Abstract
In this report, we use a proteomic strategy to identify glycoproteins on the surface of exosomes derived from myeloid-derived suppressor cells (MDSCs), and then test if selected glycoproteins contribute to exosome-mediated chemotaxis and migration of MDSCs. We report successful modification of a surface chemistry method for use with exosomes and identify 21 surface N-glycoproteins on exosomes released by mouse mammary carcinoma-induced MDSCs. These glycoprotein identities and functionalities are compared with 93 N-linked glycoproteins identified on the surface of the parental cells. As with the lysate proteomes examined previously, the exosome surface N-glycoproteins are primarily a subset of the glycoproteins on the surface of the suppressor cells that released them, with related functions and related potential as therapeutic targets. The “don’t eat me” molecule CD47 and its binding partners thrombospondin-1 (TSP1) and signal regulatory protein α (SIRPα) were among the surface N-glycoproteins detected. Functional bioassays using antibodies to these three molecules demonstrated that CD47, TSP1, and to a lesser extent SIRPα facilitate exosome-mediated MDSC chemotaxis and migration.
Keywords: exosomes, myeloid derived suppressor cells, N-glycoproteome, cell surface capture
INTRODUCTION
Proteins on cell surfaces are mediators of cell functions, serving as receptors, transporters, and adhesives. They transport nutrition and waste, respond to stimuli from the environment, and protect the integrity of the cell. They can also serve as ligands for receptors on other cells and deliver signals to surrounding cells in their locale. In the context of the tumor microenvironment, CD47 and other surface proteins are considered drug target candidates,1,2 as well as biomarkers for distinguishing cells of interest.3,4 Exosomes, nanoscale vesicles released by almost all types of cells, have been shown to carry cargos that include proteins, lipids, and RNAs and to differ according to the type of parental cells. Exosomes are bounded by a phospholipid bilayer that contains tetraspanins and other proteins. Tetraspanins CD63, CD81, CD82, and CD9 have been proposed as markers of exosomes.5–7 Electron micrographs of immunolabeled CD63 clearly show the protein on the surface of blood-derived exosomes.8 Several other studies also report visualization of selected proteins on the surfaces of exosomes.5,9,10 Recent work has highlighted the role that integrins on the surface of exosomes play in uptake by recipient cells leading to premetastatic niche formation and cancer cell migration.11,12 Here we report an unbiased study of glycoproteins on the surface of exosomes released by myeloid-derived suppressor cells (MDSCs). Our broad objectives are to identify therapeutic targets for reducing MDSC suppressive functions and to gain insight into cell-to-cell communication within the tumor microenvironment that promotes tumor progression.
MDSCs are immature myeloid cells. They accumulate in the blood and tumor microenvironment of virtually all cancer patients and tumor-bearing mice. They are an acknowledged obstacle to innate antitumor immunity and to cancer immunotherapies.13 MDSCs are multifunctional cells that use a variety of suppressive mechanisms to inactivate antitumor immunity. They (i) inhibit the activation of tumor-reactive T cells through their production of reactive oxygen and nitrogen species and by sequestering or degrading amino acids that are essential for T cell function, (ii) polarize immunity and macrophages toward a type 2 phenotype that supports tumor growth, (iii) prevent naive T cells from entering lymph nodes and becoming activated by down-regulating T cell expression of L-selectin, (iv) inhibit the cytotoxic activity of natural killer cells, (v) promote neovascularization through their production of vascular endothelial growth factor (VEGF), and (vi) promote metastasis through their production of matrix metalloproteases.14 The inflammatory milieu present in most solid tumors exacerbates both the quantity and potency of MDSCs and induces their differentiation and accumulation from hematopoietic progenitor cells.15–17 MDSCs mediate most of their immune suppressive and tumor-promoting functions within the tumor microenvironment (TME). Therefore, MDSC function requires that the cells migrate from the bone marrow where they are generated into solid tumors.14
Cell-to-cell interactions, such as those between MDSCs and their target cells, are frequently initiated by contact between plasma membrane receptors and their soluble or membrane-bound ligands. MDSCs and other cells in the tumor microenvironment release exosomes, which also facilitate communication between cells and promote tumor growth and metastasis.18–20 Glycoproteins, such as the receptor for advanced glycation endproducts (RAGE), facilitate the activation of MDSCs.21 Our previous studies using proteomic approaches demonstrated that exosomes released by MDSCs contain one of the ligands for RAGE21 and that these proteins are chemotactic for parental MDSCs and also polarize macrophages toward a tumor-promoting M2 phenotype.20 Based on these findings, we hypothesized that proteomic approaches may be useful for identifying additional MDSC plasma membrane and exosomal glycoproteins, which may regulate MDSC function and migration.
A number of methods have been developed to take advantage of the unique accessibility of molecules at the surfaces of intact cells. These include lectin affinity columns,22 alkylation of accessible lysine residues with linked biotin,23,24 nanoparticle pellicles,25,26 and alkylation of oxidized glycans with linked biotin or hydrazide.27–30 We have adopted the latter strategy for this study of surface glycoproteins on MDSC-derived exosomes and their parental cells. In particular, the milder oxy-amino alkylation method of Weekes et al.31 was followed because its compatibility with physiological pH minimizes the potential for lysing the exosomes.
We hypothesize that multiple glycoproteins reside on the surface of our exosomes, where they play critical roles in exosome–cell interactions. Here we report the first successful adaptation of cell surface methodology to examine the surface of exosomes released by tumor-induced mouse MDSC. The diameters of these exosomes average around 30 nm, on the small end of the range reported for exosomes.16 With a high radius of curvature and a small surface area, these vesicles provide an appropriate challenge for surface chemistry. We have characterized 21 glycoproteins and show that these meet several criteria for a surface origin. We have carried out a parallel identification of 93 glycoproteins on the surface of parental MDSCs and discuss similarities in composition that are consistent with the similar functions reported previously for the exosomes and their parent cells. Because MDSC chemotaxis and migration are critical for MDSC function, we have focused our biological studies on CD47, an N-glycosylated integral membrane protein identified by our proteomic studies. We report here that CD47, known for its ability to protect cancer cells from phagocytosis, is also used by MDSC-derived exosomes to chemoattract MDSCs.
