Abstract
This study used a high-throughput in vitro microassay, in vitro batch culture, and the Rumen Simulation Technique (RUSITEC) to screen recombinant fibrolytic enzymes for their ability to increase the ruminal fiber degradability of barley straw. Eleven different recombinant enzymes in combination with a crude mixture of rumen enzymes (50% recombinant enzyme:50% crude mixture of rumen enzymes) were compared with the crude mixture of rumen enzymes alone. In the microassay, all treatments were applied at 15 mg of protein load per gram barley straw glucan. Based on the microassay results, 1 recombinant endoglucanase [EGL7A, from the glycoside hydrolase (GH) family 7], 2 recombinant xylanases (XYL10A and XYL10C, from GH10), and a recombinant enzyme mixture were selected and compared with a crude mixture of fibrolytic enzymes from Aspergillus aculeatus for their ability to hydrolyze barley straw. For batch culture, enzymes were applied to barley straw at 2 dosages (100 and 500 µg of protein/g of substrate DM). All enzymes increased (P < 0.05) DM disappearance and total VFA production, but the mixture of recombinant enzymes was not superior to the use of a single recombinant enzyme. Based on positive results (P < 0.05) for total DM disappearance and VFA production in batch culture, 3 enzymes (EGL7A, XYL10A, and XYL10C) were selected and applied to barley straw at 500 µg of protein per gram for further assessment in RUSITECs fed a concentrate:barley straw diet (300:700 g/kg DM). In RUSITECs, the recombinant enzyme XYL10A increased (P < 0.05) barley straw DM, NDF, and ADF disappearance, whereas EGL7A and XYL10C had no effect. The enzymes selected based on the high-throughput in vitro microassay consistently increased barley straw degradation in ruminal batch culture, but not in the semicontinuous culture RUSITEC system.
Keywords: fiber, fibrolytic enzymes, in vitro, rumen bacteria, RUSITEC, straw
INTRODUCTION
A number of studies have examined supplementation of ruminant diets with fibrolytic enzymes, with the aim of increasing the rate and/or extent of fiber digestibility, and ultimately growth, milk yield, and feed efficiency (Meale et al., 2014; Arriola et al., 2017). Fibrolytic enzymes have the potential to improve the utilization of cereal crop residues by ruminants, but responses have been equivocal or inconsistent (Adesogan et al., 2014; Meale et al., 2014). The fact that these enzyme products are not formulated for optimal activity within the rumen is one factor that could contribute to their inconsistency (Beauchemin et al., 2003; Adesogan et al., 2014; Meale et al., 2014). Additionally, information on the types of microbes and fibrolytic enzymes in the rumen has not been used to formulate enzyme products that act synergistically with the natural enzymes produced by the rumen microbial community (Ribeiro et al., 2016).
According to rumen metagenome and metatranscriptome studies (Morgavi et al., 2013; Riley et al., 2014; Dai et al., 2015), the glycoside hydrolase (GH) family 7 is absent in the rumen, but is present in aerobic microorganisms. Our previous work (Badhan et al., 2014) showed that mixing rumen enzymes with the recombinant enzyme, EGL7A (endo-β-1,4-glucanase, GH7) from the aerobic fungi Thielavia terrestris, enhanced the digestion of alkaline peroxide-treated barley straw and alfalfa hay in an in vitro microassay. Therefore, the objective of this study was to screen new recombinant fibrolytic enzymes for their ability to increase the ruminal fiber degradability of barley straw using a series of rumen in vitro techniques. We hypothesized that enzymes selected on the basis of synergy with rumen enzymes using a high-throughput in vitro microassay would also increase barley straw degradation in ruminal batch culture and in the Rumen Simulation Technique (RUSITEC).
MATERIALS AND METHODS
All animal procedures and protocols used in this experiment were reviewed and approved by the Lethbridge Research and Development Centre Animal Care Committee (ACC number 1501) in accordance with the guidelines of the Canadian Council on Animal Care (CCAC, 2009).
High-Throughput In Vitro Microassay
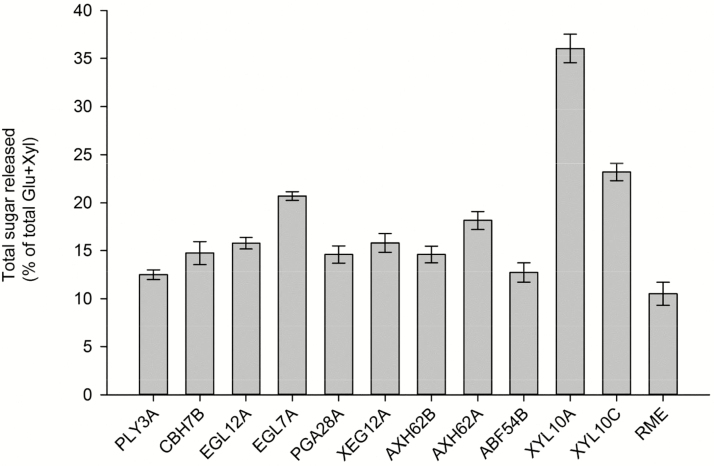
Eleven recombinant fibrolytic enzymes were produced in Aspergillus niger as previously described (Badhan et al., 2014). The recombinant enzymes tested were PLY3A (pectate lyase, EC 4.2.2.2, PL3 from Pleurotus ostreatus), CBH7B (cellobiohydrolase, EC 3.2.1.91, GH7 from Phanerochaete chrysosporium), EGL12A (endoglucanase, EC 3.2.1.151, GH12, Gloeophyllum trabeum), EGL7A (endo-β-1,4-glucanase, EC 3.2.1.4, GH7 from T. terrestris), PGA28A (endo-polygalacturonase, EC 3.2.1.15 GH28 from G. trabeum), XEG12A (xyloglucanase, EC 3.2.1.151, GH12 from A. niger), AXH62B (arabinoxylan arabinofuranosidase, EC 3.2.1.55, GH62 from Myceliophthora thermophile), AXH62A (arabinoxylan arabinofuranosidase, EC 3.2.1.55, GH62 from M. thermophile), ABF54B (alpha-arabinofuranosidase, EC 3.2.1.55, GH54 from Chaetomium thermophilum), XYL10A (1,4-β-xylanase, EC 3.2.1.8, GH10 from Rasamsonia emersonii), and XYL10C (1,4-β-xylanase, EC 3.2.1.8, GH10 from A. niger). These recombinant enzymes in combination with a mixture of rumen enzymes were compared with mixed rumen enzymes alone in a high-throughput in vitro microassay for their ability to increase the hydrolysis of barley straw. A detailed description of rumen content collection and processing for the production of the crude mixture of rumen enzymes and of the high-throughput in vitro microassay can be found in Badhan et al. (2014). In brief, rumen fluid were collected 2 h after feeding from 6 rumen-cannulated cows fed 70% barley straw and 30% concentrate (DM basis) containing corn-dried distillers grains and canola meal. Collected rumen fluids were strained through 4 layers of cheesecloth, pooled, lyophilized, and aliquoted into 15-mL flacon tubes for storage at −20 °C. Lyophilized aliquots of rumen fluid were reconstituted in 50 mM sodium citrate (pH 6.0, containing 5 µg/mL cycloheximide and 0.02% sodium azide), centrifuged at 38,300 × g for 20 min. The supernatant (S1) was stored at 4 °C. The pellets were treated with 1% triethylamine (to liberate bound enzymes into solution) and centrifuged, and the supernatant (S2) was buffer exchanged using 50 mM sodium citrate buffer (above). The supernatants S1 and S2 (after buffer exchange) were pooled to represent the crude mixture of rumen enzymes. For the in vitro microassay, ground barley straw was mixed in buffered suspension (composition as shown above) at concentration of 0.5% wt/vol, and 200 µL of this suspension was incubated with the specific enzyme treatment. All enzymes were applied at 15 mg of protein per gram glucan of 0.5% barley straw suspension in 50 mM sodium citrate (pH = 6.0). Recombinant enzyme treatments were a mixture of 50% of the crude mixture of rumen enzymes and 50% of the specific recombinant enzyme. The control treatment consisted only of the crude mixture of rumen enzymes (RME). The reaction mixtures were incubated in triplicate at 39 °C for 48 h. After incubation, glucose and xylose were determined colorimetrically using an enzyme-coupled assay kit (Megazyme International, Bray, Ireland). The recombinant enzymes that increased total sugar release (glucose + xylose) by at least 20% when compared with the crude mixture of rumen enzymes were selected for the rumen in vitro batch culture study (Fig. 1).
Figure 1.
Percent of total sugar [glucose (Glu) + xylose (Xyl)] released from barley straw. All treatments were applied at 15 mg of protein load per gram glucan of 0.5% barley straw suspension in 50 mM sodium citrate (pH = 6.0). Recombinant enzyme treatments were a mixture of 50% of rumen enzymes and 50% of the recombinant enzyme. The reaction mixture was incubated in triplicate at 39 °C for 48 h. PLY3A, pectate lyase, EC 4.2.2.2, PL3 from Pleurotus ostreatus; CBH7B, cellobiohydrolase, EC 3.2.1.91, GH7 from Phanerochaete chrysosporium; EGL12A, endoglucanase, EC 3.2.1.151, GH12, Gloeophyllum trabeum; EGL7A, endo-β-1,4-glucanase, EC 3.2.1.4, GH7 from Thielavia terrestris; PGA28A, endo-polygalacturonase, EC 3.2.1.15, GH28 from Gloeophyllum trabeum; XEG12A, xyloglucanase, EC 3.2.1.151, GH12 from Aspergillus niger; AXH62B, arabinoxylan arabinofuranosidase, EC 3.2.1.55, GH62 from Myceliophthora thermophile; AXH62A, arabinoxylan arabinofuranosidase, EC 3.2.1.55, GH62 from M. thermophile; ABF54B, alpha-arabinofuranosidase, EC 3.2.1.55, GH54 from Chaetomium thermophilum; XYL10A, 1,4-β-xylanase, EC 3.2.1.8, GH10 from Rasamsonia emersonii; XYL10C, 1,4-β-xylanase, EC 3.2.1.8, GH10 from A. niger; RME, crude mixture of rumen enzymes.
