Abstract
Hepatocyte growth factor (HGF) signals mediate mouse skeletal muscle stem cell, or satellite cell (SC), reentry into the cell cycle and myoblast proliferation. Because the athletic horse experiences exercise-induced muscle damage, the objective of the experiment was to determine the effect of HGF on equine SC (eqSC) bioactivity. Fresh isolates of adult eqSC were incubated with increasing concentrations of HGF and the initial time to DNA synthesis was measured. Media supplementation with HGF did not shorten (P > 0.05) the duration of G0/G1 transition suggesting the growth factor does not affect activation. Treatment with 25 ng/mL HGF increased (P < 0.05) eqSC proliferation that was coincident with phosphorylation of extracellular signal-regulated kinase (ERK)1/2 and AKT serine/threonine kinase 1 (AKT1). Chemical inhibition of the upstream effectors of ERK1/2 or AKT1 elicited no effect (P > 0.05) on HGF-mediated 5-ethynyl-2′-deoxyuridine (EdU) incorporation. By contrast, treatment of eqSC with 2 µm Gö6983, a pan-protein kinase C (PKC) inhibitor, blocked (P < 0.05) HGF-initiated mitotic activity. Gene-expression analysis revealed that eqSC express PKCα, PKCδ, and PKCε isoforms. Knockdown of PKCδ with a small interfering RNA (siRNA) prevented (P > 0.05) HGF-mediated EdU incorporation. The siPKCδ was specific to the kinase and did not affect (P > 0.05) expression of either PKCα or PKCε. Treatment of confluent eqSC with 25 ng/mL HGF suppressed (P < 0.05) nuclear myogenin expression during the early stages of differentiation. These results demonstrate that HGF may not affect activation but can act as a mitogen and modest suppressor of differentiation.
Keywords: myogenesis, PKCδ, proliferation, satellite cell
INTRODUCTION
Satellite cells (SC), a population of muscle stem and progenitor cells, are located adjacent to the skeletal muscle fiber under the basal lamina (Mauro, 1961). The SC typically resides in a quiescent state but becomes mitotically active in response to muscle damage. Genetic ablation of SC within the adult mouse completely abolishes skeletal muscle regeneration demonstrating its essential role in damage repair (Sambasivan et al., 2011; von Maltzahn et al., 2013). A case for their involvement in muscle hypertrophy also exists. Removal of ≥90% of SC in adult mice prevented fiber hypertrophy in response to synergist ablation (SA) of soleus and gastrocnemius muscles (Egner et al., 2016). Using a different approach, disruption of SC fusogenic capabilities prevented fiber hypertrophy during SA further supporting a role for the cell type during growth (Goh and Millay, 2017). These findings are in stark contrast to others demonstrating that SC are not required for overload-induced hypertrophy (McCarthy et al., 2011), atrophic recovery (Jackson et al., 2012), or fiber remodeling and plasticity (Lee et al., 2016; Murach et al., 2017a, 2017b). Differences may be attributed to genetic ablation models, animal age, and subtleties in hypertrophy measures.
Contribution of the SC to growth and regeneration requires exit from G0 followed by commitment to the myoblast lineage and progression through myogenesis; a subpopulation undergoes self-renewal to maintain the muscle precursor pool. Multiple growth factors, cytokines, and other signaling molecules that reside within the fiber niche work in concert to regulate myogenesis. For example, Notch signals are required for SC self-renewal and quiescence (Bjornson et al., 2012; Bi et al., 2016). Genetic ablation of Hes/Hey, Notch target genes, causes SC to undergo precocious differentiation (Fukada et al., 2011). Migration and motility of SC to sites of microdamage are controlled by spatial localization of ephrins and their ligands on myofibers and interstitial cells (Stark et al., 2011; Gu et al., 2016). Expansion of the SC pool to support fiber damage repair relies upon the mitogenic actions of fibroblast growth factor (FGF) 2 and FGF receptor signaling (Yablonka-Reuveni et al., 2015; Galimov et al., 2016; Pawlikowski et al., 2017). Fusion of SC is positively regulated by IGF-1 and multiple genetic models demonstrate the hypertrophic effects of the growth factor on skeletal muscle (Kandalla et al., 2011; Schiaffino and Mammucari, 2011). The presence and diverse actions of these niche-localized factors underscore the complexity of signals present during normal growth as well as injury repair.
Hepatocyte growth factor (HGF) is an autocrine growth factor produced by the SC that participates in multiple stages of myogenesis (Anderson, 2016). Early work established that HGF activates G0 exit of quiescent SC and inhibition of HGF signaling delays cell cycle entry in vitro (Allen et al., 1995; Miller et al., 2000; Sheehan et al., 2000). The bioavailability of HGF to the SC is controlled sequentially by nitric oxide production, which in turn causes the enzymatic release of membrane-tethered HGF allowing the ligand to dock with the HGF receptor (MET; Tatsumi et al., 2002; Wozniak et al., 2003; Wozniak and Anderson, 2007, 2009; Tatsumi et al., 2009). In addition to its role as an activator, HGF promotes SC proliferation through recruitment of GRB2-associated-binding protein (Gab1) and tyrosine-protein phosphatase nonreceptor type-11 (SHP2) to the MET kinase domain which may allow for sustained ERK1/2 phosphorylation (Leshem et al., 2002; Halevy and Cantley, 2004; Li et al., 2009). The local concentration of HGF and intracellular ERK1/2 signal intensity are critical to SC biology as elevation in either leads to cell cycle exit (Reed et al., 2007; Yamada et al., 2010). The role of HGF during myoblast differentiation remains unresolved. Satellite cell-specific ablation of MET impairs migration, motility, and myoblast fusion during muscle regeneration (Webster and Fan, 2013). Human primary myoblasts cultured in differentiation permissive conditions increase fusion and myotube formation in response to HGF treatment (Walker et al., 2015). Treatment of mice with a synthetic, bivalent HGF protein initiates MET signal transduction through AKT serine/threonine kinase 1 (AKT1) to promote muscle fiber hypertrophy (Cassano et al., 2008). By contrast, others report that HGF inhibits SC differentiation in vitro and injection of the growth factor blunts fiber formation during damage repair (Leshem et al., 2000; Miller et al., 2000).
The athletic horse, similar to humans, experiences skeletal muscle damage upon completion of strenuous sporting activities. Several growth factors are expressed in muscle during the immediate post-race recovery period including HGF (Kawai et al., 2013). Due to the importance of HGF as a regulator SC function, the objectives of the experiment were to determine its effects on equine SC (eqSC) bioactivity and to identify the signaling systems that mediate HGF effects.
MATERIALS AND METHODS
Animal Care
Animal work was reviewed and approved by the Institutional Animal Care and Use Committee at Virginia Polytechnic Institute and State University (16–074).
Satellite Cell Isolation and Culture
Adult light breed geldings (n = 6; 3–8 yr of age) were utilized for all studies. Animals were sedated with xylazine (1 mg/kg; Bimeda-MTC Animal Health, Cambridge, ON) and administered a local anesthetic (Lidocaine; Aspen Veterinary Resources, Liberty, MO) subcutaneously atop the middle gluteal muscle. The muscle was chosen for sampling due to the large amount of data available and its participation in strenuous exercise. Biopsies (~200 mg/sample) were retrieved with a vacuum-assisted biopsy device (Vacora Biopsy System, 10 G; C. R. Bard, Tempe, AZ) and maintained individually in wash buffer [WB; 5% fetal bovine serum (FBS), 5% penicillin-streptomycin, 0.2% gentamicin in PBS] on ice prior to transport and processing in the lab. Muscle samples were washed three times with WB, dissected free of fat and connective tissue, and minced using sterile dissection scissors. Minced tissue was incubated at 37 °C for 40 min in 1 mg/mL protease (Type XIV, Sigma Aldrich, St. Louis, MO) in PBS. The tissue slurry was passed through a 70 µm cell strainer and retained fiber fragments were collected in WB, vortexed 2 min, and passed through a 40 µm cell strainer. The filtrate containing eqSC was collected by centrifugation at 800 × g for 5 min. The pellet was resuspended in Dulbecco’s Modified Eagle Medium (DMEM) containing 20% FBS, 1% penicillin-streptomycin, 0.2% gentamicin, and 4 ng/mL recombinant human FGF-2 (growth media; GM). Equine SC were seeded onto entactin–collagen–laminin (ECL; EMD Millipore) coated plates and cultured at 37 °C in a humidified, 5% CO2-controlled atmosphere. All culture media and supplements were purchased from Gibco-ThermoFisher, Waltham, MA. Myogenicity (>90%) was confirmed by Pax7, myogenin, and myosin heavy chain (MyHC) immuoreactivity prior to commencement of experiments. Cells were manually dissociated from the substratum with a sterile cell scraper for passage and expansion prior to proliferation and differentiation assays. Biological specimens were maintained and treated individually throughout all assays. All assays included a minimum of four biological replicates in duplicate.