EXPERIMENTAL SECTION
Materials
Fetal bovine serum was obtained from Atlanta Biologicals (Lawrenceville, GA). Tris-HCl, PBS, aniline, iodoacetamide, dithiothreitol (DTT), urea, protease inhibitor cocktail solution, sodium(meta)periodate, Triton X-100, SDS, NH4HCO3, and glycerol were purchased from Sigma-Aldrch (St. Louis, MO). NaCl and trifluoroacetic acid was purchased from EMD chemicals (Gibbstown, NJ). Trypsin was obtained from Promega (Madison, WI). PNGaseF and G7 buffer were purchased from New England Biolabs (Ipswich, MA). High capacity streptavidin beads were obtained from Thermo Scientific (Rockford, IL). Amino-oxy-biotin was purchased from Biotium (Hayward, CA). Optima LC/MS grade acetonitrile and formic acid were purchased from Fisher Scientific. Pierce C18 spin columns were purchased from Glygen Corp (Columbia, MD). Fluorescently labeled monoclonal antibodies to murine Gr1 (clone RB6-8C5), CD11b (clone M170), CD47 (clone miap301), and isotype-matched controls were from BioLegend (San Diego, CA). Deionized water used for all experiments was obtained from a Milli-Q A10 system.
Myeloid-Derived Suppressor Cells and Exosome Harvesting
Female BALB/c mice, 6–10 weeks of age (bred in the UMBC animal facility from breeding pairs obtained from The Jackson Laboratory, Bar Harbor, ME), were injected in the mammary fat pad with 1 × 105 mammary carcinoma 4T1 cells transfected with the IL-1β gene (4T1/IL-1β). Mice were bled from the submandibular vein when tumors were 9–11 mm in diameter. MDSCs were harvested from the blood, labeled with fluorescently coupled antibodies to MDSC markers, and assessed by flow cytometry (Beckman/Coulter Cyan ADP) for purity, as reported earlier.20,32 Cell populations that were>90% pure Gr1+CD11b+ cells were used in these studies. Approximately 108 MDSCs were obtained from 2–3 mice.
Following isolation from mice, MDSCs were either frozen in 15% DMSO and stored at −80 °C (for studies with intact MDSCs) or maintained in serum-free HL-1 media overnight at 37 °C (for collection of exosomes), as previously described.20 After 16 h, each supernatant containing exosomes was centrifuged at 805g and 2090g to remove residual cells and then ultracentrifuged at 100 000g to pellet the exosomes. In a previous study, the pellet was shown to comprise homogeneous exosomes with diameters of 25–30 nm and density of 1.2–1.3 g/mL.20 Pellets containing the exosomes were resuspended in PBS, and absorbances were measured at 260 and 280 nm. Protein concentration was assessed by Bradford Quick Start assay (Biorad, Hercules, CA). Exosomes were never frozen and were used fresh within a week of isolation.
MDSC and Exosome Cell Surface Chemistry
Approximately 108 MDSCs were washed with 50 mM PBS (CaCl2, MgCl2, pH 7.4) and then incubated in 1 mM sodium periodate in 50 mM PBS (CaCl2, MgCl2, pH 7.4, 5%FBS) for 20 min at 4 °C in the dark to oxidize surface glycans. Oxidized glycans were biotinylated in 100 μM amino-oxy-biotin and 10 mM aniline (in PBS with CaCl2, MgCl2, 5% FBS, pH 6.7) for 1 h at 4 °C, washed two times with PBS (CaCl2, MgCl2, pH 7.4; 5 min at 900g), and then resuspended in lysis buffer (150 mM NaCl, 5 mM IAA, 10 mM Tris-HCl, triton X-100, and protease inhibitor cocktail) and incubated overnight at 4 °C. The following day, mechanical lysis was carried out. The nuclear material and debris were removed by two 40 min centrifugations at 2800g, followed by two 40 min centrifugations at 16 000g. During the final centrifugation, streptavidin beads were prepared in snap cap spin columns (Pierce, Rockford, IL) via two washes (1 min at 1000g) in lysis buffer. The cell lysate was incubated on-column for 2 h at 4 °C. The beads were washed with lysis buffer and PBS (pH 7.4, 0.5% SDS). Proteins were reduced on column with 100 mM DTT in PBS (pH 7.4 0.5% SDS) for 20 min at room temperature. The beads were then washed with urea containing (UC) buffer (6 M urea, 100 mM Tris-HCl) and alkylated with 50 mM iodoacetamide in UC buffer for 20 min at room temperature. The biotinylated glycoproteins were digested on-column using 5 μg of trypsin in 50 mM ammonium bicarbonate at room temperature, overnight. The tryptic peptides, released from the beads and present in the supernatant, were collected by centrifugation, and a C18 column cleanup was performed. The snap cap columns were washed with PBS, followed by Milli-Q water, and G7 buffer (50 mM sodium phosphate). The N-glycopeptides (still attached to the beads) were released via digestion with PNGase F (15 000 U) overnight in G7 buffer at 37 °C. The peptides were eluted from the beads and are present in the supernatant, which was then collected by centrifugation.
Preparation of surface proteins from exosomes was performed with starting material of exosomes collected from 1 × 109 MDSCs. The exosomes were incubated in 1 mM sodium periodate in 50 mM PBS (CaCl2, MgCl2, pH 7.4) for 20 min at 4 °C in the dark and then centrifuged at 100 000g for 70 min. Exosome pellet was washed with PBS (CaCl2, MgCl2, pH 7.4, 2% BSA), and another 100 000g spin was performed. The supernatant was removed, and the pellet was resuspended in a solution of 100 μM amino-oxy-biotin and 10 mM aniline (in PBS, w/CaCl2, MgCl2 2% BSA, pH 6.7) and incubated for 1 h at 4 °C. Intact exosomes were then washed with PBS (CaCl2, MgCl2, pH 7.4, 2% BSA) (70 min, 100 000g) and then resuspended in 100 μL of PBS (pH 7.4) and incubated overnight at 4 °C. The following day, exosomes were lysed in 8 M urea. NH4HCO3 (50 mM) was added to each sample to dilute the final urea concentration to 0.8 M. Streptavidin beads were prepared in snap cap spin columns. The exosome lysate was incubated on-column for 2 h at 4 °C. The beads were washed with ammonium bicarbonate and PBS (pH 7.4, 0.5% SDS). Proteins were reduced on column with 100 mM DTT in PBS (pH 7.4, 0.5% SDS) for 20 min at room temperature. The beads were then washed with UC buffer and alkylated with 50 mM IAA in UC buffer for 20 min at room temperature. The biotinylated glycoproteins were digested on-column using 0.75 μg of trypsin in 50 mM ammonium bicarbonate. The tryptic peptides were collected by centrifugation, and a C18 column cleanup was performed. The snap cap columns were washed twice with PBS, followed by two washes with Milli-Q water, and two more washes with G7 buffer. The N-glycopeptides (still attached to the beads) were released via digestion with PNGase F (15 000 U) overnight in G7 buffer at 37 °C, and collected by centrifugation.