Batch Culture Screening of Fibrolytic Enzymes with Barley Straw
The semiautomated in vitro gas production technique as described by Mauricio et al. (1999) was used to study the effects of exogenous fibrolytic enzymes on ruminal fermentation and digestion of barley straw and concentrate (Table 1). Recombinant enzymes that promoted greater sugar release in the microassay (EGL7A, XYL10A, and XYL10C) were selected for evaluation in batch cultures. A mixture of five of the most promising recombinant fibrolytic enzymes (20:20:20:20:20 mix of EGL7A, XYL10A, XYL10C, AXH62A, and AXE1A) was also included to test whether they synergistically promoted greater straw degradation than individual enzymes. The enzyme AXE1A (acetylxylan esterase, EC 3.1.1.72, CE1, M. thermophile), which was not tested in the microassay, was included in the recombinant enzyme mixture because it was hypothesized that it would hydrolyse acetyl side-groups that impede the access of xylanases to the core of hemicellulose, facilitating plant cell wall hydrolysis. In addition, Viscozyme L (Novozymes Inc., Copenhagen, Denmark), a commercial enzyme product from Aspergillus aculeatus containing a variety of carbohydrases (i.e., arabanase, cellulase, beta-glucanase, hemicellulose, and xylanase), was also included in the batch culture screening for comparison purposes.
Table 1.
Chemical composition of the substrates used in in vitro experiments
| Barley straw1 | Concentrate2 | Diet3 | |
|---|---|---|---|
| DM, % | 95.1 | 95.6 | 95.3 |
| OM, % of DM | 93.2 | 89.7 | 92.2 |
| CP, % of DM | 7.0 | 34.6 | 15.3 |
| NDF, % of DM | 77.7 | 36.0 | 65.2 |
| ADF, % of DM | 44.6 | 12.5 | 35.0 |
1Barley straw used in batch cultures and RUSITEC studies.
2Composition (DM basis): 66.7% corn dried distillers grains with solubles, 26.6% canola meal, 4.2% calcium carbonate, 1% urea, 0.8% dicalcium phosphate, 0.5% salt, 0.17% feedlot premix, and 0.01% vitamin E.
3Diet (70% barley straw:30% of concentrate, DM basis) fed to the RUSITEC.
The specific activity (U) per gram of total protein (expressed in µmol/min per gram) of the recombinant enzymes selected was assessed with standard conditions at pH 5 and 40 °C. The enzyme EGL7A endoglucanase activity was 19,900 U/g using 1% carboxymethylcellulose. The enzyme XYL10A xylanase activity was 242,800 U/g using 1% beechwood xylan. The enzyme XYL10C xylanase activity was 269,000 U/g using 1% birchwood xylan. The enzyme AXH62A p-nitrophenyl-a-l-arabinofuranoside activity was 280 U/g. The enzyme AXE1A activity on acetylated xylan was 48,300 U/g. Viscozyme L main activity according to the manufacturer (Novozymes Inc.) is beta-glucanase with ≥100 U/g. According to Gama et al. (2015), Viscozyme L endoglucanase activity was 263,600 U/g using 1% carboxymethylcellulose, and xylanase activity was 191,100 U/g using 1% birchwood xylan.
The enzyme dosages used were 100 and 500 µg of protein per gram of barley straw DM. The treatments were as follows: 1) Control (no enzyme); 2) 100 µg of Viscozyme L (µg/g of substrate DM); 3) 500 µg of Viscozyme L; 4) 100 µg of EGL7A; 5) 500 µg of EGL7A; 6) 100 µg of XYL10A; 7) 500 µg of XYL10A; 8) 100 µg of XYL10C; 9) 500 µg of XYL10C; 10) 100 µg of recombinant enzyme mixture (20:20:20:20:20 mix of EGL7A, XYL10A, XYL10C, AXH62A, and AXE1A); and 11) 500 µg of the same recombinant enzyme mixture. Treatments were arranged as a completely randomized design with 3 runs and 3 replicate vials per incubation time point (12, 24, and 48 h) per run. In total, 9 vials were prepared per treatment per run. Each run was conducted on separate weeks.
To prepare substrate bags, barley straw was dried at 55 °C, ground to pass through a 1-mm screen, and weighed (0.7 g/bag) into acetone-washed and preweighed filter bags (F57 ANKOM bag, Ankom Technology Corp., Macedon, NY) that were then heat sealed. Fibrolytic enzymes were diluted with 50 mM sodium citrate buffer (pH = 6) and applied (1 mL) directly onto straw in the filter bags (before sealing). Individual bags were then placed into 125-mL serum vials. After enzyme application, serum vials were kept at 39 °C for 18 h prior to incubation. Rumen fluid was collected before the morning feeding from 4 ruminally cannulated Angus × Hereford cows fed 50% grass hay, 30% barley straw, 15% corn dried distillers’ grains plus solubles, and 5% mineral/vitamin supplement (DM basis). Rumen contents were collected from 4 distinct sites in the rumen, strained through 4 layers of cheesecloth, and equal volumes from each cow were combined. The inoculum was prepared by mixing rumen fluid 1:4 with mineral buffer (Goering and Van Soest, 1970). Inoculum (65 mL) was transferred to each vial under a stream of O2-free CO2. Vials containing inoculum, but no substrate were included as blank controls. Vials were sealed with rubber stoppers and placed in a rotary shaker (120 rpm) in an incubator at 39 °C.
Gas pressure in each vial was measured at 3, 6, 12, 24, and 48 h of incubation by inserting a 23 gauge (0.6 mm) needle attached to a pressure transducer (model PX4200-015GI, Omega Engineering, Inc., Laval, QC, Canada). After 12, 24, and 48 h of incubation, 3 replicate vials from each treatment were retrieved from the incubator, and the filter bags were removed. Bags were thoroughly rinsed with cold water until the water ran clear, dried at 55 °C for 48 h, weighed, and subsequently analyzed for NDF, and ADF to determine NDF and ADF disappearance. Samples (1.5 mL) of the fermentation fluid were collected and transferred to microcentrifuge tubes (2 mL) containing 300 µL of H2SO4 (1%; vol/vol) for NH3-N analysis, and another sample (1.5 mL) was collected and acidified with 300 µL of metaphosphoric acid (25%; wt/vol) for VFA analysis.
Disappearances of DM, NDF, and ADF were calculated as the difference between the amount of those components in the substrates before and after incubation. The gas production (mL) at each time point was calculated from the measured gas pressure (psi) as described by Eun and Beauchemin (2007). Total gas production at each time point was corrected for the blank control. To estimate fermentation kinetic parameters, gas production (GP) data were fitted to the nonlinear model of Krishnamoorthy et al. (1991):
where A is the asymptotic gas production (mL/g DM incubated), c is the fractional rate of gas production (h−1), L is the lag time (h), and t is the incubation time (h). The parameters A, c, and L were estimated by an iterative least square procedure using the NLIN procedure of SAS (SAS Institute Inc., Cary, NC). Average fermentation rate (AFR, mL gas/h), that is, average gas production rate between the start of the incubation and the time at which cumulative gas production reaches half of its asymptotic value, was estimated according to the equation of Hervás et al. (2005):
RUSITEC Evaluation of Recombinant Fibrolytic Enzymes for Barley Straw-Based Diet
As the recombinant fibrolytic enzymes EGL7A, XYL10A and XYL10C increased DM disappearance and total VFA production in the batch culture study, they were selected for further assessment in the RUSITEC. The recombinant enzyme mixture was not selected for further assessment, as it did not show any improvement above that observed for individual recombinant enzymes. Five hundred microgram of enzyme dose (µg enzyme protein/g barley straw substrate DM) was selected for assessment in the RUSITEC, as it promoted greater total VFA production compared with the lower dose (100 µg/g of substrate DM) in the batch culture study.
The experiment was conducted over 15 d with an adaptation of 5 d and sample collection over 10 d. The experiment was a complete randomized block design using two 8-vessel RUSITEC apparatuses (Czerkawski and Breckenridge, 1977) with 4 treatments as follows: 1) Control (no enzyme added); 2) 500 µg of EGL7A (µg/g of substrate DM); 3) 500 µg of XYL10A; and 4) 500 µg of XYL10C. Four replicate vessels were used for each treatment. All fermenters were fed a 30% concentrate:70% barley straw diet (DM basis). The same barley straw as was used in previous experiments was used in the RUSITEC. The chemical composition of the concentrate and barley straw is reported in Table 1. The concentrate was a mixture of 66.7% of corn dried distiller grains with solubles, 26.6% of canola meal, 4.2% calcium carbonate, 1% of urea, 0.8% dicalcium phosphate, 0.5% salt, 0.17% feedlot premix, and 0.01% vitamin E (DM basis). Enzymes were applied only to the barley straw. Straw (control or enzyme treated) and concentrate (not enzyme treated) were incubated in separate polyester bags (for concentrate: R510; 50 × 100 mm; pore size = 50 µm; for straw: R1020; 100 × 200 mm; pore size = 50 µm; ANKOM, Ankom Technol. Corp., Macedon, NY). Fibrolytic enzymes were diluted with 50 mM sodium citrate buffer (pH = 6) and slowly applied (5 mL) directly onto the barley straw in the polyester bags (before sealing). As per batch cultures, straw was incubated at 39 °C for 18 h after enzyme treatment before being placed in the fermenters.