Lag Period Measurement
Immediately following eqSC isolation, cells (n = 5) were seeded equally into ECL-coated 48-well plates and allowed to attach to the matrix overnight. At 22 h post-seeding, the cells were washed repeatedly with WB and re-fed GM or placed into DMEM + 1% FBS + 1% penicillin-streptomycin + 0.2% gentamicin (basal medium; BM) supplemented with 1, 5, 10, or 25 ng/mL recombinant human HGF (HGF; R&D Systems, Minneapolis, MN), doses of HGF demonstrated to affect cell cycle or regeneration, or 0.1% BSA in PBS control (Miller et al., 2000). At 24-h intervals subsequent, eqSC were pulsed for 2 h with 10 µM 5-ethynyl-2′-deoxyuridine (EdU), a thymidine analog. Cells were fixed with 4% paraformaldehyde (PFA) in PBS for 15 min at room temperature at the end of the EdU pulse period. EdU was detected with click chemistry (Click-It EdU AlexaFluor488, Invitrogen, Carlsbad, CA) and total nuclei were visualized with Hoechst 33342 (10 µg/mL). A mitotic index was calculated as EdU (+)/total Hoechst (+) × 100.
Proliferation and Differentiation Assays
Serially passaged eqSC at 30% confluency were treated with 1, 5, 10, or 25 ng/mL HGF or 0.1% BSA PBS control in BM for 48-h proliferation assays, with EdU included during the final 2 h prior to PFA fixation. For assays involving the inhibition of signaling intermediates, chemical inhibitors or an equal amount of dimethyl sulfoxide were added for 1 h prior to and for the duration of the stimulus. Chemical inhibitors and concentrations were 25 µM PD98059 (Cell Signaling Technology, Danvers, MA), 200 nM wortmannin (Cell Signaling Technology), and 2 µM Gö6983 (Tocris Bioscience, Minneapolis, MN).
Equine SC at 80% confluency were treated with 25 ng/mL HGF or 0.1% BSA in PBS in low-glucose DMEM containing 1% FBS, 1% penicillin-streptomycin, and 0.2% gentamicin (differentiation media) for 24 or 48 h for analysis of early and late differentiation, respectively. Cells were fixed with PFA for 15 min at room temperature or acidified formalin alcohol on ice for 4 mins then washed repeatedly with PBS. Nonspecific antigen sites were blocked with 3% BSA in PBS containing 0.1% Triton ×100 for 20 min followed by overnight incubation with anti-myogenin hybridoma supernatant (F5D, Developmental Studies Hybridoma Bank, Iowa City, IA) or anti-MyHC hybridoma supernatant (MF20, Developmental Studies Hybridoma Bank) diluted 1:5 in blocking buffer. Cells were washed with PBS prior to 1-h incubation with goat anti-mouse IgG AlexaFluor568 (1:200; ThermoFisher), 10 μg/mL Hoechst 33342. Representative photomicrographs were captured at equal shutter speeds to ensure consistent intensity at 200-fold magnification with a Nikon Eclipse TS100 epifluorescent microscope (Nikon Imaging Corp., Melville, NY) connected to a CoolSNAP HQ2 camera (Photometrics, Tuscon, AZ) and digitized with NIS Elements AR Ver4.13.00 software (Nikon). As an indicator of early differentiation, an index was calculated as myogenin (+)/Hoechst (+) × 100. Fusion index was calculated as (number of Hoechst 33342 positive cells residing within the myotube)/(total number of Hoechst 33342 positive cells) × 100. Representative images are shown in black and white for ease of reading.
Small Interfering RNA Knockdown of PKC Isoforms
Semi-confluent eqSC were transiently transfected with 5 nM of scrambled or target-specific small interfering RNA (siRNA) (Lipofectamine RNAiMAX; Invitrogen), according to manufacturer’s protocols. Oligonucleotides were custom synthesized by Invitrogen (Table 1). After 24 h, the media was replaced with BM supplemented with 25 ng/mL HGF or an equivalent amount of 0.1% BSA in PBS. Cells were pulsed with 10 µM EdU 2 h prior to fixation at 24 h in treatment. Detection of EdU was performed using Click-it EdU detection kit (Click-It EdU AlexaFluor 488; Invitrogen) and nuclei were detected with Hoechst 33342 (10 μg/mL). EdU (+) and Hoechst (+) cells were visualized and enumerated for the calculation of mitotic index, as described above.
Table 1.
Small interfering RNA sequences
| Sense (5ʹ-3ʹ) | Antisense (5ʹ-3ʹ) | |
|---|---|---|
| PKCα | CGACGACUCUCUGUAGAAA | UUUCUACAGAGAUCGUCG |
| PKCα—scramble | CGACGACUGAGUGUAGAAA | UUUCUACACUCAGUCGUCG |
| PKCδ | GCAUGAACGUGCACCAUAA | UUAUGGUGCACGUUCAUGC |
| PKCδ—scramble | GCAUGAACCACCACCUAA | UUAUGGUGGUGGUUCAUGC |
| PKCε | ACCUCGAAUUAAAACCAAA | UUUGGUUUUAAUUCGAGGU |
| PKCε—scramble | ACCUCGAAAAUAAACCAAA | UUUGGUUUAUUUUCGAGG |
Quantitative Reverse-Transcription PCR
Total RNA was extracted using Trizol reagent coupled with column purification (PureLink RNA Mini Kit, Invitrogen). Genomic DNA was removed by DNAse I (Invitrogen) digestion. RNA quantification and purity were assessed by spectrophotometry (Nanodrop, ThermoFisher). One microgram of total RNA was reverse transcribed (High Capacity cDNA kit, ThermoFisher) in a final volume of 20 µL. Fifty nanograms of cDNA was amplified with DNA polymerase (GoTaq Green master mix, Promega, Madison, WI) and gene-specific primers for glyceraldehyde phosphate dehydrogenase (F, 5-CCACCCCTAACGTGTCAGTC, R, 5-AATCGCAGGAGACAACCTGG, efficiency = 0.89), pan-protein kinase C (PKC) α (F, 5-GACTCCCTGGTATGTGCTCG, R, 5-GCTC CTCACAAGACCGGAAA, efficiency = 0.84), PKCδ (F, 5-GTTCCCAACAATGAACCGCC, R, 5-CAGAAGGTGGGCTGCCTAAA, efficiency = 0.85), and PKCε (F, 5-TGTCAACATG CCCCACAAGT, R, 5-TGTCATTGCACAACAG AGG, efficiency = 0.85). Polymerase chain reaction conditions were 95 °C for 2 min followed by 40 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s. A final extension of 72 °C for 5 min was performed. Amplicons were electrophoretically separated through 1.2% TAE agarose gels impregnated with SYBR-Safe dye (Invitrogen). Amplicon size was confirmed by comparison to a molecular weight ladder (exACTGene DNA Ladder, Fisher Scientific, Waltham, MA). Quantitative PCR was performed with 5 ng cDNA and gene-specific primers using SYBR chemistry (Power SYBR Green PCR, ThermoFisher) in an Eppendorf Realplex thermocycler (Eppendorf, Hamburg, Germany). The optimum thermal cycling parameters included 95 °C for 10 min, 40 cycles of 95 °C for 15 s and 60 °C for 1 min. A melt curve was generated by 95 °C for 15 s, 60 °C for 15 s followed by 1.75 °C/min for 20 min. Glyceraldehyde phosphate dehydrogenase was used as a housekeeping gene for normalization and fold change for all the samples relative to its own scrambled control was calculated using the 2−ΔΔCt method.