HPLC-MS/MS Analysis
Tryptic peptides and glycopeptides from both MDSC and exosome samples were fractionated on a C18 analytical column (Grace Vydac, Deerfield, IL) in line with an LTQ-orbitrap XL (Thermo Scientific, San Jose, CA) with a combined linear gradient of 0–40% solvent B (97.5% ACN, 2.5% H2O, and 0.1% formic acid) over 140 min and 40–85% solvent B over an additional 25 min. The flow rate was 500 nL/min. Precursor ion scans were recorded at a resolution of 30 000 at 400 m/z. In each cycle, the nine most highly abundant precursor ions were isolated for fragmentation via collisional induced dissociation (CID) in the ion trap. Fragment ions were analyzed in the ion trap. Samples from one biological replicate each of the parental cells were also analyzed on an orbitrap fusion. In this case, peptides from parental cells were fractionated on a C18 Easy Spray column, 75 ¼m ID × 25 cm (Thermo Scientific, San Jose, CA), with a linear gradient of 5–32% acetonitrile/0.1% formic acid through 120 min at a flow rate of 500 nL/min. Spectra were acquired using a resolution of 120 000 (at 200 m/z) for full scans, followed by HCD fragmentation and detection of the fragment ions in the ion trap in the Top N mode.
In total, three biological replicates were analyzed for MDSCs and four for the MDSC exosomes. Between three and six injections were examined for each biological replicate.
Bioinformatics
Spectra were converted to peak lists and reformatted as mzXML using msconvert from the ProteoWizard project33 and uploaded to the PepArML peptide identification meta-search engine34 for searching using MS-GF+.35 Spectra were searched against the UniProtKB reviewed mouse reference proteome (August 2015). Precursor ion tolerance of 0.05 Da was required for either the monoisotopic or first 13C peak of the precursor isotope cluster, and tolerance of 0.5 Da was used for fragment ion matching. A fixed carbamidomethylation Cys modification was specified throughout. Peptides were required to match the trypsin proteolytic digest motif at both the N- and C-terminus. Up to two missed trypsin cleavages were permitted. Spectral FDR was estimated using a reversed protein sequence database.
Peptides in the fraction resulting from PNGaseF deglycosylation and release of glycan-bound tryptic glycopeptides were searched using variable deamidation of Asn residues in the N-XS/T glycosylation sequence motif and only those peptides with a deamidation mass-shift (+0.98 Da), characteristic of N-glycan cleavage, were retained. Peptides in the fraction resulting from tryptic digestion of bound glycoproteins were searched separately, without this variable modification, and the resulting peptide identifications were combined with the PNGaseF fraction deglycosylated glycopeptides. Protein parsimony analysis was applied to the combined exosome and MDSC peptide identifications, after filtering at 5% spectral FDR, to infer glycoproteins. In addition, at least one deglycosylated glycopeptide identification from the PNGaseF fractions was required for each inferred protein. Proteins inferred on the basis of a single deglycosylated glycopeptide were held to a higher standard, with the single deglycosylated glycopeptide identified required to satisfy spectral FDR at most 1% and to have a unique alignment to UniProt reviewed mouse reference sequences. Protein FDR for the unified parsimony analysis was 2.05%, estimated by tracking decoy peptides and proteins throughout the protein inference procedure.
Nine of the 21 exosome proteins were identified based on the identification of a single deglycosylated glycopeptide. Six of these are matched to more than one spectrum. In the parental MDSC cell surface analysis, 18 glycoproteins out of 93 were identified based on a single PNGaseF fraction deglycosylated glycopeptide. Stringent one-peptide identifications for the surface proteins have been considered sufficient by other laboratories exploring surface glycoproteins.36
The NetNglyc server was used to predict N-glycosylation sites on peptides identified from the PNGaseF fraction.37 Uniprot and Protter were used to make predictions on topology domains of the glycopeptides.38
MDSC Migration and Chemotaxis
MDSC chemotaxis with exosomes was assessed as previously described with the following modifications.20 Intact MDSCs from 4T1-tumor-bearing mice (1 × 106 of >90% Gr1+CD11b+ cells) were placed in the upper chamber of transwells containing an 8 μm filter and MDSC-derived exosomes were place in the lower chamber. In some experiments, MDSC-derived exosomes were premixed at 4 °C for 1 h with 5 ¼g/mL antibodies to mouse CD47 (clone miap301, Rat IgG2a, catalog no. 127501, BioLegend), mouse thrombospondin-1 (TSP-1; clone 6.1, catalog no. 14-9756-82, EBioscience), mouse signal regulatory protein-α (SIRPα; CD172a, clone P84, catalog no. 144002, BioLegend), or the irrelevant isotype matched control antibodies rat IgG2a (clone RTK2758, no. 400515, BioLegend), mouse IgG1 (clone P.3.6.2.8.15, no. 16-4714-83, EBioscience), or rat IgG1 (clone BRG1, no. 16-4301-85, EBioscience), respectively. Preincubated exosomes or medium containing no exosomes was then added to the lower chamber of the transwells, and the transwell plates were incubated at 37 °C, 5% CO2. At the end of 4 h, cells migrating to the lower chamber were counted by hemocytometer. In some experiments, parental MDSCs were pretreated at 5 µg/mL with the CD47, TSP-1, SIRPα, or corresponding irrelevant isotype matched antibodies for 1 h at room temperature, and unbound antibody was removed by washing with PBS. Samples were run in triplicate, and each sample was counted six times. Values are the average ± SD of triplicate samples per experimental condition. % migration = {experimental – control medium)/ (exosomes – control medium)} × 100%. Statistical analysis was by Student’s t test.
RESULTS AND DISCUSSION
Twenty-one surface N-glycoproteins have been identified on the exosome samples and are listed in Table 1. A recent publication speculates that a single exosome can carry about 100 proteins.39 In this context and allowing for heterogeneity within an exosome sample,4 the identification of 21 glycoproteins on the exosome surface would seem to be a reasonable first result. Twenty-one protein identifications were based on 25 N-glycopeptide identifications (Table S1) on 25 sites. All glycopeptides contain a motif for N-glycosylation in which an asparagine has been modified (by PNGaseF cleavage) to aspartic acid. Sixteen of the glycosylation sites have been previously identified in other studies using experimental methods,40 and nine are characterized here for the first time.