To begin the experiment, each fermenter was filled with 180 mL of prewarmed artificial saliva (McDougall, 1948) modified to contain 0.3 g/L of (NH4)2SO4, and 720 mL of filtered rumen fluid. Solid rumen digesta (20 g), barley straw (7 g), and concentrate (3 g) for each respective treatment were placed in 3 separate polyester bags within each fermenter. Rumen fluid and solids were obtained from the same 4 ruminally fistulated cattle in the same manner as described for the batch culture experiment. Rumen fluid and contents from different cows were kept separate. Each treatment had 4 replicates, which were randomly allocated to the fermenters and inoculated with rumen fluid from different cows. Fermenters were immersed in a water bath at 39 °C, and bags within the vessels were moved up and down within fermentation fluid at 8 cycles/min. After 24 h, the polyester bag containing solid rumen digesta was replaced with one bag containing barley straw and one containing concentrate. Thereafter, polyester bags were replaced daily, resulting in each bag remaining in the fermenter for 48 h.
Artificial saliva (pH 8.2) was infused continuously into each fermenter at 2.9% per hour, replacing 70% of the fermenter volume daily. The experiment consisted of 5 d of adaptation (day 1 to 5) and 10 d of sampling (day 6 to 15). Effluent from each fermenter was collected into a 1-L flask, and gas was collected into a reusable 2 L, vinyl collection bag (Curity, Covidien Ltd., Mansfield, MA) attached to each effluent flask. Daily total gas production and effluent volume from each fermenter were recorded at the time of feed bag exchange, and the pH of fermenter fluid was measured. During feed bag exchange, fermenters were flushed with O2-free CO2.
From day 6 to 12, the 48-h incubated feed bags were removed and washed under cold tap water until the water was clear. Bags were dried at 55 °C to a constant weight. Straw residue in the bags was kept separately for each of the 7 d, whereas the residues in concentrate bags were pooled over 3 d to ensure sufficient sample for chemical analysis. All samples were ground through a 1-mm screen prior to chemical analysis.
Daily total gas production was determined throughout the experiment using a gas meter (Alexander-Wright, London, UK). From day 9 to 15, a 20-mL gas sample was taken from the septum of each collection bag using a 26 gauge needle (Becton Dickinson, Franklin Lakes, NJ) and transferred to evacuated 6.8-mL exetainers (Labco Ltd., Wycombe, Bucks, UK) for immediate analysis of CH4.
To determine daily VFA and NH3-N production, effluent was collected daily from day 9 to 12 in flasks containing 20 mL of 3.66 M H2SO4 (20%, vol/vol; Giraldo et al., 2007b). Subsamples of fermenter effluent (2.5 mL) were taken directly from the effluent flask at the time of feed-bag exchange and placed in screw-capped vials containing 0.5 mL of 25% (wt/wt) metaphosphoric acid and stored at −20 °C until VFA analysis. At the same time, 2.5-mL subsamples of fermenter effluent were also placed in a screw-capped vial with 0.5 mL of 1% (vol/vol) H2SO4 until analyzed for NH3-N.
To estimate microbial protein synthesis, bacteria in the fermenters were labeled using 15N. On day 7, 0.3 g/L (NH4)2SO4 in McDougall’s buffer was replaced with 0.3 g/L 15N-enriched (NH4)2SO4 (Sigma Chemical Co., St. Louis, MO; minimum 15N enrichment 10.01 atom %) until the end of the experiment. On days 13, 14 and 15, 24-h accumulation of effluent in each flask was preserved with 20% (wt/vol) sodium azide (3 mL), and 50 mL was subsampled for isolation of liquid-associated bacteria as described by Ribeiro et al. (2015). Feed particle-associated (FPA) and feed particle-bound (FPB) bacterial fractions were prepared from 48-h feed residues on days 13, 14, and 15 as described by Wang et al. (2001) and Ribeiro et al. (2015).
Xylanase and endoglucanase activity from days 13, 14, and 15 was determined in the supernatant of FPA samples after centrifuging at 20,000 × g for 30 min at 4 °C. Three milliliters of sample was combined with 3-mL substrate solution of either a 2% suspension (wt/vol) beechwood or low-viscosity carboxymethylcellulose in 0.2 M phosphate buffer, pH 6.8, for measurement of xylanase and endoglucanase activity, respectively. Samples were incubated at 39 °C, with shaking, for 2 h and terminated by placing the tubes into boiling water for 10 min. Samples were centrifuged (20,000 × g; 15 min; 4 °C), and the supernatants were assayed for reducing sugars using the 3,5-dinitrosalicylic acid assay against a xylose or glucose standard (Wood and Bhat, 1988).
Protozoa counts were determined on days 9, 11, and 13 using pooled fluid samples collected daily from both the 48-h straw and concentrate feed bags from each fermenter. Bags were pressed to expel fermentation fluid and a 2.5-mL subsample was obtained and preserved using 2.5 mL of methyl green formalin-saline solution. Protozoa samples were stored in the dark at room temperature until enumerated by light microscopy with a Levy-Hausser counting chamber (Hausser Scientific, Horsham, PA).
Feed and fermentation residues were analyzed for DM (AOAC, 2006; method 930.15) and ash (AOAC, 2006; method 942.05). The NDF and ADF contents were determined by the sequential method (Van Soest et al., 1991) with the ANKOM200 Fiber Analyzer (ANKOM Technology Corp., Macedon, NY; Vogel et al., 1999). Sodium sulphite (S430-3 sodium sulfite anhydrous, Fisher Scientific, Pittsburgh, PA) and α-amylase (Termamyl 120, Sigma–Aldrich, St. Louis, MO) were used during NDF determination. Total N (AOAC, 2006; method 990.03) and atom percent excess of 15N were analyzed by combustion analysis linked to a mass spectrometer (NA 1500, Carlo Erba Instruments, Rodano, Italy).
Concentrations of VFA and NH3-N in the liquid effluent were analyzed by gas chromatography (Wang et al., 2001) and the modified Berthelot method (Rhine et al., 1998), respectively. Methane concentration in gas was determined using a Varian gas chromatograph equipped with GS-Carbon PLOT 30 m × 0.32 mm × 3 µm column and thermal conductivity detector (Agilent Technologies Canada Inc., Mississauga, ON, Canada) at an isothermal oven temperature of 35 °C with helium as the carrier gas (27 cm/s).
The effluent, FPA, and FPB microbial N (MN) production was calculated as described by Ribeiro et al. (2015) and Oss et al. (2016). True DM and N disappearance was calculated by subtracting the microbial mass from feed residues. Microbial mass in feed residues was calculated by multiplying MN production (mg) in feed residues by the microbial mass per milligram of MN (g of DM of microbial pellet/mg of MN). Microbial mass per milligram of MN was determined in FPA bacterial pellets. Ammonia-N and daily VFA production were calculated by multiplying the concentration of the fermentation end products in the effluent by the daily production of effluent.
Statistical Analyses
Data were analyzed using the MIXED procedure of SAS (SAS Institute Inc.). In the batch culture experiment, the model included the fixed effects of treatment, time (12, 24, and 48 h), treatment × time, and the random effects of run (1 to 3) and replicate vials (1 to 3), with time included as a repeated measure. Degrees of freedom were adjusted using the Kenward–Roger option, and the covariance structure for repeated measurements was selected based on the lowest Akaike and Bayesian information criteria values. Differences among treatments were tested using the PDIFF option. The sums of squares were further partitioned by orthogonal contrasts to analyze differences between specific enzyme treatments and the control (no enzyme).
For the RUSITEC, individual fermenter was the experimental unit for statistical analysis. The MIXED model used included the fixed effects of enzyme treatment, day of sampling, and enzyme treatment × day, with the day of sampling from each fermenter treated as a repeated measure, and random effects of RUSITEC system (1 to 2) and inoculum (cow 1 to 4). The minimum Akaike information criterion value was used to select the covariance structure. The sums of squares were further partitioned by orthogonal contrasts to analyze differences between specific enzyme treatments with the control (no enzyme). A significant effect was declared at P ≤ 0.05, and tendency was considered at 0.05 < P < 0.10.
RESULTS
Enzyme Screening of Recombinant Fibrolytic Enzymes
In the microassay, the enzyme treatments EGL7A, XYL10A, and XYL10C increased total sugar release from barley straw compared with the rumen enzyme mixture alone by 97%, 243%, and 206%, respectively (Fig. 1).
For gas production kinetics in batch cultures, enzymes compared with the control (Con vs. Enz) reduced (P = 0.04) L, increased (P ≤ 0.01) AFR and total gas production, but did not affect (P > 0.10) c or A (Table 2). Increasing enzyme dose (Enz 100 vs. Enz 500) decreased (P = 0.01) L and increased (P < 0.001) AFR and total gas production. Comparison of the gas production kinetics of each specific enzyme against the control revealed that Viscozyme L increased (P = 0.01) AFR, XYL10A increased (P ≤ 0.04) total gas production, A and AFR; XYL10C reduced (P = 0.001) L and increased (P ≤ 0.02) total gas production, A and AFR. The recombinant enzyme mixture reduced L (P = 0.05) and increased (P ≤ 0.01) total gas production and AFR. However, EGL7A did not affect (P > 0.05) gas production kinetics when compared with control, with only a tendency (P = 0.07) for greater AFR observed.
Table 2.