Western Blotting
Equine SC (n = 5) were lysed in ice-cold radioimmunoprecipitation assay buffer containing protease and phosphatase inhibitors (Halt Inhibitor Cocktail, ThermoFisher). Protein concentration was measured by bicinchoninic acid assay (ThermoFisher). Twenty micrograms of protein were denatured with lithium dodecyl sulfate sample buffer (ThermoFisher) at 95 °C for 5 min prior to electrophoretic separation through 10% Tris-glycine gels (Novex Wedge Well, ThermoFisher). Proteins were transferred to nitrocellulose (iBlot 2, ThermoFisher). Membranes were blocked in 0.1% Tween-20 Tris-buffered saline (TBST) containing 5% nonfat dry milk for 1 h. Membranes were washed 3 × 5 min with TBST prior to incubation with primary antibodies diluted in TBST containing 5% BSA overnight at 4 °C. Antibodies (Cell Signaling Technology) and dilutions were anti-ERK1/2, 1:1000, anti-phosphoERK1/2, 1:2000, anti-AKT1, 1:1000, anti-phosphoAKT1, 1:2000, and anti-beta tubulin, 1:1000. Membranes were washed with TBST and incubated with goat anti-rabbit IgG peroxidase (1:1000, ThermoFisher) for 1 h at room temperature. Following TBST washes, the antigen-antibody complexes were visualized by chemiluminescence (ECL, ThermoFisher) using a Chemi-Doc MP system equipped with Image Lab software (BioRad, Hercules, CA).
Statistics
The GLM procedure of SAS (SAS Enterprise Guide, Cary, NC) was used to generate a one- or two-way ANOVA. The model included horse, concentration of growth factor, chemical additive, the interaction between growth factor and chemical additive, and hours in culture where appropriate. Technical replicates were averaged for each biological replicate prior to calculation of least square means and SEM. Post-test comparisons were performed using Tukey’s adjustment to analyze pre-planned comparisons between groups. Significance was set at P < 0.05.
RESULTS
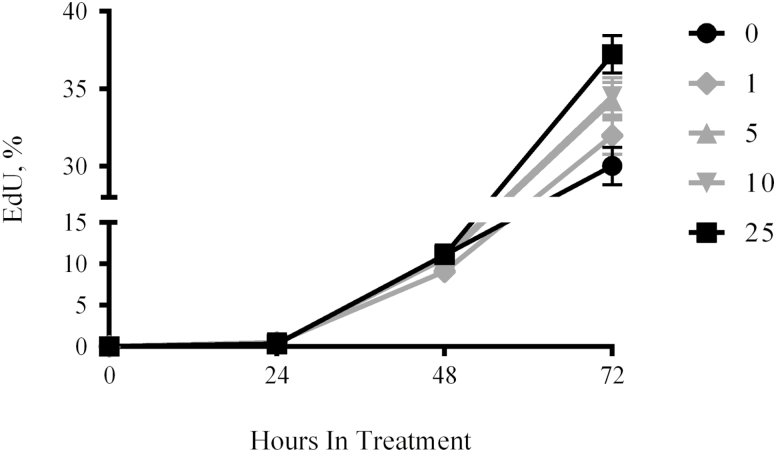
Hepatocyte growth factor is regarded as an activator of rodent SC that facilitates their exit from G0 and decreases the time to first division in vitro (Anderson, 2016). To determine if the growth factor elicits similar effects on eqSC, isolates from adult horses were treated with increasing concentrations of HGF for 72 h and the numbers of mitotic cells were enumerated daily. Results demonstrate the presence of EdU (+) cells after 48 h of treatment indicating the cells have exited G0 and transitioned into S-phase (Fig. 1). No differences (P > 0.05) in the percentage of cells incorporating the thymidine analog were evident in the presence of 0, 1, 5, 10, or 25 ng/mL HGF after 48 h. Treatment with 25 ng/mL increased (P < 0.05) proliferation at 72 h by comparison to controls receiving vehicle-only. No differences (P > 0.05) in proliferation rate were evident at 72 h for 0, 1, 5, or 10 ng/mL HGF.
Figure 1.
Hepatocyte growth factor supplementation does not alter time to S-phase. Fresh isolates of eqSC (n = 5) were allowed to attach overnight followed by treatment with 0, 1, 5, 10, and 25 ng/mL HGF for 72 h. Cells were pulsed with EdU for 2 h prior to fixation at 24-h intervals. EdU (+) and total nuclei were enumerated. Percent EdU = EdU(+)/Hoechst 33342 (+) × 100. Means and SEMs shown. Black symbols (0 and 25 ng/mL) are different (P < 0.05) from one another at 72 h only. eqSC = equine satellite cell.
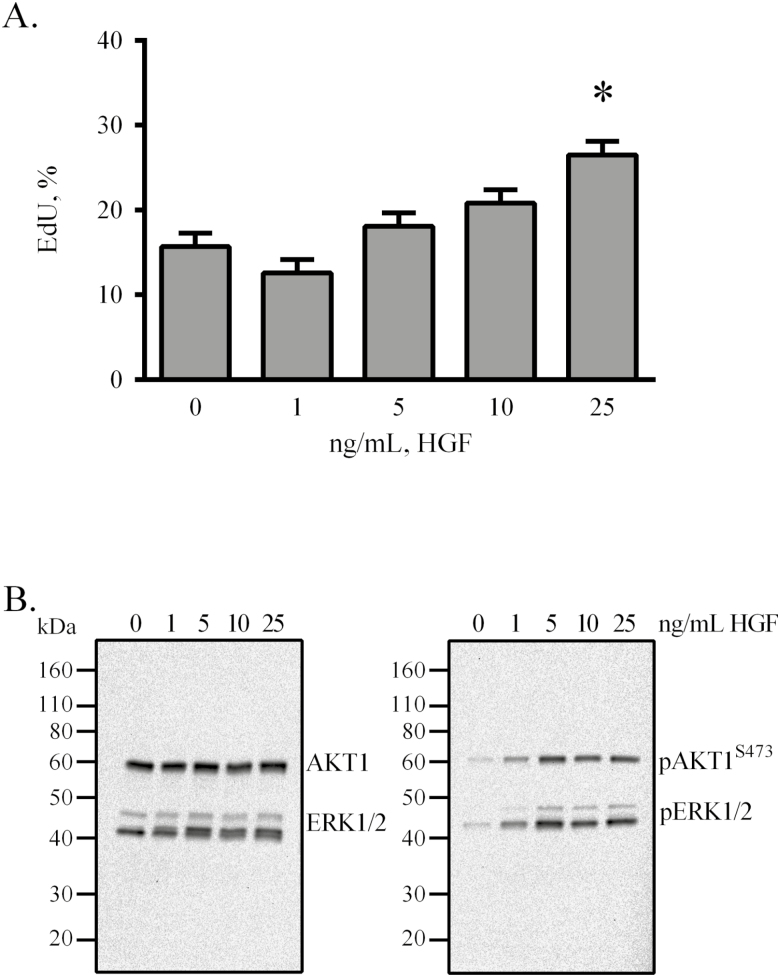
To further evaluate the effects of HGF on proliferation, serial-passaged eqSC were treated for 48 h with BM supplemented with 0, 1, 5, 10, and 25 ng/mL HGF and a mitotic index calculated. Results demonstrate that only 25 ng/mL HGF stimulated (P < 0.05) cell proliferation (Fig. 2A). Previous studies indicate that ERK1/2 and AKT1 are activated in response to HGF (Halevy and Cantley, 2004). The status of these two signaling axes was investigated by Western blot using lysates prepared from cells treated with the various concentrations of HGF. After 20 min of stimulation, eqSC exhibited phosphorylation of both AKT1 and ERK1/2 (Fig. 2B). No apparent differences in chemiluminescent band intensity for the phosphoproteins was evident by visual appraisal suggesting that 1 ng/mL was as effective as 25 ng/mL at initiating AKT1 and ERK1/2 modification. Interestingly, HGF stimulates preferentially the phosphorylation of ERK2 by comparison to ERK1.