Table 1.
N-Glycoproteins Identified on the Surface of Exosomes Released by MSDCa
| glycopeptide | Uniprot accession number | protein |
|---|---|---|
| DQCIVDDITYNVNDTFHK | P11276 | fibronectin |
| LDAPTNLQFVNETDR | P11276 | fibronectin |
| TEAADLCQAFNSTLPTMDQMK | P15379 | CD44 |
| INLTTNVVDVNRPLPLAAYNNR | Q3UZZ4 | olfactomedin-4 |
| DAMVGNYTCEVTELSR | Q61735 | leukocyte surface antigen CD47 |
| DSSGVINVMLNGSEPK | Q64277 | ADP-ribosyl cyclase/CD157 |
| AVNQTGALYQCDYSTSR | P05555 | integrin α-M/CD11b |
| LNYTLVGEPLR | P05555 | integrin α-M/CD11b |
| YLNFTASEMTSK | P05555 | integrin α-M/CD11b |
| ISENGSSVAGILSSPNMEK | Q9Z0M6 | CD97 |
| VVIRPFYLTNSTDMV | Q07797 | galectin-3-binding protein |
| VVNVSELYGTPCTK | P55772 | ectonucleoside triphosphate diphosphohydrolase 1/CD39 |
| DCIQSGPGCSWCQKLNFTGPGEPDSLR | P11835 | integrin β-2/CD18 |
| AFMNSSFTIDPK | O88792 | junctional adhesion molecule A/CD321 |
| ALMPFDSLHDDPCLLTNR(S) | P11247 | myeloperoxidase |
| KVSCPIMPCSNATVPDGECCPR | P35441 | thrombospondin-1 |
| VVNSTTGPGEHLR | P35441 | thrombospondin-1 |
| GFCEADNSTVSENNPEDWPVNTEGCMEK | P40237 | CD82 |
| FLEQQNQVLQTKWELLQQVNTSTR | Q6IFZ6 | keratin, type II cytoskeletal 1b |
| YKGTAGNALMDGASQLVGENR(T) | Q8K0E8 | fibrinogen β chain |
| GTAGNALMDGASQLVGENR(T) | Q8K0E8 | fibrinogen β chain |
| VQGCMSQPGCNLLNGTQTIGPVDVSER | Q8R2S8 | CD177 |
| AELSNVSDTVWNIR | Q9D8U6 | mast cell-expressed membrane protein 1 |
| IVDVNLTSEGK | Q9ET30 | transmembrane 9 superfamily member 3 |
| ALNASQEETGAVFLCPWK | Q9QUM0 | integrin α-IIb/CD41 |
| LLENCGFNMTAK | Q9WVG5 | endothelial lipase |
Asparagine residues in the motif N-X-S/T, which were deamidated upon release by PNGase F, are highlighted in bold.
From the surface of the parental MDSCs, 188 N-linked glycopeptides were characterized, supporting the identification of 93 glycoproteins (Table S2). On 16 of these glycopeptides, we identified glycosylation at two sites. In all, 204 glycosites are identified. One hundred and fifty-two of the 204 observed sites of N-glycosylation have already been confirmed by experimental evidence in other studies,40 while 52 are characterized for the first time here. Twenty-seven of the latter cohort are not predicted by the NetNGlyc database (Table S3).
The topological domains of the N-linked glycosites identified from the exosomal surface were also evaluated, seeking confirmation of the fidelity of oxidative alkylation on the surface of our exosomes (Table S4). Based on the chemistry, we would expect all the glycopeptides observed to originate in solvent accessible regions of the proteins. Relevant topologic information is not available for all of the proteins; however the majority of glycosites identified on the surface of the exosomes and also on the parental MDSC surface can be assigned to extracellular regions of their proteins. A few glycosites originate from proteins in the extracellular space and some from GPI anchored proteins that do not have transmembrane domains (Figure 1).
Figure 1.
Origins of N-linked glycopeptides identified on the surfaces of parental MDSCs and exosomal N-glycopeptides. The topology information was created using Uniprot and Protter.
Cluster of differentiation (CD) proteins are of particular interest because of their interactive functions at the cell surface. Ten CD proteins were identified on the exosomes, and 35 on MDSCs (Figure 2). The CD proteins found on surfaces of both parental cells and exosomes include CD11b (also known as Mac-1), which is used as a marker for MDSCs although it is also expressed on other hematopoietic cells. CD47 was also found on both surfaces and is of interest as it is reported to prevent phagocytosis by macrophages and thereby protects cancer cells from immune destruction.41 CD82 found on both surfaces is known to bind to CD4 and stimulate the T-cell receptor signaling cascade.42
Figure 2.
CD proteins identified on the surfaces of the MDSC and MDSC derived exosomes.
The similarity or difference between proteins carried by exosomes and their parental cells is of considerable interest to researchers studying the role of exosomes in intercellular communication.4– In the present study, 17 of the 21 exosome glycoproteins were found in common on the surface of parental MDSCz with N-linked glycopeptide identifications on exosomes. Another three proteins identified on the basis of N-glycopeptides in exosomes are supported by tryptic peptides in MDSCs (ADP-ribosyl cyclase (CD157), transmembrane 9 superfamily member 3, and endothelial lipase) (Table S5). Only junctional adhesion molecule A (CD321) was identified on the exosome surface but not detected at all in our current examination of parental MDSC samples. Intriguingly, ADP-ribosyl cyclase (CD157) is identified in the exosomes by virtue of one deglycosylated glycopeptide and no tryptic peptides, while in the parental MDSCs, four distinct tryptic peptides were observed, but no deglycosylated glycopeptide, suggesting it is differentially glycosylated in exosomes. The overlap between exosome and parental MDSC surface proteins demonstrated here exceeds 80% (17 of 21 proteins) and suggests that the surface of the exosomes primarily carry proteins representative of the surface of the primary cells that released them.