Parameters of gas production kinetics (L, c, A, and AFR) and total gas production (TGP) of barley straw treated with fibrolytic enzymes incubated in ruminal batch culture
| Treatments1 | Gas production parameters2 | TGP, mL/g of DM | ||||||
|---|---|---|---|---|---|---|---|---|
| Dose, µg/g | L, h | c, h−1 | A, mL/g of DM | AFR, mL/h | 12 h | 24 h | 48 h | |
| Control | 0 | 1.7 | 0.025 | 145 | 2.50 | 31.8 | 62.3 | 98.1 |
| Viscozyme L | 100 | 1.5 | 0.029 | 135 | 2.66 | 33.9 | 64.3 | 97.6 |
| 500 | 1.3 | 0.030 | 138 | 2.87 | 36.3 | 69.1 | 102.8 | |
| EGL7A | 100 | 1.5 | 0.027 | 144 | 2.68 | 33.7 | 65.8 | 100.9 |
| 500 | 1.6 | 0.032 | 124 | 2.70 | 33.9 | 63.7 | 94.4 | |
| XYL10A | 100 | 1.7 | 0.027 | 156 | 2.68 | 32.9 | 66.3 | 101.1 |
| 500 | 1.1 | 0.026 | 175 | 2.84 | 38.1 | 69.9 | 109.0 | |
| XYL10C | 100 | 1.2 | 0.025 | 173 | 2.64 | 34.8 | 65.9 | 103.2 |
| 500 | 0.5 | 0.028 | 162 | 3.16 | 44.0 | 76.6 | 117.3 | |
| Enzyme mixture | 100 | 1.3 | 0.027 | 150 | 2.73 | 35.0 | 67.4 | 103.4 |
| 500 | 1.1 | 0.031 | 140 | 3.02 | 38.7 | 71.2 | 105.7 | |
| SEM | 0.18 | 0.0032 | 15.1 | 0.177 | 4.81 | |||
| Contrasts P-value3 | ||||||||
| Con vs. Enz | 0.04 | 0.21 | 0.50 | 0.002 | 0.01 | |||
| Con vs. Enz 100 | 0.23 | 0.43 | 0.37 | 0.06 | 0.15 | |||
| Con vs. Enz 500 | 0.01 | 0.11 | 0.70 | <0.001 | 0.002 | |||
| Enz 100 vs. Enz 500 | 0.01 | 0.14 | 0.40 | <0.001 | 0.001 | |||
| Con vs. Viscozyme L | 0.20 | 0.13 | 0.35 | 0.01 | 0.16 | |||
| Con vs. EGL7A | 0.56 | 0.13 | 0.23 | 0.07 | 0.56 | |||
| Con vs. XYL10A | 0.18 | 0.64 | 0.04 | 0.02 | 0.02 | |||
| Con vs. XYL10C | 0.001 | 0.58 | 0.02 | <0.001 | <0.001 | |||
| Con vs. Enzyme mixture | 0.05 | 0.17 | 0.99 | 0.001 | 0.01 | |||
1Viscozyme L (Novozymes Inc., Copenhagen, Denmark), commercial crude enzyme preparation from Aspergillus aculeatus, EGL7A, endo-β-1,4-glucanase, EC 3.2.1.4, GH7 from Thielavia terrestris; XYL10A, 1,4-β-xylanase, EC 3.2.1.8, GH10 from Rasamsonia emersonii; XYL10C, 1,4-β-xylanase, EC 3.2.1.8, GH10 from Aspergillus niger; Enzyme mixture, 20:20:20:20:20 mix of EGL7A, XYL10A, XYL10C, AXH62A (arabinoxylan arabinofuranosidase, EC 3.2.1.55, GH62 from Myceliophthora thermophile), and AXE1A (acetylxylan esterase, EC 3.1.1.72, CE1 from M. thermophile).
2 L = lag time; c = fractional rate of gas production; A = asymptotic gas production; AFR = average fermentation rate.
3Con = control treatment; Enz = all enzyme treatments; Enz 100 = enzyme treatments with 100 µg of protein load per gram of substrate DM; Enz 500 = enzyme treatments with 500 µg of protein load per gram of substrate DM.
Compared with the control, enzymes improved (P < 0.01) DM and NDF disappearance and tended to increase (P = 0.08) ADF disappearance (Table 3). Contrasting with gas production kinetics, increasing enzyme dose (Enz 100 vs. Enz 500) did not improve DM, NDF, or ADF disappearance (P > 0.10). Compared with control, Viscozyme L and XYL10A increased (P ≤ 0.02) DM disappearance, tended to increase NDF disappearance (P ≤ 0.09), and did not affect (P > 0.10) ADF disappearance. EGL7A, XYL10C, and the recombinant enzyme mixture increased (P ≤ 0.03) DM and NDF disappearance. In addition, XYL10C and the recombinant enzyme mixture also tended to increase ADF disappearance (P ≤ 0.08) compared with control.
Table 3.
Effect of fibrolytic enzymes on DM, NDF, and ADF disappearance of barley straw after 12, 24, and 48 h of incubation in ruminal batch culture
| Treatments1 | Dose, µg/g | DMD, % | NDFD, % | ADFD, % | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 12 h | 24 h | 48 h | 12 h | 24 h | 48 h | 12 h | 24 h | 48 h | ||
| Control | 0 | 19.5 | 31.5 | 40.7 | 10.0 | 24.7 | 35.5 | 7.4 | 21.9 | 32.5 |
| Viscozyme L | 100 | 20.8 | 33.0 | 41.2 | 10.9 | 26.0 | 35.6 | 5.9 | 23.9 | 33.2 |
| 500 | 20.7 | 33.6 | 41.5 | 11.2 | 26.6 | 36.2 | 8.6 | 22.9 | 34.3 | |
| EGL7A | 100 | 21.2 | 33.6 | 41.3 | 11.9 | 26.8 | 36.1 | 7.4 | 24.1 | 33.3 |
| 500 | 20.7 | 33.8 | 40.6 | 10.8 | 26.8 | 35.4 | 7.6 | 23.8 | 33.6 | |
| XYL10A | 100 | 20.8 | 32.9 | 42.1 | 10.9 | 25.9 | 37.2 | 7.8 | 23.4 | 34.4 |
| 500 | 21.1 | 31.7 | 42.3 | 10.9 | 24.5 | 37.0 | 4.9 | 21.4 | 35.4 | |
| XYL10C | 100 | 20.4 | 33.2 | 42.7 | 10.5 | 26.3 | 37.7 | 5.9 | 24.1 | 35.5 |
| 500 | 21.3 | 32.8 | 44.0 | 11.6 | 25.7 | 38.9 | 6.6 | 22.4 | 36.4 | |
| Enzyme mixture | 100 | 21.2 | 34.0 | 41.7 | 11.0 | 27.2 | 36.0 | 5.7 | 24.7 | 33.1 |
| 500 | 20.7 | 32.8 | 43.6 | 10.9 | 25.8 | 38.9 | 8.7 | 22.5 | 35.6 | |
| SEM | 1.22 | 1.51 | 1.47 | |||||||
| Contrasts P-value2 | ||||||||||
| Con vs. Enz | <0.001 | 0.008 | 0.08 | |||||||
| Con vs. Enz 100 | 0.002 | 0.01 | 0.12 | |||||||
| Con vs. Enz 500 | 0.001 | 0.01 | 0.07 | |||||||
| Enz 100 vs. Enz 500 | 0.73 | 0.82 | 0.69 | |||||||
| Con vs. Viscozyme L | 0.02 | 0.08 | 0.18 | |||||||
| Con vs. EGL7A | 0.01 | 0.03 | 0.12 | |||||||
| Con vs. XYL10A | 0.01 | 0.09 | 0.35 | |||||||
| Con vs. XYL10C | <0.001 | 0.003 | 0.06 | |||||||
| Con vs. enzyme mixture | <0.001 | 0.007 | 0.08 | |||||||
1Viscozyme L (Novozymes Inc., Copenhagen, Denmark) = commercial crude enzyme preparation from Aspergillus aculeatus; EGL7A = endo-β-1,4-glucanase, EC 3.2.1.4, GH7 from Thielavia terrestris; XYL10A = 1,4-β-xylanase, EC 3.2.1.8, GH10 from Rasamsonia emersonii; XYL10C = 1,4-β-xylanase, EC 3.2.1.8, GH10 from Aspergillus niger; enzyme mixture = 20:20:20:20:20 mix of EGL7A, XYL10A, XYL10C, AXH62A (arabinoxylan arabinofuranosidase, EC 3.2.1.55, GH62 from Myceliophthora thermophile), and AXE1A (acetylxylan esterase, EC 3.1.1.72, CE1 from M. thermophile).
2Con = control treatment; Enz = all enzyme treatments; Enz 100 = enzyme treatments with 100 µg of protein load per gram of substrate DM; Enz 500 = enzyme treatments with 500 µg of protein load per gram of substrate DM.
In agreement with the increase in DM disappearance, enzymes also increased (P < 0.001) total VFA production when compared with the control (Table 4). Increasing enzyme dose (Enz 100 vs. Enz 500) increased total VFA (P < 0.001) despite the lack of an effect on DM disappearance. However, this increase in VFA production with increasing enzyme dose aligned with the increase in total gas production. In addition, the increase in total VFA due to enzymes was accompanied by greater (P < 0.01) proportions of acetate, and less propionate and butyrate. Consequently, the acetate:propionate ratio increased (P < 0.001) with the addition of enzymes when compared with the control. Compared with control, each individual enzyme also increased total VFA and acetate concentrations. Overall, enzymes did not affect (P > 0.10) ammonia-N concentration (Con vs. Enz). However, when comparing only XYL10C with the control, ammonia-N concentration was increased (P = 0.02).
Table 4.