Figure 2.
Hepatocyte growth factor stimulates proliferation and phosphorylation of AKT1 and ERK1/2. Semi-confluent eqSC (n = 5) were treated with HGF for 48 h with a 2-h pulse of EdU prior to fixation (A). Cells were treated with the indicated concentration of HGF for 20 min, lysed and analyzed by Western blot for total and phosphorylated forms of AKT1 and ERK1/2 (B). EdU (+) and total nuclei were enumerated. Western blot exposure time was 2 mins. Percent EdU = EdU (+)/Hoechst 33342 (+) × 100. Means and SEMs shown. *Significance between control and treatment at P < 0.05. AKT1 = AKT serine/threonine kinase 1; eqSC = equine satellite cell.
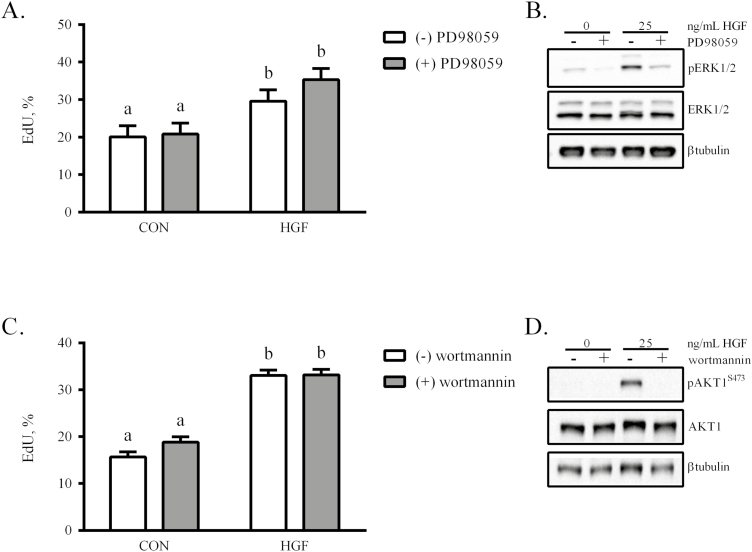
Using a dose of 25 ng/mL HGF, the necessity of ERK1/2 activation on eqSC proliferation was evaluated. Treatment with HGF increased (P < 0.05) proliferation, as measured by EdU incorporation, by comparison to control (CON; Fig. 3A). Incubation of eqSC with HGF in the presence of a MEK1/2 inhibitor (PD98059) did not suppress (P > 0.05) the proliferative effect. The concentration of MEK1/2 inhibitor was sufficient to block ERK1/2 phosphorylation (Fig. 3B). Thus, HGF initiation of ERK1/2 phosphorylation does not underlie its ability to stimulate eqSC proliferation. In a similar manner, eqSC were treated with HGF in the presence or absence of wortmannin. HGF increased proliferation (P < 0.05) that remained unaffected (P > 0.05) by inclusion of the phosphoinositide 3-kinase (PI3K) inhibitor (Fig. 3C). Western blot confirmed that 200 nM wortmannin was sufficient to prevent phosphorylation and subsequent activation of AKT1 (Fig. 3D).
Figure 3.
The mitotic actions of HGF are not mediated by phosphorylation of either ERK1/2 or AKT1. Semi-confluent eqSC (n = 4) were treated with 25 ng/mL HGF in the presence or absence of 25 µM PD98059 (A), a MEK1/2 inhibitor, or 200 nM wortmannin (B), a PI3K inhibitor. After 48 h, cells were fixed and EdU(+) and total nuclei were enumerated. Parallel plates were lysed for Western blot analysis of phosphoERK1/2 (C) and phosphoAKT1 (D). Percent EdU = EdU (+)/Hoechst 33342 (+) × 100. Means and SEMs shown. Means with different letters are significant at P < 0.05. AKT1 = AKT serine/threonine kinase 1; eqSC = equine satellite cell; PI3K = phosphoinositide 3-kinase.
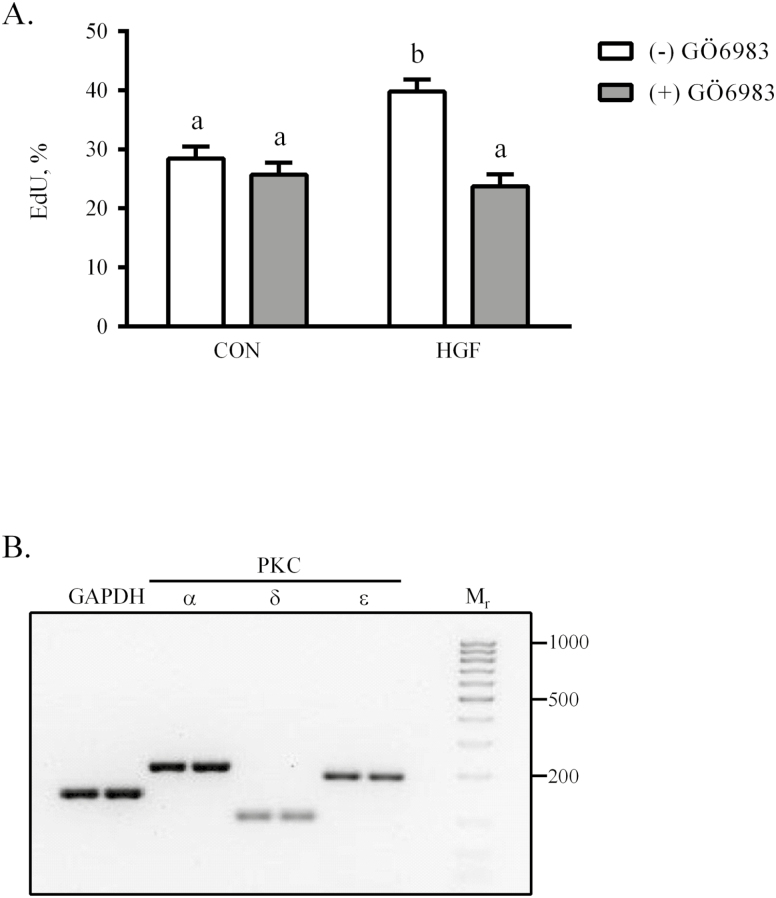
The finding that neither AKT1 nor ERK1/2 activity was required for HGF-initiated eqSC proliferation was unexpected. Because PKC isoforms can increase mitotic rates in other cells (Poli et al., 2014), we chose to examine this system as a modulator of HGF effects. Semi-confluent eqSC were treated for 48 h in basal media supplemented with 25 ng/mL HGF with or without 2 µM Gö6983, an inhibitor that preferentially targets both classical and novel PKC isoforms. HGF stimulated (P < 0.05) eqSC proliferation by comparison to CON treated with vehicle-only (Fig. 4A). Co-incubation of cells with HGF and Gö6983 diminished (P < 0.05) EdU incorporation to levels observed in CON. Thus, a PKC isoform likely is involved in HGF-driven eqSC division. To narrow the list of potential candidates, total RNA was isolated from semi-confluent eqSC, reverse transcribed and amplified with gene-specific primers for PKCα, PKCβ, PKCδ, PKCε, and PKCζ. End-point PCR reveals that eqSC express PKCα, PKCδ, and PKCε; no amplicons for PKCβ and PKCζ were detected (Fig. 4B).
Figure 4.