Exosomes are proposed to transport active biomolecules from sender cells to receiver cells, a hypothesis that raises questions about recognition and adhesion (as well as transmembrane mechanisms).46–50 We have shown that MDSC exosomes stimulate migration of MDSCs,20 and we have summarized in Table 2 glycoproteins known to be partners in receptor/ligand interactions and thus, potentially, in MDSC/exosome couplings. Leukocyte surface antigen (CD47) and CD44 on the exosome surface both bind glycoproteins identified on the MDSC surface51,52and CD29 and CD172a on the MDSC surface bind to TSP-141,53 and fibrinogen,54 respectively, identified on the exosome surface. Most of the cell surface molecules listed in Table 2 are also receptors or ligands expressed by other cells in the tumor microenvironment.55–59 Our survey appears to support adhesion between exosomes and cells, though not yet selective recognition.
Table 2.
Ligand/Substrate Binding Partners Identified on the Surface of MDSC and MDSC-Derived Exosomes
| exosome | MDSC |
|---|---|
| integrin α-IIb/CD41 | thrombospondin, fibronectin, fibrinogen |
| integrin α-M/CD11b | haptoglobin, fibrinogen, fibronectin |
| integrin β2/CD18 | haptoglobin, fibrinogen |
| leukocyte surface antigen (CD47) | thrombospondin-1, CD172a |
| CD44 | fibronectin |
| thrombospondin-1 | CD29, leukocyte surface antigen (CD47) |
MDSC-derived exosomes exhibit immunosuppressive functionalities similar to that of their parental cells,20 and proteins on their surface should similarly be considered as potential drug targets. Neutrophil granule protein is a protein of interest identified in this study on the surfaces of both MDSCs and MDSC-derived exosomes. CD47 is particularly interesting because cancer cells express elevated levels of CD47, which sends a “don’t eat me” signal when it complexes with CD172a (signal regulatory protein α or SIRPα), thereby preventing phagocytosis by macrophages.41 CD47 is also known to affect cellular aggregation and migration,60 and it is currently under investigation as a potential therapeutic target against cancer.1,2 These characteristics made CD47 an interesting candidate for functional assays.
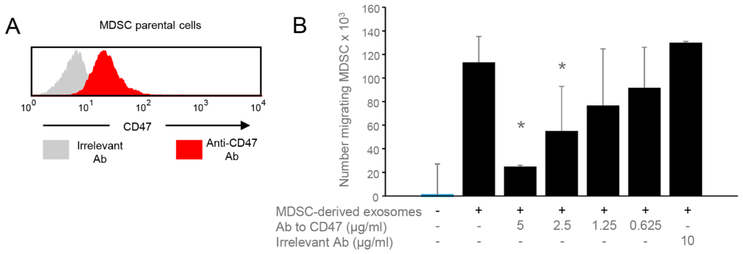
MDSCs mediate their immune suppressive functions within the tumor microenvironment. Since MDSCs are generated in the bone marrow and traffic via the circulatory system, their entry into solid tumors involves chemoattraction. Our previous studies have demonstrated that tumor-derived inflammatory factors21 and MDSC-derived exosomes facilitate this chemoattraction.20 We first confirmed by flow cytometry that parental MDSCs express cell surface CD47 by staining MDSCs with an antibody specific to CD47 or an irrelevant isotype matched antibody (Figure 3A). As expected from the proteomic analysis, MDSCs expressed high levels of CD47. To determine if CD47 impacts MDSC migration, we used a transwell system in which MDSCs were placed in the upper chamber of a transwell containing a semipermeable membrane, and MDSC-derived exosomes were placed in the lower chamber. The ability of the exosomes to chemoattract parental MDSCs was determined by quantifying the number of MDSCs migrating into the lower chamber. The role of CD47 was determined by assessing MDSC migration in the presence of an antibody that sterically blocked CD47. In the absence of exosomes, MDSCs did not migrate, confirming that exosomes are chemotactic for MDSCs. Preincubation of the exosomes with increasing quantities of anti-CD47 antibody, but not an irrelevant control antibody, significantly decreased the number of migrating MDSCs (Figure 3B). These results indicate that CD47 facilitates exosome-mediated migration of MDSCs.
Figure 3.
CD47 regulates MDSC chemotaxis and migration in response to MDSC-derived exosomes. (A) Parental MDSCs express CD47 on their cell surface. Tumor-induced MDSCs were stained with fluorescent antibody to CD47 or with irrelevant control isotype-matched antibody and analyzed by flow cytometry. (B) BALB/c tumor-induced MDSCs were placed in the upper chamber of a transwell and MDSC-derived exosomes, with or without titered quantities of antibody to CD47 or irrelevant control antibody IgG2b were placed in the lower chamber. MDSCs migrating to the lower chamber were quantified by counting. * indicates values are statistically significantly different from exosomes without antibody to CD47 or exosomes with isotype control antibody (p < 0.02). Data are from one of six independent experiments with 5 μg/mL CD47 antibody.
CD47 binds two ligands: the secreted protein TSP-1 and the integral membrane protein SIRPα. Our proteomic analysis identified TSP-1 in both parental MDSCs and MDSC-derived exosomes, while SIRPα was only detected in the parental MDSCs. Exosome driven chemotaxis of MDSCs could involve either or both of these ligands and either the MDSCs or the exosomes could express the receptor CD47 or the ligand TSP-1 or SIRPα To determine whether CD47 is used by the MDSC or the exosomes, exosomes or MDSCs were pretreated with blocking antibodies to CD47 or an irrelevant control isotype matched antibody prior to inclusion in the migration assay. Anti-CD47 antibody pretreatment of the exosomes (Figure 4A) but not the MDSCs (Figure 4B) prevented migration, while the irrelevant antibody had no effect. These results indicate that the exosomes and not the MDSCs use the integral membrane protein CD47 as the receptor. Since the receptor CD47 is functional on the exosomes, MDSC chemotaxis could involve MDSC-produced TSP-1 or SIRPα. To determine if either of these ligands are involved and to ascertain if MDSCs are the source of the ligand, exosomes or MDSCs were pretreated with blocking antibodies to TSP-1 or SIRPα prior to inclusion in the migration assay. Pretreatment of either exosomes or MDSCs with antibodies to TSP-1 significantly decreased MDSC migration (Figure 5A.B, respectively). Since antibodies cannot be removed from the exosome solution prior to the assay, these anti-TSP-1 antibodies could be neutralizing TSP-1 produced by either the MDSC or exosomes. In contrast, pretreatment of either exosomes or MDSCs with blocking antibodies to SIRPα only minimally reduced MDSC migration, and this trend was not statistically significantly different from the effect of the irrelevant antibody controls (Figure 6A,B, respectively). These results demonstrate that TSP-1 is the dominant CD47 ligand that regulates exosome-driven MDSC chemotaxis.