Effects of fibrolytic enzymes on ammonia-N and VFA profiles after 12, 24, and 48 h incubation of barley straw in ruminal batch culture
| Treatments1 | Dose, µg/g | Ammonia-N, mM | Total VFA, mM | Acetate, % | Propionate, % | Butyrate, % | Acetate: propionate | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 12 h | 24 h | 48 h | 12 h | 24 h | 48 h | 12 h | 24 h | 48 h | 12 h | 24 h | 48 h | 12 h | 24 h | 48 h | 12 h | 24 h | 48 h | ||
| Control | 0 | 8.7 | 10.2 | 14.2 | 26.5 | 40.1 | 52.8 | 64.9 | 60.8 | 59.8 | 17.4 | 20.2 | 21.8 | 7.4 | 7.3 | 7.3 | 3.71 | 3.01 | 2.75 |
| Viscozyme | 100 | 8.8 | 9.8 | 14.8 | 27.0 | 41.0 | 53.5 | 65.5 | 61.3 | 60.6 | 17.3 | 20.0 | 21.3 | 7.2 | 7.3 | 7.2 | 3.79 | 3.07 | 2.85 |
| 500 | 9.5 | 10.6 | 14.2 | 27.8 | 43.0 | 56.3 | 65.6 | 61.5 | 60.4 | 17.2 | 20.0 | 21.5 | 7.3 | 7.4 | 7.4 | 3.83 | 3.08 | 2.81 | |
| EGL7A | 100 | 8.3 | 9.6 | 14.8 | 26.7 | 42.5 | 53.8 | 65.6 | 61.7 | 60.7 | 17.6 | 19.9 | 21.2 | 7.0 | 7.2 | 7.2 | 3.74 | 3.11 | 2.86 |
| 500 | 8.6 | 10.7 | 14.0 | 27.8 | 41.4 | 54.9 | 65.8 | 61.5 | 60.5 | 17.1 | 20.0 | 21.4 | 7.0 | 7.2 | 7.2 | 3.85 | 3.07 | 2.83 | |
| XYL10A | 100 | 8.9 | 10.6 | 13.9 | 25.2 | 42.1 | 54.2 | 64.5 | 61.7 | 60.6 | 17.7 | 20.0 | 21.4 | 7.3 | 7.3 | 7.2 | 3.69 | 3.10 | 2.83 |
| 500 | 9.2 | 11.4 | 13.5 | 28.5 | 42.2 | 57.9 | 67.1 | 62.1 | 61.3 | 17.0 | 19.5 | 21.3 | 6.6 | 7.1 | 7.1 | 3.95 | 3.19 | 2.88 | |
| XYL10C | 100 | 9.3 | 9.9 | 14.8 | 27.4 | 43.6 | 55.1 | 66.8 | 61.9 | 61.0 | 16.7 | 19.9 | 21.2 | 6.8 | 7.1 | 7.2 | 4.00 | 3.15 | 2.88 |
| 500 | 9.9 | 11.9 | 15.8 | 31.4 | 46.0 | 61.1 | 69.0 | 64.0 | 62.0 | 17.1 | 19.0 | 21.5 | 5.3 | 6.1 | 6.6 | 4.05 | 3.37 | 2.88 | |
| Enzyme mixture | 100 | 8.1 | 10.0 | 14.4 | 27.1 | 43.1 | 55.4 | 66.1 | 61.6 | 60.4 | 17.1 | 20.1 | 21.4 | 7.0 | 7.2 | 7.3 | 3.82 | 3.07 | 2.82 |
| 500 | 8.6 | 11.0 | 15.2 | 28.5 | 42.3 | 57.6 | 66.8 | 62.4 | 61.3 | 16.5 | 19.5 | 21.4 | 6.7 | 7.0 | 7.1 | 4.12 | 3.21 | 2.87 | |
| SEM | 0.81 | 0.97 | 0.41 | 0.32 | 0.16 | 0.064 | |||||||||||||
| Contrasts P-value2 | |||||||||||||||||||
| Con vs. Enz | 0.37 | <0.001 | <0.001 | 0.005 | <0.001 | <0.001 | |||||||||||||
| Con vs. Enz 100 | 0.95 | 0.003 | <0.001 | 0.04 | <0.001 | 0.001 | |||||||||||||
| Con vs. Enz 500 | 0.10 | <0.001 | <0.001 | 0.001 | <0.001 | <0.001 | |||||||||||||
| Enz 100 vs. Enz 500 | 0.007 | <0.001 | <0.001 | 0.04 | <0.001 | <0.001 | |||||||||||||
| Con vs. Viscozyme L | 0.54 | 0.001 | <0.001 | 0.10 | 0.18 | 0.01 | |||||||||||||
| Con vs. EGL7A | 0.86 | 0.006 | <0.001 | 0.09 | <0.001 | 0.01 | |||||||||||||
| Con vs. XYL10A | 0.59 | <0.001 | <0.001 | 0.06 | <0.001 | <0.001 | |||||||||||||
| Con vs. XYL10C | 0.02 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | |||||||||||||
| Con vs. Enzyme mixture | 0.62 | <0.001 | <0.001 | 0.003 | <0.001 | <0.001 | |||||||||||||
1Viscozyme L (Novozymes Inc., Copenhagen, Denmark) = commercial crude enzyme preparation from Aspergillus aculeatus; EGL7A = endo-β-1,4-glucanase, EC 3.2.1.4, GH7 from Thielavia terrestris; XYL10A = 1,4-β-xylanase, EC 3.2.1.8, GH10 from Rasamsonia emersonii; XYL10C = 1,4-β-xylanase, EC 3.2.1.8, GH10 from Aspergillus niger; Enzyme mixture = 20:20:20:20:20 mix of EGL7A, XYL10A, XYL10C, AXH62A (arabinoxylan arabinofuranosidase, EC 3.2.1.55, GH62 from Myceliophthora thermophile), and AXE1A (acetylxylan esterase, EC 3.1.1.72, CE1from M. thermophile).
2Con = control treatment; Enz = all enzyme treatments; Enz 100 = enzyme treatments with 100 µg of protein load per gram of substrate DM; Enz 500 = enzyme treatments with 500 µg of protein load per gram of substrate DM.
RUSITEC Evaluation of Recombinant Fibrolytic Enzymes for Barley Straw-Based Diet
In the RUSITEC, XYL10A increased (P ≤ 0.03) true DM disappearance of straw, concentrate, and the complete diet (straw + concentrate) compared with the control (Table 5), whereas EGL7A and XYL10C had no effect on DM disappearance. Increased true DM disappearance was consistent with increased (P ≤ 0.05) NDF and ADF disappearance from straw, increased (P = 0.08) NDF disappearance from concentrate, and increased (P = 0.06) NDF and (P = 0.03) ADF disappearance from the total diet for XYL10A compared with the control. However, compared with the control, only a tendency for greater N disappearance (P = 0.08) from straw was observed with XYL10A.
Table 5.
Effect of recombinant fibrolytic enzymes on true DM, NDF, ADF, and N disappearance of a barley straw–concentrate diet in the RUSITEC
| Item | Treatments1 | SEM | Contrasts P-value | |||||
|---|---|---|---|---|---|---|---|---|
| Control | EGL7A | XYL10A | XYL10C | Con vs. EGL7A | Con vs. XYL10A | Con vs. XYL10C | ||
| True DM disappearance, % | ||||||||
| Concentrate | 64.7 | 65.4 | 68.4 | 64.0 | 3.84 | 0.67 | 0.03 | 0.70 |
| Straw | 49.8 | 49.4 | 51.1 | 50.0 | 0.95 | 0.42 | 0.01 | 0.67 |
| Total diet | 54.3 | 54.2 | 56.2 | 54.3 | 1.83 | 0.93 | 0.01 | 0.95 |
| NDF disappearance, % | ||||||||
| Concentrate | 58.4 | 59.4 | 63.2 | 58.2 | 3.83 | 0.73 | 0.08 | 0.93 |
| Straw | 42.4 | 42.7 | 43.7 | 42.6 | 1.18 | 0.61 | 0.04 | 0.85 |
| Total diet | 45.0 | 45.1 | 46.7 | 45.2 | 1.51 | 0.90 | 0.06 | 0.75 |
| ADF disappearance, % | ||||||||
| Concentrate | 44.4 | 46.2 | 46.9 | 43.6 | 3.38 | 0.53 | 0.38 | 0.78 |
| Straw | 34.9 | 34.8 | 36.4 | 35.2 | 1.37 | 0.88 | 0.05 | 0.67 |
| Total diet | 36.3 | 36.5 | 37.9 | 36.5 | 1.56 | 0.72 | 0.03 | 0.79 |
| N disappearance, % | ||||||||
| Concentrate | 65.2 | 60.4 | 62.3 | 63.6 | 6.91 | 0.15 | 0.39 | 0.63 |
| Straw | 68.7 | 69.9 | 71.4 | 71.2 | 2.28 | 0.45 | 0.08 | 0.10 |
| Total diet | 66.3 | 63.4 | 65.2 | 66.0 | 4.17 | 0.14 | 0.59 | 0.90 |
1EGL7A = endo-β-1,4-glucanase, EC 3.2.1.4, GH7 from Thielavia terrestris; XYL10A = 1,4-β-xylanase, EC 3.2.1.8, GH10 from Rasamsonia emersonii; and XYL10C = 1,4-β-xylanase, EC 3.2.1.8, GH10 from Aspergillus niger.
Recombinant enzymes did not affect (P > 0.10) fermenter pH or total VFA production (Table 6). However, XYL10A increased (P ≤ 0.02) ammonia-N production and the proportions of acetate and isovalerate, and lowered (P = 0.02) the proportion of propionate compared with the control. Consequently, acetate:propionate ratio increased (P < 0.01) with XYL10A compared with control. Total gas and CH4 production in milligram per gram of degraded DM were increased (P < 0.05), and CH4 production in milligram per gram of incubated DM tended to increase (P = 0.08) by EGL7A compared with the control, but the concentration of CH4 in the total gas and the total daily CH4 production (mL or mg) were not affected (P > 0.10). Gas and CH4 production were not affected (P > 0.10) by XYL10A or XYL10C.