Protein kinase C mediates the proliferative effects of HGF. Semi-confluent eqSC (n = 4) were treated with HGF in the presence or absence of 2 µM Gö6983 for 48 h (A). Cells were pulsed with EdU 2 h prior to fixation. Total and EdU (+) nuclei were enumerated. Isoforms of PKC expressed by eqSC were identified by RT-PCR using gene-specific primers. Amplicons were separated electrophoretically through SYBR-Safe impregnated agarose gels. (B). RNA isolates from two animals are shown per gene of interest. Molecular weight ladder ranges from 1 kb to 25 bp. Amplicon sizes are GAPDH, 150 bp, PKCα, 201 bp, PKCδ, 102 bp and PKCε, 191 bp. Percent EdU = EdU (+)/Hoechst (+) × 100. Means and SEMs shown. Means with different letters are significant at P < 0.05. eqSC = equine satellite cell; GAPDH = glyceraldehyde phosphate dehydrogenase; RT-PCR = real-time PCR.
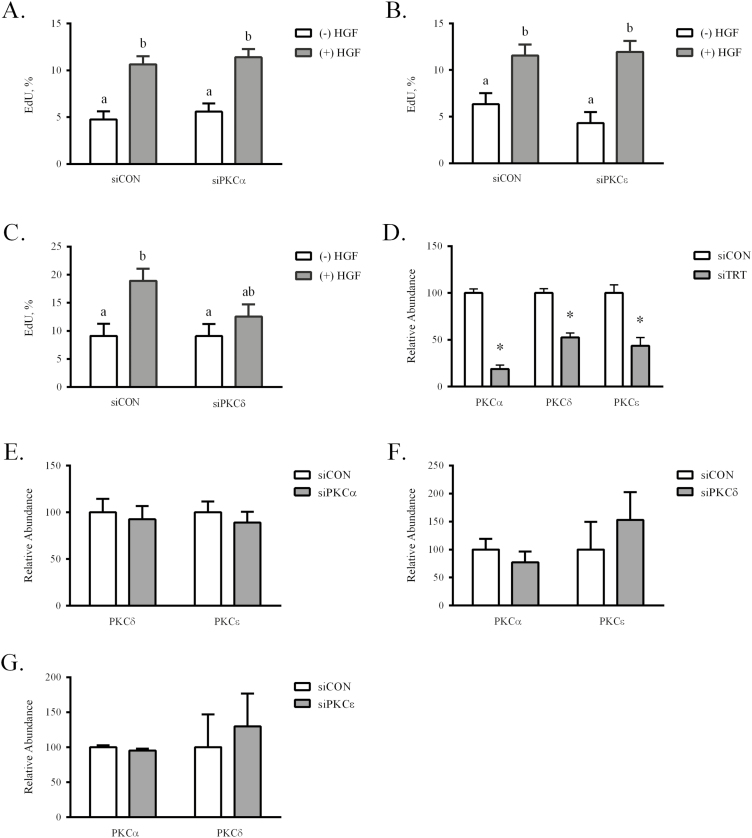
The contribution of the PKC isoforms to HGF-mediated proliferation was examined using siRNA methodology. In brief, siRNA specific for PKCα, PKCδ, and PKCε (siPKCα, siPKCδ, siPKCε) were designed and transfected into semi-confluent eqSC. Scrambled oligonucleotides for each of the PKC isoforms were included as controls (siCON) for nonspecific effects. Twenty-four hours after siRNA loading, eqSC were treated with HGF for 24 h and a mitotic index calculated. Results demonstrate that treatment of eqSC with siCON or siPKCα proliferated (P < 0.05) in response to 25 ng/mL HGF (Fig. 5A). A similar result was found for siPKCε (Fig. 5B). By contrast, eqSC ectopically expressing siPKCδ failed to respond (P > 0.05) to HGF treatment, retaining a proliferation rate similar to vehicle-treated siCON (Fig. 5C). Despite multiple attempts, antibodies against the PKC isoforms failed to detect the respective proteins by western blot. Thus, mRNA knockdown was confirmed by real-time PCR for the siPKCα, siPKCδ, and siPKCε oligonucleotides (Fig. 5D). Transfection of the isoform-specific siRNA resulted in an approximate 70, 50, and 60% reduction (P < 0.05) in PKCα, PKCδ, and PKCε, respectively, in the absence of detectable off-targeting (P > 0.05; Fig. 5E–G). Reduced PKCδ mRNA prevented eqSC response to 25 ng/mL HGF, in contrast to loss of either PKCα or PKCε. Thus, the results provide a role for PKCδ as a transducer of HGF signals that support eqSC proliferation.
Figure 5.
Hepatocyte growth factor signaling through PKCδ stimulates eqSC proliferation. Semi-confluent eqSC (n = 5) were transfected with scrambled (siCON) or siRNA specific for PKC isoforms. After 24 h, the cells were treated with HGF for 24 h with a 2 h EdU pulse prior to fixation. Total and EdU (+) nuclei were enumerated. Equine SC treated with siPKCα (A) or siPKCε (B) did not prevent HGF stimulated proliferation. Cells expressing siPKCδ did not respond to HGF (C). Means with different letters are significant at P < 0.05. Total RNA from siCON- and siPKC-treated cells was analyzed by qPCR for specific knockdown (D) and off-target (E–G) reduction of PKCα, PKCδ, and PKCε mRNA. Relative expression was calculated by 2−ΔΔCt method. *Significance at P < 0.05 within treatment. eqSC = equine satellite cell; qPCR = quantitative PCR; siRNA = small interfering RNA.
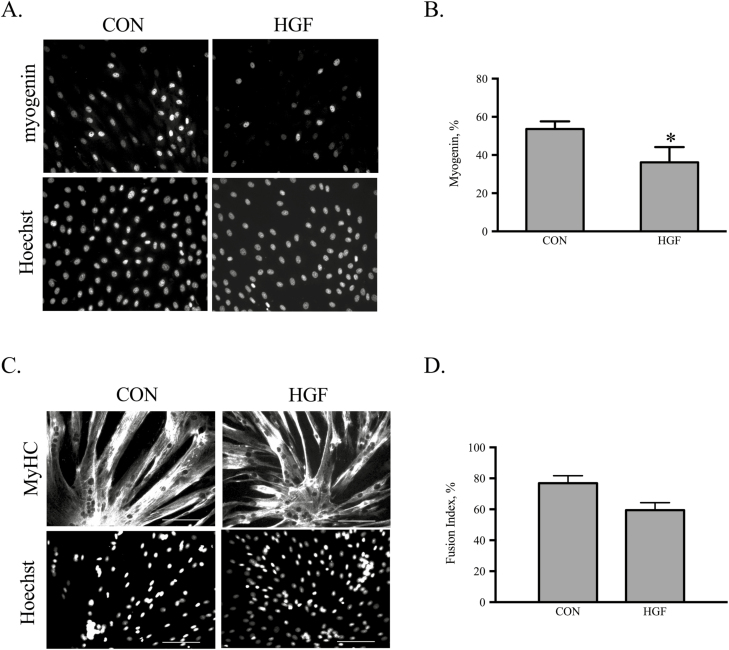
Increased differentiation was reported following treatment of human SC with HGF (Walker et al., 2015). The role of HGF during eqSC myotube formation was examined in vitro. Cultures at 80% confluency were treated with 25 ng/mL HGF for 24 h or 48 h followed by fixation. Immunocytochemical detection of myogenin revealed fewer cells express the transcription factor at 24 h (Fig. 6A). Enumeration of total and myogenin (+) nuclei indicates HGF reduces (P < 0.05) the percentage of terminally differentiated myoblasts by approximately 30% (Fig. 6B). By 48 h in culture both CON and HGF cultures contain large multinucleated myotubes (Fig. 6C) with no difference (P > 0.05) in the calculated fusion index (Fig. 6D). Thus, HGF serves to delay but not prevent myogenesis.
Figure 6.
