A., Johnston D. J. A., Perry D. B., Goddard M. E. A., and Burrow H. M. D.. 2003b. Genetic and phenotypic characterisation of animal, carcass, and meat quality traits from temperate and tropically adapted beef breeds. 2. Abattoir carcass traits. Aust. J. Exp. Agric. 54:119–134. doi: 10.1071/AR02085.0004-9409/03/020107 [DOI] [Google Scholar]
- Rezende F. M., Ferraz J. B. S., Groeneveld E., Mourão G. B., Oliveira P. S., and Eler J. P.. 2009. Estimation of genetic and phenotypic parameters for meat and carcass traits in Nellore bulls. In Books of Abstracts of the 60th Annual Meeting of the European Association of Animal Production; August 2009; Barcelona, Spain. [Google Scholar]
- Riley D. G., Chase C. C. Jr, Hammond A. C., West R. L., Johnson D. D., Olson T. A., and Coleman S. W.. 2002. Estimated genetic parameters for carcass traits of Brahman cattle. J. Anim. Sci. 80:955–962. doi: 10.2527/2002.804955x [DOI] [PubMed] [Google Scholar]
- Riley D. G., Chase C. C. Jr, Hammond A. C., West R. L., Johnson D. D., Olson T. A., and Coleman S. W.. 2003. Estimated genetic parameters for palatability traits of steaks from Brahman cattle. J. Anim. Sci. 81:54–60. doi: 10.2527/2003.81154x [DOI] [PubMed] [Google Scholar]
- Sargolzaei M., Chesnais J. P., and Schenkel F. S.. 2014. A new approach for efficient genotype imputation using information from relatives. BMC Genomics 15:478. doi: 10.1186/1471-2164-15-478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelford S., Koohmaraie M., Cundiff L., Gregory K., Rohrer G., and Savell J.. 1994. Heritabilities and phenotypic and genetic correlations for bovine postrigor calpastatin activity, intramuscular fat content, Warner-Bratzler shear force, retail product yield. J. Anim. Sci. 72:857–863. doi: 10.2527/1994.724857x [DOI] [PubMed] [Google Scholar]
- Shanks B. C., Tess M. W., Kress D. D., and Cunningham B. E.. 2001. Genetic evaluation of carcass traits in Simmental-sired cattle at different slaughter end points. J. Anim. Sci. 79:595–604. [DOI] [PubMed] [Google Scholar]
- Smith T., Domingue J. D., Paschal J. C., Franke D. E., Bidner T. D., and Whipple G.. 2007. Genetic parameters for growth and carcass traits of Brahman steers. J. Anim. Sci. 85:1377–1384. doi: 10.2527/jas.2006-653 [DOI] [PubMed] [Google Scholar]
- Tizioto P. C., Decker J. E., Taylor J. F., Schnabel R. D., Mudadu M. A., Silva F. L., Mourao G. B., Coutinho L. L., Tholon P., Sonstegard T. S.,. et al. 2013. Genome scan for meat quality traits in Nelore beef cattle. Physiol. Genomics. 45:1012–1020. doi: 10.1152/physiolgenomics.00066.2013 [DOI] [PubMed] [Google Scholar]
- Utrera A. R., and Van Vleck L. D.. 2004. Heritability estimates for carcass traits of cattle: a review. Genet. Mol. Res. 3:380–394. [PubMed] [Google Scholar]
- VanRaden P. M. 2008. Efficient methods to compute genomic predictions. J. Dairy Sci. 91:4414–4423. doi: 10.3168/jds.2007-0980 [DOI] [PubMed] [Google Scholar]
- Van Tassell C. P., and Van Vleck L. D.. 1996. Multiple-trait Gibbs sampler for animal models: flexible programs for Bayesian and likelihood-based (co)variance component inference. J. Anim. Sci. 74:2586–2597. [PubMed] [Google Scholar]
- Warner R. D., Greenwood P. L., Pethick D. W., and Ferguson D. M.. 2010. Genetic and environmental effects on meat quality. Meat Sci. 86:171–183. doi: 10.1016/j.meatsci.2010.04.042 [DOI] [PubMed] [Google Scholar]
- Wheeler T. L., Cundiff L. V., Shackelford S. D., and Koohmaraie M.. 2001. Characterization of biological types of cattle (cycle V): carcass traits and longissimus palatability. J. Anim. Sci. 79:1209–1222. doi: 10.2527/2001.7951209x [DOI] [PubMed] [Google Scholar]