Table 6.
Effect of recombinant fibrolytic enzymes on pH, ammonia-N, VFA, gas, methane (CH4) and microbial N production, protozoa numbers, and enzymatic activities of a barley straw–concentrate diet in the RUSITEC
| Treatments1 | Contrasts P-value | |||||||
|---|---|---|---|---|---|---|---|---|
| Item | Control | EGL7A | XYL10A | XYL10C | SEM | Con vs. EGL7A | Con vs. XYL10A | Con vs. XYL10C |
| pH | 6.79 | 6.78 | 6.78 | 6.78 | 0.023 | 0.27 | 0.37 | 0.25 |
| Ammonia-N, mg/d | 94.1 | 94.2 | 103.7 | 96.2 | 6.489 | 0.98 | 0.01 | 0.57 |
| Total VFA, mmol/d | 44.8 | 44.4 | 45.7 | 44.4 | 3.04 | 0.73 | 0.48 | 0.74 |
| Acetate, mol/100 mol | 66.0 | 65.9 | 67.4 | 66.4 | 1.54 | 0.80 | 0.02 | 0.47 |
| Propionate, mol/100 mol | 23.9 | 24.0 | 22.9 | 23.7 | 0.82 | 0.76 | 0.02 | 0.70 |
| Butyrate, mol/100 mol | 6.69 | 6.61 | 6.52 | 6.60 | 0.501 | 0.74 | 0.49 | 0.71 |
| Valerate, mol/100 mol | 1.57 | 1.55 | 1.48 | 1.58 | 0.122 | 0.86 | 0.38 | 0.93 |
| Isobutyrate, mol/100 mol | 0.69 | 0.73 | 0.72 | 0.71 | 0.027 | 0.09 | 0.16 | 0.53 |
| Isovalerate, mol/100 mol | 1.07 | 1.18 | 1.20 | 1.09 | 0.059 | 0.01 | 0.003 | 0.65 |
| Caproate, mol/100 mol | 0.06 | 0.06 | 0.06 | 0.06 | 0.013 | 0.86 | 0.77 | 0.63 |
| Acetate:propionate | 2.77 | 2.78 | 2.96 | 2.78 | 0.158 | 0.90 | 0.01 | 0.87 |
| Total gas production, L/d | 1.59 | 1.78 | 1.66 | 1.59 | 0.222 | 0.01 | 0.38 | 0.98 |
| CH4, % | 5.2 | 5.2 | 5.1 | 5.3 | 0.70 | 0.97 | 0.68 | 0.94 |
| CH4, mL/d | 91.5 | 106.4 | 88.5 | 91.9 | 18.83 | 0.27 | 0.82 | 0.98 |
| CH4, mg/d | 59.4 | 70.8 | 57.5 | 59.7 | 11.92 | 0.19 | 0.81 | 0.97 |
| CH4, mg/g incubated DM | 5.7 | 6.8 | 5.5 | 5.7 | 1.02 | 0.08 | 0.73 | 0.99 |
| CH4, mg/g degraded DM | 11.6 | 14.4 | 11.1 | 11.8 | 2.30 | 0.02 | 0.63 | 0.86 |
| Protozoa (× 104) | 9.6 | 10.8 | 9.5 | 12.0 | 1.67 | 0.37 | 0.92 | 0.09 |
| Production of microbial N, mg/d | ||||||||
| Effluent | 31.3 | 33.5 | 35.1 | 32.0 | 2.92 | 0.14 | 0.01 | 0.65 |
| FPA2 | 4.8 | 4.8 | 5.2 | 4.9 | 0.29 | 0.96 | 0.06 | 0.80 |
| FPB3 straw | 27.5 | 27.1 | 29.3 | 29.4 | 0.80 | 0.63 | 0.05 | 0.03 |
| FPB concentrate | 4.8 | 5.2 | 4.5 | 4.5 | 0.32 | 0.23 | 0.47 | 0.53 |
| Total | 69.5 | 70.8 | 72.4 | 70.2 | 2.30 | 0.32 | 0.03 | 0.59 |
| Enzyme activity (µmol/min per gram) | ||||||||
| Xylanase | 420.8 | 407.3 | 419.9 | 396.3 | 20.21 | 0.53 | 0.97 | 0.26 |
| Endoglucanase | 6.21 | 10.33 | 9.61 | 5.92 | 1.096 | 0.01 | 0.03 | 0.85 |
1EGL7A = endo-β-1,4-glucanase, EC 3.2.1.4, GH7 from Thielavia terrestris; XYL10A = 1,4-β-xylanase, EC 3.2.1.8, GH10 from Rasamsonia emersonii; and XYL10C = 1,4-β-xylanase, EC 3.2.1.8, GH10 from Aspergillus niger.
2FPA = feed particle-associated bacterial fraction.
3FPB = feed particle-bound bacterial fraction.
Recombinant enzymes did not affect (P > 0.10) total protozoa numbers (Table 6). However, application of XYL10A increased (P ≤ 0.05) effluent, straw FPB, and total MN production and tended to increase (P = 0.06) MN production in FPA fraction compared with the control. Addition of XYL10C also increased (P = 0.03) MN in straw FPB fraction, whereas EGL7A had no affect (P > 0.10) on MN production. Compared with control, enzymes did not (P > 0.10) increase xylanase activity. However, endoglucanase activity was increased (P ≤ 0.03) by EGL7A and XYL10A, but not (P = 0.85) by XYL10C.
DISCUSSION
Sustainable ruminant livestock production systems will require more efficient utilization of available feedstuffs to satisfy the predicted increase in global demand for meat and milk (Thornton, 2010; Alexandratos and Bruinsma, 2012). Lignocellulosic cereal crop residues, such as straw and corn stover, are widely available and presently largely discarded. However, their utilization in ruminant diets is very limited as often less than 50% of their cell wall carbohydrates are digested (Horton, 1978; Sari et al., 2015). Low fiber digestibility reduces ruminant livestock intake and performance (Jung and Allen, 1995; Oba and Allen, 2000). Consequently, improving ruminal fiber degradability of these crop residues would provide additional energy to ruminants and increase the nutritional value of these underutilized feeds.
Since the 1960s, many studies have evaluated the effects of adding fibrolytic enzymes to ruminant diets, with responses being equivocal or inconsistent as discussed by many excellent reviews (McAllister et al., 2001; Beauchemin et al., 2003; Adesogan et al., 2014; Meale et al., 2014; Arriola et al., 2017). These reviews highlight that most of these enzyme products were not formulated or designed for optimal activities under normal ruminal conditions, being originally designed for other nonfeed applications. In the present study, we used the current knowledge of ruminal GH families and our previous results on enzyme activities associated with increased fiber degradation in the rumen, to screen and select effective pure recombinant fibrolytic enzymes under ruminal conditions.
Many different endoglucanases and xylanases from a variety of GH families were first prescreened using a high-throughput in vitro microassay as described by Badhan et al. (2014; Fig. 1). Concentrations of enzymes in the microassay were applied at levels that were below the saturation point, while allowing for clear comparisons among recombinant enzymes. The most promising recombinant enzymes were then selected for screening at concentrations that were economically feasible for commercialization using the rumen in vitro batch culture technique. As many cellulases and endoglucanases act synergistically to hydrolyze plant cell walls, a mixture of 5 promising recombinant fibrolytic enzymes was included in the screening. The commercial crude enzyme preparation, Viscozyme L from a strain of A. aculeatus contained a range of carbohydrases including arabanase, cellulase, beta-glucanase, hemicellulose, and xylanase, was also assessed in the batch culture assay.
Substrate degradation by ruminal microbes in vivo or in vitro generates VFA, gas, and microbial biomass. Gas production in vitro is highly correlated to substrate degradation (Menke and Steingass, 1988; Blümmel and Ørskov, 1993); hence, rumen in vitro gas production kinetics have been widely used to indirectly measure rumen DM degradation kinetics. In the present study, compared with the control, all fibrolytic enzymes reduced L and increased AFR, but not A. This is in agreement with the general theory that enzymes increase the rate rather than the extent of feed degradation in the rumen (Beauchemin and Holtshausen, 2010). However, the recombinant enzymes XYL10A and XYL10C increased A and DM disappearance after 48 h of incubation when compared with the control, indicating that they increased the extent of feed degradation. XYL10A and XYL10C may target unique sites within lignocellulose, acting synergistically with rumen enzymes and thereby increasing overall rumen degradation and saccharification of recalcitrant lignocellulosic materials. The presence of complementary activities in exogenous fibrolytic enzymes that are inhibited, inactivated, or absent in the rumen has been proposed as a mechanism for the increase in the extent of digestion by exogenous fibrolytic enzymes (Adesogan et al., 2014). Using a multiple regression model, Blümmel and Ørskov (1993) demonstrated that A, but not c, was highly correlated with feed intake (0.88), digestible DMI (0.93), and growth rate (0.95). Consequently, enzymes that increase A are theorized to be more likely to improve animal performance than enzymes that increase only rate of gas production.
In agreement with total gas production, fibrolytic enzymes also generally increased DM and NDF disappearance and tended to increase ADF disappearance. However, an enzyme dose effect was not observed for DM, NDF, or ADF disappearance, in contrast to observations for total gas production. Compared with the control, total VFA concentration followed the same trend of total gas production with greater values observed when enzymes were applied at the higher dose. This observation was consistent with the increase in acetate concentration, acetate:propionate ratio, and fiber degradation with enzyme treatments. Microbial attachment to the straw residues may have prevented us from observing an enzyme dose effect for DM, NDF, and ADF disappearance. In addition, the gravimetric methods used for determining the disappearance of these different fractions are prone to more variation compared with total gas and VFA production methods.