Hepatocyte growth factor inhibits eqSC differentiation. Eighty percent confluent eqSC (n = 4) were treated with HGF for 24 or 48 h followed by fixation and immunocytochemical detection of myogenin or MyHC. Representative black and white photomicrographs of myogenin expression at equal shutter speeds for CON cells receiving vehicle and eqSC treated with 25 ng/mL HGF are shown (A). Percent myogenin (+) was calculated as total myogenin/total Hoechst 33342 × 100 (B). Representative black and white images of MyHC immunostaining at 48 h (C). Percent fusion was calculated as total number nuclei within a MyHC myotube/total number nuclei × 100 (D). Scale bar = 100 µm. *Significance at P < 0.05. eqSC = equine satellite cell.
DISCUSSION
The skeletal muscle of athletes performing strenuous exercise undergoes a period of post-event recovery that includes activation of SC to repair fiber microdamage (Paulsen et al., 2012; Kawai et al., 2013; Bryan et al., 2017). Increased expression of several cytokines and growth factors is observed prior to SC activation suggesting they may play a role in modulating SC bioactivity. In the horse, HGF and Pax7 mRNA expression increase nearly 2-fold at day 3 of the exercise-recovery period (Kawai et al., 2013). Because HGF plays a significant role during SC exit from G0 in rodents, we investigated the ability of the growth factor to perform a similar function in eqSC. Interestingly, our results demonstrate that supplementation of eqSC with HGF does not alter the time to S-phase following initial seeding. Thus, HGF may not be the requisite activator of eqSC. Species-specific differences are noted between human and mouse SC activation (Quarta et al., 2016). To prolong quiescence, mouse SC are cultured with both MET and FGF receptor inhibitors while only the FGF receptor inhibitor is supplemented to the human SC. FGF2 is present in the initial eqSC plating media prior to its replacement with low serum treatment medium thus, the mitogen could serve as an activator during that 24 h window. It should be noted that neither the metabolic activation status of the cells nor their residence in G1 was measured. It is possible that the eqSC are primed (Galert) and ready to progress into the cell cycle but arrested at a restriction point due to insufficient growth factors. The inability of HGF to reduce the initial lag period also may be attributed to subtle differences in ligand:receptor interactions. The human HGF used in these studies is 91% identical at the amino acid level to equine HGF. The strong amino acid homology coupled with the ability of human HGF to stimulate phosphorylation of intracellular signaling kinases supports effective ligand docking with the equine MET receptor. Importantly, SC synthesize and secrete HGF and autocrine HGF may cause local activation of the cell and prevent a response to exogenous growth factor due to receptor down-regulation (Sheehan et al., 2000). This aspect should be explored in future experiments. From the current experiments, we interpret the results to conclude that HGF-alone is unable to accelerate G1 progression as indicated by no change in the initial lag period.
Satellite cell migration and proliferation are controlled by specific intracellular signaling pathways initiated following HGF binding to MET, the cognate receptor. Ligand docking and MET kinase domain transphosphorylation lead to recruitment of the scaffold protein, Gab1. Genetic ablation of Gab1 results in compromised muscle formation due to the inability of migratory muscle progenitor cells to populate the analages (Sachs et al., 2000). The adaptor protein serves as a binding surface for multiple intracellular signaling intermediates including Grb2, SHP2, and signal transducer and activator of transcription 3 (STAT3; Barrow-McGee and Kermorgant, 2014). The mediator of Gab1 migratory effects involves SHP2, a tyrosine phosphatase. Site-directed mutagenesis of the SHP2-docking site in Gab1 results in both fewer muscle progenitor cells as well as their migration into the dorsal limb (Schaeper et al., 2007). Previous work by our group demonstrated that SHP2 is required for HGF-directed proliferation of mouse myoblasts likely through a MET/Gab1/SHP2 signalosome (Li et al., 2009). Further, conditional ablation of SHP2 in mouse SC disrupts activation and proliferation following injury (Griger et al., 2017). Extension of the cascade may include PKC isoforms. SHP2 is phosphorylated in vitro by multiple PKC isoforms including PKCδ (Strack et al., 2002). The phosphatase directs PKC to the insulin receptor complex thereby allowing PKC to phosphorylate and inactivate signal transduction (Müssig et al., 2005). Serving as a docking interface between PKCδ and MET, SHP2 may mediate the downstream signals that underlie HGF-initiated SC proliferation.
Satellite cells are required for the formation of new fibers following myotrauma and play an integral role during muscle hypertrophy through their ability to increase myonuclear content (von Maltzahn et al., 2013; Egner et al., 2016; Goh and Millay, 2017). Both functions are dependent upon SC fusion and subsequent expression of the myogenic differentiation program. In general, growth factors that act as SC mitogens (FGF2, PDGF) also suppress differentiation through mechanisms that are independent of proliferation (Pawlikowski et al., 2017). Our results indicate HGF elicits a reduction in early differentiation as determined by a lower percentage of myogenin (+) myoblasts. No differences were found in myoblast fusion or a subjective measure of apparent size of the myotubes. The ability of eqSC to form myotubes even in the presence of mitogenic concentrations of HGF may be partially attributed to ligand-induced phosphorylation of AKT1, a positive effector of biochemical and morphological myocyte differentiation (Schiaffino et al., 2013). A synthetic MET agonist with structural similarity to native HGF causes the phosphorylation and activation of AKT1 in C2C12 myoblasts that results in increased fusion and myotube size (Cassano et al., 2008; Perini et al., 2015). Injection of the MET agonist into skeletal muscle or transgenic expression of the protein in skeletal muscle increases fiber cross-sectional area and improves exercise performance (Ronzoni et al., 2017). The decreased number of myogenin positive nuclei and absence of an effect on myoblast fusion point to a delay in the differentiation program in response to HGF. The inability of HGF to completely abrogate eqSC differentiation may be attributed to the strong predisposition of primary SC to spontaneously differentiate even the presence of FBS and mitogens (Charville et al., 2015).
In summary, our results extend upon existing literature to provide novel roles for HGF during SC myogenesis. Unlike rodent SC, the initial lag period between G0 exit and S-phase is not shortened in eqSC treated with HGF. The growth factor is capable of initiating intracellular signaling cascades in eqSC that include phosphorylation of ERK1/2 and AKT1. Signals through the nonclassical PKCδ, however, are required for HGF-mediated SC proliferation. These results provide a foundation for future efforts modulating PKCδ as a means of promoting eqSC bioactivity.