Figure 4.
CD47 on MDSC-derived exosomes, and not CD47 on intact (parental) MDSCs, regulates MDSC chemotaxis and migration. Chemotaxis assay was performed as in Figure 3 except (A) exosomes or (B) MDSCs in some samples were pretreated with antibody to CD47 or an irrelevant isotype matched antibody prior to their placement in the transwells. Data are representative of one of six and three independent experiments for panels A and B, respectively.
Figure 5.
Thrombospondin, a ligand for CD47, is produced by parental MDSCs and facilitates MDSC chemotaxis and migration. Chemotaxis assay was performed as in Figure 3 except (A) exosomes or (B) MDSCs in some samples were pretreated with antibody to thrombospondin (TSP-1) or an irrelevant isotype matched antibody prior to their placement in the transwells. Data are representative of one of three and one of two independent experiments for panels A and B, respectively.
Figure 6.
SIRPα, a ligand for CD47, is produced by parental MDSCs and may contribute to MDSC chemotaxis and migration. (A) Chemotaxis assay was performed as in Figure 3 except (A) exosomes or (B) MDSCs in some samples were pretreated with antibody to SIRPα, antibody to CD47, or an irrelevant isotype matched antibody prior to their placement in the transwells. Data are representative of one of three and three independent experiments for panels A and B, respectively.
CONCLUSIONS
We have successfully established a method to identify glycoproteins on the surfaces of exosomes and demonstrated that multiple glycoproteins are present on the surface of MDSC-derived exosomes. About 80% of the surface glycoproteins on these exosomes were also identified on the surface of parental MDSCs, providing further support to the conclusion that the protein cargo in exosomes closely resembles that of the parent cells. Our studies identified numerous molecules that have the potential to regulate exosome activity and function. For example, exosomes were observed to contain CD321 (also known as junctional adhesion molecule A encoded by the F11R gene). In addition to its function in cellular tight junctions, CD321 binds to leukocyte function antigen 1 (LFA-1 or CD11a), which we detected in MDSCs. LFA-1 is also expressed by T cells raising the possibility that MDSC-derived exosomes might deliver their immune suppressive cargo to T cells by their binding of CD321 to T cell LFA-1.
The leukocyte surface antigen CD47 and its ligand TSP-1 were among eight CDs found on both cell and exosome surfaces. CD47 is well-known for its ability to protect tumor cells from phagocytosis. Our studies demonstrate that CD47 also promotes tumor progression by enhancing the trafficking of immune suppressive MDSCs and thereby inhibiting antitumor immunity. Therefore, in-progress clinical trials testing the therapeutic effect of antibodies to CD47 may not only demonstrate direct effects on tumor cells but may also facilitate the development of antitumor immunity.61
Supplementary Material
Peptides identified from tryptic and PngaseF fractions used to identify N-glycoproteins from the surface of MDSC exosomes (XLSX)
Peptides identified from tryptic and PngaseF fractions used to identify N-glycoproteins from the surface of MDSC (XLSX)
Glycosite analysis of N-glycoproteins identified on the surface of MDSC (XLSX)
Glycosite analysis of N-glycoproteins identified on the surface of MDSC derived exosomes (XLSX)
Proteins identified from parental MDSC with only tryptic peptides (XLSX)
ACKNOWLEDGMENTS
We thank Dr.Yan Wang, Director of Proteomic Core Facility, Maryland Pathogen Research Institute, University of Maryland, College Park, for advice with the LC–MS/MS analysis. This work was supported by the National Institutes of Health, Grants GM021248 and RO1CA115880.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.6b00811.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Notes
The authors declare no competing financial interest.
REFERENCES
- (1).Tseng D; Volkmer J-P; Willingham SB; Contreras-Trujillo H; Fathman JW; Fernhoff NB; Seita J; Inlay MA; Weiskopf K; Miyanishi M; et al. Anti-CD47 antibody-mediated phagocytosis of cancer by macrophages primes an effective antitumor T-cell response. Proc. Natl. Acad. Sci. U. S. A 2013, 110, 11103–11108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Unanue ER Perspectives on anti-CD47 antibody treatment for experimental cancer. Proc. Natl. Acad. Sci. U. S. A 2013, 110, 10886–10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Sun B; Hood L Protein-Centric N-Glycoproteomics Analysis of Membrane and Plasma Membrane Proteins. J. Proteomics Res 2014, 13, 2705–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Bock T; Moest H; Omasits U; Dolski S; Lundberg E; Frei A; Hofmann A; Bausch-Fluck D; Jacobs A; Krayenbuehl N; et al. Proteomic analysis reveals drug accessible cell surface N-glycoproteins of primary and established glioblastoma cell lines. J. Proteome Res 2012, 11, 4885–4893. [DOI] [PubMed] [Google Scholar]
- (5).Mathivanan S; Ji H; Simpson RJ Exosomes: Extracellular organelles important in intercellular communication. J. Proteomics 2010, 73, 1907–1920. [DOI] [PubMed] [Google Scholar]
- (6).Kowal J; Arras G; Colombo M; Jouve M; Morath JP; Primdal-bengtson B; et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. U. S. A 2016, 113, E968–E977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Andreu Z; Yáñez-Mó M Tetraspanins in extracellular vesicle formation and function. Front. Immunol 2014, 5, 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Heijnen BHFG; Schiel AE; Fijnheer R; Geuze HJ; Sixma JJ Activated Platelets Release Two Types of Membrane Vesicles. Blood J 1999, 94, 3791–3800. [PubMed] [Google Scholar]