Supplementing a commercial enzyme complex (Viscozyme L) or a mixture of recombinant fibrolytic enzymes, compared with individual recombinant fibrolytic enzymes did not further improve the in vitro ruminal degradation of barley straw, possibly suggesting a redundancy of enzyme activities within tested mixtures. Similar results were also observed by Eun and Beauchemin (2007), where combinations of pure recombinant endoglucanases and xylanases did not perform better than individual enzymes alone. The lack of synergistic effects highlights the difficulty in finding an ideal enzyme mixture that complements ruminal fibrolytic enzymes and stimulates fiber digestion. The mixture of effective fibrolytic enzymes was expected to act at a variety of active sites catalyzing more hydrolysis reactions than is possible by a single enzyme. This lack of synergism may suggest that the mechanisms by which individual enzymes improve fiber digestion in the rumen may be more indirect through promoting microbial attachment to the feed, changes in the microbial population and corresponding enzymatic activity rather than solely a direct effect on fiber hydrolysis as suggested by others (Wang et al., 2001; Giraldo et al., 2007a).
In contrast to the batch culture study, only XYL10A increased DM degradation compared with the control in the RUSITEC. Ribeiro et al. (2015) also observed that a fibrolytic enzyme product that exhibited positive effects in batch cultures, failed to augment feed degradation in the RUSITEC. As suggested by Ribeiro et al. (2015), the larger particle size of the samples incubated in the RUSITEC when compared with batch cultures may reduce the surface area and restrict enzyme access to substrates. Interestingly, both xylanases (EC 3.2.1.8) from the GH10 family (XYL10A and XYL10C) increased MN attached to barley straw compared with control, but only XYL10A increased straw degradation. An increase in ruminal microbial attachment to substrates by pretreatment with enzymes has been observed previously (Wang et al., 2001; Giraldo et al., 2007a; Ribeiro et al., 2015) and has been proposed as a possible mechanism for enzyme action in the rumen (Morgavi et al., 2000; Wang et al., 2001). The release of reducing sugars and other products of hydrolysis during the pretreatment of feeds with enzymes is proposed to cause a chemotactic response in ruminal microbes, stimulating their attachment to feed particles (Cheng and McAllister, 1997; Nsereko et al., 2000; Beauchemin et al., 2003; Giraldo et al., 2007b). Although XYL10A and XYL10C differ in their specific activities, they also seem to differ in their influence on microbial populations as XYL10A resulted in greater effluent MN production, FPA fraction MN production, and endoglucanase activity when compared with XYL10C. This suggests that XYL10A was more effective at stimulating attachment of bacterial populations that were more effective in degrading feed particles than XYL10C. The pretreatment of feed with fibrolytic enzymes may change the species profile of colonizing bacteria as suggested by Wang et al. (2001). It is interesting to note that although the recombinant enzymes were not applied to the concentrate portion of the diet, XYL10A also promoted concentrate DM disappearance. This finding may reflect a shift in the bacterial species profile in the liquid fraction of the RUSITEC fermenters, and consequently changes in the bacterial population colonizing the concentrate fraction of the diet. Similar to the rumen, microbial pools or compartments (liquid-free suspension, loosely associated to solid substrate, and firmly attached to solid substrate) in the RUSITEC are in a dynamic equilibrium where microbes are constantly flowing between compartments (Czerkawski and Cheng, 1988). This flow of microbes between the different compartments supports our theory that changes in the microbes attached to the straw can change the microbial population in the liquid fraction and consequently in the population attached to the concentrate fraction of the diet.
Pretreatment of barley straw with EGL7A resulted in only minor increases in isovalerate and total gas production, without affecting feed degradation or total VFA production. The GH family classification is based on substrate specificity, molecular mechanisms, and amino acid sequence similarities, as there is a direct relationship between sequence and protein folding (Adesogan et al., 2014; CAZy, 2018). This classification reflects the structural characteristics of the enzymes and not necessarily enzyme activity. Although GH7 have not been identified in the rumen, similar enzyme activities may be exhibited by rumen enzymes of different GH families. Total gas production, and consequently CH4 expressed as milligram per gram of degraded DM increased by EGL7A compared with the control despite no change in feed degradation, total VFA production, and proportions of acetate, propionate, and butyrate. This is hard to explain as the production of acetate and butyrate is associated with the release of H2, which is used by methanogens to form CH4 (Stewart et al., 1997). Although not measured in this study, an increase in the activity of the methanogen population by EGL7A could explain the increase in CH4 production observed.
The pretreatment of barley straw with xylanases (XYL10A and XYL10C) or with an endoglucanase (EGL7A) did not affect the xylanase activity in the FPA fraction. However, endoglucanase activity in the FPA fraction increased when EGL7A and XYL10A were added to barley straw. This increase in endoglucanase activity with the addition of XYL10A supports our hypothesis that this fibrolytic enzyme may promote a shift in the microbial population colonizing feed particles and is consistent with the increase in barley straw fiber digestion observed. The increase in the proportion of acetate and a decrease in propionate with XYL10A whens compared with the control are also consistent with higher fiber digestion. The endoglucanase activity in the FPA fraction was increased with EGL7A treatment, but fiber degradation was unaffected. Increased xylanase activity in the FPA fraction by a predominantly xylanase enzyme preparation without an increase in fiber degradation has also been observed previously (Wang et al., 2001). This suggests that this activity assay performed with a pure form of substrate (i.e., carboxymethylcellulose) does not correlate well with results generated using a recalcitrant lignocellulosic substrate such as barley straw.
In conclusion, the enzymes selected based on the high-throughput in vitro microassay consistently increased barley straw degradation in ruminal batch cultures, but not in the RUSITEC. Despite the apparent absence of GH7 in the rumen, an endoglucanase from this family did not improve the ruminal fermentation of barley straw, and a mixture of recombinant enzymes proved to be no better than a single recombinant enzyme. Only the recombinant enzyme XYL10A consistently improved substrate degradation in both batch culture and continuous rumen fermentation systems. The observed increase in the extent of digestion of barley straw with XYL10A could potentially improve the intake of straw-based diets and consequently growth or milk production. Scale-up production of this enzyme for evaluation in vivo has been accomplished and in vivo experiments using sheep and beef cattle are currently underway.
Footnotes
The financial support from this study from Elanco Animal Health and the Agricultural Innovation Program of Agriculture and Agri-Food Canada is gratefully acknowledged. The authors thank the Lethbridge Research and Development Centre (AAFC) staff for their technical and animal care assistance.
LITERATURE CITED
- Adesogan A. T., Ma Z. X., Romero J. J., and Arriola K. G.. 2014. Ruminant nutrition symposium: Improving cell wall digestion and animal performance with fibrolytic enzymes. J. Anim. Sci. 92:1317–1330. doi: 10.2527/jas.2013-7273 [DOI] [PubMed] [Google Scholar]
- Alexandratos N., and Bruinsma J.. 2012. World agriculture towards 2030/2050. FAO, United Nations, Rome. [Google Scholar]
- Arriola K. G., Oliveira A. S., Ma Z. X., Lean I. J., Giurcanu M. C., and Adesogan A. T.. 2017. A meta-analysis on the effect of dietary application of exogenous fibrolytic enzymes on the performance of dairy cows. J. Dairy Sci. 100:4513–4527. doi: 10.3168/jds.2016-12103 [DOI] [PubMed] [Google Scholar]
- Association of Official Analytical Chemists (AOAC) 2006. Official methods of analysis. 18th ed AOAC, Arlington, VA. [Google Scholar]
- Badhan A., Wang Y., Gruninger R., Patton D., Powlowski J., Tsang A., and McAllister T.. 2014. Formulation of enzyme blends to maximize the hydrolysis of alkaline peroxide pretreated alfalfa hay and barley straw by rumen enzymes and commercial cellulases. BMC Biotechnol. 14:31. doi: 10.1186/1472-6750-14-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchemin K. A., Colombatto D., Morgavi D. P., and Yang W. Z.. 2003. Use of exogenous fibrolytic enzymes to improve feed utilization by ruminants. J. Anim. Sci. 81:E37–E47. doi: 10.2527/2003.8114_suppl_2E37x [DOI] [Google Scholar]
- Beauchemin K. A., and Holtshausen L.. 2010. Developments in enzyme usage in ruminants. In: Bedford M. R. and Partridge G. G., editors, Enzymes in farm animal nutrition. 2nd ed CABI, Oxford, UK: p. 206–230. [Google Scholar]
- Blümmel M., and Ørskov E. R.. 1993. Comparison of in vitro gas production and nylon bag degradability of roughages in predicting feed intake in cattle. Anim. Feed Sci. Technol. 40:109–119. doi: 10.1016/0377-8401(93)90150-I [DOI] [Google Scholar]
- Canadian Council on Animal Care 2009. CCAC guidelines on: The care and use of farm animals in research, teaching and testing. Can. Council Anim. Care, Ottawa, Canada. [Google Scholar]
- CAZy 2018. Carbohydrate-active enzymes database [accessed March 8, 2018]. http://www.cazy.org/Glycoside-Hydrolases.html.