LITERATURE CITED
- Allen R. E., Sheehan S. M., Taylor R. G., Kendall T. L., and Rice G. M.. 1995. Hepatocyte growth factor activates quiescent skeletal muscle satellite cells in vitro. J. Cell. Physiol. 165:307–312. doi: 10.1002/jcp.1041650211 [DOI] [PubMed] [Google Scholar]
- Anderson J. E. 2016. Hepatocyte growth factor and satellite cell activation. Adv. Exp. Med. Biol. 900:1–25. doi: 10.1007/978-3-319-27511-6_1 [DOI] [PubMed] [Google Scholar]
- Barrow-McGee R., and Kermorgant S.. 2014. Met endosomal signalling: in the right place, at the right time. Int. J. Biochem. Cell Biol. 49:69–74. doi: 10.1016/j.biocel.2014.01.009 [DOI] [PubMed] [Google Scholar]
- Bi P., Yue F., Sato Y., Wirbisky S., Liu W., Shan T., Wen Y., Zhou D., Freeman J., and Kuang S.. 2016. Stage-specific effects of Notch activation during skeletal myogenesis. Elife. 5:3593 doi:10.7554/eLife.17355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornson C. R., Cheung T. H., Liu L., Tripathi P. V., Steeper K. M., and Rando T. A.. 2012. Notch signaling is necessary to maintain quiescence in adult muscle stem cells. Stem Cells. 30:232–242. doi: 10.1002/stem.773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan K., McGivney B. A., Farries G., McGettigan P. A., McGivney C. L., Gough K. F., MacHugh D. E., Katz L. M., and Hill E. W.. 2017. Equine skeletal muscle adaptations to exercise and training: evidence of differential regulation of autophagosomal and mitochondrial components. BMC Genomics. 18:595. doi: 10.1186/s12864-017-4007-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassano M., Biressi S., Finan A., Benedetti L., Omes C., Boratto R., Martin F., Allegretti M., Broccoli V., Cusella De Angelis G.,. et al. 2008. Magic-factor 1, a partial agonist of Met, induces muscle hypertrophy by protecting myogenic progenitors from apoptosis.S. Wölfl, editor. PLoS ONE. 3:e3223 doi:10.1371/journal.pone.0003223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charville G. W., Cheung T. H., Yoo B., Santos P. J., Lee G. K., Shrager J. B., and Rando T. A.. 2015. Ex vivo expansion and in vivo self-renewal of human muscle stem cells. Stem Cell Reports. 5:621–632. doi: 10.1016/j.stemcr.2015.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egner I. M., Bruusgaard J. C., and Gundersen K.. 2016. Satellite cell depletion prevents fiber hypertrophy in skeletal muscle. Development. 143:2898–2906. doi: 10.1242/dev.134411 [DOI] [PubMed] [Google Scholar]
- Fukada S., Yamaguchi M., Kokubo H., Ogawa R., Uezumi A., Yoneda T., Matev M. M., Motohashi N., Ito T., Zolkiewska A., et al. 2011. Hesr1 and hesr3 are essential to generate undifferentiated quiescent satellite cells and to maintain satellite cell numbers. Development. 138:4609–4619. doi: 10.1242/dev.067165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galimov A., Merry T. L., Luca E., Rushing E. J., Mizbani A., Turcekova K., Hartung A., Croce C. M., Ristow M., and Krützfeldt J.. 2016. MicroRNA-29a in adult muscle stem cells controls skeletal muscle regeneration during injury and exercise downstream of fibroblast growth factor-2. Stem Cells. 34:768–780. doi: 10.1002/stem.2281 [DOI] [PubMed] [Google Scholar]
- Goh Q., and Millay D. P.. 2017. Requirement of myomaker-mediated stem cell fusion for skeletal muscle hypertrophy. Elife. 6:e20007 doi:10.7554/eLife.20007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griger J., Schneider R., Lahmann I., Schöwel V., Keller C., Spuler S., Nazare M., and Birchmeier C.. 2017. Loss of Ptpn11 (Shp2) drives satellite cells into quiescence. Elife. 6:633 doi:10.7554/eLife.21552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J. M., Wang D. J., Peterson J. M., Shintaku J., Liyanarachchi S., Coppola V., Frakes A. E., Kaspar B. K., Cornelison D. D., and Guttridge D. C.. 2016. An NF-κb–ephrinA5-dependent communication between NG2(+) interstitial cells and myoblasts promotes muscle growth in neonates. Dev. Cell. 36:215–224. doi: 10.1016/j.devcel.2015.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halevy O., and Cantley L. C.. 2004. Differential regulation of the phosphoinositide 3-kinase and MAP kinase pathways by hepatocyte growth factor vs. Insulin-like growth factor-I in myogenic cells. Exp. Cell Res. 297:224–234. doi: 10.1016/j.yexcr.2004.03.024 [DOI] [PubMed] [Google Scholar]
- Jackson J. R., Mula J., Kirby T. J., Fry C. S., Lee J. D., Ubele M. F., Campbell K. S., McCarthy J. J., Peterson C. A., and Dupont-Versteegden E. E.. 2012. Satellite cell depletion does not inhibit adult skeletal muscle regrowth following unloading-induced atrophy. Am. J. Physiol. Cell Physiol. 303:C854–C861. doi: 10.1152/ajpcell.00207.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandalla P. K., Goldspink G., Butler-Browne G., and Mouly V.. 2011. Mechano growth factor E peptide (MGF-E), derived from an isoform of IGF-1, activates human muscle progenitor cells and induces an increase in their fusion potential at different ages. Mech. Ageing Dev. 132:154–162. doi: 10.1016/j.mad.2011.02.007 [DOI] [PubMed] [Google Scholar]
- Kawai M., Aida H., Hiraga A., and Miyata H.. 2013. Muscle satellite cells are activated after exercise to exhaustion in thoroughbred horses. Equine Vet. J. 45:512–517. doi: 10.1111/evj.12010 [DOI] [PubMed] [Google Scholar]
- Lee J. D., Fry C. S., Mula J., Kirby T. J., Jackson J. R., Liu F., Yang L., Dupont-Versteegden E. E., McCarthy J. J., and Peterson C. A.. 2016. Aged muscle demonstrates fiber-type adaptations in response to mechanical overload, in the absence of myofiber hypertrophy, independent of satellite cell abundance. J. Gerontol. A. Biol. Sci. Med. Sci. 71:461–467. doi: 10.1093/gerona/glv033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshem Y., Gitelman I., Ponzetto C., and Halevy O.. 2002. Preferential binding of grb2 or phosphatidylinositol 3-kinase to the met receptor has opposite effects on HGF-induced myoblast proliferation. Exp. Cell Res. 274:288–298. doi: 10.1006/excr.2002.5473 [DOI] [PubMed] [Google Scholar]
- Leshem Y., Spicer D. B., Gal-Levi R., and Halevy O.. 2000. Hepatocyte growth factor (HGF) inhibits skeletal muscle cell differentiation: a role for the bHLH protein twist and the cdk inhibitor p27. J. Cell. Physiol. 184:101–109. doi: 10.1002/(SICI)1097-4652(200007) [DOI] [PubMed] [Google Scholar]
- Li J., Reed S. A., and Johnson S. E.. 2009. Hepatocyte growth factor (HGF) signals through SHP2 to regulate primary mouse myoblast proliferation. Exp. Cell Res. 315:2284–2292. doi: 10.1016/j.yexcr.2009.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Maltzahn J., Jones A. E., Parks R. J., and Rudnicki M. A.. 2013. Pax7 is critical for the normal function of satellite cells in adult skeletal muscle. Proc. Natl. Acad. Sci. U. S. A. 110:16474–16479. doi: 10.1073/pnas.1307680110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro A. 1961. Satellite cell of skeletal muscle fibers. J. Biophys. Biochem. Cytol. 9:493–495. doi:10.1083/jcb.9.2.493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy J. J., Mula J., Miyazaki M., Erfani R., Garrison K., Farooqui A. B., Srikuea R., Lawson B. A., Grimes B., Keller C., et al. 2011. Effective fiber hypertrophy in satellite cell-depleted skeletal muscle. Development. 138:3657–3666. doi: 10.1242/dev.068858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K. J., Thaloor D., Matteson S., and Pavlath G. K.. 2000. Hepatocyte growth factor affects satellite cell activation and differentiation in regenerating skeletal muscle. Am. J. Physiol. Cell Physiol. 278:C174–C181. doi: 10.1152/ajpcell.2000.278.1.C174 [DOI] [PubMed] [Google Scholar]