- (9).Bellingham SA; Guo BB; Coleman BM; Hill AF Exosomes: Vehicles for the transfer of toxic proteins associated with neurodegenerative diseases? Front. Physiol 2012, 3, 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Chahar H; Bao X; Casola A Exosomes and Their Role in the Life Cycle and Pathogenesis of RNA Viruses. Viruses 2015, 7, 3204–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Hoshino A; Costa-Silva B; Shen T-L; Rodrigues G; Hashimoto A; Tesic Mark M; Molina H; Kohsaka S; Di Giannatale A; Ceder S; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Fedele C; Singh A; Zerlanko BJ; Iozzo RV; Languino LR The αvΛ6 integrin is transferred intercellularly via exosomes. J. Biol. Chem 2015, 290, 4545–4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Marvel D; Gabrilovich DI Myeloid-derived suppressor cells in the tumor microenvironment: expect the unexpected. J. Clin. Invest 2015, 125, 3356–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Parker KH; Beury DW; Ostrand-rosenberg S Myeloid-Deived Suppressor Cells: Critical cells driving Immune Suppression in the Tumor Microenvironment. Adv. Cancer Res 2015, 128, 95–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Ostrand-Rosenberg S; Sinha P; et al. Myeloid-derived suppressor cells: linking inflammation and cancer. J. Immunol 2009, 182, 4499–4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Sinha P; Clements VK; Fulton AM; Ostrand-Rosenberg S Prostaglandin E2 promotes tumor progression by inducing myeloid-derived suppressor cells. Cancer Res 2007, 67, 4507–4513. [DOI] [PubMed] [Google Scholar]
- (17).Parker KH; Sinha P; Horn LA; Clements VK; Yang H; Li J; Tracey KJ; Ostrand-Rosenberg S HMGB1 enhances immune suppression by facilitating the differentiation and suppressive activity of myeloid-derived suppressor cells. Cancer Res 2014, 74, 5723–5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Xiang X; Poliakov A; Liu C; Liu Y; Deng Z; Wang J; Cheng Z; Shah SV; Wang G-J; Zhang L; et al. Induction of myeloid-derived suppressor cells by tumor exosomes. Int. J. Cancer 2009, 124, 2621–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Yang C; Robbins PD The roles of tumor-derived exosomes in cancer pathogenesis. Clin. Dev. Immunol 2011, 2011, 842849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Burke M; Choksawangkarn W; Edwards N; Ostrand-Rosenberg S; Fenselau C Exosomes from Myeloid-Derived Suppressor Cells Carry Biologically Active Proteins. J. Proteome Res 2014, 13 (2), 836–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Sinha P; Okoro C; Foell D; Freeze HH; Ostrand-Rosenberg S; Srikrishna G Proinflammatory S100 Proteins Regulate the Accumulation of Myeloid-Derived Suppressor Cells. J. Immunol 2008, 181, 4666–4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Ghosh D; Krokhin O; Antonovici M; Ens W; Standing KG; Beavis RC; Wilkins JA Lectin Affinity as an Approach to the Proteomic Analysis of Membrane Glycoproteins research articles. J. Proteome Res 2004, 3, 841–850. [DOI] [PubMed] [Google Scholar]
- (23).Rybak J-N; Scheurer SB; Neri D; Elia G Purification of biotinylated proteins on streptavidin resin: a protocol for quantitative elution. Proteomics 2004, 4, 2296–2299. [DOI] [PubMed] [Google Scholar]
- (24).Shin BK; Wang H; Yim AM; Le Naour F; Brichory F; Jang JH; Zhao R; Puravs E; Tra J; Michael CW; et al. Global profiling of the cell surface proteome of cancer cells uncovers an abundance of proteins with chaperone function. J. Biol. Chem 2003, 278, 7607–7616. [DOI] [PubMed] [Google Scholar]
- (25).Choksawangkarn W; Kim S; Cannon JR; Edwards NJ; Lee SB; Fenselau C Enrichment of Plasma Membrane Proteins Using Nanoparticle Pellicles: Comparison between Silica and Higher Density Nanoparticles. J. Proteome. Res 2013, 12, 1134–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Elschenbroich S; Kim Y; Medin J.a.; Kislinger T Isolation of cell surface proteins for mass spectrometry-based proteomics. Expert Rev. Proteomics 2010, 7, 141–154. [DOI] [PubMed] [Google Scholar]
- (27).Boeggeman E; Ramakrishnan B; Kilgore C; Khidekel N; Hsieh-Wilson LC; Simpson JT; Qasba PK Direct identification of nonreducing GlcNAc residues on N-glycans of glycoproteins using a novel chemoenzymatic method. Bioconjugate Chem 2007, 18, 806–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Ramya TNC; Weerapana E; Cravatt BF; Paulson JC Glycoproteomics enabled by tagging sialic acid- or galactose-terminated glycans. Glycobiology 2013, 23, 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Gundry RL; Raginski K; Tarasova Y; Tchernyshyov I; Bausch-Fluck D; Elliott ST; Boheler KR; Van Eyk JE; Wollscheid B The mouse C2C12 myoblast cell surface N-linked glycoproteome: identification, glycosite occupancy, and membrane orientation. Mol. Cell. Proteomics 2009, 8, 2555–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Zhang H; Li X-J; Martin DB; Aebersold R Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat. Biotechnol 2003, 21, 660–666. [DOI] [PubMed] [Google Scholar]
- (31).Weekes MP; Antrobus R; Lill JR; Duncan LM; Hör S; Lehner PJ Comparative analysis of techniques to purify plasma membrane proteins. J. Biomol Technol 2010, 21, 108–115. [PMC free article] [PubMed] [Google Scholar]
- (32).Chornoguz O; Grmai L; Sinha P; Artemenko KA; Zubarev RA; Ostrand-Rosenberg S Proteomic pathway analysis reveals inflammation increases myeloid-derived suppressor cell resistance to apoptosis. Mol. Cell. Proteomics 2011, 10, M110.002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Kessner D; Chambers M; Burke R; Agus D; Mallick P ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics 2008, 24, 2534–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Edwards NJ PepArML: A Meta-Search Peptide Identification Platform for Tandem Mass Spectra. Curr. Protoc Bioinformatics 2013, 13.23.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Kim S; Pevzner PA MS-GF+ makes progress towards a universal database search tool for proteomics. Nat. Commun 2014, 5, 5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Gundry RL; Riordon DR; Tarasova Y; Chuppa S; Juhasz O; Wiedemeier O; Milanovich S; Noto FK; Tchernyshyov I; Raginski K; et al. A Cell Surfaceome Map for Immunophenotyping and Sorting Pluripotent Stem Cells. Mol. Cell. Proteomics 2012, 11, 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Gupta R; Brunak S Prediction of glycosylation across the human proteome and the correlation to protein function. Pac. Symp. Biocomput 2002, 322, 310–322. [PubMed] [Google Scholar]