- Cheng K.-J., and McAllister T. A.,. 1997. Compartmentation in the rumen. In: Hobson P. N. and Stewart C. S., editors, The rumen microbial ecosystem. 2nd ed Blackie Academic & Professional, London, UK: p. 492–522. [Google Scholar]
- Czerkawski J. W., and Breckenridge G.. 1977. Design and development of a long-term Rumen Simulation Technique (Rusitec). Br. J. Nutr. 38:371–384. doi: 10.1079/BJN19770102 [DOI] [PubMed] [Google Scholar]
- Czerkawski J. W., and Cheng K-J.. 1988. Compartmentation in the rumen. In: Hobson P. N., editor, The rumen microbial ecosystem. Elsevier Sci. Publ, New York, NY: p. 361–385. [Google Scholar]
- Dai X., Tian Y., Li J., Luo Y., Liu D., Zheng H., Wang J., Dong Z., Hu S., and Huang L.. 2015. Metatranscriptomic analyses of plant cell wall polysaccharide degradation by microorganisms in the cow rumen. Appl. Environ. Microbiol. 81:1375–1386. doi: 10.1128/AEM.03682-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eun J.-S., and Beauchemin K. A.. 2007. Assessment of the efficacy of varying experimental exogenous fibrolytic enzymes using in vitro fermentation characteristics. Anim. Feed Sci. Technol. 132:298–315. doi: 10.1016/j.anifeedsci.2006.02.014 [DOI] [Google Scholar]
- Gama R., Van Dyk J. S., and Pletschke B. I.. 2015. Optimisation of enzymatic hydrolysis of apple pomace for production of biofuel and biorefinery chemicals using commercial enzymes. 3 Biotech. 5:1075–1087. doi: 10.1007/s13205-015-0312-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo L. A., Ranilla M. J., Tejido M. L., and Carro M. D.. 2007a. Influence of exogenous fibrolytic enzymes and fumarate on methane production, microbial growth and fermentation in Rusitec fermenters. Br. J. Nutr. 98:753–761. doi: 10.1017/S0007114507744446 [DOI] [PubMed] [Google Scholar]
- Giraldo L. A., Tejido M. L., Ranilla M. J., and Carro M. D.. 2007b. Effects of exogenous cellulase supplementation on microbial growth and ruminal fermentation of a high-forage diet in Rusitec fermenters. J. Anim. Sci. 85:1962–1970. doi: 10.2527/jas.2006-318 [DOI] [PubMed] [Google Scholar]
- Goering H. K., and Van Soest P. J.. 1970. Forage fiber analyses (apparatus, reagents, procedures, and some applications). Agric. Handbook Number 379. ARS-USDA, Washington, DC. [Google Scholar]
- Hervás G., Frutos P., Giraldez F. J., Mora M. J., Fernandez B., and Mantecon A. R.. 2005. Effect of preservation on fermentative activity of rumen fluid inoculum for in vitro gas production techniques. Anim. Feed Sci. Technol. 123–124:107–118. doi: 10.1016/j.anifeedsci.2005.05.004 [DOI] [Google Scholar]
- Horton G. 1978. The intake and digestibility of ammoniated cereal straws by cattle. Can. J. Anim. Sci. 58:471–478. doi: 10.4141/cjas78-060 [DOI] [Google Scholar]
- Jung H., and Allen M. S.. 1995. Characteristics of plant cell walls affecting intake and digestibility of forages by ruminants. J. Anim. Sci. 73:2774–2790. doi: 10.2527/1995.7392774x [DOI] [PubMed] [Google Scholar]
- Krishnamoorthy U., Soller H., Steingass H., and Menke K. H.. 1991. A comparative study on rumen fermentation of energy supplements in vitro. J. Anim. Physiol. Anim. Nutr. 65:28–35. doi: 10.1111/j.1439-0396.1991.tb00237.x [DOI] [Google Scholar]
- Mauricio R. M., Mould F. L., Dhanoa M. S., Owen E., Channa K. S., and Theodorou M. K.. 1999. A semi-automated in vitro gas production technique for ruminant feedstuff evaluation. Anim. Feed Sci. Technol. 79: 321–330. doi: 10.1016/S0377-8401(99)00033-4 [DOI] [Google Scholar]
- McAllister T. A., Hristov A. N., Beauchemin K. A., Rode L. M., and Cheng K. J.. 2001. Enzymes in ruminant diets. In: Bedford M. R. and Partridge G. G., editors, Enzymes in farm animal nutrition. CABI Publishing, Wallingford, UK: p. 273–289. [Google Scholar]
- Mcdougall E. I. 1948. The composition and output of sheep’s saliva. Biochem. J. 43:99–109. doi: 10.1042/bj0430099 [DOI] [PubMed] [Google Scholar]
- Meale S. J., Beauchemin K. A., Hristov A. N., Chaves A. V., and McAllister T. A.. 2014. Board-Invited Review: Opportunities and challenges in using exogenous enzymes to improve ruminant production. J. Anim. Sci. 92:427–442. doi: 10.2527/jas.2013-6869 [DOI] [PubMed] [Google Scholar]
- Menke K. H., and Steingass H.. 1988. Estimation of the energetic feed value obtained from chemical analysis and in vitro gas production using rumen fluid. Anim. Res. Dev. 28:7–55. [Google Scholar]
- Morgavi D. P., Beauchemin K. A., Nsereko V. L., Rode L. M., Iwaasa A. D., Yang W. Z., McAllister T. A., and Wang Y.. 2000. Synergy between ruminal fibrolytic enzymes and enzymes from Trichoderma longibrachiatum. J. Dairy Sci. 83:1310–1321. doi: 10.3168/jds.S0022-0302(00)74997-6 [DOI] [PubMed] [Google Scholar]
- Morgavi D. P., Kelly W. J., Janssen P. H., and Attwood G. T.. 2013. Rumen microbial (meta)genomics and its application to ruminant production. Animal 7(Suppl 1):184–201. doi: 10.1017/S1751731112000419 [DOI] [PubMed] [Google Scholar]
- Nsereko V. L., Morgavi D. P., Rode L. M., Beauchemin K. A., and McAllister T. A.. 2000. Effects of fungal enzyme preparations on hydrolysis and subsequent degradation of alfalfa hay fiber by mixed rumen microorganisms in vitro. Anim. Feed Sci. Technol. 88:153–170. doi: 10.1016/S0377-8401(00)00225-X [DOI] [Google Scholar]
- Oba M., and Allen M. S.. 2000. Effects of brown midrib 3 mutation in corn silage on productivity of dairy cows fed two concentrations of dietary neutral detergent fiber: 3. Digestibility and microbial efficiency. J. Dairy Sci. 83:1350–1358. doi: 10.3168/jds.S0022-0302(00)75002-8 [DOI] [PubMed] [Google Scholar]
- Oss D. B., Ribeiro G. O. Jr, Marcondes M. I., Yang W., Beauchemin K. A., Forster R. J., and McAllister T. A.. 2016. Synergism of cattle and bison inoculum on ruminal fermentation and select bacterial communities in an artificial rumen (Rusitec) fed a barley straw based diet. Front. Microbiol. 7:2032. doi: 10.3389/fmicb.2016.02032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhine E. D., Mulvaney R. L., Pratt E. J., and Sims G. K.. 1998. Improving the Berthelot reaction for determining ammonium in soil extracts and water. Soil Sci. Soc. Am. J. 62:473–480. doi: 10.2136/sssaj1998.03615995006200020026x [DOI] [Google Scholar]
- Ribeiro G. O. Jr, Goncalves L. C., Pereira L. G. R., Chaves A. V., Wang Y., Beauchemin K. A., and McAllister T. A.. 2015. Effect of fibrolytic enzymes added to a Andropogon gayanus grass silage-concentrate diet on rumen fermentation in batch cultures and the artificial rumen (Rusitec). Animal 9:1153–11162. doi: 10.1017/S1751731115000221 [DOI] [PubMed] [Google Scholar]
- Ribeiro G. O., Gruninger R. J., Badhan A., and McAllister T. A.. 2016. Mining the rumen for fibrolytic feed enzymes. Anim. Front. 6:20–26. doi: 10.2527/af.2016-0019 [DOI] [Google Scholar]
- Riley R., Salamov A. A., Brown D. W., Nagy L. G., Floudas D., Held B. W., Levasseur A., Lombard V., Morin E., Otillar R.,. et al. 2014. Extensive sampling of basidiomycete genomes demonstrates inadequacy of the white-rot/brown-rot paradigm for wood decay fungi. Proc. Natl. Acad. Sci. U.S.A. 111:9923–9928. doi: 10.1073/pnas.1400592111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sari M., Ferret A., and Calsamiglia S.. 2015. Effect of pH on in vitro microbial fermentation and nutrient flow in diets containing barley straw or non-forage fiber sources. Anim. Feed Sci. Technol. 200:17–24. doi: 10.1016/j.anifeedsci.2014.11.011 [DOI] [Google Scholar]
- Stewart C. S., Flint H. J., and Bryant M. P.. 1997. The rumen bacteria. In: P. N. Hobson and C. S. Stewart, editors. The Rumen Microbial Ecosystem. Dordrecht (Netherlands): Springer; p. 10–72. [Google Scholar]
- Thornton P. K. 2010. Livestock production: Recent trends, future prospects. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 365:2853–2867. doi: 10.1098/rstb.2010.0134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Soest P. J., Robertson J. B., and Lewis B. A.. 1991. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 74:3583–3597. doi: 10.3168/jds.S0022-0302(91)78551-2 [DOI] [PubMed] [Google Scholar]
- Vogel K. P., Pedersen J. F., Masterson S. D., and Toy J. J.. 1999. Evaluation of a filter bag system for NDF, ADF, and IVDMD forage analysis. Crop Sci. 39:276–279. doi: 10.2135/cropsci1999.0011183X003900010042x [DOI] [Google Scholar]
- Wang Y., McAllister T. A., Rode L. M., Beauchemin K. A., Morgavi D. P., Nsereko V. L., Iwaasa A. D., and Yang W.. 2001. Effects of an exogenous enzyme preparation on microbial protein synthesis, enzyme activity and attachment to feed in the Rumen Simulation Technique (Rusitec). Br. J. Nutr. 85:325–332. doi: 10.1079/BJN2000277 [DOI] [PubMed] [Google Scholar]
- Wood T. M., and Bhat K. M.. 1988. Methods for measuring cellulase activities. Methods Enzymol. 160:87–112. doi: 10.1016/0076-6879(88)60109-1 [DOI] [Google Scholar]