- Murach K. A., Confides A. L., Ho A., Jackson J. R., Ghazala L. S., Peterson C. A., and Dupont-Versteegden E. E.. 2017a. Depletion of pax7+ satellite cells does not affect diaphragm adaptations to running in young or aged mice. J. Physiol. 595:6299–6311. doi: 10.1113/JP274611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murach K. A., White S. H., Wen Y., Ho A., Dupont-Versteegden E. E., McCarthy J. J., and Peterson C. A.. 2017b. Differential requirement for satellite cells during overload-induced muscle hypertrophy in growing versus mature mice. Skelet. Muscle. 7:14. doi: 10.1186/s13395-017-0132-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müssig K., Staiger H., Fiedler H., Moeschel K., Beck A., Kellerer M., and Häring H. U.. 2005. Shp2 is required for protein kinase C-dependent phosphorylation of serine 307 in insulin receptor substrate-1. J. Biol. Chem. 280:32693–32699. doi: 10.1074/jbc.M506549200 [DOI] [PubMed] [Google Scholar]
- Paulsen G., Mikkelsen U. R., Raastad T., and Peake J. M.. 2012. Leucocytes, cytokines and satellite cells: what role do they play in muscle damage and regeneration following eccentric exercise?Exerc. Immunol. Rev. 18:42–97. [PubMed] [Google Scholar]
- Pawlikowski B., Vogler T. O., Gadek K., and Olwin B. B.. 2017. Regulation of skeletal muscle stem cells by fibroblast growth factors. Dev. Dyn. 246:359–367. doi: 10.1002/dvdy.24495 [DOI] [PubMed] [Google Scholar]
- Perini I., Elia I., Lo Nigro A., Ronzoni F., Berardi E., Grosemans H., Fukada S., and Sampaolesi M.. 2015. Myogenic induction of adult and pluripotent stem cells using recombinant proteins. Biochem. Biophys. Res. Commun. 464:755–761. doi: 10.1016/j.bbrc.2015.07.022 [DOI] [PubMed] [Google Scholar]
- Poli A., Mongiorgi S., Cocco L., and Follo M. Y.. 2014. Protein kinase C involvement in cell cycle modulation. Biochem. Soc. Trans. 42:1471–1476. doi: 10.1042/BST20140128 [DOI] [PubMed] [Google Scholar]
- Quarta M., Brett J. O., DiMarco R., De Morree A., Boutet S. C., Chacon R., Gibbons M. C., Garcia V. A., Su J., Shrager J. B., et al. 2016. An artificial niche preserves the quiescence of muscle stem cells and enhances their therapeutic efficacy. Nat. Biotechnol. 34:752–759. doi: 10.1038/nbt.3576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed S. A., Ouellette S. E., Liu X., Allen R. E., and Johnson S. E.. 2007. E2F5 and LEK1 translocation to the nucleus is an early event demarcating myoblast quiescence. J. Cell. Biochem. 101:1394–1408. doi: 10.1002/jcb.21256 [DOI] [PubMed] [Google Scholar]
- Ronzoni F., Ceccarelli G., Perini I., Benedetti L., Galli D., Mulas F., Balli M., Magenes G., Bellazzi R., De Angelis G. C., et al. 2017. Met-activating genetically improved chimeric factor-1 promotes angiogenesis and hypertrophy in adult myogenesis. Curr. Pharm. Biotechnol. 18:309–317. doi: 10.2174/1389201018666170201124602 [DOI] [PubMed] [Google Scholar]
- Sachs M., Brohmann H., Zechner D., Müller T., Hülsken J., Walther I., Schaeper U., Birchmeier C., and Birchmeier W.. 2000. Essential role of Gab1 for signaling by the c-Met receptor in vivo. J. Cell Biol. 150:1375–1384. doi:10.1083/jcb.150.6.1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambasivan R., Yao R., Kissenpfennig A., Van Wittenberghe L., Paldi A., Gayraud-Morel B., Guenou H., Malissen B., Tajbakhsh S., and Galy A.. 2011. Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development. 138:3647–3656. doi: 10.1242/dev.067587 [DOI] [PubMed] [Google Scholar]
- Schaeper U., Vogel R., Chmielowiec J., Huelsken J., Rosario M., and Birchmeier W.. 2007. Distinct requirements for Gab1 in met and EGF receptor signaling in vivo. Proc. Natl. Acad. Sci. U. S. A. 104:15376–15381. doi: 10.1073/pnas.0702555104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiaffino S., Dyar K. A., Ciciliot S., Blaauw B., and Sandri M.. 2013. Mechanisms regulating skeletal muscle growth and atrophy. Febs J. 280:4294–4314. doi: 10.1111/febs.12253 [DOI] [PubMed] [Google Scholar]
- Schiaffino S., and Mammucari C.. 2011. Regulation of skeletal muscle growth by the IGF1-Akt/PKB pathway: insights from genetic models. Skelet. Muscle 1:4. doi: 10.1186/2044-5040-1-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan S. M., Tatsumi R., Temm-Grove C. J., and Allen R. E.. 2000. HGF is an autocrine growth factor for skeletal muscle satellite cells in vitro. Muscle Nerve. 23:239–245. doi:10.1002.(SCI)1097-4598 [DOI] [PubMed] [Google Scholar]
- Stark D. A., Karvas R. M., Siegel A. L., and Cornelison D. D.. 2011. Eph/ephrin interactions modulate muscle satellite cell motility and patterning. Development. 138:5279–5289. doi: 10.1242/dev.068411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack V., Krützfeldt J., Kellerer M., Ullrich A., Lammers R., and Häring H. U.. 2002. The protein-tyrosine-phosphatase SHP2 is phosphorylated on serine residues 576 and 591 by protein kinase C isoforms alpha, beta 1, beta 2, and eta. Biochemistry. 41:603–608. doi:10.1021/bi011327v CCC [DOI] [PubMed] [Google Scholar]
- Tatsumi R., Hattori A., Ikeuchi Y., Anderson J. E., and Allen R. E.. 2002. Release of hepatocyte growth factor from mechanically stretched skeletal muscle satellite cells and role of pH and nitric oxide. Mol. Biol. Cell. 13:2909–2918. doi: 10.1091/mbc.E02-01-0062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsumi R., Wuollet A. L., Tabata K., Nishimura S., Tabata S., Mizunoya W., Ikeuchi Y., and Allen R. E.. 2009. A role for calcium-calmodulin in regulating nitric oxide production during skeletal muscle satellite cell activation. Am. J. Physiol. Cell Physiol. 296:C922–C929. doi: 10.1152/ajpcell.00471.2008 [DOI] [PubMed] [Google Scholar]
- Walker N., Kahamba T., Woudberg N., Goetsch K., and Niesler C.. 2015. Dose-dependent modulation of myogenesis by HGF: implications for c-Met expression and downstream signalling pathways. Growth Factors 33:229–241. doi: 10.3109/08977194.2015.1058260 [DOI] [PubMed] [Google Scholar]
- Webster M. T., and Fan C-M.. 2013. c-MET regulates myoblast motility and myocyte fusion during adult skeletal muscle regeneration.V. Mouly, editor. PLoS ONE. 8:e81757 doi:10.1371/journal.pone.0081757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak A. C., and Anderson J. E.. 2007. Nitric oxide-dependence of satellite stem cell activation and quiescence on normal skeletal muscle fibers. Dev. Dyn. 236:240–250. doi: 10.1002/dvdy.21012 [DOI] [PubMed] [Google Scholar]
- Wozniak A. C., and Anderson J. E.. 2009. The dynamics of the nitric oxide release-transient from stretched muscle cells. Int. J. Biochem. Cell Biol. 41:625–631. doi: 10.1016/j.biocel.2008.07.005 [DOI] [PubMed] [Google Scholar]
- Wozniak A. C., Pilipowicz O., Yablonka-Reuveni Z., Greenway S., Craven S., Scott E., and Anderson J. E.. 2003. C-met expression and mechanical activation of satellite cells on cultured muscle fibers. J. Histochem. Cytochem. 51:1437–1445. doi: 10.1177/002215540305101104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yablonka-Reuveni Z., Danoviz M. E., Phelps M., and Stuelsatz P.. 2015. Myogenic-specific ablation of Fgfr1 impairs FGF2-mediated proliferation of satellite cells at the myofiber niche but does not abolish the capacity for muscle regeneration. Front. Aging Neurosci. 7:85. doi: 10.3389/fnagi.2015.00085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M., Tatsumi R., Yamanouchi K., Hosoyama T., Shiratsuchi S., Sato A., Mizunoya W., Ikeuchi Y., Furuse M., and Allen R. E.. 2010. High concentrations of HGF inhibit skeletal muscle satellite cell proliferation in vitro by inducing expression of myostatin: a possible mechanism for reestablishing satellite cell quiescence in vivo. Am. J. Physiol. Cell Physiol. 298:C465–C476. doi: 10.1152/ajpcell.00449.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]