- (38).Omasits U; Ahrens CH; Muller S; Wollscheid B Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 2014, 30, 884–886. [DOI] [PubMed] [Google Scholar]
- (39).Ung TH; Madsen HJ; Hellwinkel JE; Lencioni AM; Graner MW Exosome proteomics reveals transcriptional regulator proteins with potential to mediate downstream pathways. Cancer Sci 2014, 105, 1384–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Bausch-Fluck D; Hofmann A; Bock T; Frei AP; Cerciello F; Jacobs A; Moest H; Omasits U; Gundry RL; Yoon C; et al. A mass spectrometric-derived cell surface protein atlas. PLoS One 2015, 10, e0121314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Lv Z; Bian Z; Shi L; Niu S; Ha B; Tremblay A; Li L; Zhang X; Paluszynski J; Liu M; et al. Loss of Cell Surface CD47 Clustering Formation and Binding Avidity to SIRPα Facilitate Apoptotic Cell Clearance by Macrophages. J. Immunol 2015, 195, 661–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Nagira M; Imai T; Ishikawa I; Uwabe K-I; Yoshie O Mouse Homologue of C33 Antigen (CD82), a member of the transmembrane 4 superfamily: Complementary DNA, genomic structure, and expression. Cell. Immunol 1994, 157, 144–157. [DOI] [PubMed] [Google Scholar]
- (43).Valadi H; Ekström K; Bossios A; Sjöstrand M; Lee JJ; Lötvall JO Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol 2007, 9, 654–659. [DOI] [PubMed] [Google Scholar]
- (44).Taylor DD; Gercel-Taylor C MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol 2008, 110, 13–21. [DOI] [PubMed] [Google Scholar]
- (45).Rabinowits G; Gercel-Taylor C; Day JM; Taylor DD; Kloecker GH Exosomal microRNA: a diagnostic marker for lung cancer. Clin. Lung Cancer 2009, 10, 42–46. [DOI] [PubMed] [Google Scholar]
- (46).Segura E; Guerin, C; Hogg N; Amigorena, S; Thery, C CD8+ Dendritic Cells Use LFA-1 to Capture MHC-Peptide Complexes from Exosomes In Vivo. J. Immunol 2007, 179, 1489–1496. [DOI] [PubMed] [Google Scholar]
- (47).Clayton A; Mitchell JP; Court J; Linnane S; Mason MD; Tabi Z Human Tumor-Derived Exosomes Down-Modulate NKG2D Expression. J. Immunol 2008, 180, 7249–7258. [DOI] [PubMed] [Google Scholar]
- (48).Clayton A; Turkes A; Dewitt S; Steadman R; Mason MD; Hallett MB Adhesion and signaling by B cell-derived exosomes: the role of integrins. FASEB J 2004, 18 (9), 977–979. [DOI] [PubMed] [Google Scholar]
- (49).Feng D; Zhao WL; Ye YY; Bai XC; Liu RQ; Chang LF; Zhou Q; Sui SF Cellular internalization of exosomes occurs through phagocytosis. Traffic 2010, 11, 675–687. [DOI] [PubMed] [Google Scholar]
- (50).Singh R; Pochampally R; Watabe K; Lu Z; Mo Y-Y Exosome-mediated transfer of miR-10b promotes cell invasion in breast cancer. Mol. Cancer 2014, 13, 256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Fujimoto TT; Katsutani S; Shimomura T; Fujimura K Thrombospondin-bound integrin-associated protein (CD47) physically and functionally modifies integrin αIIbβ3 by its extracellular domain. J. Biol. Chem 2003, 278, 26655–26665. [DOI] [PubMed] [Google Scholar]
- (52).Jalkanen S; Jalkanen M Lymphocyte CD44 binds the COOH-terminal heparin-binding domain of fibronectin. J. Cell Biol 1992, 116, 817–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Short SM; Derrien A; Narsimhan RP; Lawler J; Ingber DE; Zetter BR Inhibition of endothelial cell migration by thrombospondin-1 type-1 repeats is mediated by beta1 integrins. J. Cell Biol 2005, 168, 643–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Smiley ST; King JA; Hancock WW Fibrinogen Stimulates Macrophage Chemokine Secretion Through Toll-Like Receptor 4. J. Immunol 2001, 167, 2887–2894. [DOI] [PubMed] [Google Scholar]
- (55).Poncz M; Eisman R; Heidenreich R; Silver SM; Vilaire G; Surrey S; Schwartz E; Bennett JS Structure of the platelet membrane glycoprotein IIb. J. Biol. Chem 1987, 262, 8476–8482. [PubMed] [Google Scholar]
- (56).Moroi M; Jung SM A mechanism to safeguard platelet adhesion under high shear flow: Von Willebrand factor-glycoprotein Ib and integrin α2 β1-collagen interactions make complementary, collagen-type-specific contributions to adhesion. J. Thromb. Haemostasis 2007, 5, 797–803. [DOI] [PubMed] [Google Scholar]
- (57).Saumet A; Legrand C; Dubernard V; de Jesus N Association of thrombospondin-1 with the actin cytoskeleton of human thrombin-activated platelets through an αIIbβ3- or CD36-independent mechanism mechanism. Biochem. J 2002, 363, 473–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).El Ghmati SM; Van Hoeyveld EM; Van Strijp JG; Ceuppens JL; Stevens EA Identification of haptoglobin as an alternative ligand for CD11b/CD18. J. Immunol 1996, 156, 2542–2552. [PubMed] [Google Scholar]
- (59).Rubel C; Gómez S; Fernández GC; Isturiz MA; Caamaño J; Palermo MS Fibrinogen-CD11b/CD18 interaction activates the NF-κB pathway and delays apoptosis in human neutrophils. Eur. J. Immunol 2003, 33, 1429–1438. [DOI] [PubMed] [Google Scholar]
- (60).Rebres RA; Kajihara K; Brown EJ Novel CD47-dependent intercellular adhesion modulates cell migration. J. Cell. Physiol 2005, 205, 182–193. [DOI] [PubMed] [Google Scholar]
- (61).Oxford University, Stanford University. CAMELLIA: Anti-CD47 Antibody Therapy in Relapsed/Refractory Acute Myeloid Leukaemia. ClinicalTrials.gov [Internet]. Bethesda (MD): National Library of Medicine (US) 2016- [cited 2016 Aug 22]. Available from: https://clinicaltrials.gov/ct2/show/NCT02678338?term=CD47&rank=1. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Peptides identified from tryptic and PngaseF fractions used to identify N-glycoproteins from the surface of MDSC exosomes (XLSX)
Peptides identified from tryptic and PngaseF fractions used to identify N-glycoproteins from the surface of MDSC (XLSX)
Glycosite analysis of N-glycoproteins identified on the surface of MDSC (XLSX)
Glycosite analysis of N-glycoproteins identified on the surface of MDSC derived exosomes (XLSX)
Proteins identified from parental MDSC with only tryptic peptides (XLSX)